Abstract
RuvABC and RecG are thought to provide alternative pathways for the late stages of recombination in Escherichia coli. Inactivation of both blocks the recovery of recombinants in genetic crosses. RuvABC resolves Holliday junctions, with RuvAB driving branch migration and RuvC catalyzing junction cleavage. RecG also drives branch migration, but no nuclease has been identified that might act with RecG to cleave junctions, apart from RusA, which is not normally expressed. We searched for an alternative nuclease using a synthetic lethality assay to screen for mutations causing inviability in the absence of RuvC, on the premise that a strain without any ability to cut junctions might be inviable. All the mutations identified mapped to polA, dam, or uvrD. None of these genes encodes a nuclease that cleaves Holliday junctions. Probing the reason for the inviability using the RusA Holliday junction resolvase provided strong evidence in each case that the RecG pathway is very ineffective at removing junctions and indicated that a nuclease component most probably does not exist. It also revealed new suppressors of recG, which were located to the ssb gene. Taken together with the results from the synthetic lethality assays, the properties of the mutant SSB proteins provide evidence that, rather than promoting recombination, a major function of RecG is to curb potentially pathological replication initiated via PriA protein at sites remote from oriC.
THE early stages of genetic recombination in Escherichia coli associated with initiation of homologous DNA pairing and with strand exchange are well established and can be described in terms of enzymology and reaction pathways (Dillingham and Kowalczykowski 2000, 2008; Singleton et al. 2004; Cox 2007a,b). However, later stages associated with resolution of Holliday junction intermediates have proven more difficult to pin down because of what appears at first sight to be a functional overlap between the RuvABC and RecG proteins. RecG also appears to have multiple roles in DNA metabolism that obscure the nature and extent of its involvement in recombination.
The RuvA and RuvB proteins together catalyze branch migration of Holliday junction intermediates and form a complex with RuvC protein that enables the latter to resolve these intermediates by a dual strand cleavage reaction (van Gool et al. 1998). RecG is a dsDNA translocase and, like RuvAB, catalyses branch migration of Holliday junctions (Lloyd and Sharples 1993; McGlynn and Lloyd 2001; Singleton et al. 2001). Its elimination from ruv mutants blocks the recovery of recombinants in genetic crosses and confers extreme sensitivity to genotoxic agents (Lloyd 1991). The strong synergism observed led to the idea that RuvABC and RecG provide partially overlapping pathways for the resolution of Holliday junctions. However, RecG proved to have no intrinsic ability to cleave junctions (Lloyd and Sharples 1993), which raised the possibility that some unidentified nuclease could act with RecG to promote Holliday junction resolution in the way RuvC acts with RuvAB. The RusA protein was a possible candidate (Sharples et al. 1994). This homodimeric endonuclease resolves Holliday junctions by a dual strand cleavage mechanism that targets specific DNA sequences (Sharples et al. 1994; Bolt and Lloyd 2002). Its expression compensates very effectively for the absence of RuvABC and in a RecG-dependent manner (Mandal et al. 1993; Mahdi et al. 1996). However, RusA is encoded by a cryptic prophage gene (rusA) and is not normally expressed because the gene lacks a promoter. Furthermore, its deletion does not reduce recombination in ruv mutant strains (Mahdi et al. 1996). Therefore, RusA cannot be the resolvase that operates in strains lacking RuvABC, although it can act as such when activated by a promoter inserted upstream of rusA (Mandal et al. 1993; Mahdi et al. 1996).
To date, our screens for mutations blocking recombination in ruv mutants failed to identify an alternative nuclease that could act with RecG, revealing only knockouts of the RecA, RecB, or RecC proteins needed to initiate exchanges or of RecG (our unpublished work). This failure could be explained if the requisite activity is needed to maintain viability, at least in the absence of RuvABC or is provided by more than one nuclease. Alternatively, there may be no such nuclease, with RecG alone able to eliminate Holliday junctions simply by driving branch migration and enabling them to merge with replication forks, as has been suggested (Wardrope et al. 2009). This possibility would suffice to explain why eliminating RecG has such a strong synergistic effect on ruv strains. But recent studies indicate that there may be an additional and perhaps more radical explanation.
RecG unwinds a variety of branched DNA molecules, at least in vitro (McGlynn and Lloyd 2000, 2001, 2002b). These include the D loops and R loops that PriA protein could otherwise exploit to initiate stable DNA replication (SDR), a form of chromosome replication that is independent of oriC and of the initiator protein DnaA, and which also includes the replication primed by recombination during repair of chromosome breaks (Vincent et al. 1996; Fukuoh et al. 1997; Kogoma 1997; McGlynn et al. 1997). SDR is elevated constitutively in cells lacking RecG (Hong et al. 1995) and triggers severe overreplication of the chromosome when increased even further by damage to DNA. This sets in motion a pathological cascade that interferes with the cell cycle and which results in the formation of extraordinarily long filaments that bud off a small cell capable of normal growth and division only after a very long delay (Rudolph et al. 2009a,b). Without RuvABC, this problem is likely to be much exacerbated by the failure to resolve Holliday junctions. Thus, ruv recG double mutants may be exceptionally sensitive to UV light and other DNA-damaging agents because they are simply overwhelmed by pathological consequences resulting from the increase in SDR.
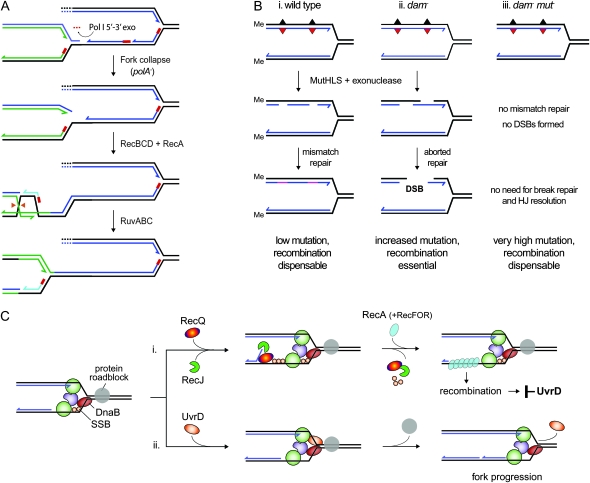
In this article, we present evidence supporting this hypothesis. The evidence stems from studies in which we had initially exploited a synthetic lethality assay to screen for mutations incompatible with a deletion of ruvC. The seven mutations identified inactivated dam, polA, or uvrD (supporting information, File S1 and Figure S1), which previous studies had reported to be inviable with ruv (Ishioka et al. 1998; Marinus 2000; Flores et al. 2005; Magner et al. 2007). The polA gene encodes Pol I, a DNA polymerase and exonuclease associated with nucleotide excision repair and the processing of Okazaki fragments during DNA replication (Moolenaar et al. 2000), whereas the dam gene encodes a deoxyadenosine methylase that directs the MutHLS mismatch repair system to the newly synthesized strands (Modrich 1991). Without efficient means to remove RNA primers and complete synthesis, it is thought that polA mutants retain gaps in the nascent strands, gaps that trigger fork collapse during the next round of replication (Figure 1A). With no means to distinguish parental strands from nascent strands, the MutHLS proteins initiate mismatch repair indiscriminately in dam mutants, which may result in chromosome breakage when repair tracks overlap (Figure 1B). Thus, the viability of both polA and dam mutants depends on recombination proteins to repair DNA breaks— RecBCD and RecA to initiate exchanges at the broken DNA ends and RuvABC to cleave Holliday junctions (Figure 1, A and B) (Lloyd et al. 1974; Hong et al. 1995; Kuzminov 1995; Ishioka et al. 1998; Marinus 2000).
Figure 1.—
Models of how chromosome replication is affected by polA, dam, and uvrD mutations. (A) Replication fork collapse following incomplete processing of Okazaki fragments in the absence of the polymerase activity of DNA polymerase I. The fork is rescued by recombination, enabling replication to be completed. (B, panel i) MutHLS-mediated mismatch repair in wild-type cells is directed by Dam methylation to the transiently unmethylated nascent strands, enabling replication errors to be eliminated. (panel ii) Chromosome breakage and repair following undirected initiation of mismatch repair in a dam mutant. (panel iii) Inactivation of MutHLS prevents chromosome breakage in a dam mutant strain and thus eliminates the need for recombination to maintain viability. (C) UvrD prevents recombination during chromosome replication by either (panel i) removing RecA filaments assembled at a stalled fork and/or by (panel ii) helping the replisome to drive through obstacles that might block fork progression.
The uvrD gene encodes a DNA helicase associated with both mismatch repair and UvrABC-dependent nucleotide excision repair (Matson and Kaiser-Rogers 1990; Modrich 1991). Recent studies indicate that it also acts to limit recombination when replication forks stall (Flores et al. 2005). UvrD has been shown to displace RecA filaments assembled on ssDNA in vitro and is thought to do so in vivo when RecA is loaded on a region of the nascent lagging strand template exposed at a stalled fork through the actions of RecQ helicase and RecJ exonuclease. With no UvrD present, the RecFOR proteins establish a stable RecA filament on the exposed template (Figure 1C), thus presumably provoking recombination even though this recombination is not essential, as indicated by the fact that inactivating RecA or preventing RecA loading restores viability to uvrD ruv cells (Flores et al. 2005; Veaute et al. 2005; Lestini and Michel 2007; Magner et al. 2007). Although UvrD is thought to reduce such pathology by displacing RecA, it might also limit fork stalling by providing a second helicase motor at the fork to help DnaB drive through obstacles (Figure 1C).
What is particularly significant about these observations is that the presence of RecG itself is clearly not sufficient to maintain the viability of polA, dam, and uvrD strains lacking RuvABC. The possibility that the products of these three genes are all essential components of a RecG recombination pathway needed to maintain viability is highly unlikely. None has any activity that might resolve Holliday junctions. Our studies provide strong genetic evidence that the inviability observed when polA, dam, and uvrD strains lack an intact RuvABC system is due in all three cases to the accumulation of Holliday junctions. More importantly, they indicate that RecG is not at all effective in removing Holliday junctions and suggest there is probably no nuclease expressed in wild-type (WT) E. coli cells that enables RecG to provide an effective alternative to RuvABC. Taken together, the results presented support the notion that a major function of RecG is to limit PriA-mediated overreplication of the chromosome and its pathological consequences.
MATERIALS AND METHODS
Strains:
Bacterial strains are listed in Table S1. All constructs used for synthetic lethality assays are based on E. coli K-12 MG1655 ΔlacIZYA (Bernhardt and de Boer 2004) carrying pRC7 derivatives. Chromosomal genes were inactivated using Tn10 or Tn10kan insertions, conferring resistance to tetracycline (Tcr) and kanamycin (Kmr), respectively, or with deletions tagged with resistance to chloramphenicol (Cmr), kanamycin (Kmr), trimethoprim (Tmr), or apramycin (Aprar). Unless referenced otherwise, tagged deletions removed the entire coding sequence of the genes concerned and were generated as described (Datsenko and Wanner 2000). Further details of the polA, dam, and uvrD mutations are presented in File S1, Figure S1, and Figure S2.
Plasmids:
The pRC7 construct is a low-copy number, mini-F derivative of pFZY1. It carries the bla gene encoding resistance to ampicillin (Apr) and the lacZYA operon under control of lacIq (Figure S1A) (Bernhardt and de Boer 2004). pAM372 carries the ruvC+ coding sequence inserted at the multiple cloning site (MCS) in pRC7 under control of the lac promoter. pAM390 carries the wild-type ruvAB operon under control of its native promoter inserted at the ApaI site in pAM372. pJJ100 and pJJ103 carry the entire recG+ gene and some upstream sequences inserted at the ApaI site in pRC7 and pAM372, respectively. pAM408 is a derivative of pAM372 carrying the entire recG+ gene, some upstream sequences, and a downstream BamHI site inserted at the ApaI site. pAM409 is a derivative of pAM408 carrying the ruvAB operon under control of its native promoter inserted at the BamHI site downstream of recG. The ruv+ and/or recG+ genes cloned in these pRC7 derivatives restore full resistance to mitomycin C and UV light in strains carrying the relevant ruv or recG null allele.
pGB066 was constructed by cloning the coding sequence for priA+ into the expression vector pT7-7, as described for the recG+ construct pAM210 (Mahdi et al. 2003). pAM423, pAM425, pAM426, pDIM024, and pDIM025 encode WT SSB protein, or K44E, N14D, R97C or Δ115–144 derivatives, respectively. They carry the coding sequences for ssb+, ssb[A130G], ssb[A40G], ssb[C289T], or ssb[Δ345-434], respectively, cloned at the NdeI site in pET22b. pCC178 and pCC180 are pET22b derivatives encoding SSB with a P176S substitution of the penultimate residue (ssb113) or a deletion of the last 10 residues (ΔC10) (Cadman and McGlynn 2004).
Media and general methods:
Growth media, and methods for monitoring cell growth, P1vir transduction, and for determining sensitivity to UV and mitomycin C have been cited (Al-Deib et al. 1996; McGlynn and Lloyd 2000; Trautinger et al. 2005).
Synthetic lethality assay:
Cultures of strains carrying pRC7 derivatives were grown overnight in LB broth containing ampicillin to maintain plasmid selection, diluted 80-fold in LB broth, and grown without ampicillin selection to an A650 of 0.4 before spreading dilutions on LB agar supplemented with X-gal and IPTG. Plates were photographed and scored after 48 hr at 37°. Where 56/2 minimal salts agar was substituted for LB agar (as indicated in Figure S4B, panel i), the plates were photographed after 72 hr (56/2 agar). Plasmid-free cells forming small white colonies were restreaked on LB agar, to see whether they could be subcultured, and were photographed after 24 hr at 37°. The assay is described in further detail in File S1 and illustrated in Figure S1, A–C.
Mapping priA and ssb suppressors of recG dam:
Suppressors were isolated as shown in Figure 3B, panel i. Mutations in priA were identified by linkage (∼90%) to metB followed by PCR sequencing. Mutations in ssb were identified by first restoring recG+ and dam+ to the initial strain isolates (JJ1329 and JJ1331), generating strains JJ1489 and JJ1490, respectively (Table S1), both of which remained sensitive to UV light. These were transduced with P1 phage grown on pools of cells carrying random kan insertions in the chromosome, generated in strain MG1655 using the EZ-Tn5 〈kan-2〉 Tnp transposome system (Epicentre Technologies). Kmr transductants that were also UVr were identified. PCR sequencing identified insertions in yjcB and yjbQ, very close to ssb. Further sequencing revealed an A to G transition at bp 130 of the ssb coding sequence of JJ1489 and at bp 40 in that of JJ1490. These changes were verified as the suppressors by engineering the same substitution into the chromosome, as described (Datsenko and Wanner 2000), and demonstrating that they conferred sensitivity to UV.
Figure 3.—
Synthetic lethality assays showing how PriAK230R confers viability on polA, dam, and uvrD cells lacking RecG or improves their viability. (A and D) polA constructs. (B) dam constructs. (C) uvrD constructs. The overlay inset in C, panel i, is a magnification of a section of the streak underneath showing a large colony variant.
Proteins:
Wild-type RecG was purified as described (Mahdi et al. 2003). Wild-type and mutant SSB proteins were expressed using the relevant ssb plasmid transformed into the nuclease-depleted BL21(DE3) derivative, STL5827, or JJ1634 and JJ1635 in the case of N14D and K44E, respectively, and purified broadly as described (Cadman and McGlynn 2004). The molecular mass of the purified SSB proteins was determined by gel filtration on a Superdex 200 10/300 column in 20 mm Tris–HCl 7.5, 150 mm NaCl, and corresponded to that of a tetramer in every case.
For wild-type PriA, cultures of STL5827 transformed with pGB066 were grown to an A650 of ∼0.6 in Mu broth supplemented with ampicillin before adding IPTG to induce expression of priA. After 3 hr of further incubation, the induced cells were collected by centrifugation, broken open by sonication, and PriA purified from the supernatant by passage through heparin- and SP-sepharose columns, followed by gel filtration on a Hiprep 16/60 sephacryl S-200 column, before storing at −80°.
Physical interaction of SSB with PriA and RecG:
Equimolar mixtures of SSB (tetramer form) and PriA or of SSB and RecG in 20 mm Tris-HCl pH 7.5 were kept on ice for 15 min before adding 2.5 μl 750 g/liter ammonium sulfate solution (final volume, 10 μl). After a further 15 min on ice, the mixtures were centrifuged for 10 min at 14,000 rpm, the supernatants removed, and pellets resuspended in 10 μl 20 mm Tris–HCl. Fractions were then analyzed by SDS–PAGE, using 15% polyacrylamide gels. Controls containing PriA, RecG, or SSB alone, or equimolar mixtures of PriA and SSB, or RecG and SSB proteins without ammonium sulfate precipitation, were analyzed in parallel.
DNA binding and unwinding assays:
DNA binding was measured by a gel retardation assay using a 50-mer oligonucleotide (5′-ATTCGGCAGCGTTAGCTATCAGAGATCTGTCGTTACAGG-3′) labeled with 32P at the 5′ end. Binding reactions (20 μl) contained 0.2 nm of labeled oligonucleotide and the indicated SSB proteins at final concentrations of 0.5, 5, and 50 nm in low ionic strength buffer, and were analyzed by electrophoresis on 4% polyacrylamide gels (Lloyd and Sharples 1993). DNA unwinding by PriA helicase, and the effect of SSB on this activity was measured using Fork 2, essentially as described (Cadman and McGlynn 2004), except the ATP and MgCl2 were both at 5 mm.
RESULTS
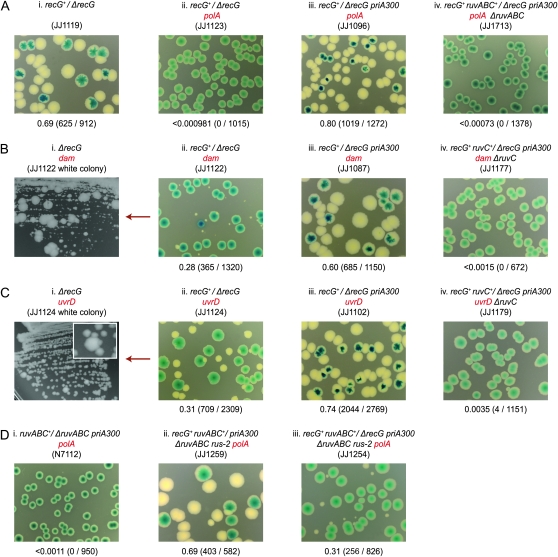
Previous studies demonstrated that polA, dam, and uvrD strains lacking RuvABC are inviable and assumed the cells accumulate Holliday junctions that interfere with growth and division (Ishioka et al. 1998; Marinus 2000; Flores et al. 2005; Magner et al. 2007). If true, it would imply that RecG does not provide an efficient alternative resolution pathway. We exploited RusA to investigate whether this is indeed the case, examining its effect on the viability of the relevant double mutant. We used a synthetic lethality assay based on pRC7 for this purpose, a mini-F derivative that lacks F stabilization systems and which is therefore easily lost (Bernhardt and de Boer 2004). It carries the lac+ genes and its loss is revealed in a Δlac background by segregation of Lac− clones. On plates containing the β-galactosidase indicator, X-gal, these clones form white colonies or white sectors within blue (Lac+) colonies, depending on whether plasmid loss occurred before or after plating (Figure S1, A and B) (Bernhardt and de Boer 2004). A ruv+ derivative of pRC7 was used to cover a ruv deletion in the chromosome (ruv+/Δruv) before introducing a deletion or insertion inactivating polA, dam, or uvrD. Inviability between the covered ruv mutation and the uncovered polA, dam, or uvrD allele is revealed by the absence of Lac− clones and the formation of uniformly blue colonies, or in the case of uvrD ruv, by the appearance of rare and rather sickly Lac− clones (Figure 2, A–D, panels i). Clearly, only those cells retaining the plasmid and therefore expressing RuvABC are capable of robust growth.
Figure 2.—
Synthetic lethality assays showing how RusA confers viability on polA, dam, and uvrD cells lacking RuvABC. (A) Control strains. (B) polA constructs. (C) dam constructs. (D) uvrD constructs. The synthetic lethality assay exploited in A–D and in subsequent figures is described in detail in materials and methods and further illustrated in Figure S1. The relevant genotype of the synthetic lethality construct used is shown above each photograph. In each case the relevant plasmid genotype/relevant chromosome genotype (e.g., ruvABC+/ΔruvABC) is indicated, along with the strain number in parentheses. The fraction of white colonies is shown below with the number of white colonies/total colonies analyzed in parentheses.
RusA restores viability to polA, dam, and uvrD cells lacking RuvABC:
Parallel constructs were made in which expression of RusA had been activated by rus-2, an IS10 insertion upstream of the rusA coding sequence (Mahdi et al. 1996). Ample growth of plasmid-free, Lac− clones was detected in each case, demonstrating that RusA confers robust viability on the double mutant cells (Figure 2, A–D, panels ii). Given RusA cleaves Holliday junctions with high specificity and efficiency, and has comparatively little activity on other forms of branched DNAs unless these can adopt a four-way branched configuration mimicking a Holliday junction (Bolt and Lloyd 2002), these observations leave little doubt that unresolved Holliday junctions are responsible for the inviability of polA, dam, and uvrD cells lacking RuvABC.
Previous studies revealed that the ability of RusA to promote recombination and DNA repair in the absence of RuvABC depends on RecG (Mandal et al. 1993; Mahdi et al. 1996). Synthetic lethality constructs based on recG+ derivatives of pRC7 revealed that the same is true for RusA's ability to confer viability on polA, dam, and uvrD cells lacking RuvABC (Figure 2, A–D, panels iii). This dependence on RecG, coupled with the robust viability observed with RecG present, points very clearly toward the conclusion that any nuclease wild-type E. coli may have that is able to act in concert with RecG to resolve Holliday junctions must operate very inefficiently indeed compared with RusA.
RecG is required to limit the activity of PriA:
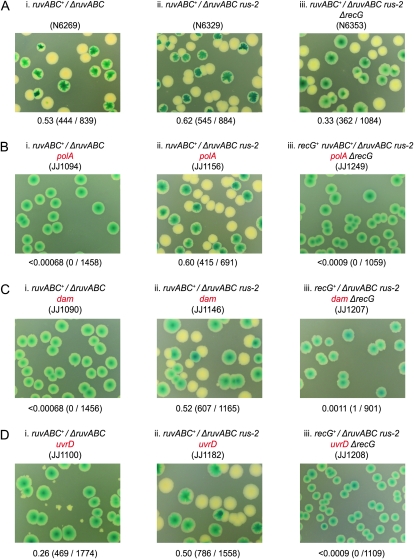
Drawing the conclusion from the data in Figure 2 that RecG is needed to support junction resolution by RusA has to be tempered by evidence that RecG itself is required to maintain viability in the case of polA and dam cells (Hong et al. 1995; Marinus 2000). Synthetic lethality assays based on a recG+ derivative of pRC7 confirmed this was so for polA recG cells (Figure 3A, panels i and ii). With dam recG cells, the assays revealed that viability is much reduced. The double mutant cells form tiny white colonies without the covering plasmid and although these colonies can be subcultured they accumulate suppressors with high frequency, as evident from the emergence of large colony variants (Figure 3B, panels i and ii). The low viability of the dam recG cells explains the previous failure to construct the double mutant by conventional crosses (Marinus 2000).
Previous studies demonstrated that it is possible to construct a recG uvrD double mutant (Mendonca and Matson 1995). However, a synthetic lethality assay based on a recG+/ΔrecG uvrD construct revealed that eliminating both RecG and UvrD reduces cell viability. This is clear from the smaller size of the colonies formed by plasmid-free segregants. These segregants also accumulate suppressors that appear as larger colony variants on subculture (Figure 3C, panels i and ii). However, neither effect is as extreme as with dam recG cells. The reduced viability of polA, dam, and uvrD cells lacking RecG is surprising given RuvABC is available and our evidence that any RecG pathway for the resolution of Holliday junctions is at best highly inefficient. A recent report also revealed that recG cells show little evidence of accumulating Holliday junctions following DNA breakage (Wardrope et al. 2009). So, why should RecG be needed to maintain viability?
Previous studies showed that the sensitivity of recG cells to mitomycin C and to other DNA damaging agents can be suppressed by mutations reducing or eliminating the helicase activity of PriA, demonstrating that this activity can be detrimental to cell viability when the DNA is damaged and RecG is absent (Al-Deib et al. 1996). We considered whether a similar explanation might also account for the reduced viability of polA, dam, and uvrD cells lacking RecG and tested this possibility using recG+/ΔrecG priA300 constructs. The priA300 allele encodes helicase-defective PriAK230R (Zavitz and Marians 1992) and is an effective suppressor of recG (Jaktaji and Lloyd 2003). With this allele present, polA, dam, and uvrD derivatives proved quite viable, as evident from the robust growth of plasmid-free Lac− colonies (Figure 3, A–C, panels iii). However, ΔruvC and ΔruvABC derivatives revealed that this requires the RuvABC system to be intact (Figure 3, A–C, panels iv; data not shown). Furthermore, priA300 fails to restore viability to equivalent ΔruvABC recG+ and ΔruvC recG+ constructs inactivated for polA, dam, or uvrD (Figure 3D, panel i; data not shown), consistent with previous studies demonstrating that it does not suppress the ruv mutant phenotype (Jaktaji and Lloyd 2003). Thus, whereas RecG is dispensable once the helicase activity of PriA is eliminated, RuvABC is most certainly not.
This dependence on RuvABC indicates that Holliday junctions still accumulate. We tried to test this directly by building a ΔrecG ΔruvABC priA300 polA construct in which the recG and ruvABC deletions were covered by a recG+ ruv+ plasmid, and in which RusA was activated by rus-2. The construct made segregates white colonies, but these are tiny and grow very poorly on subculture, consistent with RusA being effective only in the presence of RecG (Figure 3D, panels ii and iii; data not shown). Nevertheless, the fact that white colonies do appear supports the notion that the cells accumulate Holliday junctions in the absence of RuvABC, despite the presence of priA300.
From these data, it would appear that the viability of polA, dam, and uvrD cells is reduced in the absence of RecG simply because they lack means to counter some deleterious effect of PriA helicase activity. When this is eliminated, RuvABC maintains viability very effectively without any assistance from RecG.
The absence of RecG provokes recombination:
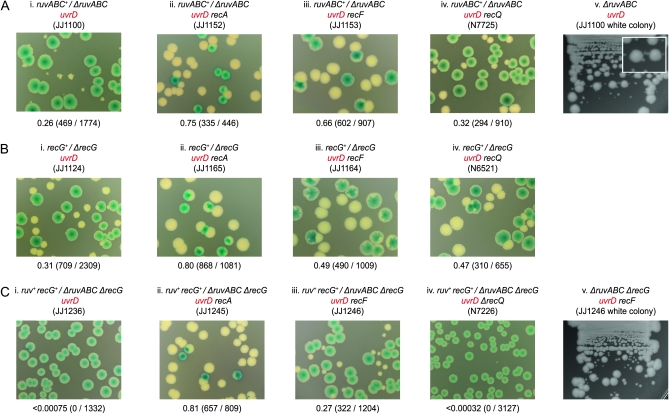
The plasmid-free cells segregated by a ruv+/Δruv uvrD construct form very sickly colonies that accumulate suppressors, which is not surprising given the mutator phenotype associated with uvrD. These suppressors appear as large colony variants on subculture (Figure 4A, panels i and v). Genetic analysis revealed that mutations inactivating RecA, RecFOR, RecJ, or RecQ restore robust viability and account for the suppressors observed (Figure 4A, panels ii–iv; Figure S3, A and B; data not shown). This analysis confirmed previous studies and supports the model outlined in Figure 1C, panel i (Flores et al. 2005; Magner et al. 2007).
Figure 4.—
Synthetic lethality assays demonstrating how RecA, RecF, and RecQ affect the viability of uvrD cells lacking RuvABC and/or RecG. (A) Cells lacking RuvABC. (B) Cells lacking RecG. (C) Cells lacking both RuvABC and RecG.
Synthetic lethality assays based on recG+/ΔrecG uvrD constructs revealed a similar improvement in the growth of plasmid-free recG uvrD cells following the inactivation of RecA, RecFOR, or RecQ (Figure 4B, panels i–iv; data not shown), suggesting perhaps there might be a common basis for the reduced viability of ruv uvrD and recG uvrD cells. We tested this by eliminating RecA from uvrD cells lacking both RuvABC and RecG. With RecA present, these cells fail to form any colonies without a covering recG+ ruv+ plasmid (Figure 4C, panel i). Removing RecA restores robust viability (Figure 4C, panel ii). Significantly, removing RecF or RecO is much less effective in this case. It allows plasmid-free cells to form colonies, but these are small relative to those formed by cells retaining the plasmid and accumulate suppressors, demonstrating that their viability is still compromised (Figure 4C, panels iii and v; data not shown). Removing RecQ does not help at all, but neither does it interfere with the improved viability observed on removing RecA (Figure 4C, panel iv; data not shown). If one accepts that the viability of a strain lacking RuvABC depends on the incidence of recombination mediated by RecA, then these data would suggest that such recombination is more frequent in ΔruvABC cells lacking both UvrD and RecG than in ΔruvABC cells lacking only UvrD. This in turn implies that eliminating RecG itself provokes recombination, and for a reason distinct from that provoked by the absence of UvrD. Since eliminating RecQ does not improve viability at all, and inactivating the RecF or RecO component of RecFOR is only partially effective, we suspect this additional recombination is not triggered by the exposure of ssDNA at stalled forks.
Identification of ssb suppressors of recG:
The feeble growth of ΔrecG dam cells facilitates the identification of suppressors as large colony variants (Figure 3B, panels i and ii). Among the ≥30 independent suppressors analyzed, we identified 11 with mutations in priA and 2 with mutations in ssb. Others were strong mutators, and reconstructions confirmed that inactivating MutH, MutL, or MutS restores viability (Figure 5, A and B; data not shown). Our analyses demonstrated that the undirected initiation of mismatch repair creates a problem for both recG and ruv cells.
Figure 5.—
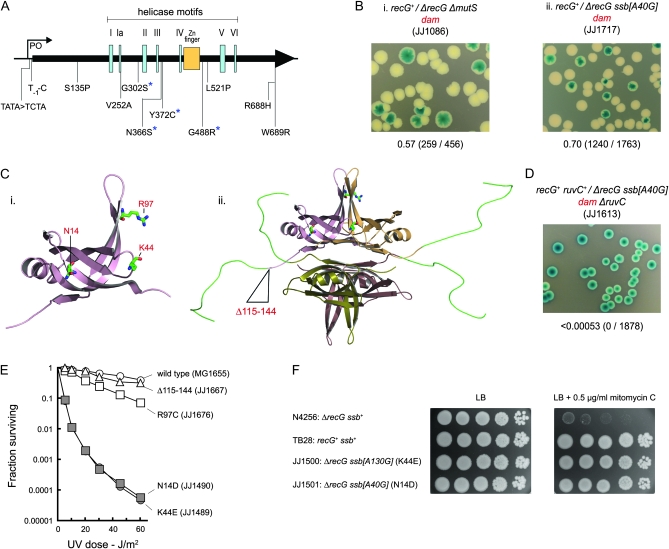
Suppressors of the low viability of dam recG cells. (A) Map showing location and identity of priA suppressors. Coding mutations are indicated in terms of the changes to PriA. The asterisks identify changes that provide for particularly strong suppression of the mitomycin C sensitivity of a recG strain. (B) Synthetic lethality assays illustrating restoration of viability to recG dam cells by mutS and ssb mutations. (C) Structure of an SSB monomer (i) and tetramer (ii) showing the amino acid residues affected by the ssb mutations identified. The models were generated using Pymol and SSB crystal coordinates from the PDB database (Matsumoto et al. 2000; Raghunathan et al. 2000; Savvides et al. 2004). (D) Synthetic lethality assay showing that the viability conferred on recG dam cells by ssb[A40G] depends on RuvC. (E) Effect of ssb mutations on sensitivity to UV light. (F) Restoration of mitomycin C resistance to a recG strain by ssb[A40G] and ssb[A130G]. Cultures of the strains indicated were grown in LB broth to an A650 of 0.4, diluted in 10-fold steps, and 10-μl aliquots spotted on LB agar without and with mitomycin C, as indicated. The plates were photographed after 24 hr at 37°.
Given the already established effect of priA300 (Figure 3B, panel iii), the priA suppressors were no surprise. The spectrum of alleles was broader than observed in a previous study that selected directly for recG suppressors restoring resistance to mitomycin C (Al-Deib et al. 1996). Indeed two were promoter mutations (Figure 5A), indicating that a reduction in the overall level of PriA suffices to improve viability in this case.
The ssb mutations proved quite robust suppressors. Both carry A to G transitions in the ssb gene (one at position 40 and one at position 130), encoding either an N14D or a K44E substitution in SSB (Figure 5C). In each case, the ability of the altered SSB to confer viability depends on having an intact RuvABC system. The same loss of viability is observed on removing RuvC alone or all three Ruv proteins (Figure 5D and data not shown). Furthermore, ruv+/Δruv dam constructs carrying these ssb mutations revealed that neither is a suppressor of ruv dam inviability (data not shown). This ability of both priA and ssb mutations to confer viability on recG dam but not on either recG dam ruv or ruv dam is significant. It demonstrates very clearly that the depletion of RecG and RuvABC creates very different problems for cells initiating mismatch repair indiscriminately during chromosome replication.
The N14D and K44E derivatives of SSB specifically suppress recG:
Both of the ssb suppressor strains proved moderately sensitive to UV light. Introducing wild-type alleles for recG or dam, or both recG and dam, established that this is a property of the ssb alleles (Figure 5E and materials and methods). It also revealed that these mutations specifically suppress recG. Despite the sensitivity conferred to UV, these mutations confer no sensitivity to mitomycin C and strongly suppress the mitomycin C sensitivity conferred by a ΔrecG allele (Figure 5F and data not shown). These findings were confirmed by reconstruction of the relevant genotypes (data not shown). The ssb mutations also improve the viability of recG polA and recG ΔpriA cells, consistent with being suppressors of recG (Figure S4B). Previous studies had demonstrated that recG ΔpriA cells have a low efficiency of plating on LB agar (McCool and Sandler 2001; Gregg et al. 2002). Neither of the ssb mutations was able to suppress the mitomycin C sensitivity conferred by a Δruv allele (data not shown), reinforcing the fact that both specifically suppress the recG mutant phenotype.
Two other ssb mutations were identified previously among suppressors of the very poor viability of ΔpriA dnaC812 strains lacking the DNA binding protein, RdgC (Moore et al. 2003). One is a C to T transition (ssb[C289T]) encoding an R97C substitution, the other an in-frame deletion of 90 bp (ssb[Δ345–434]) removing 30 amino acids from the long C-terminal arms extending from the core of the SSB tetramer (Figure 5C) (Matsumoto et al. 2000; Raghunathan et al. 2000; Savvides et al. 2004). These mutations also improve the viability of recG dam cells. However, they are less effective, at least as judged by colony size, consistent with being less effective suppressors of the mitomycin C sensitivity of recG (Figure S4A and data not shown). They also confer little or no sensitivity to UV (Figure 5E). Thus, in the case of SSB mutations, the ability to strongly suppress recG and, consequently to restore robust viability to recG dam cells, seems to come at the price of a reduced ability to survive UV irradiation.
The SSB mutants retain the ability to bind DNA, PriA, and RecG:
SSB has been shown to interact with several proteins associated with DNA repair and the rescue of replication forks stalled on the template DNA, including RecG and PriA (Curth et al. 1996; Cadman and McGlynn 2004; Lecointe et al. 2007; Shereda et al. 2007; Buss et al. 2008). Given priA mutations that reduce or eliminate PriA helicase activity suppress recG, it is particularly significant that SSB not only binds PriA but also stimulates its helicase activity (Cadman and McGlynn 2004). It raises the possibility that the N14D and K44E mutations eliminate the SSB/PriA interaction. We purified wild-type and mutant SSB proteins to test this directly.
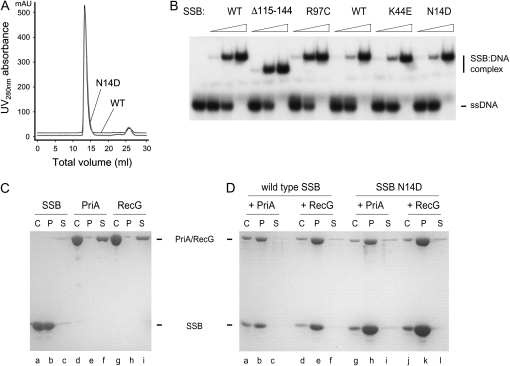
Gel filtration revealed that both mutant proteins migrate with the same molecular mass as wild-type SSB, which forms a stable tetramer in solution (Figure 6A and data not shown). Band-shift assays revealed that they also retain the ability to bind ssDNA, as do the R97C and Δ115-114 proteins (Figure 6B). This is not surprising given that strains expressing these proteins are viable. SSB is an essential protein, being required to bind the unwound lagging strand template during replication of the chromosome.
Figure 6.—
Properties of SSB suppressor proteins. (A) Gel filtration of SSB wild-type and N14D proteins. (B) ssDNA binding activity. Reactions contained 0.2 nm labeled oligonucleotide and the indicated proteins at final concentrations of 0.5, 5, and 50 nm. (C) and (D) SDS–PAGE analysis showing coprecipitation of PriA and RecG with SSB wild-type and N14D proteins, as indicated. C, no ammonium sulfate control; P and S, pellet and supernatant fractions respectively after ammonium sulfate precipitation.
To examine the ability to interact with PriA, we exploited the insolubility of SSB in low concentrations of ammonium sulfate that do not precipitate PriA (Figure 6C, lanes a–f) (Shereda et al. 2007). When the same low level of ammonium sulfate is added to an equimolar mixture of PriA and wild-type SSB (tetramer), most of the PriA is precipitated along with the SSB, with little of either protein remaining in solution (Figure 6D, lanes a–c). Similar coprecipitation was observed when wild-type SSB was replaced with either the N14D or K44E mutant (Figure 6D, lanes g–i; data not shown). These studies also confirmed that SSB interacts with RecG, as reported (Buss et al. 2008; Lecointe et al. 2007), and demonstrated that the mutants retain this property (Figure 6C, lanes g–i; Figure 6D, lanes d–f and j–l).
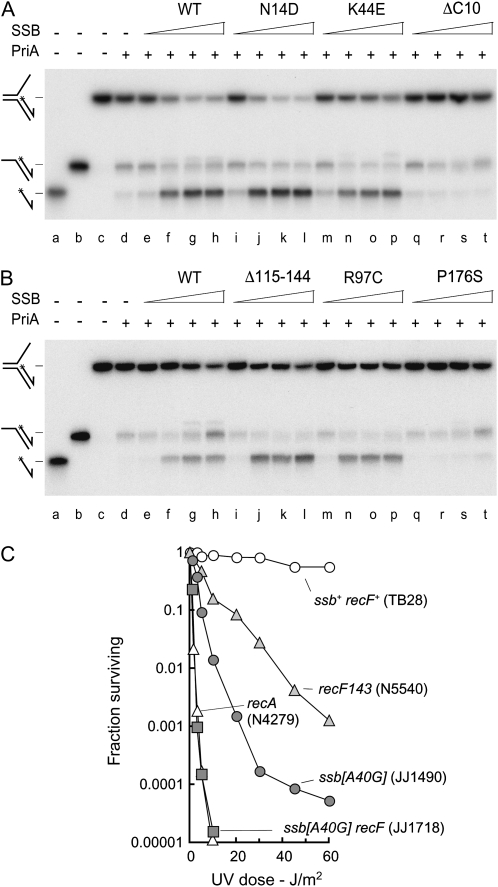
Next, we examined the ability of the mutant SSB proteins to stimulate the 3′–5′ DNA helicase activity of PriA. McGlynn and co-workers demonstrated that wild-type SSB stimulates PriA to unwind the lagging strand at a fork lacking a leading strand (Cadman and McGlynn 2004). Furthermore, they showed that this activity is eliminated by a deletion removing the last 10 residues from the C terminus of SSB or by a P176S substitution of the penultimate residue, consistent with the idea that the extended C-terminal projection is associated specifically with protein–protein interactions (Shereda et al. 2008). We recapitulated these observations using the same fork (Figure 7, A and B, lanes e–h and q–t). We also found that the N14D, K44E, R97C, and Δ115-144 mutants retain the ability to stimulate PriA (Figure 7, A and B, lanes i–p).
Figure 7.—
Effect of SSB proteins on the helicase activity of PriA and effect of SSB N14D on UV repair in recF cells. (A and B) Unwinding of a DNA fork by PriA in the presence of SSB. (Lanes a–c) Labeled oligonucleotide, DNA partial duplex, and fork markers. (Lane d) A total of 0.2 nm labeled fork DNA plus 5 nm PriA. (Lanes e–t) A total of 0.2 nm labeled fork DNA, 5 nm PriA, and the indicated SSB protein at 0.2, 2, 20, or 200 nm. (C) Effect of ssb[A40G] on the UV sensitivity of a recF strain. The strains used are as identified.
SSB protein has a very high affinity for ssDNA and therefore rapidly sequesters any single strands exposed during chromosome replication or repair, preventing loading of RecA and thus establishing a primary defense against unnecessary recombination. When ssDNA is exposed, and recombination called for, the RecFOR proteins are recruited to help load RecA, displacing SSB and stabilizing the RecA nucleoprotein filament (Umezu et al. 1993; Morimatsu and Kowalczykowski 2003; Cox 2007a,b). Given the UV sensitivity of cells expressing N14D or K44E derivatives of SSB, we considered the possibility these two proteins might modify one or more interactions needed by RecFOR to load RecA and maintain a stable nucleoprotein filament. Figure 7C shows that an ssb[A40G] strain expressing the N14D mutant is substantially more sensitive to UV than a recF mutant. The double mutant is exceedingly sensitive, essentially as sensitive as a recA null strain at higher doses (Figure 7C), indicating perhaps that the combination of these two mutations effectively blocks RecA-dependent reactions. A similar synergism was observed between recF and the ssb[A130G] mutation expressing the K44E derivative of SSB (data not shown). Thus it is possible these two mutant SSB proteins are more resistant to displacement by RecA. However, the fact that SSB interacts with so many proteins means we cannot exclude the possibility of some other defect.
DISCUSSION
The accumulation of unresolved recombination intermediates has been offered previously as an explanation for the inviability or poor viability of polA, uvrD, and dam cells lacking the RuvABC Holliday junction resolvase. However, given the reported redundancy between RecG and RuvABC (Lloyd 1991), it raised the question of why the postulated RecG pathway is unable to cope. We exploited a synthetic lethality assay to address this question, comparing the relative viabilities of polA, uvrD, and dam cells lacking either RuvABC or RecG and examining the ability of the RusA resolvase and PriA helicase deficiency to overcome any reduction in viability. During this work, we also identified mutations in ssb that suppress the recG phenotype, but not that associated with ruv mutations. The main findings are summarized in Table 1.
TABLE 1.
Viability of uvrD, dam, and polA strains lacking RuvABC or RecGa
|
uvrD, dam, polA, recG genotype |
||||||||
|---|---|---|---|---|---|---|---|---|
| Other genotype |
uvrD+ dam+ polA+ |
uvrD+ dam+ polA− |
uvrD+ dam− polA+ |
uvrD− dam+ polA+ |
||||
| recG+ | ΔrecG | recG+ | ΔrecG | recG+ | ΔrecG | recG+ | ΔrecG | |
| None | + | + | + | − | + | * | + | * |
| ΔruvABC | + | + | − | − | − | − | * | − |
| ΔruvABC rus-2 | + | + | + | − | + | − | + | − |
| rus-2 | + | + | + | − | + | * | + | * |
| priA300 | + | + | + | + | + | + | + | + |
| priA300 ΔruvABC | + | + | − | − | − | − | − | − |
| priA300 ΔruvABC rus-2 | + | + | + | −b | + | − | + | − |
| ssb[A40G]c | + | + | ND | + | + | + | ND | + |
| ssb[A40G] Δruv(AB)C | + | + | −d | − | −d | − | * | − |
+, plasmid-free segregants account for >20% of the colonies observed under the conditions employed and form healthy colonies that can be subcultured without difficulty; −, colonies of plasmid-free segregants not detected or form <0.2% of the colonies observed; *, plasmid-free segregants form small or tiny colonies that tend to accumulate suppressors that allow formation of larger colonies on subculture.
As determined using a synthetic lethality assay based on ability of plasmid-free (Lac−) segregants to form colonies on LB agar.
Very tiny colonies of plasmid-free cells detected, but these could not be subcultured, consistent with the idea that RusA needs the presence of RecG to function efficiently.
Identical results were recorded using ssb[A130G].
Only ΔruvC tested.
The synthetic lethality assays revealed that the RusA resolvase confers robust viability on polA, uvrD, and dam cells lacking RuvABC, provided RecG is available (Table 1; Figure 2). Given the reported high specificity of RusA for Holliday junctions (Bolt and Lloyd 2002), this establishes that unresolved junctions are indeed the reason for the inviability, as suspected. However, it also demonstrates very clearly that if the postulated RecG pathway involves some nuclease that acts to resolve Holliday junctions by junction cleavage, then this nuclease must operate very inefficiently compared with RusA. Indeed, it strongly suggests that such a nuclease does not exist.
Evidence against the existence of such a nuclease emerged from our analysis of how the viability of polA, uvrD, and dam cells is compromised without RecG (Table 1). Ishioka et al. (1997) proposed that the inviability of recG polA cells is due to the accumulation of unresolved recombination intermediates and a consequent failure of chromosome segregation. A similar argument was put forward to explain the failure to construct a recG dam double mutant (Marinus 2000). However, it is not obvious why RuvABC alone should not suffice to maintain viability in these cases. Our studies with constructs carrying priA300 proved very informative. They revealed that RuvABC alone is sufficient, provided the helicase activity of PriA is reduced or eliminated (Table 1; Figure 3). Thus, recG cells may simply lack means to curb a potentially deleterious effect of PriA helicase activity rather than being partially defective in Holliday junction resolution. Once PriA helicase activity is eliminated, RecG is quite dispensable as far as the viability of polA, uvrD, and dam cells is concerned. RuvABC is then able to cope very well despite the fact that recombination is known to be increased in these mutants (Arthur and Lloyd 1980; Kuzminov 1995; Marinus 2000).
A clue as to why PriA helicase activity might be so detrimental in the absence of RecG emerged from comparisons of how RecA, RecFOR, and RecQ affect the viability of uvrD ruv, uvrD recG, and uvrD ruv recG cells (Figure 4). Our studies confirmed previous reports demonstrating that the absence of UvrD provokes RecA-mediated recombination via a mechanism that could be countered by eliminating RecFOR, RecJ, or RecQ (Flores et al. 2005; Magner et al. 2007). They revealed in addition that the absence of RecG might also provoke recombination, but for a very different reason as this effect could not be countered efficiently in a recG ruv uvrD genetic background by removing RecFOR or RecQ. However, this conclusion requires the assumption that the viability of a strain lacking RuvABC depends on the incidence of recombination mediated by RecA. Since we did not measure recombination directly, we cannot exclude alternative explanations.
Evidence of increased recombination in recG cells was reported previously during assays of DNA double strand break repair and of the frequency of exchanges between tandem duplications (Lovett et al. 1993; Grove et al. 2008). The increased recombination evident from this study is clearly triggered independently of RecQ and RecFOR, indicating that it most probably has little to do with replication fork stalling and exposure of ssDNA as described in Figure 1C. It could be accounted for instead by an increase in RecBCD-mediated loading of RecA at dsDNA ends. As with recombination initiated at ssDNA gaps, UvrD would be expected to limit such exchanges by dissociating the RecA nucleoprotein filaments assembled by RecBCD (Dillingham and Kowalczykowski 2008). However, this begs the question of why the incidence of dsDNA ends should be increased in the absence of RecG.
The ability of RecG to catalyze DNA branch migration is well documented (Lloyd and Sharples 1993; Whitby et al. 1993; Whitby and Lloyd 1995; McGlynn and Lloyd 2001), as is its ability to facilitate the recovery of recombinants in strains lacking RuvABC (Lloyd 1991; Ryder et al. 1994; Mahdi et al. 1996). It has been suggested that RecG also promotes replication of damaged DNA (McGlynn and Lloyd 2000, 2002a,b), but this has proven more contentious (Donaldson et al. 2004; Michel et al. 2007). What is not in doubt is the ability of RecG to limit the incidence of DnaA-independent SDR, which is mediated via PriA-dependent loading of DnaB and subsequent replisome assembly at D loops and R loops (Asai and Kogoma 1994a,b; Masai et al. 1994; Hong et al. 1995). Without RecG to dissociate these branched structures (Vincent et al. 1996; Fukuoh et al. 1997), replication may initiate wherever they arise, rather than being restricted to DnaA-dependent events at oriC. Such initiations may set up replication forks that travel toward oriC, breaking the replichore arrangement that otherwise directs fork movement from oriC toward the terminus (Reyes-Lamothe et al. 2008). This might result in more frequent head-on collisions with RNA polymerase complexes (Rudolph et al. 2007), increasing the likelihood of fork breakage and recombination and causing additional difficulties for polA, uvrD, or dam cells. It would also be expected to increase collisions between replication forks as the new forks meet those coming from oriC (Rudolph et al. 2009b). The replichore arrangement normally restricts fork encounters to a single event within the terminus area each cycle of cell growth and division. An increase in the number of encounters may cause even more difficulties, especially if they take place outside of the control normally exerted by the Tus terminator protein, which appears to limit rereplication (Hiasa and Marians 1994; Krabbe et al. 1997; Markovitz 2005).
The ability of priA300 to restore viability to polA, uvrD, and dam cells lacking RecG is consistent with this idea. Although helicase-deficient PriA proteins retain the ability to assemble a primosome and to complement the DNA repair and growth defects associated with a priA null allele (Zavitz and Marians 1992), they reduce SDR quite substantially (Tanaka et al. 2003), a fact consistent with genetic data indicating that the K230R derivative may not be able to initiate replication at D loops (Mahdi et al. 2006). In doing so, we believe they reduce the chromosome pathology arising as a consequence of unscheduled DNA replication (Rudolph et al. 2009a,b).
A reduction in SDR may also account for the ssb suppressors of recG we identified during this work. Replication is initiated at a D loop, and presumably at an R loop, via the action of PriA, which recognizes and binds to a DNA branch point (McGlynn et al. 1997; Liu and Marians 1999; Nurse et al. 1999). This leads to the recruitment of a primosome complex composed of PriB, DnaT, DnaB, and DnaG, and finally the assembly of a DNA polymerase III holoenzyme complex that initiates replication in an SSB-dependent manner (Liu and Marians 1999; Liu et al. 1999). Strains expressing the mutant SSB proteins we identified as suppressors of recG grow well, indicating that normal DNA replication is impaired very little, if at all. Furthermore, the mutant proteins bind ssDNA, interact with PriA, and stimulate its ability to unwind the lagging strand at a fork (Figures 6 and 7). At first sight, this last property would seem to exclude a simple explanation for their ability to suppress recG, namely that they lack the ability to stimulate a potentially toxic helicase activity of PriA. However, the most effective ssb mutations confer sensitivity to UV light and in a manner that is synergistic with a recF mutation (Figure 7C). This could be explained if the mutant SSB proteins reduce the efficiency of PriA-mediated replisome assembly. If true, it might be expected to reduce the efficiency of replication restart and of inducible SDR after UV irradiation. It might also reduce the basal level of SDR in unirradiated recG cells. This leads to the idea that it is perhaps some downstream consequence of PriA-mediated unscheduled replication (SDR) that is toxic for recG cells, not the helicase activity of PriA per se.
Could increased chromosome pathology associated with SDR explain the much-reduced recovery of recombinants from conjugational and transductional crosses with ruv recG double mutants? The idea that E. coli has two partially overlapping pathways for the resolution of Holliday junctions, one dependent on RuvABC and the other on RecG, came from studies demonstrating that although ruv and single recG mutants are reasonably proficient in conjugational recombination, the double mutant is quite deficient (Lloyd 1991). The two new replication forks postulated to be established as each end of the linear donor DNA invades the recipient chromosome (Smith 1991) would likely exacerbate existing difficulties with replication caused by the absence of RecG and may trigger sufficient pathology for the zygotic cell to be no longer viable without RuvABC. The fact that inactivation of RecFOR, which reduces SDR (Kogoma 1997; Rudolph et al. 2008), improves the recovery of recombinants by ∼10- to 15-fold, lends support to this argument (Ryder et al. 1994).
Pathological replication might also provide an explanation for the strong synergism between ruv and recG mutations with respect to radiation sensitivity and DNA double strand break repair (Lloyd 1991; Meddows et al. 2004; Grove et al. 2008). However, Grove et al. (2008) reported that the RecG-dependent repair pathway produces both crossover and noncrossover products, and in the same proportions as the RuvABC-dependent pathway, consistent with Holliday junction cleavage in either of the two possible orientations. If we accept there is no nuclease that acts with RecG to cleave junctions, then whatever is responsible for the productive repair (recombination) seen without RuvABC would also have to account for the generation of these two alternative resolution products.
Although the results presented argue against a nuclease that could act with RecG to resolve Holliday junctions, we do not rule out the existence of a RecG resolution pathway. Indeed, while the yield of recombinants observed in crosses with ruv recG strains lacking RecFOR is much increased, it is still ∼20-fold lower than with the equivalent recG+ construct (Ryder et al. 1994), indicating that RecG may play a part in promoting recombination, at least in the absence of RuvABC. The dsDNA translocase activity of RecG might be sufficient in this case to eliminate a junction, perhaps by driving it to merge with replication forks, as has been suggested (Wardrope et al. 2009). However, such activity may fail to cope when the incidence of Holliday junctions is higher, as for instance after UV irradiation or in mutant strains in which DNA repair is compromised and where recombination is therefore elevated. This may not matter in wild-type cells as RuvABC has clearly evolved to resolve junctions with high efficiency. As suggested here, and elsewhere (Rudolph et al. 2009a), RecG may have a more basic housekeeping role to limit initiation of replication by PriA, thus reducing the incidence of pathological events that might result from such replication.
Acknowledgments
We thank those who provided E. coli strains and plasmids and Tim Moore for constructing pDIM024 and pDIM025. We are particularly grateful to Carol Buckman and Lynda Harris for excellent technical support. We also thank Christian Rudolph and Amy Upton for stimulating discussions on the possible functions of RecG. This work was supported by the Medical Research Council.
Note added in proof: Fonville et al. [N. C. Fonville, M. D. Blankschien, D. B. Magner and S. M. Rosenberg 2010 RecQ-dependent death-by-recombination in cells lacking RecG and UvrD. DNA Repair (in press; doi:10.1016/j.dnarep.2009.12.019)] reported that recG uvrD is inviable, contrary to our results (Figure 3C). We find that transduction of strain MG1655 ΔrecG∷apra to Tcr with P1 phage from a metE∷Tn10 uvrD1∷kan donor produces ∼17% small Kmr (i.e., recG uvrD) colonies that can be subcultured on LB agar. The cotransduction frequency was 16% with MG1655 recG+ as a recipient. We do not know the basis of the difference between our observations and those of Fonville et al. (2010).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.114413/DC1.
References
- Al-Deib, A. A., A. A. Mahdi and R. G. Lloyd, 1996. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J. Bacteriol. 178 6782–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur, H. M., and R. G. Lloyd, 1980. Hyper-recombination in uvrD mutants of Escherichia coli K-12. Mol. Gen. Genet. 180 185–191. [DOI] [PubMed] [Google Scholar]
- Asai, T., and T. Kogoma, 1994. a D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J. Bacteriol. 176 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai, T., and T. Kogoma, 1994. b Roles of ruvA, ruvC and recG gene functions in normal and DNA damage-inducible replication of the Escherichia coli chromosome. Genetics 137 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, T. G., and P. A. de Boer, 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt, E. L., and R. G. Lloyd, 2002. Substrate specificity of RusA resolvase reveals the DNA structures targeted by RuvAB and RecG in vivo. Mol. Cell 10 187–198. [DOI] [PubMed] [Google Scholar]
- Buss, J. A., Y. Kimura and P. R. Bianco, 2008. RecG interacts directly with SSB: implications for stalled replication fork regression. Nucleic Acids Res. 36 7029–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman, C. J., and P. McGlynn, 2004. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 32 6378–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, M. M., 2007. a Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell. Biol. 8 127–138. [DOI] [PubMed] [Google Scholar]
- Cox, M. M., 2007. b Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 42 41–63. [DOI] [PubMed] [Google Scholar]
- Curth, U., J. Genschel, C. Urbanke and J. Greipel, 1996. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 24 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko, K. A., and B. L. Wanner, 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham, M. S., and S. C. Kowalczykowski, 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72 642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, J. R., C. T. Courcelle and J. Courcelle, 2004. RuvAB and RecG are not essential for the recovery of DNA synthesis following UV-induced DNA damage in Escherichia coli. Genetics 166 1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores, M. J., N. Sanchez and B. Michel, 2005. A fork-clearing role for UvrD. Mol. Microbiol. 57 1664–1675. [DOI] [PubMed] [Google Scholar]
- Fukuoh, A., H. Iwasaki, K. Ishioka and H. Shinagawa, 1997. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 16 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg, A. V., P. McGlynn, R. P. Jaktaji and R. G. Lloyd, 2002. Direct rescue of stalled DNA replication forks via the combined action of PriA and RecG helicase activities. Mol. Cell 9 241–251. [DOI] [PubMed] [Google Scholar]
- Grove, J. I., L. Harris, C. Buckman and R. G. Lloyd, 2008. DNA double strand break repair and crossing over mediated by RuvABC resolvase and RecG translocase. DNA Repair 7 1517–1530. [DOI] [PubMed] [Google Scholar]
- Hiasa, H., and K. J. Marians, 1994. Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 269 26959–26968. [PubMed] [Google Scholar]
- Hong, X., G. W. Cadell and T. Kogoma, 1995. Escherichia coli RecG and RecA proteins in R-loop formation. EMBO J. 14 2385–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishioka, K., H. Iwasaki and H. Shinagawa, 1997. Roles of the recG gene product of Escherichia coli in recombination repair: effects of the delta recG mutation on cell division and chromosome partition. Genes Genet. Syst. 72 91–99. [DOI] [PubMed] [Google Scholar]
- Ishioka, K., A. Fukuoh, H. Iwasaki, A. Nakata and H. Shinagawa, 1998. Abortive recombination in Escherichia coli ruv mutants blocks chromosome partitioning. Genes Cells 3 209–220. [DOI] [PubMed] [Google Scholar]
- Jaktaji, R. P., and R. G. Lloyd, 2003. PriA supports two distinct pathways for replication restart in UV-irradiated Escherichia coli cells. Mol. Microbiol. 47 1091–1100. [DOI] [PubMed] [Google Scholar]
- Kogoma, T., 1997. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski, S. C., 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25 156–165. [DOI] [PubMed] [Google Scholar]
- Krabbe, M., J. Zabielski, R. Bernander and K. Nordstrom, 1997. Inactivation of the replication-termination system affects the replication mode and causes unstable maintenance of plasmid R1. Mol. Microbiol. 24 723–735. [DOI] [PubMed] [Google Scholar]
- Kuzminov, A., 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16 373–384. [DOI] [PubMed] [Google Scholar]
- Lecointe, F., C. Serena, M. Velten, A. Costes, S. McGovern et al., 2007. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 26 4239–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestini, R., and B. Michel, 2007. UvrD controls the access of recombination proteins to blocked replication forks. EMBO J. 26 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and K. J. Marians, 1999. PriA-directed assembly of a primosome on D loop DNA. J. Biol. Chem. 274 25033–25041. [DOI] [PubMed] [Google Scholar]
- Liu, J., L. Xu, S. J. Sandler and K. J. Marians, 1999. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc. Natl. Acad. Sci. USA 96 3552–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. G., 1991. Conjugational recombination in resolvase-deficient ruvC mutants of Escherichia coli K-12 depends on recG. J. Bacteriol. 173 5414–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. G., and G. J. Sharples, 1993. Dissociation of synthetic Holliday junctions by E. coli RecG protein. EMBO J. 12 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R. G., K. B. Low, G. N. Godson and E. A. Birge, 1974. Isolation and characterization of an Escherichia coli K-12 mutant with a temperature-sensitive RecA− phenotype. J. Bacteriol. 120 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett, S. T., P. T. Drapkin, V. A. Sutera, Jr. and T. J. Gluckman-Peskind, 1993. A sister-strand exchange mechanism for recA-independent deletion of repeated DNA sequences in Escherichia coli. Genetics 135 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magner, D. B., M. D. Blankschien, J. A. Lee, J. M. Pennington, J. R. Lupski et al., 2007. RecQ promotes toxic recombination in cells lacking recombination intermediate-removal proteins. Mol. Cell 26 273–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi, A. A., G. J. Sharples, T. N. Mandal and R. G. Lloyd, 1996. Holliday junction resolvases encoded by homologous rusA genes in Escherichia coli K-12 and phage 82. J. Mol. Biol. 257 561–573. [DOI] [PubMed] [Google Scholar]
- Mahdi, A. A., G. S. Briggs, G. J. Sharples, Q. Wen and R. G. Lloyd, 2003. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 22 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdi, A. A., C. Buckman, L. Harris and R. G. Lloyd, 2006. Rep and PriA helicase activities prevent RecA from provoking unnecessary recombination during replication fork repair. Genes Dev. 20 2135–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, T. N., A. A. Mahdi, G. J. Sharples and R. G. Lloyd, 1993. Resolution of Holliday intermediates in recombination and DNA repair: indirect suppression of ruvA, ruvB and ruvC mutations. J. Bacteriol. 175 4325–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus, M. G., 2000. Recombination is essential for viability of an Escherichia coli dam (DNA adenine methyltransferase) mutant. J. Bacteriol. 182 463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovitz, A., 2005. A new in vivo termination function for DNA polymerase I of Escherichia coli K12. Mol. Microbiol. 55 1867–1882. [DOI] [PubMed] [Google Scholar]
- Masai, H., T. Asai, Y. Kubota, K. Arai and T. Kogoma, 1994. Escherichia coli PriA protein is essential for inducible and constitutive stable DNA replication. EMBO J. 13 5338–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson, S. W., and K. A. Kaiser-Rogers, 1990. DNA helicases. Annu. Rev. Biochem. 59 289–329. [DOI] [PubMed] [Google Scholar]
- Matsumoto, T., Y. Morimoto, N. Shibata, T. Kinebuchi, N. Shimamoto et al., 2000. Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis. J. Biochem. 127 329–335. [DOI] [PubMed] [Google Scholar]
- McCool, J. D., and S. J. Sandler, 2001. Effects of mutations involving cell division, recombination, and chromosome dimer resolution on a priA2:kan mutant. Proc. Natl. Acad. Sci. USA 98 8203–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn, P., A. A. Al-Deib, J. Liu, K. J. Marians and R. G. Lloyd, 1997. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J. Mol. Biol. 270 212–221. [DOI] [PubMed] [Google Scholar]
- McGlynn, P., and R. G. Lloyd, 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101 35–45. [DOI] [PubMed] [Google Scholar]
- McGlynn, P., and R. G. Lloyd, 2001. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Nat. Acad. Sci. USA 98 8227–8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn, P., and R. G. Lloyd, 2002. a Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18 413–419. [DOI] [PubMed] [Google Scholar]
- McGlynn, P., and R. G. Lloyd, 2002. b Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 3 859–870. [DOI] [PubMed] [Google Scholar]
- Meddows, T. R., A. P. Savory and R. G. Lloyd, 2004. RecG helicase promotes DNA double-strand break repair. Mol. Microbiol. 52 119–132. [DOI] [PubMed] [Google Scholar]
- Mendonca, V. M., and S. W. Matson, 1995. Genetic analysis of ΔhelD and ΔuvrD mutations in combination with other genes in the RecF recombination pathway in Escherichia coli: suppression of a ruvB mutation by a uvrD deletion. Genetics 141 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, B., H. Boubakri, Z. Baharoglu, M. LeMasson and R. Lestini, 2007. Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst.) 6 967–980. [DOI] [PubMed] [Google Scholar]
- Modrich, P., 1991. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25 229–253. [DOI] [PubMed] [Google Scholar]
- Moolenaar, G. F., C. Moorman and N. Goosen, 2000. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J. Bacteriol. 182 5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T., P. McGlynn, H. P. Ngo, G. J. Sharples and R. G. Lloyd, 2003. The RdgC protein of Escherichia coli binds DNA and counters a toxic effect of RecFOR in strains lacking the replication restart protein PriA. EMBO J. 22 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu, K., and S. C. Kowalczykowski, 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11 1337–1347. [DOI] [PubMed] [Google Scholar]
- Nurse, P., J. Liu and K. J. Marians, 1999. Two modes of PriA binding to DNA. J. Biol. Chem. 274 25026–25032. [DOI] [PubMed] [Google Scholar]
- Raghunathan, S., A. G. Kozlov, T. M. Lohman and G. Waksman, 2000. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 7 648–652. [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe, R., X. Wang and D. Sherratt, 2008. Escherichia coli and its chromosome. Trends Microbiol. 16 238–245. [DOI] [PubMed] [Google Scholar]
- Rudolph, C. J., P. Dhillon, T. Moore and R. G. Lloyd, 2007. Avoiding and resolving conflicts between DNA replication and transcription. DNA Repair 6 981–993. [DOI] [PubMed] [Google Scholar]
- Rudolph, C. J., A. L. Upton and R. G. Lloyd, 2008. Maintaining replication fork integrity in UV-irradiated Escherichia coli cells. DNA Repair (Amst.) 7 1589–1602. [DOI] [PubMed] [Google Scholar]
- Rudolph, C. J., A. L. Upton, L. Harris and R. G. Lloyd, 2009. a Pathological replication in cells lacking RecG DNA translocase. Mol. Microbiol. 73 352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, C. J., A. L. Upton and R. G. Lloyd, 2009. b Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol. Microbiol. 74 940–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder, L., M. C. Whitby and R. G. Lloyd, 1994. Mutation of recF, recJ, recO, recQ, or recR improves Hfr recombination in resolvase-deficient ruv recG strains of Escherichia coli. J. Bacteriol. 176 1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides, S. N., S. Raghunathan, K. Futterer, A. G. Kozlov, T. M. Lohman et al., 2004. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 13 1942–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples, G. J., S. C. Chan, A. A. Mahdi, M. C. Whitby and R. G. Lloyd, 1994. Processing of intermediates in recombination and DNA repair: identification of a new endonuclease that specifically cleaves Holliday junctions. EMBO J. 13 6133–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereda, R. D., D. A. Bernstein and J. L. Keck, 2007. A central role for SSB in Escherichia coli RecQ DNA helicase function. J. Biol. Chem. 282 19247–19258. [DOI] [PubMed] [Google Scholar]
- Shereda, R. D., A. G. Kozlov, T. M. Lohman, M. M. Cox and J. L. Keck, 2008. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43 289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, M. R., S. Scaife and D. B. Wigley, 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 107 79–89. [DOI] [PubMed] [Google Scholar]
- Singleton, M. R., M. S. Dillingham, M. Gaudier, S. C. Kowalczykowski and D. B. Wigley, 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432 187–193. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., 1991. Conjugational recombination in E. coli: myths and mechanisms. Cell 64 19–27. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., C. Taniyama, K. Arai and H. Masai, 2003. ATPase/helicase motif mutants of Escherichia coli PriA protein essential for recombination-dependent DNA replication. Genes Cells 8 251–261. [DOI] [PubMed] [Google Scholar]
- Trautinger, B. W., R. P. Jaktaji, E. Rusakova and R. G. Lloyd, 2005. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell 19 247–258. [DOI] [PubMed] [Google Scholar]
- Umezu, K., N. Chi and R. D. Kolodner, 1993. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl. Acad. Sci. USA 90 3875–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gool, A. J., R. Shah, C. Mezard and S. C. West, 1998. Functional interactions between the Holliday junction resolvase and the branch migration motor of Escherichia coli. EMBO J. 17 1838–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute, X., S. Delmas, M. Selva, J. Jeusset, E. Le Cam et al., 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, S. D., A. A. Mahdi and R. G. Lloyd, 1996. The RecG branch migration protein of Escherichia coli dissociates R-loops. J. Mol. Biol. 264 713–721. [DOI] [PubMed] [Google Scholar]
- Wardrope, L., E. Okely and D. Leach, 2009. Resolution of joint molecules by RuvABC and RecG following cleavage of the Escherichia coli chromosome by EcoKI. PLoS One 4 e6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, M. C., and R. G. Lloyd, 1995. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 14 3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby, M. C., L. Ryder and R. G. Lloyd, 1993. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75 341–350. [DOI] [PubMed] [Google Scholar]
- Zavitz, K. H., and K. J. Marians, 1992. ATPase-deficient mutants of the Escherichia coli DNA replication protein PriA are capable of catalyzing the assembly of active primosomes. J. Biol. Chem. 267 6933–6940. [PubMed] [Google Scholar]