Abstract
The waved with open eyes (woe) locus is a spontaneous recessive mouse mutation that exhibits wavy fur, eyelids open at birth, and enlarged heart and esophagus. In this study, we confirmed the previously identified woe phenotypes and additionally identified anterior eye segment defects, absence of the meibomian glands, and defects in the semilunar cardiac valves. Positional cloning identified a C794T substitution in the Adam17 gene that ablates a putative exonic splicing enhancer (ESE) sequence in exon 7 resulting in aberrant Adam17 splicing. The predominant woe transcript, Adam17Δexon7, lacks exon 7 resulting in an in-frame deletion of 90 bp and a putative Adam17Δ252-281 protein lacking residues 252–281 from the metalloprotease domain. Western blot analysis in woe identified only the precursor form of Adam17Δ252-281 protein. Absence of cleavage of the prodomain renders Adam17Δ252-281 functionally inactive; however, constitutive and stimulated shedding of Adam17 substrates was detected in woe at significantly reduced levels. This residual Adam17 shedding activity in woe most likely originates from full-length Adam17T265M encoded by the Adam17C794T transcript identified expressed at severely reduced levels. These results show that even small amounts of functional Adam17 allow woe mice to survive into adulthood. In contrast to Adam17−/− mice that die at birth, the viability of woe mice provides an excellent opportunity for studying the role of Adam17 throughout postnatal development and homeostasis.
THE waved with open eyes (woe) mouse was identified via a screen for mouse models of syntenic human ocular disorders (Chang et al. 2005). Initially the locus was named waved3 (wa3) (Chang et al. 2005) and has since been renamed woe. Phenotypic analysis of woe mice identified “eyelids open at birth” (EOB), wavy coat, and enlargements of the heart and esophagus. The woe mutation arose spontaneously on the C57BL/6 background and exhibits recessive inheritance; coarse linkage analysis assigned woe to mouse chromosome 12 (Chang et al. 2005).
The wavy coat observed in woe mice has been previously described in mice with altered epidermal growth factor receptor (Egfr) signaling (Schneider et al. 2008). Egfr belongs to a family of tyrosine kinase receptors. Following ligand binding to the extracellular domain, Egfr is dimerized and autophosphorylated, which subsequently induces an intracellular signaling cascade (Schneider et al. 2008). Mice carrying different mutations in Egfr such as wa2, wa5, and velvet, as well as mice carrying mutations in the transforming growth factor α (Tgfa) all exhibit wavy fur (Luetteke et al. 1993, 1994; Miettinen et al. 1995; Thaung et al. 2002; Fitch et al. 2003; Du et al. 2004). Tgfa is a member of the Egfr family of ligands that binds to Egfr (Harris et al. 2003). Egfr signaling has been implicated in the differentiation and maturation of the hair follicle (Hansen et al. 1997). Interestingly Egfr and Tgfa mouse mutants, in addition to the wavy coat phenotype, also demonstrate the EOB phenotype (Luetteke et al. 1993, 1994; Miettinen et al. 1995; Thaung et al. 2002; Fitch et al. 2003; Du et al. 2004). During mammalian embryonic eye development, at the tip of the newly formed eyelids, epithelial sheets extend over the cornea and move toward the center of the eye resulting in eyelid closure (Li et al. 2003). In mice with Egfr signaling defects there is a failure of the migration of the leading edges and consequent failure of embryonic eyelid closure. Egfr signaling has been established as one of the essential signaling pathways required for leading eyelid edge migration and the formation of actin stress fibers (Xia and Karin 2004).
Even though woe mice phenotypically resemble mice with mutations in Tgfa and Egfr, linkage assignment of the woe locus to chromosome 12 excluded the possibility that woe is allelic with either Tgfa on chromosome 6 or Egfr on chromosome 11. While initial mapping of woe demonstrated linkage to chromosome 12, the exact position within the chromosome was not determined (Chang et al. 2005). Therefore, as a part of this study, we positionally cloned and identified the mutation responsible for the woe phenotype. Our results show that woe is a mutation in Adam17, a member of a family of membrane-anchored metalloproteinase disintegrins that plays an essential role in the proteolytic cleavage and release of membrane bound precursors termed “shedding” (Blobel 2005). Adam17 has been implicated in the shedding of numerous proteins involved in multiple signaling mechanisms (Huovila et al. 2005; Mezyk et al. 2003). However, the primary role of Adam17 has been established as a mediator of Egfr signaling via shedding of membrane-bound precursor forms of epiregulin, Tgfa, amphiregulin, and Hbegf (Peschon et al. 1998; Sahin et al. 2004; Horiuchi et al. 2007). Functional analysis of the woe allele in this study provides evidence that woe is a hypomorphic mutation in Adam17. Since woe mice are viable, whereas null Adam17 mice die at birth, the woe mutation provides an excellent opportunity for studying the role of Adam17 in postnatal development and, specifically, has helped uncover a critical role for Adam17 in the development of the anterior segment of the eye and meibomian glands, most likely caused by defects in Adam17-dependent Egf-signaling.
MATERIALS AND METHODS
Mice:
C57BL/6, woe, and C3H/HeJ mice were obtained from Jackson Laboratories (Bar Harbor, ME). Adam17ΔZn/+ mice were obtained from Roy Black, Amgen (Seattle, WA). All mice exhibited normal breeding patterns. Progeny were genotyped utilizing PCR protocols (primer sequences are summarized in Table 1) as previously described (Talamas et al. 2006).
TABLE 1.
PCR primers used to amplify genomic and cDNA segments
| Target | Primer name | Primer sequence (5′ → 3′) | Annealing temp. |
|---|---|---|---|
| Adam17 | Adam17 1.2 F | CCCAATGTGAGCAGTTTCCCGAAC | 58° |
| Exon 1 | Adam17 1.2 R | CGCGTCCCTCCAATCACTCTGG | |
| Adam17 | Adam17 2.1 F | TGAGCAGCGAGTTAATGCTCTT | 58° |
| Exon 2 | Adam17 2.2 R | CATCAAGTACAAATCTTTACGA | |
| Adam17 | Adam17 3 F | TGGGTGGCTATTGTTCATCT | 58° |
| Exon 3 | Adam17 3 R | TTCCATCATTGCAGAAAGTG | |
| Adam17 | Adam17 4 F | GGTCAGCATGGGCTATAAGA | 58° |
| Exon 4 | Adam17 4 R | CCTGCTGATTCATTTTCCTG | |
| Adam17 | Adam17 5 F | TGTTTAGAAGGTGGTCTTGG | 60° |
| Exon 5 | Adam17 5 R | GCCAGAAACAGTATGATGTGC | |
| Adam17 | Adam17 6 F | AAAACCCTCCTTGTTTTGATG | 60° |
| Exon 6 | Adam16 6 R | AAATCTCTGCAGCCTTCTCTT | |
| Adam17 | Adam17 7 F | CCACTAGATGCCTATTCTAGTAGGTT | 60° |
| Exon 7 | Adam17 7 R | TCAGGAAGGTTTTGGAAGAA | |
| Adam17 | Adam17 8 F | TCAGTGTCTTAGGATGTTTTGG | 60° |
| Exon 8 | Adam17 8 R | AAGGTTTCTACCATGTTCTTTCA | |
| Adam17 | Adam17 9/10 F | GTAATGTTGAGGTGCCCTTG | 58° |
| Exon 9–10 | Adam17 9/10 R | ATGCTTATGTGCATGTGGTG | |
| Adam17 | Adam17 11 F | CCCAACTGGCAAAAATAACT | 62° |
| Exon 11 | Adam17 11 R | TTTCTGTTAACATGGCATCG | |
| Adam17 | Adam17 12 F | GACTGCTTGACACAGTGGTG | 58° |
| Exon 12 | Adam17 12 R | CTGCATTTGCAATCTCCTGT | |
| Adam17 | Adam17 13 F | AACCATAGCATTTGATGTTGG | 58° |
| Exon 13 | Adam17 13 R | CACCTGAAGGAAACTTGACC | |
| Adam17 | Adam17 14 F | GCTGGGTCCACATTTTTATG | 58° |
| Exon 14 | Adam17 14 R | CCCTATTTGGCTTCCTCACT | |
| Adam17 | Adam17 15 F | ATCTGGGAGTAAGGCCAAAG | 58° |
| Exon 15 | Adam17 15 R | AGACCAAGGCTGCTTAAGTG | |
| Adam17 | Adam17 16 F | TGGTTACTTGCCCATTGTTT | 58° |
| Exon 16 | Adam17 16 R | GTCATAACGGGAATCAGCTT | |
| Adam17 | Adam17 17 F | CCAGGAGACTCAGGAGAGGT | 62° |
| Exon 17 | Adam17 17 R | GAGAAGCTGTCTTGAACACTCA | |
| Adam17 | Adam17 18.2 F | GATGGCTGGGTTCCTTTAAAAT | 58° |
| Exon 18 | Adam17 18.3 R | CAGATCCTTTTAACTTCCACTAGCC | |
| Adam17 | Adam17 19 F | CATAGCCCAGGGTTACTGT | 58° |
| Exon 19 | Adam17 19 R | TCATTATAATCTATGTTTTGATTCAGG | |
| Adam17 | Adam17−/− F | CTCTGCAAGTGCAGTGACACTC | 67° |
| neo | Adam17−/− R | GGCCACTTGTGTAGCGCCAAGT | |
| Adam17 | Adam17+/+ F | TGCGTCATTCCACTCCAACC | 67° |
| Without neo | Adam17+/+ R | GCATCGCCTTCTTTCCTACCA | |
| Adam17 intron 5 | Adam17 intron 5–1 F | TTGCAGTCTCCAAAAGTATGTGG | 58° |
| Adam17 intron 5–1 R | TTTTTCCAGACAGGGTTTCTGTATATC | ||
| Adam17 intron 5–2 F | CGCAGGCATTTAACCAGCAC | ||
| Adam17 intron 5–2 R | CCAGGGTATTTGGAGGCATCTCT | ||
| Adam17 intron 5–3 F | CTCCATAAACACACTTTTGCTTG | ||
| Adam17 intron 5–3 R | AGTAAAAAGTAATATAAGGGGCTAGAGAG | ||
| Adam17 intron 5–4 F | CCATTGGTTTTGGATTCAGCAAT | ||
| Adam17 intron 5–4 R | TTCTCCACGGCCCATGTATTTAT | ||
| Adam17 intron 6 | Adam17 intron 6–1 F | ACGAGCTGAACCTAACCCCTTGA | 58° |
| Adam17 intron 6–1 R | GGAGAGAATGGAAAGACGGCAAT | ||
| Adam17 intron 6–2 F | TCCCTGGAATTGTTTTGTTTCTCA | ||
| Adam17 intron 6–2 R | GGGGAACACGTGTATGTATGTGC | ||
| Adam17 intron 6–3 F | TTCAAAGTGCATTTCTCACTGCTA | ||
| Adam17 intron 6–3R | ACCCTTTAAACCCTGCATTATCC | ||
| Adam17 intron 7 | Adam17 intron 7–1 F | CCGAGTTGATGACATATACCGGAAC | 58° |
| Adam17 intron 7–1 R | CCACTGAGGATCAGAAGGAGAGAAG | ||
| Adam17 intron 7–2 F | GGGGTTCTGATGGAATCCTACAGTT | ||
| Adam17 intron 7–2 R | AGTCCTTGGGGAAGGGGTTAGAG | ||
| Adam17 intron 7–3 F | CACTGAGGATGAATTGGAGGGTGT | ||
| Adam17 intron 7–3R | CGACAGTTCTTCCGAAGGTCCA | ||
| Adam17 intron 7–3.2F | CAGAATTAACCTTGACCTTCTGCTT | ||
| Adam17 intron 7–3.2R | CCGACAGTTCTTCCGAAGGT | ||
| Adam17 intron 7–4 F | CAAATGTGGAGGTTGTGAGAATATA | ||
| Adam17 intron 7–4 R | TGCAAGTTCATAACCTTCCATAAC | ||
| Adam17 intron 7–5 F | TTTGAGCTAAACAATTTCAGTTGC | ||
| Adam17 intron 7–5 R | TCTCTTCGTTTGGGAAACTTTTT | ||
| Adam17 cDNA | RTAdam17 F 5′-UTR 67 | CCCAATGTGAGCAGTTTCCC | 58° |
| RTAdam17 R568 | CCTTCGTCGAGAAGTGATAG | ||
| RTAdam17 F377 | GGGTTCTAGCCCACATAGGAGA | ||
| RTAdam17 R937 | CCCAAGCATCCTTCTCTTCGTTT | ||
| RTAdam17 F768 | CCGAGTTGATGACATATACCGGAA | ||
| RTAdam17 R1461 | ACTCCAGGGTGGATGAAGGAGAG | ||
| RTAdam17 F1305 | GTATCCCATAGCTGTGAGCG | ||
| RTAdam17 R2190 | AAAGGGCTTGATGATGCGAA | ||
| RTAdam17 F2015 | TCGTTGGGTCTGTTCTGGTTT | ||
| RTAdam17 3'-UTR 82 | TCATTATAATCTATGTTTTGAT | ||
| semi-quantitative RT–PCR | RT-Adam17 F377 | GGGTTCTAGCCCACATAGGAGA | 58° |
| RT-Adam17 R937 | CCCAAGCATCCTTCTCTTCGTTT | ||
| Gapdh | GAPDH F | CTTTGGCATTGTGGAAGGG | 58° |
| GAPDH R | CCTCTCTTGCTGCAGTGTC |
Histology:
For embryo analysis, embryonic day 0.5 (E0.5) was defined as the morning of the day that a vaginal plug was first observed. Collected tissues were fixed in 4% paraformaldehyde, Zn-formalin, or Davidson's solution (Miething et al. 2006), embedded in paraffin, serially sectioned to 4 μm, and stained with H&E. The sections were photographed with a DXM1200 camera (Nikon) on a BX50 microscope (Olympus) and Nikon DS-Fi1 camera on Nikon Eclipse 80i microscope.
Cell culture:
Primary mouse embryonic fibroblasts (mEFs) were generated from C57BL/6, woe, and Adam17ΔZn/ΔZn embryos at E13.5 and cultured as described previously (Weskamp et al. 2002).
Linkage mapping:
The woe locus is on the congenic C57BL/6 background. The woe/woe mice were outcrossed to C3H/HeJ and F1 mice were backcrossed to woe/woe to generate 138 progeny. At 3 weeks of age, F2 mice were scored for the wavy coat appearance/small eye phenotype, killed, and genotyped with microsatellite markers: D12Mit12, D12Mit147, D12Mit52, D12Mit263 as previously described (Talamas et al. 2006). The resulting linkage data were analyzed with Map Manager QTX (http://www.mapmanager.org/mmQTX.html).
Sequence analysis of Adam17:
Exon scanning primers were designed to anneal 50 bp from intron/exon boundaries (Table 1) and analyzed as described previously (Talamas et al. 2006). Comparative sequence analysis was performed using DNAStar software (Madison, WI). For cDNA evaluation RNA was isolated from woe mEFs, reverse transcribed, and amplified as previously described (Talamas et al. 2006) using primers in Table 1. The PCR products were electophoresed, gel extracted, and sequenced as previously described (Talamas et al. 2006). For semiquantitative analysis of Adam17 RT–PCR products were generated while in the exponential phase of amplification and Gapdh was used as an internal control (Table 1). PCR band intensities were quantified using ImageJ software (http://rsbweb.nih.gov/ij/) and are expressed relative to Gapdh. The results represent at least three independent experiments performed in triplicates. Comparison between wild-type and woe groups was analyzed by Student's t-test and data are expressed as mean ± SEM. A value of P < 0.05 was taken as statistically significant.
Western blot analysis:
Wild-type and woe mEFs were lysed for 15 min on ice in PBS containing 1% TritonX-100 10 mm phenantroline, 4 μm Marimastat, and 1× protease inhibitor cocktail. Following centrifugation at 15,000g for 10 min to remove nuclei and cell debris, glycoproteins from the cleared lysates were concentrated with Concanavalin A sepharose beads at 4° for 4 hr. Bound glycoproteins were eluted in Laemmli sample loading buffer with DTT for western blotting followed by alkylation with 20 mm iodoacetamide for 30 min at room temperature. Following electrophoresis on 9% SDS-polyacrylamide gels, the samples were transferred to nitrocellulose membranes (Biotrace, Pall Corporation, Port Washington, NY), blocked in 3% dry milk reconstituted in PBS 0.05% Tween-20 (PBS-T), and then incubated with an antiserum against the cytoplasmic domain of Adam17 (Schlondorff et al. 2000) in PBS-T for 1 hr at room temperature. The membrane was washed three times with PBS-T followed by incubation with horseradish peroxidase-conjugated affinity purified anti-rabbit IgG (H+L) (Promega, Madison, WI) for 45 min at room temperature. Bound antibodies were detected by enhanced chemiluminescence using an ECL detection reagent (GE Healthcare–Amersham Biosciences, Piscataway, NJ) and a Bio-Rad Geldoc (Bio-Rad, Hercules, CA) instrument.
Adam17 activity assay:
Endogenous Adam17 activity in wild-type, woe, and Adam17ΔZn/ΔZn mEF cell lysates was evaluated using the SensoLyte 520 TACE activity assay protocol (AnaSpec, San Jose, CA) following the manufacturer's protocol. A Michaelis–Menten curve was constructed for FRET–Adam17 peptide substrate at 0, 2.5, 5, 10, 20, 40, 80 μm concentrations following the manufacturer's protocol. Fluorescence intensity was monitored every 5 min over a period of 6 hr (excitation, 490 nm, 6-nm slit; emission, 520 nm, 6-nm slit) and measured using an LS55 Luminescence Spec (Perkin-Elmer, Waltham, MA). Samples were normalized to the FAM standard and protein concentration. Data from at least three independent experiments were compiled and the Km and Vmax values were calculated using GraphPad Prism software (La Jolla, CA). Comparison between groups was analyzed by Student's t-test and data are expressed as mean ± SEM.
Ectodomain shedding assay:
These assays were performed as previously described (Le Gall et al. 2009). Briefly, primary wild-type and woe mEFs were transiently transfected by GenJet transfection reagent (SignaGen, MD) with a vector containing Tgfa, tumor necrosis factor α (Tnfa), selectin (Cd62l), or betacellulin (Btc) with an alkaline phosphatase tag (AP-tag) (Sahin et al. 2004). For shedding experiments, transfected cells were washed with OptiMEM that was subsequently replaced after 1 hr with fresh OptiMEM with or without 25 ng/ml phorbol myristate acetate (PMA), 100 μm pervanadate (PV), or 2.5 μm ionomycin (IM). Both PMA and PV have been previously shown to stimulate Adam17-dependent shedding activity whereas iononmycin stimulates both Adam10 and Adam17 shedding activity (Le Gall et al. 2009). To evaluate stimulated and constitutive shedding, cells were incubated for 30 min and 6 hr, respectively. AP activity in the supernatant and the cell lysates was measured by colorimetry (Sahin et al. 2004). The ratio of total AP activity in the supernatant and total AP activity in the cell lysate and supernatant was calculated from two identically prepared wells and averaged with all experiments repeated at least three times. This ratio reflects the activity of Adam17 toward the AP-tagged proteins (Le Gall et al. 2009). Shedding in wild-type cells is set to 1 to allow a comparison of constitutive shedding levels between wild-type and woe mEFs. Comparison between wild-type and woe groups was analyzed by unpaired t-test and data are expressed as mean ± SEM. A value of P < 0.05 was taken as statistically significant.
RESULTS
The woe phenotype:
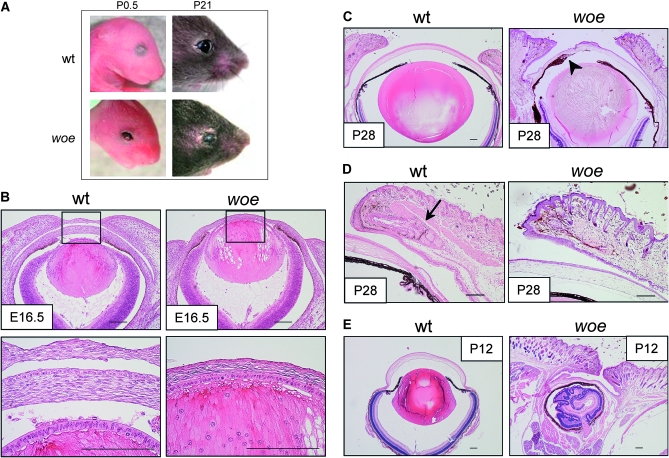
At birth, woe mice exhibit fully penetrant EOB, with eyelids remaining open throughout postnatal development (Figure 1A); slit-lamp examination revealed corneal opacities, corneal inflammation, and neovascularization. Histological analysis at E16.5 confirmed failure of eyelid fusion in woe eyes when compared to age-matched wild-type mice (Figure 1B). The woe cornea appeared much thinner with a less differentiated corneal epithelium, absent endothelial layer, and a vacuolated lens (Figure 1B) suggesting that woe eyes have defects in the development of anterior segment structures. Therefore, we proceeded to evaluate the woe ocular phenotypes at P28, following completion of anterior structure development (Smith 2002). The woe eyes exhibited corneal scarring, stromal kerititis, and neovascularization. Although the anterior chamber was formed, there were variable anterior synechiae, with variable absence of the Descemet's membrane and a small or absent Schlemm's canal. The lens showed cataracts, while the retina and the optic nerve remained normal. Histological analysis of P28 woe eyelid skin revealed evidence of chronic dermatitis with variable development of the hair follicles (Figure 1D). At the eyelid margin, there was excess pigment dispersal within the dermis and the complete absence of the meibomian glands (Figure 1D). We also examined the progression of the woe ocular phenotype. We did not identify any difference between woe eyes at P28 and at 12 months (not shown). Although the external woe evaluation identified microphthalmia in most woe mice, about 20% of woe mice exhibited unilateral or bilateral anophthalmia (not shown). Histological sections of the presumed anophthalmic eyes revealed aphakic eyes with highly disorganized retinas suggesting severe microphthalmia (Figure 1E).
Figure 1.—
Eye phenotypes in woe mice. (A) All woe mice are born with open eyelids that remain open throughout postnatal development (bottom row); woe eyes appear smaller compared to age-matched wild-type eyes (top row). (B) Histological analysis at E16.5 confirmed failure of embryonic eyelid closure in woe mice compared to the age-matched wild-type mice. Boxes indicate regions of higher magnification showing that in woe mice the corneal endothelium has not developed and the cornea is much thinner than wild-type mice. (C) At P28, woe eyes exhibit defects consistent with anterior segment dysgenesis. The arrowhead indicates the anterior synechiae observed in woe mice. (D) Histological analysis of woe eyelids shows that the meibomian glands (arrow) do not develop in woe eyelids. (E) Histological analysis of a subset of woe mice, which exhibit severe microphthalmia (right) consequent to aphakea. (Scale bars, 100 μm).
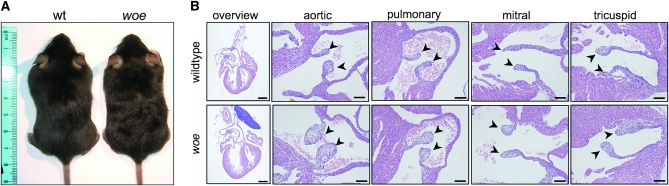
External evaluation of woe mice confirmed the wavy fur phenotype (Chang et al. 2005) (Figure 2A), although we did not observe any obvious defect of the vibrissae (not shown). Histological analysis of the skin identified variable development of the hair follicle without any abnormalities of the skin (not shown). Analysis of woe hearts at P0.5 did not consistently identify any significant size differences; however, the analysis identified hypertrophic and thickened pulmonary and aortic valves as compared to wild-type mice (Figure 2B). In contrast, the tricuspid and mitral valve thicknesses were within normal limits (Figure 2B). Evaluation of the esophagus did not identify any differences between woe and wild-type animals (not shown).
Figure 2.—
Fur and cardiac valve phenotypes in woe mice. (A) woe mice develop wavy fur. (B) Histological evaluation of the cardiac valves (arrowheads) of P0.5 wild-type and woe hearts identified thickening of the semilunar (aortic and pulmonary) valves in woe mice. Atrioventricular (mitral and tricuspid) valves in woe did not differ from wild-type controls. (Scale bars: overview images scale bars, 100 μm; valve images scale bars, 50 μm).
Positional cloning of the woe locus:
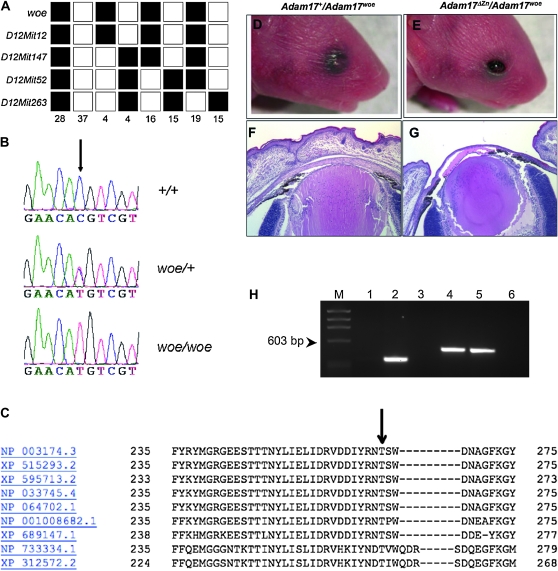
Our mapping analysis confirmed linkage to mouse chromosome 12 (Chang et al. 2005) and further mapped woe to a region of 4 cM between the centromere and D12Mit147. No recombinants were identified between woe and D12Mit12 (Figure 3A). The woe critical region encompassed 36.4 Mb containing ∼130 candidate genes. In parallel to our positional cloning efforts, we searched MGI phenotype database (http://www.informatics.jax.org/phenotypes.shtml) for genetically altered mice with a similar phenotype that may map within the woe critical region. In silico evaluation identified two Adam17−/− mouse models that exhibited developmental defects similar to woe mice including failure of eyelid development, hair, and heart defects (Peschon et al. 1998; Horiuchi et al. 2007). Thus, we proceeded to evaluate Adam17 as a candidate gene.
Figure 3.—
Genetic analysis of the woe locus. (A) Utilizing 138 meioses, the woe locus was mapped between the centromere of chromosome 12 and D12Mit147. The woe locus cosegregated with D12Mit12. (B) Sequence analysis of woe DNA identified a C794T substitution in Adam17. (C) The C794 substitution results in a T265M substitution. T265 (arrow) shows a high level of conservation between human (NP_003174.3), chimpanzee (XP_515293.2), cattle (XP_595713.2), mouse (NP_033745.4), rat (NP_064702.1), chicken (NP_001008682.1), zebrafish (XP_689147.1), fruit fly (NP_733334.1), and mosquito (XP_312572.2). (D–G) Complementation breeding of Adam17ΔZn/+ to woe mice resulted in progeny that exhibited the wild-type phenotype (D and F) and progeny that exhibited the open eyelid phenotype (E and G). (H) Genotyping of mice from the complementation breeding: lanes 1–3 depicts PCR products amplifying to the neo cassette and lanes 4–6 depict PCR products for exon 7 that were subsequently sequenced to confirm the presence of the woe allele. Lanes 1 and 4 contain genomic DNA from the mouse depicted in D and lanes 2 and 5 contain DNA from the mouse depicted in E. Lanes 3 and 6 are no template controls and M = ΦX174 DNA–HaeIII Digest. The results show that affected mice depicted in E and G are compound heterozygotes Adam17ΔZn/Adam17woe indicating failure of the Adam17ΔZn allele to complement the woe allele.
Sequence analysis identified a C794T substitution in exon 7 (Figure 3B) that leads to a T265M amino acid change in the metalloproteinase domain of Adam17. Threonine 265 is a highly evolutionarily conserved amino acid in Adam17 suggesting conservation of function (Figure 3C). Evaluation of the SNP database did not reveal any polymorphisms for Adam17 exon 7 (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=11491). These findings collectively suggested that Adam17C794T is likely not a common polymorphism. To exclude a possibility that Adam17C794T is a rare previously unidentified polymorphism and to unequivocally prove that the Adam17C794T is the woe-causing mutation, we utilized a genetic approach. Although Adam17ΔZn/ΔZn mice die at birth, Adam17ΔZn/+ mice are viable and phenotypically normal (Peschon et al. 1998). We set up breedings between Adam17ΔZn/+ and woe/woe mice. About 50% of F1 progeny appeared phenotypically normal (Figure 3D) and 50% progeny exhibited the EOB phenotype (Figure 3E) confirmed by histological analysis (Figure 3F and 3G). Genotyping revealed that unaffected progeny carried the Adam17 wild-type and woe alleles and the progeny exhibiting EOB were compound heterozygotes Adam17ΔZn/Adam17C794T indicating a failure to complement (Figure 2E). This finding provided genetic evidence that the C794T substitution in Adam17 is the woe phenotype causing mutation. The Adam17ΔZn/Adam17C794T compound heterozygotes survived into adulthood and exhibited similar phenotypes to woe mice (not shown).
Functional analysis of the woe mutation:
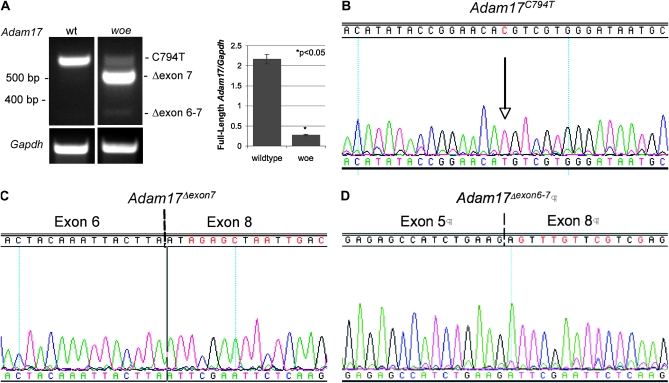
RT–PCR analysis of wild-type mEFs identified a single Adam17 transcript matching the Adam17 ORF sequence (NM_009615). In contrast, RT–PCR in woe mEFs identified three Adam17 transcripts: full-length Adam17C794T, Adam17Δexon7 lacking exon 7, and Adam17Δexons6-7 lacking exons 6 and 7 (Figure 4, A–D). To further evaluate the mechanism of aberrant Adam17 splicing in woe, we sequenced introns 5, 6, and 7. Sequence comparison between C57BL/6 and woe did not identify any large genomic deletions, insertions, or rearrangements in these three introns. The sequence differences between C57BL/6 and woe closest to the intron/exon junctions were identified in intron 5 at IVS5+107A–T and IVS5-266G–A, in intron 6 at IVS6+80A–T and IVS6-821C–T and in intron 7 at IVS7+117-2 and IVS-20A–G. None of these intronic sequence differences seemed likely to be responsible for Adam17 aberrant splicing in woe mEFs. Therefore, we explored the possibility that the Adam17 C794 nucleotide in exon 7 may be a part of a putative exonic splicing enhancer (ESE) sequence. To this aim, wild-type and woe exon 7 Adam17 sequences were analyzed by a web-based program called ESEfinder (Cartegni et al. 2003). The results showed that the C794T substitution eliminated a putative binding site for the splicing factor SRp55 and decreased the score for a binding site for another splicing factor SF2/ASF (data not shown).
Figure 4.—
Aberrant splicing of Adam17 in woe mEFs. (A) RT–PCR identified a single Adam17 transcript and three Adam17 transcript variants in wild-type and woe mEFs respectively. RT–PCR of glyceraldehyde-3-phosphate dehydrogenase (Gapdh) was examined as a control. Right": the semi-quantitative analysis of the amount of full-length Adam17 expressed in wild-type mEFs and full-length Adam17C794T expressed in woe mEFs as the ratio of the sample to the internal standard (Gapdh). (B–D) sequence analysis of the three Adam17 transcripts identified in woe mEFs: (B) sequence of the full-length Adam17C794T transcript indicating C to T substitution at nucleotide 794 (arrow), (C) sequence analysis showing that the predominant Adam17Δexon7 transcript in woe mEFs exhibits skipping of exon 7, (D) sequence of Adam17Δexon6-7 transcript exhibiting skipping of exons 6 and 7.
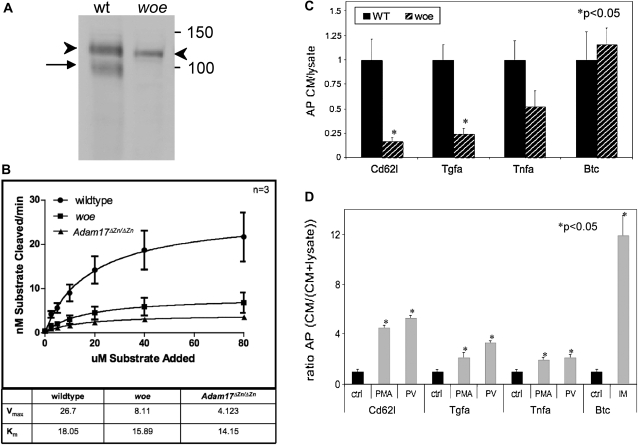
To assess the levels of precursor and mature forms of Adam17 in wild-type and woe mEFs, we performed Western blots. In cell lysates from wild-type mEFs, Adam17 was detected as a pro-form of ∼120 kDa and a mature form of ∼100 kDa (Figure 5A), similarly to previous reports for Adam17 (Schlondorff et al. 2000). In contrast, in woe mEFs an Adam17 immunoreactive band of an apparent mass of 110 kDa was detected, most likely reflecting the precursor Adam17Δ252-281 protein encoded by the Adam17Δexon7 transcript (Figure 5A). We did not detect Adam17 immunoreactive bands corresponding to the precursor and mature protein forms encoded by Adam17C794T.
Figure 5.—
Functional analysis of the woe allele. (A) Western blot analysis shows presence of the Adam17 precursor form of 120 kDa (left arrowhead) and mature form of 100 kDa (arrow) in wild-type mEFs; in woe mEFs a 110-kDa Adam17 immunoreactive band was identified (right arrowhead). Protein molecular mass markers (kDa) are indicated on the right. (B) Proteolytic cleavage of the FRET-peptide cleaved at different concentrations following incubation with cell lysates from wild-type, woe, and Adam17ΔZn/ΔZn mEFs. Vmax and Km values were calculated from the Michaelis–Menten curve as indicated in materials and methods. (C) Constitutive shedding analysis in wild-type (solid bars) and woe (lined bars) mEFs following transfection with AP-tagged l-selectin (Cd62l), Tgfa, Tnfa, and Btc. (D) Stimulated shedding analysis in untransfected woe mEFs (solid bars) and woe mEFs transfected with l-selectin (Cd62l), Tgfa, Tnfa, and Btc (shaded bars) following stimulation with 25 ng/ml PMA, 100 μM PV, or 2.5 μM IM.
To determine if woe mEFs exhibited any defects in shedding activity, we evaluated the cleavage of a fluorogenic FRET-peptide Adam17 substrate derived from the 10 amino acid sequence (LAQAVRSSSR) surrounding the scissile bond of the Adam17 cleavage site from the Tnfa precursor (Jin et al. 2002). The Tnfa precursor is a well-established substrate of Adam17 (Black et al. 1997), and cleavage between the A-V scissile bond of the Tnfa LAQAVRSSSR peptide has been previously established as a reliable measure of the Adam17 activity (Jin et al. 2002). Although the Adam17 fluorogenic FRET-peptide was established as highly specific for Adam17, some proteolytic activity was also reported for other metalloproteinases and this substrate (Jin et al. 2002). To establish if any of the FRET-peptide cleavage occurs independent of Adam17, we also evaluated the cell lysates from Adam17ΔZn/ΔZn mEFs. Under the conditions of the assay, cell lysates from Adam17ΔZn/ΔZn and woe mEFs showed a 5.0-fold and a 2.8-fold Vmax decrease respectively relative to the Vmax from the wild-type mEFs (Figure 5B). No major differences were observed in Km between the three cell lysates (Figure 5B).
To further evaluate the Adam17 shedding activity, we transfected both wild-type and woe mEFs with alkaline phosphatase-tagged Adam17-substrates: l-selectin, Tgfa, and Tnfa. Our results showed a significant decrease in the constitutive shedding of all Adam17 substrates in woe mEF cells when compared to wild-type controls (Figure 5C). To determine whether the woe mutation may have had an epigenetic effect on the function of Adam10, we also evaluated shedding of Adam10 substrate Btc. No difference in the constitutive shedding of Btc was noted between wild-type and woe mEFs (Figure 5C). The results from woe mEFs showed shedding of all three Adam17 substrates indicating that woe mEFs still respond to stimulation (Figure 5D), even though the levels of unstimulated shedding of these Adam17 substrates were significantly reduced compared to wild-type controls (Figure 5C).
DISCUSSION
In this study, we describe a spectrum of phenotypes in woe mice resulting from a C794T substitution in Adam17 that disrupts a putative ESE sequence required for binding of SRp55 and SF2/ASF proteins. The role of ESE sequences is to provide binding sites for serine/arginine (SR) proteins (Cartegni et al. 2002). Both SRp55 and SF2/ASF belong to a family of SR proteins that when bound to ESE promote exon inclusion by recruiting components of the splicing machinery. Numerous reports have shown that disruption of ESE sequences can cause the splicing machinery to skip the mutant exon with dramatic effects on protein structure of the gene product (Cartegni et al. 2002). In woe, the C794T substitution results in a predominant Adam17Δexon7 transcript that lacks exon 7 resulting in an in-frame deletion and a putative protein with residues 252–281 missing from the metalloprotease domain. Western blot analysis identified only the precursor form of the truncated Adam17Δ252-281 protein. Similar to other members of the Adam family of proteins, prodomain removal is required for Adam17 to exhibit shedding activity (Schlondorff et al. 2000). The prodomain acts as an inhibitor of protease activity via a cysteine-switch mechanism where a free cysteine residue from the prodomain interacts with the zinc ion from the active site (Schlondorff et al. 2000). Thus, presence of the prodomain would render the precursor form of Adam17Δ252-281 functionally inactive.
Even though the predominant Adam17Δexon7 transcript most likely leads to a functionally inactive precursor Adam17Δ252-281 protein, our shedding analysis experiments showed that woe mEFs exhibit residual shedding of Adam17 substrates. We cannot exclude the possibility that trace amounts of Adam17Δ252-281, below detection via Western blotting, may undergo maturation and may exhibit shedding activity. This, however, is unlikely since a loss of amino acid residues 252–281 presumably would also prevent proper folding and intracellular degradation before the protein can enter the secretory pathway en route to the cell surface (Suzuki et al. 1998). The residual Adam17 shedding observed in woe mEFs most likely originates from the full-length Adam17C794T transcript that encodes a putative full length Adam17T265M protein. In woe mEFs, Adam17C794T is expressed at severely reduced levels when compared to Adam17 expressed in wild-type mEFs. Our inability to detect precursor and mature Adam17T265M protein likely is due to the levels of Adam17T265M protein below detection via Western blotting. Nevertheless, constitutive and stimulated shedding of Adam17 substrates such as Tgfa, Tnfa, and l-selectin can be detected in woe mEFs, albeit at significantly reduced levels compared to wild-type controls. Our results are further supported by the observation that woe mice survive into adulthood, in contrast to Adam17−/− mice (Peschon et al. 1998; Horiuchi et al. 2007). These results imply that T265M substitution does not significantly affect shedding activity of Adam17T265M. Evidently, even the small amount of functional Adam17 is sufficient to allow woe mice to survive.
Adam17−/− mice exhibit enlargement of the semilunar and atrioventricular cardiac valves (Jackson et al. 2003; Horiuchi et al. 2007). Histological analysis of woe hearts identified enlargement only in the semilunar valves (Figure 2B). Previous in vivo and in vitro studies have established Adam17 as the principal sheddase of Hbegf and subsequent Egfr transactivation essential for valvulogenesis (Jackson et al. 2003; Sahin et al. 2004). Differences in valve phenotypes between woe and null Adam17 mice may be due to the reduced, but not abolished, Hbegf-mediated Egfr signaling in woe. It has been hypothesized that reduced Egfr signaling is sufficient for normal atrioventricular valve development, but insufficient for semilunar valve development (Jackson et al. 2003). As a part of this study, we evaluated shedding activity of three Adam17 substrates (Tgfa, Tnfa, and l-selectin) in woe, but we did not evaluate shedding of pro-Hbegf. Given that Adam17 is the principal sheddase of Hbegf, we expect that reduced pro-Hbegf shedding and consequent reduced Egfr signaling is responsible for the woe cardiac phenotype.
The milder cardiac phenotype observed in woe mice is most likely responsible for the postnatal survival of woe when compared to Adam17−/− mice. This is especially relevant for the evaluation of ocular phenotypes. Although formation of the anterior segment is initiated at E10, the differentiation and maturation of most anterior segment structures is not complete until P21 (Smith 2002; Cvekl and Tamm 2004). The woe anterior segment dysgenesis phenotype reported here shares features previously noted for the anterior segment dysgenesis phenotypes of Tgfa−/− and Egfr−/− mice (Luetteke et al. 1993; Miettinen et al. 1995). The role of Tgfa/Egfr signaling has been implicated in the formation of the corneal endothelium (Reneker et al. 1995). Whether Adam17 plays an essential role in mediating only the Tgfa activation of Egfr or Adam17 mediates signaling of other ligands/molecular pathways essential for anterior segment development is unclear. Histological analysis of woe eyelids showed an absence of the meibomian glands, thus identifying a novel role of Adam17 in normal eyelid development and function. Meibomian glands are specialized sebaceous glands located at the rim of the eyelid that secrete sebum, a lipid substance that prevents evaporation of the precorneal tearfilm. Absence of meibomian glands has been used as one of the most common ophthalmic clinical features in patients affected with ectodermal displasia, a genotypically and phenotypically heterogeneous group of inherited disorders that are characterized by primary defects in the development of two or more tissues derived from embryonic ectoderm (Kaercher 2004). Meibomian glands are ectodermally derived appendages that develop as a result of the interaction between the ectoderm and the underlying mesoderm in the skin. Recently, the role of Egfr signaling has also been proposed as the promoter of the intrafollicular epidermal fate at the expense of hair follicle development (Richardson et al. 2009). It is possible that Egfr signaling also promotes the development of the meibomian glands in the eyelids; mice with a mutation in Tgfa exhibit absence of the meibomian glands (Luetteke et al. 1993). However, absence of meibomian glands was not reported for Egfr−/−, but that phenotype may have been overlooked due to prenatal and perinatal lethality in null Egfr mice (Miettinen et al. 1995). Thus, we cannot exclude the possibility that the role of Adam17 in the meibomian gland development may be independent from Egfr signaling. Therefore, further analyses will address the role of Adam17 in the meibomian gland formation either dependent or independent from the Egfr signaling pathway.
In summary, we describe characterization of the woe mouse mutant. Genetic analysis identified a C794T substitution in Adam17 resulting in aberrant Adam17 splicing and rendering the majority of Adam17 protein nonfunctional. However, a small amount of functional Adam17 is produced in woe animals, sufficient to support shedding of Adam17 substrates, albeit at significantly reduced levels. The characterization of the different forms of Adam17 in the woe mutant mouse and their functional properties in woe mEFs provides a plausible explanation for the observed hypomorphic Adam17 phenotype. Since woe mice are viable, whereas Adam17−/− mice die at birth, the woe mutation provides an excellent opportunity for studying the role of Adam17 in postnatal development, and specifically helped uncover a critical role for Adam17 in the development of the anterior segment of the eye and meibomian glands, most likely caused by defects in Adam17-dependent Egf signaling.
Acknowledgments
We thank Roy Black at Amgen, Seattle, for generously providing Adam17ΔZn/+ mice. This work was supported in part by National Institutes of Health grant EY15173 and EY018872 (D.J.S.), and EY15719 and GM64750 (C.P.B.).
References
- Black, R. A., C. T. Rauch, C. J. Kozlosky, J. J. Peschon, J. L. Slack et al., 1997. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 385 729–733. [DOI] [PubMed] [Google Scholar]
- Blobel, C. P., 2005. ADAMs: key components in EGFR signalling and development. Nat. Rev. Mol. Cell Biol. 6 32–43. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., S. L. Chew and A. R. Krainer, 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3 285–298. [DOI] [PubMed] [Google Scholar]
- Cartegni, L., J. Wang, Z. Zhu, M. Q. Zhang and A. R. Krainer, 2003. ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31 3568–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, B., N. L. Hawes, R. E. Hurd, J. Wang, D. Howell et al., 2005. Mouse models of ocular diseases. Vis. Neurosci. 22 587–593. [DOI] [PubMed] [Google Scholar]
- Cvekl, A., and E. R. Tamm, 2004. Anterior eye development and ocular mesenchyme: new insights from mouse models and human diseases. Bioessays 26 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X., K. Tabeta, K. Hoebe, H. Liu, N. Mann et al., 2004. Velvet, a dominant Egfr mutation that causes wavy hair and defective eyelid development in mice. Genetics 166 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, K. R., K. A. McGowan, C. D. van Raamsdonk, H. Fuchs, D. Lee et al., 2003. Genetics of dark skin in mice. Genes Dev. 17 214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, L. A., N. Alexander, M. E. Hogan, J. P. Sundberg, A. Dlugosz et al., 1997. Genetically null mice reveal a central role for epidermal growth factor receptor in the differentiation of the hair follicle and normal hair development. Am. J. Pathol. 150 1959–1975. [PMC free article] [PubMed] [Google Scholar]
- Harris, R. C., E. Chung and R. J. Coffey, 2003. EGF receptor ligands. Exp. Cell Res. 284 2–13. [DOI] [PubMed] [Google Scholar]
- Horiuchi, K., T. Kimura, T. Miyamoto, H. Takaishi, Y. Okada et al., 2007. Cutting edge: TNF-alpha-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 179 2686–2689. [DOI] [PubMed] [Google Scholar]
- Huovila, A. P., A. J. Turner, M. Pelto-Huikko, I. Karkkainen and R. M. Ortiz, 2005. Shedding light on ADAM metalloproteinases. Trends Biochem. Sci. 30 413–422. [DOI] [PubMed] [Google Scholar]
- Jackson, L. F., T. H. Qiu, S. W. Sunnarborg, A. Chang, C. Zhang et al., 2003. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 22 2704–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, G., X. Huang, R. Black, M. Wolfson, C. Rauch et al., 2002. A continuous fluorimetric assay for tumor necrosis factor-alpha converting enzyme. Anal. Biochem. 302 269–275. [DOI] [PubMed] [Google Scholar]
- Kaercher, T., 2004. Ocular symptoms and signs in patients with ectodermal dysplasia syndromes. Graefes Arch. Clin. Exp. Ophthalmol. 242 495–500. [DOI] [PubMed] [Google Scholar]
- Le Gall, S. M., P. Bobe, K. Reiss, K. Horiuchi, X. D. Niu et al., 2009. ADAMs 10 and 17 represent differentially regulated components of a general shedding machinery for membrane proteins such as transforming growth factor alpha, l-selectin, and tumor necrosis factor alpha. Mol. Biol. Cell 20 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G., C. Gustafson-Brown, S. K. Hanks, K. Nason, J. M. Arbeit et al., 2003. c-Jun is essential for organization of the epidermal leading edge. Dev. Cell 4 865–877. [DOI] [PubMed] [Google Scholar]
- Luetteke, N. C., T. H. Qiu, R. L. Peiffer, P. Oliver, O. Smithies et al., 1993. TGF alpha deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell 73 263–278. [DOI] [PubMed] [Google Scholar]
- Luetteke, N. C., H. K. Phillips, T. H. Qiu, N. G. Copeland, H. S. Earp et al., 1994. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 8 399–413. [DOI] [PubMed] [Google Scholar]
- Mezyk, R., M. Bzowska and J. Bereta, 2003. Structure and functions of tumor necrosis factor-alpha converting enzyme. Acta Biochim. Pol. 50 625–645. [PubMed] [Google Scholar]
- Miettinen, P. J., J. E. Berger, J. Meneses, Y. Phung, R. A. Pedersen et al., 1995. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376 337–341. [DOI] [PubMed] [Google Scholar]
- Miething, F., S. Hering, B. Hanschke and J. Dressler, 2006. Effect of fixation to the degradation of nuclear and mitochondrial DNA in different tissues. J. Histochem. Cytochem. 54 371–374. [DOI] [PubMed] [Google Scholar]
- Peschon, J. J., J. L. Slack, P. Reddy, K. L. Stocking, S. W. Sunnarborg et al., 1998. An essential role for ectodomain shedding in mammalian development. Science 282 1281–1284. [DOI] [PubMed] [Google Scholar]
- Reneker, L. W., D. W. Silversides, K. Patel and P. A. Overbeek, 1995. TGF alpha can act as a chemoattractant to perioptic mesenchymal cells in developing mouse eyes. Development 121 1669–1680. [DOI] [PubMed] [Google Scholar]
- Richardson, G. D., H. Bazzi, K. A. Fantauzzo, J. M. Waters, H. Crawford et al., 2009. KGF and EGF signalling block hair follicle induction and promote interfollicular epidermal fate in developing mouse skin. Development 136 2153–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin, U., G. Weskamp, K. Kelly, H. M. Zhou, S. Higashiyama et al., 2004. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J. Cell Biol. 164 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlondorff, J., J. D. Becherer and C. P. Blobel, 2000. Intracellular maturation and localization of the tumour necrosis factor alpha convertase (TACE). Biochem. J. 347(1): 131–138. [PMC free article] [PubMed] [Google Scholar]
- Schneider, M. R., S. Werner, R. Paus and E. Wolf, 2008. Beyond wavy hairs: the epidermal growth factor receptor and its ligands in skin biology and pathology. Am. J. Pathol. 173 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R., 2002. Systematic Evaluation of the Mouse Eye. CRC Press, Cleveland, OH.
- Suzuki, T., Q. Yan and W. J. Lennarz, 1998. Complex, two-way traffic of molecules across the membrane of the endoplasmic reticulum. J. Biol. Chem. 273 10083–10086. [DOI] [PubMed] [Google Scholar]
- Talamas, E., L. Jackson, M. Koeberl, T. Jackson, J. L. McElwee et al., 2006. Early transposable element insertion in intron 9 of the Hsf4 gene results in autosomal recessive cataracts in lop11 and ldis1 mice. Genomics 88 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaung, C., K. West, B. J. Clark, L. McKie, J. E. Morgan et al., 2002. Novel ENU-induced eye mutations in the mouse: models for human eye disease. Hum. Mol. Genet. 11 755–767. [DOI] [PubMed] [Google Scholar]
- Weskamp, G., H. Cai, T. A. Brodie, S. Higashyama, K. Manova et al., 2002. Mice lacking the metalloprotease-disintegrin MDC9 (ADAM9) have no evident major abnormalities during development or adult life. Mol. Cell. Biol. 22 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y., and M. Karin, 2004. The control of cell motility and epithelial morphogenesis by Jun kinases. Trends Cell Biol. 14 94–101. [DOI] [PubMed] [Google Scholar]