Abstract
During growth under selection, mutant types appear that are rare in unselected populations. Stress-induced mechanisms may cause these structures or selection may favor a series of standard events that modify common preexisting structures. One such mutation is the short junction (SJ) duplication with long repeats separated by short sequence elements: AB*(CD)*(CD)*E (* = a few bases). Another mutation type, described here, is the tandem inversion duplication (TID), where two copies of a parent sequence flank an inverse-order segment: AB(CD)(E′D′C′B′)(CD)E. Both duplication types can amplify by unequal exchanges between direct repeats (CD), and both are rare in unselected cultures but common after prolonged selection for amplification. The observed TID junctions are asymmetric (aTIDs) and may arise from a symmetrical precursor (sTID)—ABCDE(E′D′C′B′A′)ABCDE—when sequential deletions remove each palindromic junction. Alternatively, one deletion can remove both sTID junctions to generate an SJ duplication. It is proposed that sTID structures form frequently under all growth conditions, but are usually lost due to their instability and fitness cost. Selection for increased copy number helps retain the sTID and favors deletions that remodel junctions, improve fitness, and allow higher amplification. Growth improves with each step in formation of an SJ or aTID amplification, allowing selection to favor completion of the mutation process.
IN general, genetic mutations are seen as discontinuous changes in base sequence whose origin can be explained by a single event. This view is based on laboratory genetics in which mutants are isolated using stringent selection for large discontinuous phenotypic changes or using screens that involve no growth limitation. These standard genetic procedures often miss the most common of all rearrangement types—gene copy-number changes, which may be extremely important to genetic adaptation during growth under selection. Because copy number increases are deleterious and unstable (Reams et al. 2010), they may often escape detection. However, selective conditions that detect copy-number increases can favor cells with secondary changes that stabilize and reduce the cost of the underlying structures. Prolonged selection can thus contribute to the formation and detection of mutations without affecting the molecular mechanisms that create them. This can happen without an increase in mutation rate.
Formation of mutations under selective conditions has been extensively studied in a system developed by Cairns and Foster (1991). The system employs a bacterial tester strain whose mutant lac operon limits the ability to use lactose. The mutant lac allele produces 2% of the β-galactosidase (LacZ) level found in revertant lac+ cells. During nonselective growth this lac allele reverts at a rate of 10−8/cell/division. Cells of this strain (108) are plated on minimal lactose medium and give rise (over several days) to ∼100 Lac+ revertant colonies that appear above a lawn of nongrowing parent cells. Since the reversion rate of the lac mutation during nonselective growth is 10−8/cell/division, the 100 colonies accumulated from 108 nongrowing cells suggested the possibility that stress might induce in nongrowing cells a mutagenic mechanism that evolved under selection for its ability to create beneficial mutations (Hall 1998; Foster 2007; Galhardo et al. 2007). We have argued that a mechanism for stress-induced general mutagenesis would be maladaptive in view of the vast excess of deleterious over beneficial mutations (Roth et al. 2003).
An alternative model, which we favor, uses selection (without mutagenesis) to explain behavior of the Cairns system and other related systems (Roth et al. 2006; Andersson et al. 2010). In this model, extremely common duplication types that are normally deleterious and unstable (Reams et al. 2010) are detected by selection for increased levels of lac expression. This is possible because the original mutant lac allele retains substantial activity (2% of the revertant β-galactosidase level). Under selection, these copy-number variants initiate colonies in which successive mutant types arise and improve growth progressively until one dominates the colony. Selection contributes to mutation formation by favoring progressive growth improvement without any increase in mutation rate. In some clones, amplification provides sufficient lac target copies that a normally rare reversion event (to lac+) occurs, permitting loss of mutant alleles and overgrowth to produce a colony populated primarily by stably Lac+ revertant cells (stable-rich). In other colonies, deletions arise that reduce the fitness cost of an amplified lac region, allowing higher amplification and faster growth. In these colonies, improvement is achieved by remodeling the original duplication structure (not by point mutations). This course of events leads to colonies rich in unstable Lac+ cells with high lac copy number (Kugelberg et al. 2006). The duplications described here were found in these unstable-rich Lac+ colonies during prolonged growth under selection.
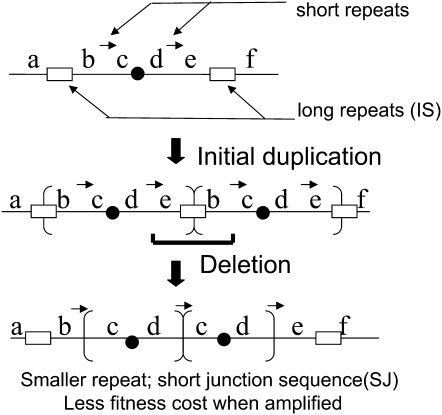
Two types of amplifications have been observed in the Cairns system (Kugelberg et al. 2006). In the first type (short junction, or SJ), directly repeated copies of the lac region are separated by short junction sequences (3–12 bp) (Kugelberg et al. 2006; Slack et al. 2006). We have proposed that these arise by remodeling of a frequent large precursor duplication formed between IS3 sequences (1.25 kb) (see Figure 1). This precursor duplication is carried by 1/500 of the unselected parent culture prior to plating, but seldom appears in selected unstable revertant colonies. We propose that deletions remove the common IS3 junction to reduce the repeat size and create a shorter duplication whose junction point is defined by the deletion end points. These modifying deletions reduce fitness cost and allow higher lac amplification. We will propose here that SJ duplications can also form from the symmetrical tandem inversion-duplication (sTID) precursors described below.
Figure 1.—
Formation of SJ duplications under selection. The short arrows between b–c and d–e in the parent sequence are repeats of 3–12 bp. The boxes are the 1.25-kb elements IS3A and IS3C that flank lac on the F′128 plasmid. The duplicated region between IS3 copies is ∼125 kb, and duplications of this type are carried by 0.1% of cells prior to selection. Deletion between identical short sequences leaves a smaller duplication, typically ∼20 kb. Shortening reduces the reversion rate and fitness cost of a duplication and thereby allows higher amplification (Kugelberg et al. 2006).
A second amplification class is the tandem inversion-duplication (TID) type described here (Kugelberg et al. 2006). In this class, the underlying duplication has direct-order sequence repeats that flank a central inverse-order segment. For example, an asymmetric inversion-duplication (aTID) rearrangement of the parent sequence ABCDEF might have the form AB(CD)(E′D′C′B′)(CD)EF, where each letter designates a multi-gene sequence. Note that the two junctions between inverse-order repeats are not extended palindromes, hence the name, asymmetric or aTID. The basic aTID unit can amplify by unequal exchanges between the flanking direct repeats, generating more copies of a basic repeat unit that includes the central region plus one flanking sequence. One example of this was described previously for a revertant derived from the Escherichia coli version of the Cairns strain (Slack et al. 2006), and its formation was attributed to unknown mechanisms brought into play by growth limitation.
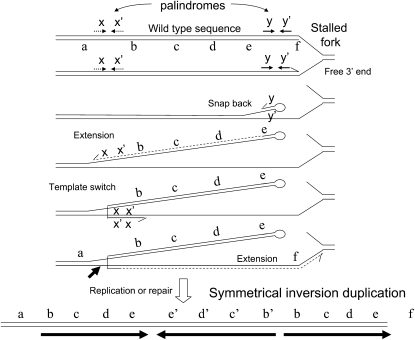
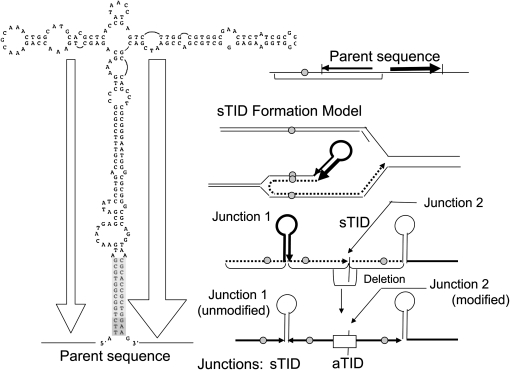
We propose that the aTID arises stepwise from a common precursor, the symmetrical sTID, whose long inverse-order repeats extend from a quasi-palindromic junction. These rearrangements arise frequently but are rapidly lost because they are both deleterious and unstable. Figure 2 diagrams a proposal for frequent formation of the initial sTID structures at quasi-palindromic sites in the parent sequence. This model will be described in more detail below with supporting evidence. These sTID structures may be deleterious due to their copy-number increases and to the tendency of their extended quasi-palindromic sequences to form a hairpin structure during replication. The sTIDs are expected to be unstable due to the demonstrated frequent deletions of hairpin structures and to the possibility of simple exchanges between the flanking direct repeats.
Figure 2.—
Proposal for the formation of symmetrical TID structures. Short quasi-palindromic sequences are frequent and can support formation of snap-back structures that initiate repair synthesis, which can switch templates and be redirected toward the fork. The resulting branched structure can be broken at arrow and repaired or can be replicated to produce the sTID structure proposed to initiate the aTID structures described here. These events may also underlie recombination-independent formation of simple duplications.
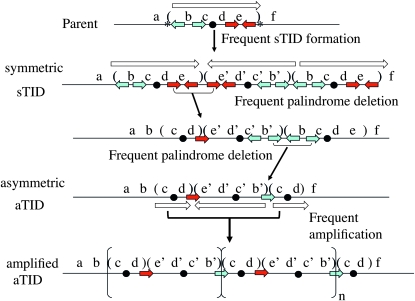
Under selection for higher expression of some gene (e.g., lac), increased gene copy number offsets the fitness cost and instability of the sTID by a process suggested in Figure 3. During prolonged slow growth under selection, deletions (frequent at palindromic junctions) render the junctions asymmetric (less unstable) and reduce repeat size (lower fitness cost). Thus selection favors the conversion of frequent unstable structures into more stable duplications that are easier to amplify under selection. Any amplification of the initial sTID duplication increases the remodeling rate by providing more sites for remodeling changes.
Figure 3.—
Conversion of an sTID precursor to an aTID duplication. The sTID forms and is lost frequently. Asterisks in the parent sequence indicate small palindromic sequences that promote sTID formation. Prolonged selection for higher expression (e.g., lac, the black dot) favors retention of the sTID and sequential accumulation of deletions that render junctions asymmetric (forming the aTID). As fitness cost is reduced, higher amplification allows faster growth and ultimately provides sufficient lac targets so that point mutations can stably alter the gene under selection. Short colored arrows are repeated oligonucleotide sequences present in the parent that serve as endpoints for deletions and junctions of the final aTID duplication. Uninvolved repeats are omitted after the first two lines.
Genetic adaptation can be extremely rapid whenever selection detects copy-number changes, which arise frequently under all growth conditions. The speed of adaptation reflects both the high frequency of contributing mutation types and the exponential increases in mutant frequency that occur during growth under selection. Ultimately, the growth of cells with more copies of a particular gene enhances the frequency of point mutations within repeats by providing more targets to the expanding clone. The process described here seems likely to contribute to human copy-number polymorphisms, to evolution of malignant cells, and to the many systems for which “stress-induced” mutation has been suggested.
MATERIALS AND METHODS
Strains, media, and chemicals:
Strains used are derivatives of Salmonella enterica (Serovar Typhimurium, strain LT2). The tester strain (TT18302) has a chromosomal leu deletion and a proB∷Tn10 insertion and carries the E. coli-derived plasmid F′128 with a triply mutant lac region. A deletion (Ω) fuses the lacI and lacZ genes, a point mutation improves the lacI promoter (IQ), and a +1 frameshift mutation (lacI33) is located within the lacI portion of the hybrid lacI-lacZ gene. The minimal medium was no-citrate E salts (NCE), containing added nutrients as recommended previously (Slechta et al. 2002). Carbon sources were used at a concentration of 0.2% (w/v). Rich medium was nutrient broth (NB; Difco Laboratories) with 5 g/liter NaCl. The chromogenic β-galactosidase substrate X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Diagnostic Chemicals, Oxford, CT) was used at 25 μg/ml in minimal media and at 40 μg/ml in NB plates.
Isolation of strains with a selected lac amplification:
Independent cultures grown in NCE glycerol were plated (2 × 108 cells) on minimal lactose plates containing X-gal and leucine. Scavenger cells (109) were added to consume contaminating carbon present in the agar. Plates were monitored for blue lac revertants every day. New independent revertant colonies appearing on day 5 were plugged and resuspended in NCE and diluted and plated for single colonies on NB medium containing X-gal. After 3 days, unstably Lac+ cells form colonies that are blue with many white (Lac−) sectors; such colonies are known to carry a tandem array of lac copies (Tlsty et al. 1984; Andersson et al. 1998; Hastings et al. 2000). These unstably Lac+ cells were used as conjugational donors and their plasmids were transferred into a recA recipient strain (DA7700) to stabilize the amplification array (Kugelberg et al. 2006).
Cloning, PCR, and sequencing:
The sequences of all primers for PCR and sequencing are available upon request. Genomic DNA was isolated using Wizard Genomic DNA purification kit (Promega). For sequencing, PCR products were purified from solution using QIAquick purification kit (Qiagen). Purified PCR product was used as template in a sequencing reaction using BigDye Terminator v3.1 cycle sequencing reaction kit (Applied Biosystems). TOPO TA cloning kit (Invitrogen) was used whenever it was necessary to clone a PCR product.
Experiments to test the structure of an aTID:
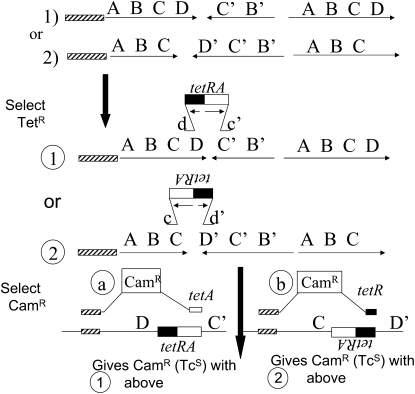
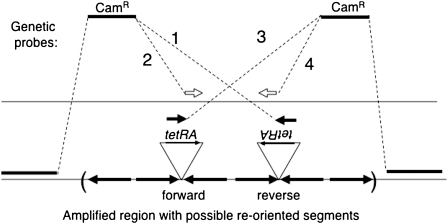
A tetracycline resistance cassette (TetRA) was added to the TID junction of strain TT25771 by linear transformation (Slechta et al. 2003). Plasmid F′128 from this strain was conjugated into TT4632 [pro-47(del:AB) recA1 srl+], which lacks drug markers. To determine the orientation of the TetRA-marked junction sequence, a second linear transformation was performed using a chloramphenicol resistance (CamR) marker as donor. Four different CamR probes were used. Two probes had a CamR determinant flanked on one side by the recombining sequence from the left side of the amplified region and on the other side by the TetRA sequence. Two other probes had a CamR determinant flanked on one side by the recombining sequence from the left side of the amplified region and on the other by the TetRA sequence in one orientation or the other. The design of these four probes is described in Figure 7 and Table 2. The specificity of these crosses was demonstrated by control transformations in which the recipient had a Tn10 insertion in an unamplified lac operon. In these controls, inheritance of CamR depended on both the presence and the proper orientation of the Tn10 insertion. All four probes were designed so that inheritance of CamR would delete the TetR determinant from the recipient sequence involved in the exchange. Transformants were selected on LB + Cam plates. Ten single transformants from each of the four different transformations were restreaked for single colonies on LB + Cam. Ten single colonies from each transformant were then tested for tetracycline resistance by patching to LB + Cam and LB + Cam + Tet plates.
Figure 7.—
Genetic testing of TID amplification structure. To determine whether an aTID junction is in orientation CD′ or DC′, a resistance determinant (tetRA) was added to the sequenced junction of strain TT25771 (see Table 1). The chromosome of the resulting strain is expected to be as diagrammed for strain 1 or strain 2 (circled). In a second transformation cross, a donor CamR determinant is introduced with one flanking sequence matching a reference region at the left and the other matching the tetRA sequence. The CamR determinant can be inherited only if the orientation of the recipient junction (tetRA or ARtet) matches that of the donor recombining sequence (right side of CamR). The donor fragments are designed such that inheritance of CamR creates a deletion of the region between the reference sequence and the right side of the TetR determinant in the recipient. The recipient strain is recA+ and was grown nonselectively before transformation.
TABLE 2.
Demonstrating the rearrangement of an amplified TID array
Whole-genome sequencing and mapping:
Whole-genome sequencing was performed with the Illumina GAII Sequencing System (Illumina). Purified genomic DNA (5 μg/sample) was fragmented to a targeted average size of 150–250 bp with a Covaris S2 System (Covaris). The DNA fragment libraries were end-polished, 3′-adenylated, and adapter-modified according to standard Illumina paired-end protocols. Reamplified library fragments of ∼200 bp were selected and purified from a 3% agarose gel for subsequent paired-end sequencing. Basecalling of raw sequencing data was performed by Illumina software. Sequence reads in the form of two 36-base sequences (mated end pairs) from each DNA fragment were mapped to both the reference S. typhimurium genome from the National Center for Biotechnology Information and the F′128 plasmid sequences with MAQ (Li et al. 2008). Mapping was done in two passes. In the first pass, each read pair was mapped with the restriction that the distance between the mapped locations is constrained to the original selected DNA fragment size. Because read pairs spanning the duplication/deletion boundary violate this constraint, they cannot be mapped in this pass. In the second pass, the remaining reads were mapped without any distance constraints. The number of reads finally mapped to each position (read-depth) was counted and used for later analysis.
Determination of junction sequences:
Determination of junction sequences of tandem duplications has been described previously (Kugelberg et al. 2006). Briefly, 77 primers across 150 kb of the F′128 plasmid were designed with 3′ ends directed away from lacZ. Seven pools of PCR primers were assembled; four pools contained primers directing replication clockwise from lac and three in a counterclockwise direction on F′128. Each of the counterclockwise pools was used in combination with each of the clockwise pools. A unique PCR product will be produced when a clockwise and counterclockwise primer cross a duplication join point. One inverse-order junction of a TID structure was identified when primers from a single pool (clockwise or counterclockwise) generated a unique PCR fragment. Hence, at TID junctions primers in the same orientation (in the parent sequence) yield a PCR product. Once a unique band was found, the PCR product was cloned into a TOPO vector and sequenced using vector-specific primers.
Numbering of base sequence from F′128 lac:
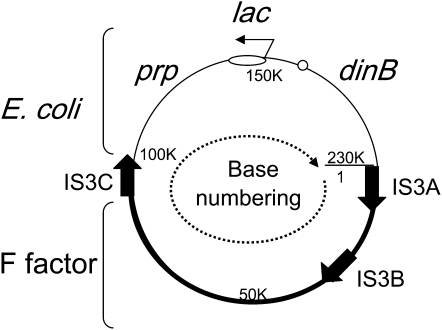
In the Cairns system, selection is imposed for reversion of a lac point mutation. Reversion involves intermediate duplications and amplifications of the lac region, which is carried on plasmid F′128. This plasmid includes the entire F element and a segment (150 kb) of the E. coli chromosome (see Figure 4). The rearrangements described here affect the E. coli-derived sequences in the top half of the F′128 lac as seen in Figure 4. In the text, “left” and “right” will refer to the position in relation to lac in this figure with “left” meaning decreasing coordinate numbers and “right” meaning increasing coordinate numbers.
Figure 4.—
Structure of the F′128 plasmid on which rearrangements occur. The base numbering conventions are the following: Bases 1–100,413 are derived from the F factor (bold arc of circle). Bases 100414–150235 are from the E. coli chromosome extending from IS3C to include all of the lac operon up to base 150,235, which is immediately adjacent to the promoter-proximal end of the lacZ coding sequence. Bases 150,236–231,638 of the plasmid are also from the E. coli chromosome and extend from lacZ to beginning of F factor sequence (at base 1). The origin and structure of this plasmid have been described in detail (Kofoid et al. 2003).
RESULTS
Characterizing selected lac amplifications:
The strain used in the Cairns selection system carries a partially functional mutant lac allele on a conjugative F′ plasmid and has no chromosomal lac region (Cairns and Foster 1991). In the original Cairns experiment, this plasmid was carried in E. coli cells. Experiments described here are done with cells of S. enterica that harbor the same mutant F′lac plasmid and have been shown to behave similarly with regard to lac reversion. Cells with this plasmid (phenotypically Lac−) are plated on solid minimal lactose medium to select revertants (see materials and methods). Over the course of 6 days new revertant colonies appeared each day. New colonies appearing on day 5 were picked and single cells were plated on rich medium with X-gal, where cells with a lac amplification form sectored (blue-white) colonies. Sectors (white) are initiated whenever some cell in the (Lac+, blue) colony experiences a segregation event that removes lac copies and therefore the Lac+ phenotype. This behavior contrasts with that of haploid revertant lac+ cells from the same revertant colony, which form solid blue (stably Lac+) colonies under the same conditions. Sectored colonies were used as conjugational donors to transfer the Lac+ phenotype into a recipient lac deletion strain with a recombination defect (recA). The RecA defect prevents copy loss and stabilizes the amplified array (Slechta et al. 2003).
Once stabilized, these high-copy amplifications were analyzed by PCR, using pools of primers designed to identify the junctions between copies repeated in tandem. To identify simple junction sequences, PCR primers were assembled into pools containing multiple primers whose 3′ ends initiate replication away from the lac locus in the same direction on the F′ lac plasmid. Primer pairs in any one pool are not expected to produce a PCR product since their replication tracks proceed in the same direction. Combinations of primer pools that initiate synthesis in opposite directions (away from lac) are not expected to generate products from a wild-type template sequence because their replication tracks diverge. However, when the template sequence includes tandem repeats, some pairs of normally divergent primers can initiate replication tracts that converge across a duplication junction; the sequence of these fragments revealed the nature of the junction for standard tandem-repeat amplifications. The primer sets used are able to detect the junction of any simple lac duplication <175 kb (Kugelberg et al. 2006).
Ninety-nine amplification strains were isolated from unstable-rich Lac+ revertant colonies that arose during prolonged growth under selection. Of these 99, two-thirds (64 revertants) had a simple tandem duplication structure described previously (Kugelberg et al. 2006). Of these 64 strains, 48 had a 3- to 12-bp SJ, 15 had a 30-base repetitive extragenic palindromic element (Gilson et al. 1984) at the junction, and one had a copy of IS3, suggesting that it arose by replicative transposition of the IS3C element. None of the 64 amplifications arising under selection had a join point formed by an exchange between IS3A and IS3C, a duplication type found in >0.1% of cells in the unselected parent culture and diagrammed in the center of Figure 1. We believe that our PCR methods can detect junctions of any simple tandem duplication. The remaining 35 (of the initial 99 amplifications) lacked a simple duplication junction and appeared to be of a distinct type.
The nature of the new amplification type was first suggested when template DNA from one unstably Lac+ revertant yielded a PCR product generated by a single primer. Initially, a single pool of primers oriented in the same direction produced a product, and later a single primer from this pool was shown to be sufficient. That is, the rearrangement included two oppositely oriented copies of a single sequence, such that one primer type could initiate converging replication tracks across the repeat junction. The sequence of this PCR fragment included our first example of a TID join point. Thirteen TID junctions are described here.
For 29 of the unstably Lac+ strains, no junctions could be identified by PCR, and the nature of the amplification in these “recalcitrant” strains is not known. These may carry new amplification types or they may be TID amplifications whose junctions resisted detection by PCR. We suspect that many are of the TID type that simply eluded our PCR attempts. TID junctions were harder to identify by PCR than SJ junctions, and one recalcitrant junction (identified as a TID type by genome sequence) has been difficult to demonstrate by PCR even when its sequence was known. To clarify the methods used to identify these join points and the model proposed to explain their formation, the formal properties of tandem inversion duplications are listed below.
Formal structure of a TID:
The 13 junctions described below collectively define the TID as a new amplification type and suggest a model for its formation. The formal properties of a TID are the following and are described in Figures 2 and 3.
The basic unit has two extended direct-order sequence repeats flanking a central inverted sequence, e.g., AB(CD)(E′D′C′B′)(CD)E. Unequal recombination exchanges between the flanking repeats (CD) of the basic unit can generate an amplified tandem array that carries repeats of the [(CD)(E′D′C′B′)] sequence with the overall structure AB[(CD)(E′D′C′B′)]nCDE.
In an aTID duplication, sequences can be present in two or three copies. The example above—AB(CD)(E′D′C′B′)(CD)E—has two copies of B and E and three copies of C and D. This distinguishes the TID from a conventional inversion, which has no repeated sequences.
A TID has two alternating asymmetric inversion junctions (tail–tail, head–head) that together provide duplication status and lead to direct-order sequence repeats. This distinguishes the TID from conventional tandem duplications, which have a single tail-to-head sequence junction (the D–A junction of ABCD–ABCD).
The TID amplification junctions described here are aTIDs. That is, the inverse-order sequences on either side of the two junctions are not an extended palindrome and thus are not prone to form extended hairpin structures. In the third and fourth lines of Figure 3, the left-hand aTID junction is the asymmetric d–e′ (not the symmetric possibilities dd′ or ee′). This contrasts with the hypothetical sTIDs in the second line of Figure 3 or the last line of Figure 2, which have extended inverse-order sequences (b c d e–e′ d′ c′ b′) and might be expected to form hairpins. Such hairpin structures have been shown to be deleterious and unstable in other systems (Leach 1994; Cromie et al. 2000), especially when the junction is perfectly palindromic.
The asymmetrical aTID structures described here are proposed to form from frequent symmetrical precursor sTID structures (bottom of Figure 2). The symmetry (or quasi-symmetry) proposed for the sTID may allow inverse-order repeats to form hairpin structures that are deleterious when extended. An initial quasi-palindromic junction can be made asymmetric by a deletion that removes unequal amounts of sequence from either side. These deletions leave extensive sequences between the inverse-order sequence repeats and minimize the chances of a hairpin extension.
Between inverse-order repeats, a unique short sequence element (4–31 bp) marks each asymmetric junction (aTID). Two copies of each junction element are present in inverse order at widely separated positions in the parent sequence (up to 24 kb apart; red and blue short arrows in Figure 3). The deletion that remodels a junction extends between such repeats and leaves one copy of the element at the remodeled TID junction. Each aTID junction sequence element characterized here is unique: no two TID amplifications characterized so far have the same junction sequence. Thirteen of these TID junction elements are described below.
Determining the sequence of TID junction elements:
To identify TID amplifications among the unstable Lac+ revertants, pools of similarly directed PCR primers were assembled. Each pool was tested for the ability to produce a PCR product using template DNA from 43 uncharacterized nonstandard amplifications—35 selected from the standard Cairns strain and 8 from a strain lacking IS3 (see materials and methods). Screening of these DNA PCR samples revealed 12 inversion-duplication junctions—both junctions from each of 4 strains (8 total) and one junction from each of 4 additional strains. A 13th junction was described later by genome sequencing.
The first six lines of Table 1 describe amplifications from 6 of the original 35 revertants that showed an unstable Lac+ phenotype but lacked standard duplication junctions. Our PCR methods showed that these six are all aTID types. Sequencing of PCR fragments revealed both junctions from 4 strains (lines 1–4: DA8097, DA7948, TT25721, TT25773) and one junction for each of two strains (lines 5 and 6: TT25790 and TT25772). Later use of genome sequencing (see below) revealed the missing left junction for the strain in line 5 (TT25790). Sequence numbering conventions are diagrammed in Figure 4.
TABLE 1.
Sequence of inversion-duplication junctions
The strains described in lines 7 and 8 were isolated from parents lacking IS3A and IS3C. The strain described in line 7 (TT25542) was isolated under selection like those above and shows an aTID amplification. The strain described in line 8 (TT25561) was trapped nonselectively (see below) and has an aTID duplication with a somewhat longer junction element at its one determined junction (a 31-bp REP element).
Verifying amplification structure by genome sequencing:
Two types of amplifications (SJ and aTID) have been inferred from the sequence of PCR fragments that include a junction. Amplifications based on SJ tandem duplications were described previously (Kugelberg et al. 2006) and those based on TID duplications are described above (Table 1). To examine the association of junction sequences and lac amplification, a full-genome sequence was determined for one previously described strain with an SJ amplification (TT24815) and a second strain inferred to carry an aTID amplification (TT25790 in Table 1).
The SJ amplification strain sequence confirmed the single junction element (AGGGCAGG) determined previously by PCR fragment sequencing (Kugelberg et al. 2006). This junction is proposed to arise when a deletion event removes the material between two copies of the small sequence element (see Figure 1). A deletion between the short elements in different duplication copies (Figure 1) removes the IS3 sequence at the junction of the original duplication and generates the observed SJ junction of a shortened duplication. Read-depth of Illumina sequencing demonstrated that the region between the two short elements in the wild-type genome was amplified in the selected Lac+ revertant. The original lac mutation was present in all copies in the amplification.
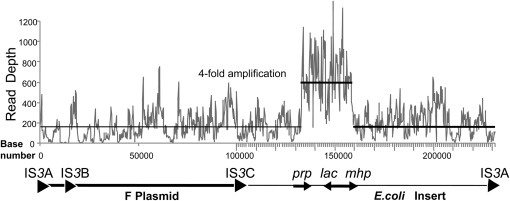
In the case of the TID-based amplification strain (TT25790), the right (divergent) junction had been characterized by sequencing a PCR product (Table 1; line 5). The genome sequence confirmed the known junction sequence and revealed the previously unknown left (convergent) join point, which had escaped identification by PCR. This junction is described below. The lac amplification carried by strain TT25790 was demonstrated by examining the number of sequence reads (see Figure 5) mapping to each genome position (read-depth). That is, the chromosome was sequenced with an average coverage of 3.2 reads/nucleotide (∼90-fold coverage at 30 bases/read). The bulk of the F′128 plasmid (outside the lac region) was sequenced at ∼5.2 reads/base, suggesting that the plasmid was present at a 1.6 excess or nearly 2 copies/cell. This low number seems reasonable since DNA was prepared from stationary phase cells in which plasmid copy number is expected to be low. The plasmid region between TID junction points was read 19.3 times/nucleotide, suggesting an amplification of approximately sixfold vis-à-vis the chromosome (fourfold vis-à-vis the rest of the plasmid). While this complete genome sequence revealed the junction sequences and the amplification of the affected region, it cannot demonstrate the structure of the amplified array (see below).
Figure 5.—
Depth of Illumina reads from the F′128 lac plasmid of a strain carrying a TID amplification. The plasmid genotype is diagrammed below the graph with the F factor plasmid sequence indicated by a bold line and the E. coli material by a finer line. Strains are recA and were grown on glycerol with no selection for lac amplification. The read depth for chromosomal genes (not shown) was ∼90-fold. The nearly 200-fold coverage seen for the plasmid (plotted horizonal line) suggests that the strain has several copies of the F′128 plasmid. The lac region is amplified ∼4-fold above the rest of the plasmid.
Even a complete genome sequence is insufficient to define the structure of a TID amplification:
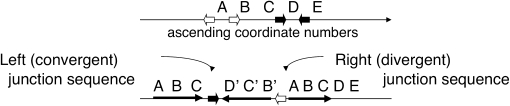
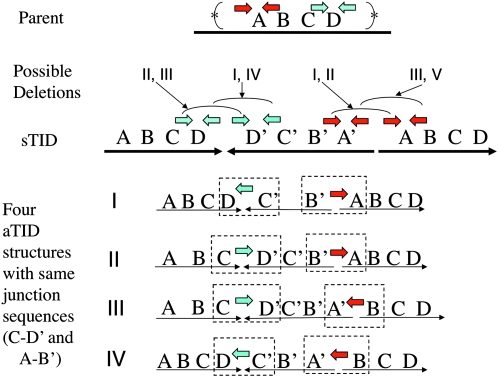
Each TID junction carries a short sequence element at which an exchange appears to have occurred to generate the inversion (see colored short arrows in Figure 3). The parent sequence has two copies of each junction element in opposite orientation. In the sTID, the sequence on opposite sides of the junction is inverted, which brings copies of the repeat element into direct orientation, such that they can serve as sites for deletion formation. As seen in Figure 6, there are two ways of using these repeat pairs to form a deletion, and each deletion type leads to a different array structure. However, the local junction sequence is identical regardless of which pair is chosen. That is, one cannot tell from local sequence information how that junction is oriented in the chromosome: a C–D′ junction is indistinguishable from a D–C′ junction. As diagrammed in Figure 6, one pair of junction sequences, C–D′(or C′–D) and A–B′(or B–A′), is consistent with any of four different TID structures. The uncertainty reflects the fact that sequence determination requires the assembly of short reads. When the parent region includes repeats of a sequence that is many kilobases in length (substantially longer than the sequence genomic fragment), the sequence reads cannot be unambiguously assembled.
Figure 6.—
Multiple structures are consistent with a single pair of junction sequences. In the examples listed (I–IV), the leftmost (converging) junction is D–C′ (equivalent to C–D′) and the rightmost (diverging) junction is B′A (equivalent to A′B). If the asymmetric inversion junctions are formed by deletions in a symmetrical precursor, then four possible structures could be generated by the various combination of deletions described in the bottom line. The colored arrows indicate repeated sequences present in inverse order in the parent chromosome and how they contribute to aTID formation.
A genetic method for testing the structure of an inversion duplication:
One can, in principle, decide which of the four structures in Figure 6 is correct by determining the orientation of each junction sequence with respect to an unamplified outside reference sequence. To make this test, a known junction of an aTID amplification was marked with an added sequence. This strain was used as the recipient for a transformation cross that revealed the orientation of the marker. First, a DNA fragment encoding the tetracycline resistance determinant (tetRA of Tn10) was added to a sequenced aTID junction (orientation unknown) by Red-mediated transformation (Figure 7, middle). Orientation of the inserted tetRA sequence was then determined by a second transformation for which the constructed donor fragment included a CamR marker flanked on the left by an outside reference sequence and on the right by a sequence from the tetRA fragment (one orientation or the other). The donated drug resistance can be inherited only if the recombining sequences (40 bp long) on the right side (in donor and recipient) are in the same orientation. The tetRA recombining sequence is chosen so that inheritance of the CamR marker deletes part of the TetR determinant. The orientation of the recipient tetRA sequence dictates which of the donor fragments (Ref-Cam-tetRA or Ref Cam-ARtet) allows introduction of CamR and removal of the recipient TetR phenotype. Control transformations using simple lac∷Tn10 insertions with no amplified array showed that formation of a CamR transformant requires that the recipient has a tetRA sequence in the orientation concordant with the donor fragment.
Figure 7 describes this cross as applied to the left junction of the strains diagrammed in Figure 6. The cross reveals whether the recipient junction resembles that of strains I and IV (in Figure 6) or strains II and III. Learning the complete structure of the duplication would require determining the orientation of the right junction by a similar series of crosses.
The procedure described above gave unanticipated results. It was applied to the convergent junction of the aTID amplification strain TT25771 (see Table 1, line 3). This junction has a known sequence but an unknown orientation. A tetRA cassette was added to the junction (Figure 7, middle), and a second transformation cross was done using two donor fragments with oppositely oriented tetRA sequences (Figure 7, bottom). Only one of the two donor fragments was expected to yield CamR transformants because only one should have a TetRA sequence whose orientation matches that of the recipient junction. When the procedure was repeated using a reference sequence on the other side of the amplified region, congruent results were expected, i.e., only one of the two donor fragments was expected to yield CamR transformants. The arrangement and results of the crosses are in Table 2. These crosses were done in RecA+ strains.
Contrary to expectation, all four donor fragments gave CamR transformants, regardless of the orientation of their tetRA recombining sequence. This suggested that the recipient population contained multiple tetRA sequence elements, some in each orientation. Either some cells have tetRA in one orientation and some in the other or multiple orientations are present within a single amplified array. Also contrary to expectation, roughly half of the CamR transformants retained TetR instead of losing it as predicted by the design of the cross. It had been expected that the TetR resistance determinant (tetRA) would be added to only one junction and the deletion made by the second cross would remove this TetR determinant. The result obtained suggested that the arrays have multiple copies of TetRA, even though only one was introduced by transformation into the amplification strain. The results suggested that the repeats of the TID amplification were rearranged during growth. This is possible because the junctions were marked and cells for this cross were grown in RecA+ cells. It is interesting that crosses involving the “forward”-oriented Tn10 element gave more transformants than those involving a “reversed” Tn10. This result suggests the presence of more forward-oriented Tn10 elements in the array than reverse-oriented ones.
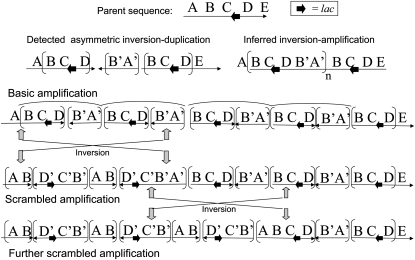
Taken together, the transformation crosses suggested that the arrays were expanding and contracting such that the introduced TetRA sequence was subject to amplification and loss during subsequent (nonselective) growth. It also suggested that the orientation of some of the introduced TetRA sequences changed during growth. Thus, it appears that the amplified arrays generated from a tandem inversion duplication have a structure that is complex. The array is being scrambled during growth, both in terms of copy number and join point orientation. A look at the sequence of an aTID array shows why this might have been expected (see Figure 8). This figure diagrams the consequences of an inversion arising between inverse sequence repeats. The end result is an amalgam of the four types diagrammed in Figure 6.
Figure 8.—
Effects of inversions on structure of amplified aTID array. One initial aTID structure is depicted with the changes that are possible due to amplification followed by recombination between inverse-order sequence repeats.
The arrays have direct and inverse-order repeats of extensive sequence blocks that are subject to recombination events that can change copy number and orientation. The copy-number changes seem reasonable (in retrospect), but were not expected to occur so frequently. Part of the explanation may be the intense recombination known to occur on the F′ plasmid (Seifert and Porter 1984; Syvanen et al. 1986). This recombination may continuously scramble the array during growth and thereby break down the formal distinction between the TID types diagrammed in Figure 6, which are expected only for a simple expansion without any inversion events.
Regardless of the nature of the initial duplication, the amplified array is predicted to become a heterogeneous series of segments containing, in random order, all of the possible inverse and direct-order sequence blocks diagrammed in Figure 6. If this structure were not scrambled, one would expect that some TID arrays would have no copies of lac in inverse order. Given that scrambling occurs, all TID arrays are predicted to have some copies of lac in inverse order once the array expands and inversions occur. This may provide a diagnostic test for TID amplifications.
A model for multi-step formation of asymmetric inversion duplications:
The defining properties of aTID duplications make it difficult to explain their origin by a single event. If they were to form by simple break-and-join events, then at least three parental copies would be required to acquire four simultaneous breaks and two rejoining events with very short junction sequence elements. The shortness of junction sequences (4–31 bp) makes it unlikely that homologous recombination is involved since the strand exchange protein (RecA) requires more extensive substrates. To make these junctions by single-strand annealing would require both multiple breaks and production of extremely long (10–20 kb) single-stranded regions. It has been suggested that junction sequence elements might appear at a broken 3′ end and initiate replication on a sister chromosome (Hastings et al. 2009). However, formation of an aTID duplication in this way would require two simultaneous breaks, each providing an end for RecA-mediated strand invasion and replication initiation at extremely short primer sequences.
The alternative model diagrammed in Figure 2 is described here in more detail. The model suggests that duplications form by multiple sequential events occurring at different times (in different cell generations). This requires simpler genetic events and may explain the formation of both standard tandem (SJ) and aTID duplications without RecA function, which has recently been reported (Reams et al. 2010). It also explains why both duplication types are more frequent following growth under selection. Each successive step in their formation provides an additional fitness advantage under selection. The first event is the formation of a symmetrical inversion duplication sTID with three copies of lac. The sTID is predicted to form at a high rate, assuring that the structures are common in any population despite being deleterious and unstable. When selection favors more lac copies, the inherent fitness cost and instability of the duplication is offset leading to an increased frequency in the population (Reams et al. 2010). As cells grow under selection, secondary deletion mutations reduce fitness cost, increase stability, and therefore allow higher lac amplification (see Figure 3). The underlying events are frequent with or without selection, but the intermediates in the process are usually lost by reversion and counterselection. Selection helps maintain the unstable intermediates in the population and favors completion of the rearrangement process.
Details of the model for formation of an aTID amplification:
The symmetrical inversion duplication (sTID) serves as precursor for an aTID:
The initial step in forming an aTID is generation of the symmetrical precursor (the sTID), which has extended palindromic sequences at each of its two junctions (see Figure 2). These junctions are prone to extrusion as a hairpin structure, whose likelihood is dictated by the size of the loop at the junction, i.e., the deviation of the center of the palindrome from perfect symmetry. Perfect palindromes of several hundred base pairs are prone to hairpin formation and are virtually lethal in bacteria (Leach 1994), and even small increases in loop size are known to increase stability (Warren and Green 1985). The proposed sTID junctions are not perfectly symmetrical, but are sufficiently stable to be replicated and amplified (by exchanges between the flanking direct repeats). Fitness cost may result from occasional hairpin formation and also from simple increases in gene dosage, which are known to impair the growth of copy-number variants (Reams et al. 2010).
Formation of the symmetrical precursor:
It is proposed that imperfect palindromic sequences in the normal sequence initiate sTID formation by the events described in Figure 2. Events of the type proposed here have been described by others in a variety of organisms (Persky and Lovett 2008). After replication fork stalling, single strands of nascent DNA are released and contribute to chromosome rearrangement (Lovett 2004). Short (imperfect) palindromic sequences are frequent (Stern et al. 1984; Bachellier et al. 1999; Vasconcelos et al. 2000) and can form “snap-back” structures in single-stranded DNA (see Figure 2).
A snap-back structure can prime repair synthesis using its own strand as template. This synthesis must open the template duplex and is likely to be prone to template switching, which has been observed in many situations, especially for the PolA repair polymerase (Ross et al. 1979; Ahmed and Podemski 1998; Pinder et al. 1998; Lovett 2004; Lee et al. 2007). Strand extension after this template switch generates a branched structure that can be resolved either by chromosome replication or by repair of the three-way junction to yield a symmetrical inversion duplication (see nick indicated in Figure 2). Formation of a symmetrical inversion duplication in this way is predicted to be independent of RecA function since it involves no strand-exchange event. These series of events may provide a way of circumventing hairpins in the template strand without requiring template breaking, homology seaches, and fork rebuilding.
Secondary deletion events remodel the sTID to form an aTID:
The initial symmetrical TID is expected to form at a high rate. The sTID is expected to be deleterious due to copy-number increase and stem formation. However, this cost can be reduced by deletions that remove material and reduce junction symmetry to produce an aTID. Such remodeling deletions (diagrammed in Figures 3 and 6) are also expected to be frequent. There are several reasons for this. A large region is available on either side of the symmetrical junction that can be removed without fitness penalty. Deletion formation at symmetrical junctions is enhanced by the known propensity of PolA to switch templates, which creates deletions as seen by Leach and coworkers in reversion of toxic perfect palindrome structures (Sharp and Leach 1996; Leach et al. 1997) and by Kleckner and co-workers in “imprecise excision” of Tn10 insertions (Foster et al. 1981). Frequent asymmetric deletions remove the extensive (51 bp) inverse repeat near one end of transposable “Mud” elements (Zieg and Kolter 1989); highly asymmetric deletions of this stem in the Salmonella chromosome were recently measured at 10−6/cell/division (S. Quinones-Soto and J. R. Roth, unpublished results) and are made more common by prior amplification of the sTID because modifying deletions can have endpoints in any pair of previous repeats.
The role of selection in aTID formation and amplification:
During selection for lac amplification, the benefits conferred by extra lac copies can compensate for the fitness cost and instability of the initial sTID and thereby favor its retention in the population. As a clone expands under selection, join point deletions improve fitness, increase stability, and allow higher lac amplification (as aTID). Amplifications can occur by unequal exchanges between direct-order sequences, which have been found to occur at rates that approximate 10−2/cell/division (Reams et al. 2010) and thus provide a frequent source of variants that improve growth rate under selection. In this way, selection favors rapid completion of a process that involves unstable deleterious intermediates.
A fulfilled prediction of the model for aTID formation:
If the aTID amplifications described here form as proposed above, one might expect to find occasional amplifications in which only one of the two junction types has been remodeled and the other retains the initial snap-back structure. A TID junction revealed by Illumina genome sequencing (Table 1, line 5, left column) fits this description. A complex hairpin structure at this junction (left side of Figure 9) is present in the parental sequence and appears at the convergent junction of the TID (junction 1 in Figure 9). The divergent junction (junction 2 in Figure 9) is of the deletion-modified aTID type and was characterized by PCR analysis (and verified by the Illumina sequence). We suggest that the stem-loop structure in Figure 9 supported a snap-back pairing that primed the formation of a more extended inverse-order repeat, and this junction was not further modified by deletion (junction 1 in Figure 9).
Figure 9.—
A TID amplification with one unmodified symmetrical junction. The diagram at left and the solid arrows at top right indicate a parental quasi-palindromic sequence that might generate snap-back pairing. The events at right describe how the snap-back structure might prime repair synthesis, leading to the TID junction sequences found in strain TT25790 (Table 1). The quasi-palindromic junction is extended without modification. The second junction forms by template switching and is later modified by deletion.
The inverse-order sequence found at this junction extends the base of the parental structure. The imperfections of the parental snap-back structure may be sufficient to make the extended palindrome (formed by replication from the snap-back structure) resistant to hairpin formation without further modification. We cannot eliminate the possibility that this junction was formed by a deletion arising in a larger sTID, but this alternative would require a deletion that fortuitously ended exactly at the base of an extensive quasi-palindromic stem-loop. It seems more likely that a quasi-palindromic snap-back structure initiated the formation of a structure that was not modified by deletion. The frequency of such junctions is likely to be revealed by genome sequences of more amplifications with “recalcitrant” junctions (see below).
Explaining why many “recalcitrant” junctions were missed by PCR:
Analysis of an initial set of 99 amplification strains by PCR revealed first the simple SJ junctions described previously (Kugelberg et al. 2006). The remaining junctions were then tested using PCR primer sets designed to identify TID junctions as described above in Table 1. In some strains, only one junction could be identified. Many other strains proved to be completely “recalcitrant” in that PCR yielded neither an SJ nor any aTID join point. The recalcitrant junctions could have escaped detection for mundane reasons; i.e., our primer sets may have been too limited. Alternatively, the recalcitrant junctions may reflect a third type of join point—one that standard PCR fails to reveal.
The junction described in Figure 9 was recalcitrant to PCR analysis, but was shown by Illumina sequencing to be part of a TID amplification. The junction is different from those found by PCR: an extensive quasi-symmetrical sequence was present in the wild-type sequence and may have been extended by repair replication from a parental snap-back structure. It seems possible that the imperfect symmetry of this junction makes it resistant to standard PCR. That is, this sequence may self-anneal when made single-stranded in the PCR process.
To test this idea, the genome sequence of this amplification strain (TT25790) was used to design primers that flank the symmetrical structure in Figure 9. So far we have been unable to recover a PCR product that includes the suggested junction sequence. If this interpretation is correct, junctions of this type may be common and genome sequencing of more strains with recalcitrant junctions may reveal many unmodified amplification junctions that are missed by PCR. More examples of this type would provide support for the multi-step model for aTID amplification formation.
sTID duplications may also give rise to SJ amplifications:
The sTIDs proposed to initiate formation of aTID junctions, can, in principle, also lead to SJ duplications. This idea is diagrammed in Figure 10 in the context of the overall process of reversion under selection in the Cairns system. Each detected aTID junction is proposed to form by a separate deletion event that removes one symmetrical junction of the parent sTID, rendering that junction asymmetric (right side of Figure 10). The endpoints of each deletion dictated the junction sequence elements for each TID junction listed in Table 1. These separate deletions can occur independently and may be frequent because palindromic sequences are known to be subject to frequent deletion events (discussed later).
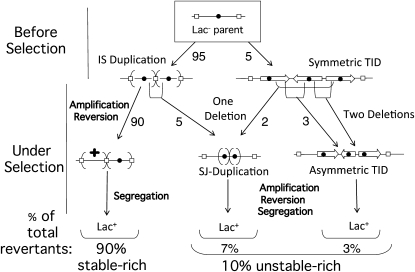
Figure 10.—
The sTID can also contribute to formation of an SJ duplication. The parental sequence is diagrammed at the top (boxed) in which open rectangles designate a copy of IS3 and the solid circle designates the lac operon. An SJ duplication can form when a deletion removes the junction of an IS3 duplication (left middle). An SJ duplication also forms when a single deletion removes both sTID junctions (right middle). Two deletions convert an sTID to an aTID duplication. Some IS3-bounded lac duplications may form through sTID intermediates.
However, the two symmetrical junctions of an sTID duplication can also be removed by a single deletion that removes the entire central repeat segment (Figure 10, middle). Such a deletion would lead to an SJ duplication, identical to that proposed to arise from a large parent IS duplication in Figure 1 and Figure 10 (left). An SJ duplication can be highly amplified under selection, regardless of how it formed. One diagrammed route for SJ duplication (Figure 10, left) can be blocked by removing IS3 repeats from the parent strain. This allows a test of the possibility of other routes to SJ duplications.
Effect of IS3 removal on the relative frequency of SJ and aTID amplifications:
Following prolonged selection for improved growth on lactose, the standard Cairns strain (with IS3 copies flanking lac) generates unstably Lac+ revertants with either SJ or aTID amplifications. It was suggested above that some SJ types arise when deletions remove the join point from an IS3 duplication (see Figure 1) or when a deletion removes the entire center segment from an sTID join point (Figure 10). In either case, fitness is improved and higher amplification is allowed, which is favored by selection. Without selection, both the SJ and aTID duplication types were rare and the recovered duplications were primarily those with IS3 junctions.
Removal of IS3A and -C from the Salmonella version of the Cairns assay strain reduced the revertant number about fourfold (Table 3). The amplifications recovered from unstable-rich revertant colonies showed fewer SJ types relative to aTID types. The fact that any SJ types were found at all suggested that their formation does not require IS3 duplications as a precursor (consistent with their formation from sTID precursors). The effects of removing IS3 sequences suggest that 2/3 of the observed SJ amplifications come from precursors with an IS3 duplication and 1/3 arise by another mechanism (e.g., an sTID).
TABLE 3.
Effect of removing IS3 repeats on the frequency of SJ and aTID duplications
| Duplication/amplification type (join-point type)a |
||||||
|---|---|---|---|---|---|---|
| Source of strains with repeat structures | SJ short join | REP junctions | IS3A/IS3C recombination | Transposition of some IS | aTID | Probable aTID |
| Amplifications arising under prolonged selection on lactose | ||||||
| Standard Cairns strain | 48% (48) | 15% (15) | 0 | 1% (1) | 6% (6) | 29% (29) |
| Strain without IS3 (4-fold fewer revertants) | 18% (3) | 12% (2) | 0 | 24% (4) | 6% (1) | 41% (7) |
| Duplications trapped following nonselective growth on glycerol | ||||||
| Standard Cairns strain | 0 | 20% (4) | 80% (20) | 0 | 0 | 0 |
| Strain without IS3 | 0 | 95% (16) | 0 | 0 | 5% (1) | 0 |
Percentages describe frequency of each duplication type among those from source strain. Parentheses enclose the actual number of duplications of that type used to determine percentage.
A rough estimate of the breakdown of revertant types in the Cairn system is shown in Figure 10. Most revertant colonies (90%) contain predominantly stably Lac+ cells, reflecting a reversion event early in the history of one of the many plated cells with an unmodified low-copy IS3 amplification (left side of Figure 10). Unstable-rich revertants (10%) have amplifications with either an SJ or an aTID junction. Seventy percent of these (7% of total) have an SJ amplification and 30% (3% of the total) have an aTID amplification. This estimate suggests that sTID duplications generate amplifications of which 1/3 are the SJ type and 2/3 are the aTID type. Unselected duplications isolated from strains lacking IS3 showed predominately REP element junctions among the reduced number of duplications found. However, one aTID type was recovered. This shows that (as for SJ types) aTID can occur in the absence of selection.
Role of F′ plasmid in aTID formation:
All of the aTID structures reported here arose on the F′ plasmid, whose transfer replication is known to stimulate RecA-dependent recombination (Seifert and Porter 1984; Syvanen et al. 1986). In our Salmonella strains, the genes for plasmid transfer (tra) are repressed. Several lines of evidence suggest that TIDs form in the chromosome and that the plasmid contributes primarily to the RecA-dependent amplification events that underlie the reversion in the Cairns experiment. Chromosomal duplications (like those on F) can form without RecA and would be explained by the model presented in Figure 10 for conversion of an sTID to an SJ duplication.
DISCUSSION
The TID rearrangements described here and the SJ tandem duplications described previously are rare in unselected cultures but common among amplifications isolated after prolonged selection for increased lac copy number. Both duplication types are proposed to arise from a very common, but normally short-lived precursor, the sTID. The initial rearrangement is proposed to form at a high rate but disappear rapidly from the population due to fitness cost and instability. Only when cells are placed under selection for increased gene copy number are the precursor structures maintained such that they can be remodeled and lead to a detectable amplification. How could these rearrangements have escaped detection for so long?
The standard practice of bacterial genetics is to use stringent positive selection to detect mutants that arise during preceding nonselective growth. These restrictive conditions have made laboratory bacterial genetics possible. Their efficacy was demonstrated by Luria, Delbrück, and Lederberg (Luria and Delbruck 1943; Lederberg and Lederberg 1952). Although these stringent conditions detect rare preexisting cells with large-effect mutations, they prevent common small-effect mutants (such as sTID duplications) from contributing to the yield of selected colonies. They systematically eliminate detection of most copy-number variants.
The Cairns selection system uses less stringent conditions than standard laboratory positive selection. The level of β-galactosidase in the parent strain is 2% of that in revertants. Selection is just stringent enough to prevent growth of the parent strain, but allows slow growth of mutants with a few extra copies of the mutant lac allele. These copy-number variants are common: ∼1 in 500 cells of the unselected plated culture carries a lac duplication. Selective conditions favor copy-number increases, which occur at a very high rate (10−2/cell/division)—1 million-fold higher than the reversion rate of the Cairns frameshift mutation. The nonstringent Cairns selection conditions (which more closely approximate natural selection) detect common copy-number variants and ultimately reveal the SJ and aTID amplifications described here. The low stringency of this selection may allow this system to reveal aspects of genetic adaptation that have been missed by stringent laboratory selection conditions and mutant screens.
Formation of duplications by a RecA-independent pathway:
Duplications have been assumed to arise by unequal recombination between extensive sequence repeats in sister chromosomes. Recent measurements suggest that homologous (RecA-mediated) recombination is not required for duplication formation even when exchanges appear to form between substantial sequence repeats (Reams et al. 2010). The model presented here may explain the RecA independence of duplication formation. That is, a common precursor (the sTID) may form in a RecA-independent manner and then be converted to either an SJ or an aTID duplication by RecA-independent deletions. These deletions are expected to be RecA-independent even when they arise between extensive sequence elements. Thus many simple tandem duplications (including some of the frequent IS3-bounded duplications) may form without recombination by multi-step pathways initiated by sTID rearrangements (see Figure 10).
The gene amplification processes described here may be major contributors to adaptive evolution because they allow extremely frequent genetic variants to be detected by selection. Whenever cellular growth is restricted (but not eliminated) by external conditions, copy-number differences may be the most common source of improvement. Copy-number variants can contribute to subsequent sequence change by providing more copies of the growth-limiting gene and by allowing expansion of a subpopulation in which point mutations arise. Later adaptive mutations may occur within the amplified genes or in unrelated genomic regions, whose likelihood is enhanced only by growth (Roth et al. 2006; Andersson et al. 2010).
For example, selective gene amplification and subsequent adaptive mutation have been shown to occur during selection for improved growth (i) on limiting carbon sources (Horiuchi et al. 1963; Sonti and Roth 1989; Andersson et al. 1998; Hendrickson et al. 2002; Slechta et al. 2003; Kugelberg et al. 2006), (ii) in the presence of toxic compounds such as antibiotics (Sandegren and Andersson 2009; Sun et al. 2009) with defective translation machinery (Nilsson et al. 2006; Lind et al. 2010), or with impaired biosynthetic pathways. The process may underlie evolution of new genes under continuous selection (Bergthorsson et al. 2007). The frequent initiation and rapid modification and expansion of gene amplifications make them challenging to study, but suggest that their contribution to genetic adaptation may have been seriously underestimated.
Acknowledgments
We thank the West Quad Computing Group at Harvard Medical School for computational resources and support. F.P.R. was supported by grants from the National Institutes of Health (NIH; G00423, HG004756, HG003224 and HG0017115) and by the Canadian Institute for Advanced Research. J.M. was supported by an Individual National Research Service Award from the NIH/National Human Genome Research Institute (HG004825). This work was supported in part by NIH grant GM27068 (J.R.R.) and grants from the Swedish Research Council to D.I.A. Elisabeth Kugelberg was supported by a fellowship from Wenner–Gren Foundation.
References
- Ahmed, A., and L. Podemski, 1998. Observations on template switching during DNA replication through long inverted repeats. Gene 223 187–194. [DOI] [PubMed] [Google Scholar]
- Andersson, D. I., E. S. Slechta and J. R. Roth, 1998. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science 282 1133–1135. [DOI] [PubMed] [Google Scholar]
- Andersson, D., D. Hughes and J. Roth, 2010. The origin of mutants under selection: interactions of mutation, growth and selection, in EcoSal: Escherichia coli and Salmonella, Cellular and Molecular Biology. ASM Press (in press). [DOI] [PubMed]
- Bachellier, S., J. M. Clement and M. Hofnung, 1999. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol. 150 627–639. [DOI] [PubMed] [Google Scholar]
- Bergthorsson, U., D. I. Andersson and J. R. Roth, 2007. Ohno's dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. USA 104 17004–17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, J., and P. L. Foster, 1991. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics 128 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie, G. A., C. B. Millar, K. H. Schmidt and D. R. Leach, 2000. Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics 154 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, P. L., 2007. Stress-induced mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 42 373–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, T. J., V. Lundblad, S. Hanley-Way, S. M. Halling and N. Kleckner, 1981. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell 23 215–227. [DOI] [PubMed] [Google Scholar]
- Galhardo, R. S., P. J. Hastings and S. M. Rosenberg, 2007. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 42 399–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson, E., J. M. Clement, D. Brutlag and M. Hofnung, 1984. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 3 1417–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, B. G., 1998. Adaptive mutagenesis: a process that generates almost exclusively beneficial mutations. Genetica 102–103 109–125. [PubMed] [Google Scholar]
- Hastings, P. J., H. J. Bull, J. R. Klump and S. M. Rosenberg, 2000. Adaptive amplification: an inducible chromosomal instability mechanism. Cell 103 723–731. [DOI] [PubMed] [Google Scholar]
- Hastings, P. J., J. R. Lupski, S. M. Rosenberg and G. Ira, 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson, H., E. S. Slechta, U. Bergthorsson, D. I. Andersson and J. R. Roth, 2002. Amplification-mutagenesis: evidence that “directed” adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc. Natl. Acad. Sci. USA 99 2164–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi, T., S. Horiuchi and A. Novick, 1963. The genetic basis of hyper-synthesis of β-galactosidase. Genetics 48 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid, E., U. Bergthorsson, E. S. Slechta and J. R. Roth, 2003. Formation of an F′ plasmid by recombination between imperfectly repeated chromosomal Rep sequences: a closer look at an old friend (F′(128) pro lac). J. Bacteriol. 185 660–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelberg, E., E. Kofoid, A. B. Reams, D. I. Andersson and J. R. Roth, 2006. Multiple pathways of selected gene amplification during adaptive mutation. Proc. Natl. Acad. Sci. USA 103 17319–17324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, D. R., 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16 893–900. [DOI] [PubMed] [Google Scholar]
- Leach, D. R., E. A. Okely and D. J. Pinder, 1997. Repair by recombination of DNA containing a palindromic sequence. Mol. Microbiol. 26 597–606. [DOI] [PubMed] [Google Scholar]
- Lederberg, J., and E. M. Lederberg, 1952. Replica plating and indirect selection of bacterial mutants. J. Bacteriol. 63 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. A., C. M. Carvalho and J. R. Lupski, 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131 1235–1247. [DOI] [PubMed] [Google Scholar]
- Li, H., J. Ruan and R. Durbin, 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind, P., C. Tobin, O. G. Berg, C. G. Kurland and D. I. Andersson, 2010. Compensatory gene amplification restores fitness after inter-species gene replacements. Mol. Microbiol. 75 1078–1089. [DOI] [PubMed] [Google Scholar]
- Lovett, S. T., 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol Microbiol 52 1243–1253. [DOI] [PubMed] [Google Scholar]
- Luria, S. E., and M. Delbruck, 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, A. I., A. Zorzet, A. Kanth, S. Dahlstrom, O. G. Berg et al., 2006. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc. Natl. Acad. Sci. USA 103 6976–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persky, N. S., and S. T. Lovett, 2008. Mechanisms of recombination: lessons from E. coli. Crit. Rev. Biochem. Mol. Biol. 43 347–370. [DOI] [PubMed] [Google Scholar]
- Pinder, D. J., C. E. Blake, J. C. Lindsey and D. R. Leach, 1998. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 28 719–727. [DOI] [PubMed] [Google Scholar]
- Reams, A. B., E. Kofoid, M. Savageau and J. R. Roth, 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184 1077–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, D. G., J. Swan and N. Kleckner, 1979. Nearly precise excision: a new type of DNA alteration associated with the translocatable element Tn10. Cell 16 733–738. [DOI] [PubMed] [Google Scholar]
- Roth, J. R., E. Kofoid, F. P. Roth, O. G. Berg, J. Seger et al., 2003. Regulating general mutation rates: examination of the hypermutable state model for Cairnsian adaptive mutation. Genetics 163 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, J. R., E. Kugelberg, A. B. Reams, E. Kofoid and D. I. Andersson, 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Microbiol. 60 477–501. [DOI] [PubMed] [Google Scholar]
- Sandegren, L., and D. I. Andersson, 2009. Bacterial gene amplification: implications for the evolution of antibiotic resistance. Nat. Rev. Microbiol. 7 578–588. [DOI] [PubMed] [Google Scholar]
- Seifert, H. S., and R. D. Porter, 1984. Enhanced recombination between lambda plac5 and mini-F-lac: the tra regulon is required for recombination enhancement. Mol. Gen. Genet. 193 269–274. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., and D. R. Leach, 1996. Palindrome-induced deletion in enterobacterial repetitive sequences. Mol. Microbiol. 22 1055–1056. [DOI] [PubMed] [Google Scholar]
- Slack, A., P. C. Thornton, D. B. Magner, S. M. Rosenberg and P. J. Hastings, 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2 e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta, E. S., J. Liu, D. I. Andersson and J. R. Roth, 2002. Evidence that selected amplification of a bacterial lac frameshift allele stimulates Lac(+) reversion (adaptive mutation) with or without general hypermutability. Genetics 161 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slechta, E. S., K. L. Bunny, E. Kugelberg, E. Kofoid, D. I. Andersson et al., 2003. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc. Natl. Acad. Sci. USA 100 12847–12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonti, R. V., and J. R. Roth, 1989. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics 123 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, M. J., G. F. Ames, N. H. Smith, E. C. Robinson and C. F. Higgins, 1984. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37 1015–1026. [DOI] [PubMed] [Google Scholar]
- Sun, S., O. G. Berg, J. R. Roth and D. I. Andersson, 2009. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics 182 1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syvanen, M., J. D. Hopkins, T. J. Griffin IV, T. Y. Liang, K. Ippen-Ihler et al., 1986. Stimulation of precise excision and recombination by conjugal proficient F′plasmids. Mol. Gen. Genet. 203 1–7. [DOI] [PubMed] [Google Scholar]
- Tlsty, T. D., A. M. Albertini and J. H. Miller, 1984. Gene amplification in the lac region of E. coli. Cell 37 217–224. [DOI] [PubMed] [Google Scholar]
- Vasconcelos, A. T., M. A. Maia and D. F. de Almeida, 2000. Short interrupted palindromes on the extragenic DNA of Escherichia coli K-12, Haemophilus influenzae and Neisseria meningitidis. Bioinformatics 16 968–977. [DOI] [PubMed] [Google Scholar]
- Warren, G. J., and R. L. Green, 1985. Comparison of physical and genetic properties of palindromic DNA sequences. J. Bacteriol. 161 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieg, J., and R. Kolter, 1989. The right end of MudI(Ap,lac). Arch. Microbiol. 153 1–6. [DOI] [PubMed] [Google Scholar]