Abstract
Nonallelic gene conversion has been proposed as a major force in homogenizing the sequences of paralogous genes. In this work, we investigate the extent and characteristics of gene conversion among gene families in nine species of the genus Drosophila. We carried out a genome-wide study of 2855 gene families (including 17,742 genes) and determined that conversion events involved 2628 genes. The proportion of converted genes ranged across species from 1 to 9% when paralogs of all ages were included. Although higher levels of gene conversion were found among young gene duplicates, at most 1–2% of the coding sequences of these duplicates were affected by conversion. Using a second approach relying on gene family size changes and gene-tree/species-tree reconciliation methods, we estimate that only 1–15% of gene trees are misled by gene conversion, depending on the lineage considered. Several features of paralogous genes correlate with gene conversion, such as intra-/interchromosomal location, level of nucleotide divergence, and GC content, although we found no definitive evidence for biased substitution patterns. After considering species-specific differences in the age and distance between paralogs, we found a highly significant difference in the amount of gene conversion among species. In particular, members of the melanogaster group showed the lowest proportion of converted genes. Our data therefore suggest underlying differences in the mechanistic basis of gene conversion among species.
IN every species, the vast majority of new genes derive from the duplication of genes already present in the genome. The duplication, loss, and sequence divergence of genes is thought to be largely responsible for the diversification of living organisms (Ohno 1970; Conant and Wolfe 2008; Hahn 2009). In most cases, the nucleotide sequences of duplicate genes tend to diverge over time. However, as long as paralogous genes share regions of sequence similarity, they can be involved in recombination events. In evolutionary terms, the crossover between homologous chromosomes during meiosis in sexually reproducing organisms represents the most relevant outcomes of recombination. But recombination can also result in the unidirectional transfer of DNA sequences, a process known as gene conversion, which has been shown to occur between both allelic and paralogous sequences.
Gene conversion and crossover differ in some essential aspects. Gene conversion implies the replacement of an acceptor DNA sequence with a donor sequence that usually does not exceed a few kilobases. On the other hand, crossing-over between two homologous chromosomes leads to an exchange of sequences that will be delimited by another crossover event or by the physical end of the chromosome. Both crossover and gene conversion ultimately result from the repair process of double-strand breaks of DNA, but conversion can derive from a broader array of repair pathways (Chen et al. 2007). Gene conversion between alleles influences several aspects of population variation, including patterns of linkage disequilibrium (Langley et al. 2000). In this article, however, we focus on the influence of nonallelic, or “ectopic” gene conversion between paralogous genes. A number of studies have suggested that concerted evolution driven by gene conversion is widespread among gene families such as rRNA (Stage and Eickbush 2007), heat-shock proteins (Bettencourt and Feder 2002), globins (Storz et al. 2007), and histones (Galtier 2003). This view has been challenged by the discovery of high rates of gene gain and loss driving the evolution of clustered gene families (Nei and Rooney 2005). The ultimate contribution of gene conversion to the evolutionary patterns among duplicates is therefore still an open question.
Genome-wide studies have shown that the proportion of duplicate genes with evidence of conversion is relatively low, from ∼2% in Caenhorhabditis elegans to ∼8% in Saccharomyces cerevisiae (in families with more than two genes; it is higher in families of size two), and ∼8–10% in rice (Semple and Wolfe 1999; Drouin 2002; Wang et al. 2007; Xu et al. 2008). In humans, different genome-wide surveys have reported from less than 1% (Benovoy and Drouin 2009) to ∼13% (McGrath et al. 2009) of paralogs affected by gene conversion. The fraction of converted young paralogs in mouse has been also reported to be between 13 and 15% (Ezawa et al. 2006; McGrath et al. 2009). The length of converted tracts between paralogous genes varies from ∼10 bp up to a few kilobases in S. cerevisiae (Drouin 2002), plants (Mondragon-Palomino and Gaut 2005; Wang et al. 2007; Xu et al. 2008), C. elegans (Semple and Wolfe 1999), humans (Jackson et al. 2005; Benovoy and Drouin 2009; McGrath et al. 2009), and Drosophila melanogaster (Gloor et al. 1991).
Recently, growing attention has been dedicated to studying the influence of conversion on patterns of nucleotide substitution, in particular the increased rate of AT → GC substitutions due to allelic gene conversion (Marais 2003; Duret and Galtier 2009). This process, known as biased gene conversion (BGC), is thought to be a major factor in shaping nucleotide composition across the genomes of vertebrates and other organisms (Birdsell 2002; Axelsson et al. 2005; Berglund et al. 2009). In Drosophila, evidence for BGC is more controversial, with some studies suggesting that this bias could be present (Galtier et al. 2006; Haddrill and Charlesworth 2008) and others that it is not (Ko et al. 2006). While the above data all come from allelic gene conversion, BGC has been also suggested to be involved in ectopic conversion events in mammals, birds, S. cerevisiae, and Arabidopsis (Galtier 2003; Kudla et al. 2004; Backstrom et al. 2005; Benovoy et al. 2005), although we did not observe any such pattern in a survey of recent duplicates in four mammalian genomes (McGrath et al. 2009).
A number of studies carried out in Drosophila have detected instances of gene conversion between paralogs, including in the α-amylase gene family (Brown et al. 1990; Hickey et al. 1991; Shibata and Yamazaki 1995), trypsin (Wang et al. 1999), antibacterial peptide attacins (Lazzaro and Clark 2001), esterase (King 1998), and engrailed transcription factors (Peel et al. 2006). Surprisingly, only a few, partial surveys have attempted to address the occurrence of gene conversion in multiple gene families in fruit flies (Thornton and Long 2005; Osada and Innan 2008). Thornton and Long (2005) focused on 13 genes from five families in D. melanogaster and found a very low proportion of converted genes using two different approaches, one relying on the number of shared polymorphisms among paralogs and the second using the GENECONV software package (Thornton and Long 2005). In contrast, the recent study by Osada and Innan (2008) detected gene conversion in 24 out of 28 pairs of paralogs in D. melanogaster using a comparative phylogenetic method (Osada and Innan 2008).
Despite the long history of studies of concerted evolution in Drosophila, the impact of nonallelic gene conversion on the evolutionary history of duplicate genes in this genus remains unknown at a genome-wide scale. The whole-genome sequencing of several Drosophila species provides the opportunity to define the role of ectopic gene conversion in a large-scale context within a well-studied, phylogenetically diverse taxonomic group. We took advantage of these resources to perform a computational survey of conversion among >17,700 paralogous genes in nine Drosophila species using two different methods.
MATERIALS AND METHODS
Data sets:
Drosophila gene duplicate sequences and gene families were obtained as described elsewhere (Hahn et al. 2007). Briefly, gene models from the nine species were obtained from the consensus gene set established by the Drosophila Genome Sequencing and Analysis Consortium (Clark et al. 2007). Gene families were built using the fuzzy reciprocal BLAST (FRB) method, which relies on all-by-all comparisons between the genomes using BLASTP; gene families are formed in the clustering step of FRB by traversing the graph of pairwise similarities to find the maximally connected clusters that are disjoint from one another while discarding nonreciprocal relationships (Clark et al. 2007). Paralogs from each family were separated according to the species they belonged to and reprocessed to obtain a better species-specific multialignment as follows. Protein sequences corresponding to these paralogs were aligned and reverse translated into their coding sequences using transAlign (Bininda-Emonds 2005) implemented with MUSCLE (Edgar 2004) to produce the nucleotide multiple-sequence alignments. To reduce the probability of false positives in the gene conversion analysis, regions of poor alignment quality were removed following a recently described procedure (Han et al. 2009). Gene conversion can be very hard or impossible to infer among identical or nearly identical sequences. Therefore, alignments with fewer than three mismatches were not screened for the GENECONV analyses. In addition, a few large histone gene families with problematic assignment to mapped contigs in non-melanogaster species were excluded from this study. Major gene conversion features, including the number of gene pairs analyzed, number of conversion events, and length of conversion tracts are summarized in supporting information, Table S1 and Table S2.
The quality of a genome assembly influences the nucleotide sequence and length of predicted genes, potentially introducing biases in the detection of gene conversion. Low sequencing coverage of a gene can lead to miscalled base pairs, likely introducing a single-nucleotide difference between paralogous genes. Because we find that duplicated sequences are mostly not converted, the result of these errors is to increase the power of GENECONV to detect conversion events. However, given that the nine species' genomes we analyzed were sequenced at deep coverage (Clark et al. 2007)—and that the low-coverage Drosophila genomes were not included in our analyses—we expect that such a bias might affect only a very limited number of paralogs. Low-quality genomes are also characterized by smaller contigs and a higher number of sequence gaps, which decrease the length of annotated gene-coding regions by loss of exon sequences and splitting of genes in more than one contig. Indeed, D. melanogaster has the best assembly and the longest coding sequences on average among the nine species. We noted that converted genes have shorter coding regions than nonconverted genes (data not shown); therefore, we should expect that genomes with lower sequence quality and overall shorter genes would have higher levels of gene conversion. However, there is no correlation between levels of conversion and average length of coding regions in the nine species (R2 = 0.0054). These observations indicate that the assembly quality most likely is not affecting the observed levels of gene conversion.
Detection of gene conversion events:
To detect gene conversion events among paralogs we used GENECONV v.1.81 [http://www.math.wustl.edu/∼sawyer/geneconv (Sawyer 1989)], which establishes significance of highly similar tracts (representing conversion events) using permutation. GENECONV can recognize conversion tracts comparing all the sequences in the alignments or by single pairwise comparisons. For these tracts, called global and pairwise fragments, respectively, GENECONV calculates P-values corrected for sequence length and also the number of sequences in the case of global comparison. In this work, however, we used GENECONV pairwise P-values to minimize possible biases introduced by families with very different number of paralogs.
GENECONV was run with default settings except for the options required to display pairwise P-values (–ListPair) and to include monomorphic sites in the calculation for alignments of only two sequences (–Include-monosites). The latter option allows the program to take into account constant sites and is required to examine alignments with only two paralogs. All fragments identified by pairwise comparisons with P < 0.05 were regarded as gene conversion events. Therefore, we expect to see 5% of all comparisons to be “significant” even when there is no conversion (see below). Tracts including one or more mismatches were not searched by GENECONV given the chosen settings. However, we noted that at least some putative ancestral converted regions with one or more mismatches were retrieved as multiple shorter tracts separated by one mismatch.
Gene conversion features:
We calculated the proportion of converted genes as the ratio of gene pairs with conversion over the total number of screened pairs per species. The genetic divergence (number of synonymous substitutions per synonymous site or dS) between paralogs was estimated from the Nei–Gojobori method obtained with the codeml package in PAML (Yang 2007). Similar divergence values were obtained from maximum-likelihood dS estimates using the same package. To correct for the decreased genetic divergence estimated between converted pairs, we multiplied the original dS value by the ratio of the alignment length and the length of the alignment minus the conversion tract. Average tract length was calculated using all pairs or only pairs where the tract is not delimited by any exon–intron boundaries or the 5′− or 3′-end of the coding sequence. Scaffolds in non-melanogaster species were mapped to Müller elements in a recent study (Schaeffer et al. 2008), allowing us to estimate the proportion of converted and nonconverted pairs residing on the same Müller elements. Chromosome organization and names differ among Drosophila species, whereas chromosomal arms (Müller elements) identify common units across these species. Therefore, Müller elements represent better physical references for analyses of interspecies, chromosome-wide properties of converted and nonconverted genes.
Effect of gene conversion on phylogenetic trees:
As previously described (McGrath et al. 2009), the effects of gene conversion on phylogenetic trees can be inferred by comparing different methods used for reconstructing the timing of gene duplication events. This is because gene conversion will cause tree-based methods to infer recent duplication events, but the results from copy-number-based methods (such as is implemented in the software package CAFE (Hahn et al. 2005; De Bie et al. 2006)) are not affected by conversion. We inferred lineage-specific duplications in each gene family using two methods. The program NOTUNG (Chen et al. 2000) was employed to calculate the number and timing of duplications by reconciliation of gene and species trees, using the gene trees for each Drosophila family in a previous study (Hahn et al. 2007). We also used the program CAFE (Hahn et al. 2005; De Bie et al. 2006) to estimate the timing of duplications by comparing the size of gene families across different genomes. For each lineage, the proportion of possible trees (gene families) affected by gene conversion is given by the number of families with more duplicates inferred by NOTUNG than by CAFE divided by the total number of families with more than two members (on that lineage).
Statistical analyses:
To determine the significant factors affecting the level of gene conversion, we used an ANOVA, implemented in R (http://www.r-project.org/). Analyzed variables included genetic divergence (dS) and physical distance between pairs, GC content, chromosome location (genes of each pair on the same or different chromosome arms, i.e., Müller elements), and species. We compared two pairs of nested general linear models, with or without the variable “species,” using a likelihood ratio test (LRT).
All Drosophila assemblies, except in the D. melanogaster and D. pseudoobscura genomes, are composed of scaffolds, many of which have not been mapped yet onto specific Müller elements. Moreover, even when two scaffolds are mapped onto the same element, their respective distance is unknown. Therefore, in our analyses of physical distance between intraelement pairs we used only pairs with genes on the same scaffold.
RESULTS
Amount of nonallelic gene conversion in Drosophila genomes:
We investigated the extent of nonallelic gene conversion among paralogous genes in nine Drosophila species. Our analysis was carried out on 17,742 genes from 2855 gene families. A total of 2040 conversion events were detected across 2628 genes from 700 families in these nine Drosophila genomes. Some of these events involved the same orthologous genes in different genomes, and—on the basis of divergence between converted pairs—we estimated that about 200 “ancestral” gene conversion events occurred before the split of different Drosophila species. Because the signature of gene conversion degrades quickly, we are able to detect only those cases where two species split very recently. However, most of the conversion events that we were able to detect occurred after the divergence of species used in our analysis and are therefore unique (see below). The number of gene conversion events, converted pairs, and converted genes varied more than twofold between Drosophila species (Table 1 and Table S1). D. ananassae exhibited the lowest extent of conversion in terms of number of events and number of converted pairs or genes, while at the other end of the spectrum D. grimshawi showed >10% higher values compared to any other Drosophila species (Table 1 and Table S1).
TABLE 1.
Levels of conversion in each species
| Dmel | Dyak | Dere | Dana | Dpse | Dwil | Dvir | Dmoj | Dgri | |
|---|---|---|---|---|---|---|---|---|---|
| Converted genes | 217 | 319 | 213 | 192 | 399 | 261 | 286 | 276 | 465 |
| Genes analyzed | 1723 | 2451 | 1797 | 1848 | 2173 | 1868 | 1731 | 1731 | 2420 |
| % converted pairs | 7.47 | 9.01 | 7.89 | 6.41 | 12.53 | 9.75 | 12.28 | 11.77 | 14.15 |
% converted pairs represents the proportion of converted pairs over all screened pairs. Dmel, D. melanogaster; Dyak, D. yakuba; Dere, D. erecta; Dana, D. ananassae; Dpse, D. pseudoobscura; Dwil, D. willistoni; Dvir, D. virilis; Dmoj, D. mojavensis; Dgri, D. grimshawi.
Levels of nonallelic gene conversion in Drosophila genomes from GENECONV analysis:
We estimated the proportion of converted genes in each species by dividing the number of gene pairs with conversion by the total number of gene pairs that we were able to screen with GENECONV. When pairs of paralogs of any age are considered, the proportion of converted pairs varies from 6.4% in D. ananassae to 14.2% in D. grimshawi. In D. melanogaster, 7.5% of paralogs showed evidence of conversion (Table 1). Note that under the null hypothesis of no conversion we expect to observe 5% of pairs with P < 0.05; therefore, the proportion of true positives likely varies from only 1–9%. These levels of conversion are comparable to what has been observed in S. cerevisiae (for families with more than two genes) and in rice (Drouin 2002; Wang et al. 2007; Xu et al. 2008), whereas only 0.88% of all human and 2% of all C. elegans paralogous pairs, respectively, appeared to have been converted among paralogs of approximately the same level of divergence (Semple and Wolfe 1999; Benovoy and Drouin 2009).
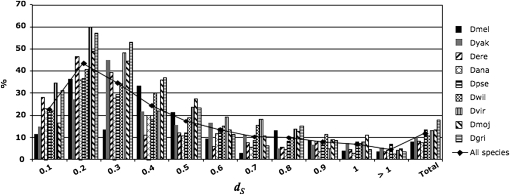
When the proportion of Drosophila converted pairs is plotted against their divergence, we observe that conversion levels are relatively low for very recent duplicates (dS < 0.1). This is most likely due to a higher proportion of gene conversion false negatives in young paralogs, as conversion events between sequences that are already very similar are extremely difficult to detect. Observed conversion levels reach a peak for 0.1 ≤ dS < 0.3 in all species and slowly decrease for paralogs with higher divergence (Figure 1). In species of the Drosophila subgenus the proportion of converted pairs is higher than in members of the Sophophora subgenus for most divergence intervals (Figure 1).
Figure 1.—
Levels of gene conversion (proportion of converted pairs over all analyzed pairs) in nine Drosophila species binned by divergence (dS) values. The line highlights the trend for the average of all nine species.
Other studies have shown that the level of conversion tends to decrease when dS increases (Semple and Wolfe 1999; Xu et al. 2008; Benovoy and Drouin 2009), although a more complicated relationship between divergence and proportion of converted pairs can emerge when only young gene duplicates are examined (McGrath et al. 2009). However, given that different methods and gene duplicates data have been used in these studies, a straightforward comparison between the levels of conversion in different organisms is not always meaningful. Given this perspective, our results offer some clues as to the features affecting variation in the level and patterns of ectopic gene conversion in species with various levels of phylogenetic relatedness (see section Factors affecting gene conversion).
The effect of gene conversion on phylogenetic trees:
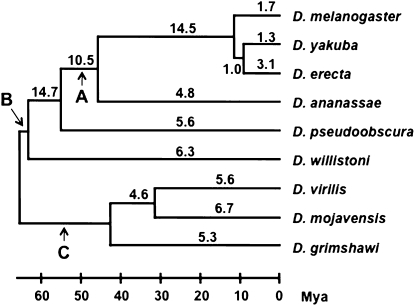
Because very high levels of gene conversion may not be detectable by GENECONV—especially in cases where the entire coding regions of two paralogs have been homogenized—we used a second measure of the effect of gene conversion by applying a procedure we recently developed (McGrath et al. 2009). Because gene conversion decreases the divergence between paralogs, it can increase the number of apparently young duplicates in a given genome; therefore, gene trees constructed from families affected by gene conversion will show evidence for recent duplications. However, gene conversion will not affect the total number of gene copies in a genome, so that methods that infer the timing of duplication based on copy number are unaffected by this bias. To examine the extent to which gene conversion affects phylogenetic reconstruction, we compared the number of recent duplicates calculated by two different methods, one affected and one unaffected by gene conversion. In the first approach the number of branch-specific duplicates is determined by gene-tree/species-tree reconciliation (see materials and methods). In the second method, gene losses and gains are estimated on the species tree using CAFE (Hahn et al. 2005; De Bie et al. 2006), which relies only on the number of paralogs in each genome. A higher number of branch-specific duplications estimated by reconciliation rather than by CAFE indicates either the occurrence of gene conversion or independent gains and losses of genes on different lineages (McGrath et al. 2009). A higher number of duplicates inferred by gene-tree methods can also occur when the species tree used in the reconciliation procedure contains a polytomy (Hahn 2007). Using this procedure we found that for families containing two genes, gene conversion affects from 1 to ∼15% of trees, with levels varying across different branches of the Drosophila species tree (Figure 2). Rates tend to be particularly low in short tip branches, such as in the melanogaster subgroup, which includes D. melanogaster, D. yakuba, and D. erecta (Figure 2). The rate appears to be especially high on the branch leading to the ancestor of the melanogaster subgroup—a known polytomy (Pollard et al. 2006)—which is consistent with the idea that reconciliation methods infer excess duplication events on these branches.
Figure 2.—
Percentage of gene trees that show more duplications on a given branch than are inferred by changes in copy number alone. The phylogram of the nine Drosophila species analyzed in this work is depicted according to Tamura et al. (2004). (A) melanogaster group. (B) Sophophora subgenus. (C) Drosophila subgenus. Mya, million years ago. Species abbreviations as in Table 1.
Length of converted regions:
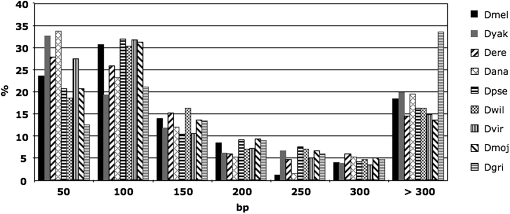
The length of converted regions ranged from ∼10 bp to more than 3 kb. D. pseudoobscura showed the smallest range with a maximum converted tract length of 1287 bp, compared to a longest tract of 3079 in D. mojavensis (Table S2). The average and median length of converted tracts varied about twofold between the nine Drosophila genomes, with particularly high values in D. grimshawi (Table S2). Approximately 49–59% of tracts are shorter than 100 bp in eight species, while in D. grimshawi only one tract out of three is <100 bp. Furthermore, 33% of tracts in D. grimshawi are longer than 300 bp, compared to less than 20% in the other species (Figure 3). Longer tracts in D. grimshawi are most likely a by-product of the low divergence between converted paralogs with respect to other species (Figure 1 and Table S2). Indeed, detectable conversion tracts tend to shorten in increasingly divergent pairs of duplicated genes, and the least divergent paralogs (dS < 0.1) have the longest tracts in every species (Figure S1). Other factors that could affect the length of observed conversion tracts seem to be less important. For instance, longer converted regions can derive from longer exons, assuming that these tracts mostly do not overlap introns. However, we found that D. grimshawi exons in converted genes have comparable length with exons in other species (Table S2). Among the nine genomes, the longest exons have an average of 675 bp (in D. ananassae), a feature that could explain the elevated average tract length in this species compared to other fruit flies (Table S2).
Figure 3.—
Length distribution of conversion tracts in nine Drosophila species. bp, base pair. Species abbreviations as in Table 1.
In the majority of Drosophila genomes, converted tracts covered ∼9–10% of the genes' coding sequences, a percentage remarkably similar between species with very different average and median tract length. D. grimshawi once again represents an outlier in this sense, with 13.5% of the coding sequences occupied by transferred tracts in converted paralogs, a consequence of the longer tracts observed in this Hawaiian species (Table S2 and Figure 3). Nevertheless, when all the analyzed pairs are considered, converted regions correspond to only ∼1–2% of the coding sequence of gene duplicates (Table S2), similarly to what has been found in mammals (McGrath et al. 2009). Note that this number includes the length of converted tracts in the 5% of expected false-positive pairs and therefore is an overestimate of the total converted sequence.
Given that we searched for conversion events in the coding regions of genes, some of the converted regions we detected may overlap with introns or go beyond the coding region boundaries at the 5′− and/or 3′-end of genes. Because this could not only affect the estimate of tract length, but also lead to different conclusions in the analyses discussed above, we examined the features of converted regions that were contained within only a single exon and do not extend to the exon–intron boundaries. Most regions (71–88%) satisfied this condition in the nine species. While the average tract length dropped by 37–93 bp, the length and age distribution of these regions were comparable to the distributions of all tracts (data not shown), suggesting that this is not a major factor affecting our results.
Factors affecting gene conversion:
Several aspects of gene structure and gene family organization have been proposed to influence levels of nonallelic conversion among paralogous genes. In our analysis, we examined the impact of these features on ectopic gene conversion in Drosophila.
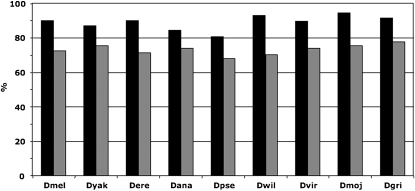
In the nine Drosophila genomes surveyed, 70–80% of paralogous pairs reside on the same Müller element. The proportion of intra-Müller element converted pairs is significantly higher than nonconverted pairs (P < 0.001, Fisher's exact test; Figure 4 and Table S1). Moreover, intraelement converted pairs tend to be physically closer than intraelement nonconverted pairs (significant support for all species except D. yakuba; t-test, P-value < 0.05; Figure S2 and Table S1). This is particularly striking for gene pairs separated by less than a few kilobases (Figure S3). Both patterns are consistent with previous studies (Semple and Wolfe 1999; Benovoy and Drouin 2009; McGrath et al. 2009). Genes in converted pairs also tend to have lower divergence than nonconverted paralogs (t-test, P-value < 0.05 for all species; Table S1). A different picture emerges from the comparison of GC content between converted and nonconverted pairs. In the four species of the melanogaster subgroup, D. melanogaster, D. yakuba, D. erecta, and D. ananassae, the proportion of G and C bases is higher in converted pairs (54.3% vs. 53.1% in converted and nonconverted pairs, respectively; see also Table S1), whereas the opposite is true for the remaining five species (50.3% vs. 51.4% in converted and nonconverted pairs, respectively; see also Table S1). These trends are significant in seven species (t-test, P-value < 0.05).
Figure 4.—
Proportion of converted pairs that are on the same Müller element (solid) compared to the proportion of nonconverted pairs (shaded). Species abbreviations as in Table 1.
Given that the physical distance between paralogs, the sequence divergence between paralogs, and the GC content within paralogs all affect levels of gene conversion, it may be that differences in these factors also determine differences in apparent levels of gene conversion among species. To test whether there is an effect of “species” independent of differences in the age, location on the same or different Müller element, and GC content among paralogs within each genome, we performed a series of nested ANOVAs. For each data set, pairs of paralogs from all the Drosophila species were combined together, and two nested models, one including and the other excluding a “species” variable, were compared. For the species variable, species were simply distinguished by assigning a different integer value to each of them. These models also included the factors we previously showed to be important predictors of gene conversion: nucleotide divergence, Müller element location, and GC content. All these variables were significant in this analysis as well (Table 2). The difference in the explanatory power of each model (with and without “species”) can be obtained using a LRT. The results of the LRT indicate that species is a highly significant variable affecting levels of gene conversion (Table 2). We also compared a similar pair of models in which we replaced the chromosomal location variable with the physical distance between intrachromosomal paralogs. Again, all variables were significant predictors of gene conversion, and the LRT between the two models showed that species membership is a significant factor after taking into account the physical distance between gene duplicates (Table S3).
TABLE 2.
Analysis of variance and likelihood ratio test (LRT) for Drosophila gene pairs excluding (M1) and including (M2) the species variable
| Variables | d.f. | MS | F | P | |
|---|---|---|---|---|---|
| M1 | dS | 1 | 24.59 | 313.042 | <2.2e − 16 *** |
| ME | 1 | 24.59 | 309.221 | <2.2e − 16 *** | |
| %GC | 1 | 3.20 | 40.736 | 1.799e − 10 *** | |
| ln(Likelihood) = −3712.529 | |||||
| M2 | dS | 1 | 24.59 | 313.042 | <2.2e − 16 *** |
| ME | 1 | 24.59 | 309.221 | <2.2e − 16 *** | |
| %GC | 1 | 3.20 | 40.736 | 1.762e − 10 *** | |
| Species | 1 | 1.14 | 14.553 | 1.368e − 04 *** | |
| ln(Likelihood) = −3704.332 | |||||
| LRT: −2Δ ln(L) = 16.4 P = 5.15e − 05 | |||||
MS: mean square. ME: Müller element.
Biased gene conversion:
The repair mechanisms involved in correcting mismatches on converted strands can introduce a biased toward G and C nucleotides (Marais 2003). We looked for evidence of BGC in our data sets using two different approaches. First, we compared the GC content of all tracts vs. nonconverted flanking regions in converted genes. We found a significantly higher percentage of GC in the tracts in all species except D. grimshawi (paired t-test, P < 0.05). The trend also holds when only tracts ≥50 bp were used (paired t-test, P < 0.05). However, this pattern could be created if GC content was a significant cause of gene conversion, rather than an effect. Therefore, we also asked whether GC content was higher in conversion tracts compared to the same region within paralogs of the same family with no conversion, whenever those paralogs were available. Our analysis revealed no significant difference between converted and nonconverted paralogs in these regions, except in D. ananassae (paired t-test, P < 0.05). This suggests that GC content does not increase upon conversion.
DISCUSSION
The dichotomy between concerted evolution and birth-and-death processes has been at the core of the debate around gene duplication for more than 20 years. We investigated the extent of gene conversion, one of the main drivers of concerted evolution in gene families, in a large genome-wide set of gene duplicates. Our survey of >17,700 paralogous genes in nine Drosophila species showed that gene conversion affects 1–9% of all paralogs, after subtracting false positives (Table 1). However, when gene duplicates are grouped by their divergence levels, the proportion of converted pairs shows a skewed pattern, with most conversion events occurring in relatively young paralogs (Figure 1). This pattern derives from two main features of gene conversion and the evolution of gene families. First, gene duplicates that diverged a long time ago share few regions of high similarity, which are the substrate of recombination, and are therefore less likely to be converted. Gene conversion tracts between old duplicates are also more difficult to detect given that they are broken up by mutations into smaller pieces and GENECONV, as well as other programs, has a limited sensitivity to detect short conversion tracts (McGrath et al. 2009). Second, conversion events between recently diverged paralogs tend to be underestimated as a consequence of the small number of substitutions between them, which are used to identify converted regions (McGrath et al. 2009). Therefore, very young paralogs are likely subjected to the highest levels of ectopic gene conversion, but these events are mostly undetectable with current methods and, more importantly, they have little evolutionary consequence given that these genes already share a very high sequence identity. On the contrary, gene conversion affecting less-similar paralogs could have a profound impact on gene families' evolution by homogenizing the coding sequences of those genes. Indeed, our data indicate that gene conversion is a relevant factor in the evolution of Drosophila gene duplicates with sequence divergence between 0.1 < dS < 0.3, where levels of conversion can vary between 30 and 60% (Figure 1).
We used an alternative approach to GENECONV to obtain an independent estimate of the effect of gene conversion on phylogenetic trees. This method asks whether reconciled gene and species trees disagree with changes in the size of gene families (see materials and methods). We found that between 1 and 3% of gene trees in the melanogaster subgroup were possibly affected by conversion, and up to ∼15% in older branches of the Drosophila genus phylogeny, particularly in the Sophophora subgenus (Figure 2). These estimates can be affected by a number of processes—not just gene conversion—including high numbers of gene gains and losses, and species trees that contain polytomies. The high level of disagreement on the branch leading to the melanogaster subgroup is most likely explained by the polytomy at the root of this group, as large numbers of duplications are incorrectly placed there by current reconciliation methods (Hahn 2007). While results from the comparison of reconciliation and copy-number methods provide a lower estimate of the effects of gene conversion than do results from GENECONV, given the short length of conversion tracts this outcome should not be too surprising. The average conversion tract contains only 9–13% of the coding region of any gene, which may have little or no effect on the genealogy of the genes considered.
Levels of gene conversion as high as 80% were reported in a recent study of young gene duplicates in D. melanogaster (Osada and Innan 2008). However, estimates of gene conversion from their method could be inflated by extensive parallel gene gains and losses, as they assumed that any families with two genes in both D. melanogaster and either D. simulans or D. sechellia were duplicated before the split of these species. Analysis of gene families across 12 Drosophila genomes revealed high levels of gene duplication and loss in all lineages (Hahn et al. 2007). Moreover, a survey of copy-number variation (CNVs) in 15 lines of D. melanogaster showed 133 entirely duplicated and 27 entirely deleted genes (Emerson et al. 2008). These observations suggest that methods relying only on phylogenetic relationships between orthologous/paralogous genes to detect gene conversion—as was used by previous authors (Osada and Innan 2008)—could be strongly affected by rapid turnover in Drosophila gene families. The results from our two different approaches indicate that gene conversion can be quite common among recent gene duplicates in Drosophila, although D. melanogaster showed an upper limit of 36% converted pairs. In addition, the GENECONV analysis indicated that gene conversion affects only ∼9–13% of the coding region of converted genes and a mere 1–2% of the coding regions of all paralogs (Table S2).
Some features of converted genes have been found to stand out when compared to genes with no conversion. Several authors have described a negative correlation between nonallelic gene conversion and both physical distance and nucleotide divergence (Semple and Wolfe 1999; Drouin 2002; Ezawa et al. 2006; Benovoy and Drouin 2009), but the relative contribution of each variable remains elusive. We recently demonstrated that when divergence is taken into account, chromosome location is still significant, but physical distance is not, in converted pairs from four mammalian genomes (McGrath et al. 2009). Here we find that these three features (intra-/interchromosomal location, physical distance within chromosomes, and sequence divergence) are significantly associated with pairs of converted genes in all Drosophila species (except physical distance in D. yakuba; Table S1).
We also found differences among species in the GC content between converted and nonconverted gene pairs, with the four species of the melanogaster group having higher average GC content in converted pairs, whereas the remaining five species show the opposite trend (Table S1). To our knowledge, this trend has not been described before and suggests possible differences in the recombination mechanisms and/or substitution patterns among species of the Drosophila genus. Analyses of substitution patterns in several organisms indicate an AT → GC mutational bias in converted sequences (Marais 2003). Our data set shows a significant enrichment in G and C nucleotides in conversion tracts compared to flanking coding sequence of converted genes in eight species, which could be the result of BGC. However, no significant GC bias was found between conversion tracts and regions corresponding to the tracts in nonconverted paralogs except in D. ananassae (t-test, P = 0.037). Together, our results suggest that G and C nucleotides play a causal role in nonallelic gene conversion and are not the result of biased substitution processes. Note that this effect of GC content may cause there to be higher levels of conversion in coding sequences in Drosophila, where the proportion of G and C nucleotides is higher.
One of the most interesting findings in our analysis concerns interspecific variation in the amount of gene conversion. Two main features emerged across species of Drosophila. First, lower levels of gene conversion were found in the melanogaster group compared to the rest of the genus. Second, D. grimshawi showed consistently higher values of conversion in terms of converted genes, amount of converted coding sequence, and length of converted tracts (Tables 1, S1 and S2; Figure 3). Indeed, our multivariate analyses revealed that species membership is a significant predictor of gene conversion levels, even after taking into account differences among species in the age, physical distance, Müller element location, and GC content of paralogs (Tables 2, S3). While it is known that Drosophila species differ in several aspects of recombination (e.g., male recombination in D. ananassae (Kikkawa 1937; Moriwaki 1937)), our results suggest an underlying difference in the machinery involved in double-strand break repair. The effects of these differences on other aspects of genome evolution may be revealed only by more thorough genetic and molecular experiments.
Acknowledgments
We thank Mira Han for assistance, A. Michelle Lawing for support with the statistical analyses, and Casey McGrath for helpful discussions. We also thank two anonymous reviewers for their helpful comments. This work was supported by a grant from the National Science Foundation (DBI-0543586) to M.W.H.
Supporting information available online at http://www.genetics.org/cgi/content/full/genetics.110.115444/DC1.
References
- Axelsson, E., M. T. Webster, N. G. Smith, D. W. Burt and H. Ellegren, 2005. Comparison of the chicken and turkey genomes reveals a higher rate of nucleotide divergence on microchromosomes than macrochromosomes. Genome Res. 15 120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backstrom, N., H. Ceplitis, S. Berlin and H. Ellegren, 2005. Gene conversion drives the evolution of HINTW, an ampliconic gene on the female-specific avian W chromosome. Mol. Biol. Evol. 22 1992–1999. [DOI] [PubMed] [Google Scholar]
- Benovoy, D., and G. Drouin, 2009. Ectopic gene conversions in the human genome. Genomics 93 27–32. [DOI] [PubMed] [Google Scholar]
- Benovoy, D., R. T. Morris, A. Morin and G. Drouin, 2005. Ectopic gene conversions increase the G + C content of duplicated yeast and Arabidopsis genes. Mol. Biol. Evol. 22 1865–1868. [DOI] [PubMed] [Google Scholar]
- Berglund, J., K. S. Pollard and M. T. Webster, 2009. Hotspots of biased nucleotide substitutions in human genes. PLoS Biol. 7 e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt, B. R., and M. E. Feder, 2002. Rapid concerted evolution via gene conversion at the Drosophila hsp70 genes. J. Mol. Evol. 54 569–586. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds, O. R., 2005. transAlign: using amino acids to facilitate the multiple alignment of protein-coding DNA sequences. BMC Bioinformatics 6 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell, J. A., 2002. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 19 1181–1197. [DOI] [PubMed] [Google Scholar]
- Brown, C. J., C. F. Aquadro and W. W. Anderson, 1990. DNA sequence evolution of the amylase multigene family in Drosophila pseudoobscura. Genetics 126 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. M., D. N. Cooper, N. Chuzhanova, C. Ferec and G. P. Patrinos, 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8 762–775. [DOI] [PubMed] [Google Scholar]
- Chen, K., D. Durand and M. Farach-Colton, 2000. NOTUNG: a program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 7 429–447. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., M. B. Eisen, D. R. Smith, C. M. Bergman, B. Oliver et al., 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450 203–218. [DOI] [PubMed] [Google Scholar]
- Conant, G. C., and K. H. Wolfe, 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9 938–950. [DOI] [PubMed] [Google Scholar]
- De Bie, T., N. Cristianini, J. P. Demuth and M. W. Hahn, 2006. CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22 1269–1271. [DOI] [PubMed] [Google Scholar]
- Drouin, G., 2002. Characterization of the gene conversions between the multigene family members of the yeast genome. J. Mol. Evol. 55 14–23. [DOI] [PubMed] [Google Scholar]
- Duret, L., and N. Galtier, 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10 285–311. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson, J. J., M. Cardoso-Moreira, J. O. Borevitz and M. Long, 2008. Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster. Science 320 1629–1631. [DOI] [PubMed] [Google Scholar]
- Ezawa, K., OOta, S., and N. Saitou, 2006. Genome-wide search of gene conversions in duplicated genes of mouse and rat. Mol. Biol. Evol. 23 927–940. [DOI] [PubMed] [Google Scholar]
- Galtier, N., 2003. Gene conversion drives GC content evolution in mammalian histones. Trends Genet. 19 65–68. [DOI] [PubMed] [Google Scholar]
- Galtier, N., E. Bazin and N. Bierne, 2006. GC-biased segregation of noncoding polymorphisms in Drosophila. Genetics 172 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor, G. B., N. A. Nassif, D. M. Johnson-Schlitz, C. R. Preston and W. R. Engels, 1991. Targeted gene replacement in Drosophila via P element-induced gap repair. Science 253 1110–1117. [DOI] [PubMed] [Google Scholar]
- Haddrill, P. R., and B. Charlesworth, 2008. Non-neutral processes drive the nucleotide composition of non-coding sequences in Drosophila. Biol. Lett. 4 438–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W., 2007. Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol. 8 R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W., 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 100 605–617. [DOI] [PubMed] [Google Scholar]
- Hahn, M. W., T. De Bie, J. E. Stajich, C. Nguyen and N. Cristianini, 2005. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 15 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, M. W., M. V. Han and S. G. Han, 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3 e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M. V., J. P. Demuth, C. L. McGrath, C. Casola and M. W. Hahn, 2009. Adaptive evolution of young gene duplicates in mammals. Genome Res. 19 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey, D. A., L. Bally-Cuif, S. Abukashawa, V. Payant and B. F. Benkel, 1991. Concerted evolution of duplicated protein-coding genes in Drosophila. Proc. Natl. Acad. Sci. USA 88 1611–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, M. S., K. Oliver, J. Loveland, S. Humphray, I. Dunham et al., 2005. Evidence for widespread reticulate evolution within human duplicons. Am. J. Hum. Genet. 77 824–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa, H., 1937. Spontaneous crossing-over in the male of Drosophila ananassae. Zool. Mag. 49 159–160. [Google Scholar]
- King, L. M., 1998. The role of gene conversion in determining sequence variation and divergence in the Est-5 gene family in Drosophila pseudoobscura. Genetics 148 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, W. Y., S. Piao and H. Akashi, 2006. Strong regional heterogeneity in base composition evolution on the Drosophila X chromosome. Genetics 174 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla, G., A. Helwak and L. Lipinski, 2004. Gene conversion and GC-content evolution in mammalian Hsp70. Mol. Biol. Evol. 21 1438–1444. [DOI] [PubMed] [Google Scholar]
- Langley, C. H., B. P. Lazzaro, W. Phillips, E. Heikkinen and J. M. Braverman, 2000. Linkage disequilibria and the site frequency spectra in the su(s) and su(w(a)) regions of the Drosophila melanogaster X chromosome. Genetics 156 1837–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro, B. P., and A. G. Clark, 2001. Evidence for recurrent paralogous gene conversion and exceptional allelic divergence in the Attacin genes of Drosophila melanogaster. Genetics 159 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais, G., 2003. Biased gene conversion: implications for genome and sex evolution. Trends Genet. 19 330–338. [DOI] [PubMed] [Google Scholar]
- McGrath, C. L., C. Casola and M. W. Hahn, 2009. Minimal effect of ectopic gene conversion among recent duplicates in four mammalian genomes. Genetics 182 615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragon-Palomino, M., and B. S. Gaut, 2005. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 22 2444–2456. [DOI] [PubMed] [Google Scholar]
- Moriwaki, D., 1937. A high ratio of crossing over in Drosophila ananassae. Z. Indukt. Abstamm. Vererbungsl. 74 17–23. [Google Scholar]
- Nei, M., and A. P. Rooney, 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, Berlin.
- Osada, N., and H. Innan, 2008. Duplication and gene conversion in the Drosophila melanogaster genome. PLoS Genet. 4 e1000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel, A. D., M. J. Telford and M. Akam, 2006. The evolution of hexapod engrailed-family genes: evidence for conservation and concerted evolution. Proc. Biol. Sci. 273 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, D. A., V. N. Iyer, A. M. Moses and M. B. Eisen, 2006. Widespread discordance of gene trees with species tree in Drosophila: evidence for incomplete lineage sorting. PLoS Genet. 2 e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer, S., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6 526–538. [DOI] [PubMed] [Google Scholar]
- Schaeffer, S. W., A. Bhutkar, B. F. McAllister, M. Matsuda, L. M. Matzkin et al., 2008. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179 1601–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple, C., and K. H. Wolfe, 1999. Gene duplication and gene conversion in the Caenorhabditis elegans genome. J. Mol. Evol. 48 555–564. [DOI] [PubMed] [Google Scholar]
- Shibata, H., and T. Yamazaki, 1995. Molecular evolution of the duplicated Amy locus in the Drosophila melanogaster species subgroup: concerted evolution only in the coding region and an excess of nonsynonymous substitutions in speciation. Genetics 141 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage, D. E., and T. H. Eickbush, 2007. Sequence variation within the rRNA gene loci of 12 Drosophila species. Genome Res. 17 1888–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, J. F., M. Baze, J. L. Waite, F. G. Hoffmann, J. C. Opazo et al., 2007. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics 177 481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., S. Subramanian and S. Kumar, 2004. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21 36–44. [DOI] [PubMed] [Google Scholar]
- Thornton, K., and M. Long, 2005. Excess of amino acid substitutions relative to polymorphism between X-linked duplications in Drosophila melanogaster. Mol. Biol. Evol. 22 273–284. [DOI] [PubMed] [Google Scholar]
- Wang, S., C. Magoulas and D. Hickey, 1999. Concerted evolution within a trypsin gene cluster in Drosophila. Mol. Biol. Evol. 16 1117–1124. [DOI] [PubMed] [Google Scholar]
- Wang, X., H. Tang, J. E. Bowers, F. A. Feltus and A. H. Paterson, 2007. Extensive concerted evolution of rice paralogs and the road to regaining independence. Genetics 177 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., T. Clark, H. Zheng, S. Vang, R. Li et al., 2008. Gene conversion in the rice genome. BMC Genomics 9 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24 1586–1591. [DOI] [PubMed] [Google Scholar]