Abstract
The ClpA chaperone combines with the ClpP peptidase to perform targeted proteolysis in the bacterial cytoplasm. ClpA monomer has an N-terminal substrate-binding domain and two AAA+ ATPase domains (D1 and D2). ClpA hexamers stack axially on ClpP heptamers to form the symmetry-mismatched protease. We used cryo-electron microscopy to visualize the ClpA-ATPγS hexamer, in the context of ClpAP complexes. Two segments lining the axial channel show anomalously low density, indicating that these motifs, which have been implicated in substrate translocation, are mobile. We infer that ATP hydrolysis is accompanied by substantial structural changes in the D2 but not the D1 tier. The entire N-domain is rendered invisible by large-scale fluctuations. When deletions of 10 and 15-residues were introduced into the linker, N-domain mobility was reduced but not eliminated and changes were observed in enzymatic activities. Based on these observations, we present a pseudo-atomic model of ClpAP holoenzyme, a dynamic proteolytic nanomachine.
Keywords: chaperone, AAA+ ATPase, unfoldase, pseudo-atomic model, image reconstruction, conformational change
Introduction
Protein degradation, a vital function shared by all kingdoms of life, serves in the programmed removal of regulatory factors and to maintain the integrity of the cellular proteome by eliminating defective proteins (Wickner et al., 1999). In the prokaryotic cytoplasm, proteolysis is performed by several related protein complexes, members of the Clp family: ClpAP, ClpXP, ClpYQ (HslUV), Lon, and the membrane-anchored FtsH. Each complex has a distinct range of substrate specificities (Gottesman, 2003). Similar complexes are found in archaea and in the mitochondria and chloroplasts of eukaryotes. They share two fundamental properties. First, as they operate intracellularly, their activity has to be stringently controlled in order to avoid damaging unintended targets. Second, they are processive; once degradation is initiated on a given substrate, it continues until the entire polypeptide chain has been reduced to peptides of 7–8 residues (Choi and Licht, 2005). These functional requirements are met by combining two oligomeric components: a peptidase whose active sites are sequestered inside the complex, thus having access only to cleavage sites that are specifically presented to them; and an unfoldase that recognizes bona fide substrates, unfolds them, and feeds them into the peptidase for degradation (Baker and Sauer, 2006; Striebel et al., 2009).
The unfoldases belong to the AAA+ family of mechanoenzymes (Ammelburg et al., 2006; Erzberger and Berger, 2006; Hanson and Whiteheart, 2005; Snider and Houry, 2008). ClpA, a Type II enzyme with two AAA+ modules, and ClpX, a Type I enzyme with a single AAA+ module, form hexameric rings in the active state; both partner the ClpP peptidase, which consists of two apposing heptameric rings. As the two components mount coaxially, there is a 6:7 symmetry mismatch at their interface (Beuron et al., 1998; Kessel et al., 1995). ClpY/HslU is a hexameric Type I unfoldase that pairs with a double hexamer of the peptidase ClpQ/HslV - hence, no symmetry mismatch in this system (Sousa et al., 2000). In Lon, the ATPase and the peptidase are co-linear segments of the same polypeptide chain so that there is necessarily no symmetry mismatch in these oligomers (Rotanova et al., 2006).
In terms of overall architecture and other key properties, these proteases resemble the proteasome, a complex that degrades proteins tagged with polyubiquitin chains in the cytoplasm and nucleus of eukaryotic cells, thus playing key roles in numerous processes (Goldberg, 2003). The proteasome also features a 6:7 pseudo-symmetry mismatch between its ATPase ring and the peptidase. However, the elaborate subunit composition of its unfoldase component – the 19S regulatory complex – complicates analysis. For this reason, the Clp enzymes whose unfoldases are relatively simple homomeric rings afford valuable model systems for the proteasome.
In the present study, we have focused on the structure of the ClpA unfoldase of E. coli (Katayama et al., 1988) in the context of the fully assembled protease. Its subunit has three domains, an α-helical N-domain connected by a flexible linker to the D1 and D2 domains which have canonical AAA+ folds (Guo et al., 2002). In the hexamer, D1 and D2 form two stacked ring-like tiers (Beuron et al., 1998; Ishikawa et al., 2004). The D2 tier engages ClpP by means of flexible loops containing a conserved IGL motif (Kim et al., 2001). The N-domains undergo large-scale fluctuations about positions distal to the D1 tier (Ishikawa et al., 2004), on whose surface substrates such as RepA initially bind, and are then unfolded and translocated axially into the degradation chamber of ClpP (Ishikawa et al., 2001; Reid et al., 2001; Weber-Ban et al., 1999). Substrate binding does not suppress N-domain mobility (Ishikawa et al., 2004). Loops lining the axial channel of ClpA were identified by cross-linking and mutagenesis experiments as important for this process (Hinnerwisch et al., 2005). Of these, the so-called diaphragm loop in the D2 tier, which contains a conserved GYVG motif, has been proposed to promote substrate translocation by adopting different conformations, depending on the protein’s nucleotide state, alternating between an “up” position closer to D1 and a “down” position closer to ClpP (Bohon et al., 2008; Farbman et al., 2008; Hinnerwisch et al., 2005).
The symmetry mismatch and the property that ClpA can bind to one or both ends of ClpP have thwarted attempts to obtain crystal structures for ClpAP. Several structures have been determined for ClpP, all of which are very similar except for the conformation of the N-terminal 17 residues (Yu and Houry, 2007), which are envisaged to be dynamic loops (Bewley et al., 2006; Gribun et al., 2005; Szyk and Maurizi, 2006). A crystal structure has also been determined for the ClpA monomer in the ADP-bound state (Guo et al., 2002). Here, we have analyzed ClpAP complexes by cryo-electron microscopy and image reconstruction to ~12 Å resolution, and by fitting in known crystal structures, derived a pseudo-atomic model for the ATP-containing ClpA hexamer. By comparison with a model of the ADP-containing hexamer (Guo et al., 2002), we infer that ATP hydrolysis is accompanied by substantial (20°) rigid-body rotations of the D2 domains, relative to a stable platform of tightly coupled D1 domains. From their diminished density in the cryo-EM density map, the “diaphragm” loops that line the axial channel in the D2 tier are seen to be mobile, a property consistent with an imputed role in substrate translocation. Even greater mobility is exhibited by the distally protruding N-domains, and this mobility is reduced but not eliminated by a 15-residue deletion in the D1:N-domain linker. Overall, this analysis reveals novel aspects of mobility of likely functional significance in a complex already known to be highly dynamic.
Results
Cryo-EM reconstruction of ClpP-bound ClpA hexamers
At ~500 kDa, ClpA is a relatively small protein complex for cryo-EM analysis and its hollow nature (Beuron et al., 1998) gives it an approximately donut-like appearance when projected in any direction. This property raises the risk of misidentifying axial views as side-views and vice-versa, particularly for micrographs recorded close to focus. To avoid this difficulty, we analyzed ClpA complexed with ClpP, because side-views of ClpAP are readily identifiable as such (Fig. 1) and they surely present side-views of ClpA (Kessel et al., 1995). With these data, the range of orientations to be searched for each particle when calculating a reconstruction is limited to the angular setting around its symmetry axis and, eventually, a small out-of-plane tilt. Moreover, a complete set of side-views affords a full sampling of the 3D Fourier transform and thus a reconstruction that is not compromised by missing data (Roseman et al., 1996; Ortega et al., 2005).
Figure 1.

A field of ClpAP complexes visualized by cryo-EM. Examples of 1:1 and 2:1 complexes are highlighted in black and white respectively. Scale bar, 300Å.
ClpA was assembled into hexamers by adding ATPγS, and then mixed with ClpP at a 1:1 molar ratio of ClpA hexamers to ClpP tetradecamers. This condition favors the formation of 1:1 complexes, while some 2:1 complexes are also produced (Fig. 1). Cryo-micrographs were recorded as focal pairs and the ClpA-containing portions were masked out from side-views of 1:1 and 2:1 complexes. These data were combined in the ensuing analysis. In brief, the reconstruction was performed by projection matching, assuming 6-fold symmetry. Two quite different starting models (see Methods) were used to confirm that the final reconstruction was not biased by the choice of initial template. According to the FSC criterion, the reconstruction, which included 6000 focal pairs, has a resolution of 12.5 Å (Fig. S1).
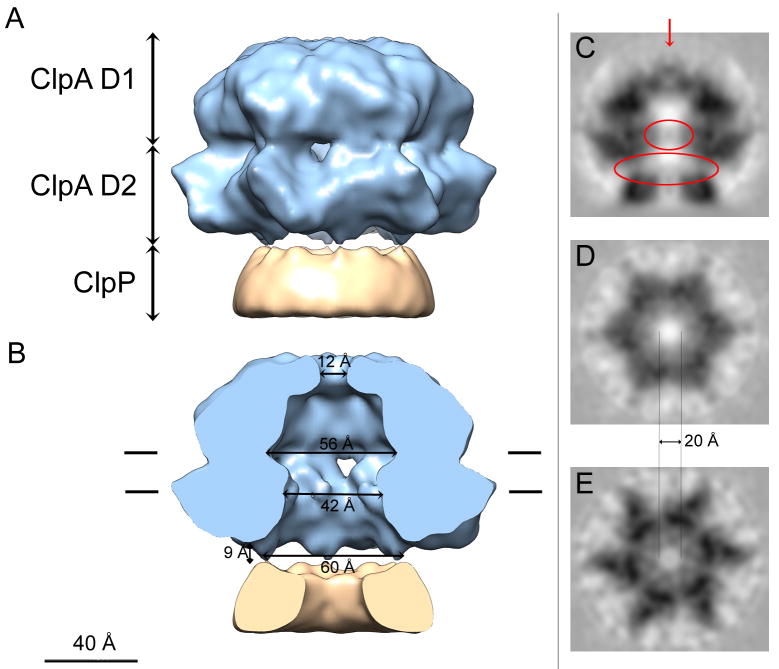
The density map depicts a 3-ring structure (Fig. 2A). The bottom ring represents ~ 50% of a ClpP heptamer. This part of ClpP (~ 70 kDa), which was included in the excised images to ensure that the experimental volume would contain all of ClpA, is too small to subvert angular determination of the ClpA hexamers. As ClpP is heptameric and six-fold symmetry was applied, the representation of this ring is almost perfectly cylindrical. Of the other two rings, the one in contact with ClpP comprises the D2 domains of ClpA, and the distal ring, its D1 domains. They have the same dimensions and a generally similar appearance as in the reconstruction of Ishikawa et al. (2004); i.e. the D2 ring has a slightly greater diameter than D1 (145 Å vs. 130 Å). However, the side-ports that were seen between the D1 and D2 tiers when the latter reconstruction was surface-rendered are much smaller. With the possible exception of a small density at the axial channel entrance on the apical surface of D1 (Fig. 2C – red arrow), the six 16-kDa N-domains are invisible, reflecting their high mobility (Ishikawa et al., 2004). This density apart, ClpA has a continuous axial channel that widens into large internal cavities between the D1 and the D2 tiers, and at the interface between D2 and ClpP (the “vestibule”) – Fig. 2B. However, when the axial region is viewed in gray-scale sections (Figs. 2C–E), the channel appears considerably narrower, as some lower but nevertheless significant density is visible (upper red oval in Fig. 2C). The diameter of the fully unobstructed channel is given in Figs. 2D & E. This observation implies that some elements lining the axial channel are locally mobile or that they depart from strict 6-fold symmetry; either eventuality would lead to their being down-weighted in the reconstruction.
Figure 2.
Cryo-EM 3D reconstruction of ClpA. (A) Surface rendering of the side-view and (B) cut away view exposing the interior. The ClpA hexamer is in blue and a portion of ClpP (~25%) is in ochre: the latter appears almost perfectly cylindrically symmetric because 6-fold symmetrization has been applied to its 7-fold-symmetric structure. The D1 and D2 tiers of ClpA are labeled. Interior dimensions at various locations are marked. In A and B, a small diffuse density plugging the apical entrance to the axial channel (red arrow in panel C) has been removed for clarity. (C–E) Grayscale sections; (C) Central longitudinal section for the plane contoured in B. D and E are transverse sections, looking towards ClpP. D is at the level of the “56 Å” channel in B, and F is at the “42 Å” mark. The upper red oval in C marks low but significant axial densities visualized within the ClpA channel that represent mobile elements. Although the surface rendering in B depicts cavity diameters of ~ 56 Å in D1 and ~ 42 Å in D2, these measures are reduced to ~ 20 Å for the completely open channel in the corresponding grayscale sections (D and E respectively). The bottom red oval encloses densities bridging between the ClpA and ClpP rings that have been diluted by applying 6-fold symmetry to the ClpP heptamer in the reconstruction.
Faint protein density bridging the gap between D2 and ClpP is seen at a peripheral site, ~ 30 Å out from the axis - Fig. 2C, bottom red oval. (The surface rendering suggests a separation of ~ 10Å between the two rings at this site - Fig. 2B). It is likely that contact between ClpP and ClpA takes place at this radius. The 7-to-6 symmetry mismatch means that each interaction is likely unique and its density is correspondingly diluted in the reconstruction as a result of the applied symmetry (Fig. 2C).
A pseudo-atomic model of the ClpA hexamer
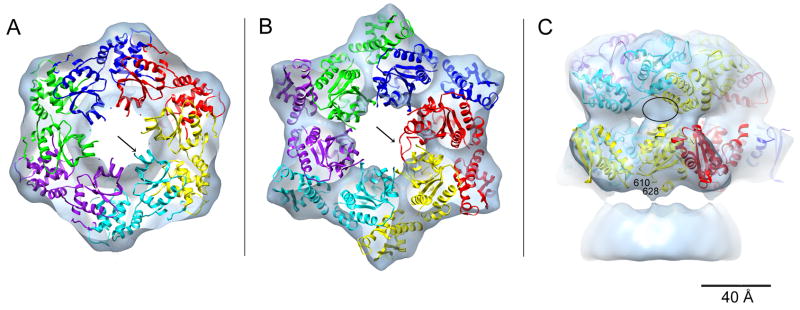
The crystal structure of ClpA has been solved as a monomer in the ADP state (Guo et al., 2002). We used this information to develop a pseudo-atomic model of the observed hexamer in the ATPγS state. To do so, we initially assumed that the folds of D1 and D2 are preserved in the hexamer and used these structures as separate rigid bodies in the modeling. In refinement, we allowed some relative movement of the large and small subdomains of each AAA+ module, which may rotate between nucleotide states (Wang et al., 2001b). D1 and D2 are quite similar to the AAA+ modules of some other family members (HslU/ClpY, NSF, p97), whose crystal structures were solved as hexamers (Bochtler et al., 2000; Davies et al., 2008; Lenzen et al., 1998). On testing the consistency of these hexamers with our density map, we found that NSF D2 solved in the AMP-PNP state (PDB 1D2N; Lenzen et al., 1998) and HslU/ClpY in the ATP state (PDB 1G31; Sousa et al., 2000) fitted quite well into the D1 and D2 tiers, respectively; accordingly, they were used as templates to build an initial model for ClpA. This model was then refined to optimize the goodness of fit in both tiers of the density map.
The model for the D1 tier fits snugly into the density map (Fig. 3A). The only discrepancies involve a short α-helix (residues 324–333) connected to the sensor 1 motif (arrow in Fig. 3A) and part of a short loop (residues 292–300) near the channel entrance (not shown). The latter was poorly defined in the crystal structure and might represent a mobile segment of D1. In the D2 tier, the initial model already fitted quite well into the EM density (Fig. S2A), but agreement could be improved by slightly rotating the small domain relative to the large domain (Figs. S2A & B). The resulting model agrees well with the density map except for the D2 “diaphragm” loop (residues 523–547), which protrudes (Fig. 3B – arrow) into the central channel. Gray-scale sections (Figs. 2C–E) show lower densities that we take to represent the D2 diaphragm loop at exactly this position. We conclude that this region is mobile. Consistent with this finding, the corresponding loops are disordered in crystal structures of ClpB (Lee et al., 2003), ClpX (Kim and Kim, 2003), and some HslU/ClpY structures (Sousa et al., 2000).
Figure 3.
Pseudo-atomic model of the ClpA hexamer. The D1 (A) and D2 (B) tiers were built separately. Each monomer is colored differently. A and B show axial views looking from ClpA towards ClpP. The arrows in A and B point to two segments of ClpA lying outside the EM density; the segment in A corresponds to an α-helix linked to the sensor 1 motif and that in B to the D2 diaphragm loop (for clarity, only one loop from the red monomer is shown). Panel C shows a side view, the unoccupied density (black oval) corresponds to the location of the 10 amino acid linker between D1 and D2 that is not shown. Residues 610 and 628 (marked) are the two ends of the ClpA motif that interacts with ClpP. The model places these two residues in the immediate vicinity of the bump of unoccupied ClpP density shown at the bottom of the panel.
As stated above, we modeled the D1 and D2 tiers separately. In the resulting structure, the relative positions of D1 and D2 are markedly different from those seen in the crystal structure (cf. in Fig. S3). We infer that this discrepancy reflects the differing nucleotide states (ATPγS vs. ADP) and, possibly, an effect of crystal contacts. In modeling, the linker between D1 and D2 (res. 435–443) was omitted; however, there is a small unoccupied region in the density map near the D1-D2 interface (Fig. 3C – black oval) that may accommodate this linker peptide. It follows from the relative positions of the C-terminus of D1 and the N-terminus of D2 in the model that the D2 domain of a given monomer is azimuthally offset relative to its D1 domain (see Fig. 3C). This arrangement is quite different from that in p97 where the D1 and D2 domains of a given monomer stack on top of each other (Davies et al., 2008), but is similar in this respect to the pseudo-atomic model proposed for ClpB (Diemand and Lupas, 2006; Lee et al., 2007).
A further validation of this pseudo-atomic model concerns the ClpA residues adjacent to ClpP. ClpA is known to interact with ClpP via a loop containing a conserved “IGL” motif that was not resolved in the ClpA crystal structure. In our model, the two ends (residues 610 and 628) of this motif are appropriately positioned in the interface region (Fig. 3C).
Structural effect of deletions in the linker connecting the N-domain to D1
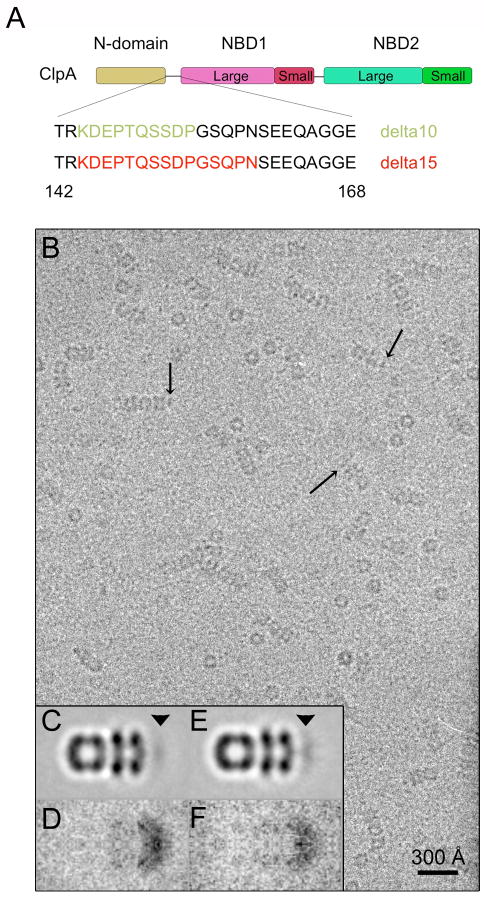
The linker connecting the N-domain to D1 is ~ 25 amino acids long (Guo et al., 2002). Anticipating that shortening it would reduce the mobility of the N-domains and hence enhance their visibility, we engineered two deletion mutants, lacking 10 and 15 amino acids (Fig. 4A). Both constructs, ClpA-Δ10 and -Δ15, hexamerize in the presence of ATPγS and bind to ClpP. Side-views of the resulting 1:1 and 2:1 complexes (Fig. 4B) are essentially undistinguishable from wild-type except for a higher incidence of molecules that have visible densities on the apical surface of ClpA (Fig. 4B – arrows). After averaging, these side-views exhibit more density in this region than does wild-type ClpA (Figs. 4C & E; cf. Ishikawa et al. (2004)). This band of apical density is more pronounced for Δ15 than for Δ10, in line with the expectation of a greater degree of immobilization in this mutant. However, it is still under-represented compared to the D1 and D2 tiers, when the respective masses are taken into account (Figs. 4C & E – arrowhead), implying that the N-domains remain mobile, albeit less so. The same trend is evidenced in the respective variance maps, which are more intense in Δ10 than in Δ15 (Figs. 4D & F), indicative of greater mobility.
Figure 4.
Study of deletion mutants in the linker region joining the N-domain to D1. A shows a schematic domain map of ClpA with the amino acid sequence of the N-domain/D1 linker. The amino acids deleted in the Δ10 and Δ15 mutants are colored in green and red respectively. B, Cryo-electron micrograph of ClpA-Δ10/ClpP complexes. Arrows point to some complexes on which N-domain densities are visible at the apical surface of ClpA. C & E, averaged side-views of 1:1 complexes of ClpP with the Δ10 and Δ15 mutants of ClpA. The arrowhead in each panel marks the location occupied by the still-mobile N-domains. D & F, side-view variance maps of complexes with the Δ10 and Δ15 mutants.
A mutant ClpA with a more drastic 20-residue deletion (Δ20) was also produced. However, this protein turned out to be more difficult to assemble into hexamers and thus to obtain a preponderance of side-views. Preliminary analysis of such cryo-EM data as were obtained revealed a similar trend as with Δ10 and Δ15 i.e. the N-domains are more visible than on the wild-type protein but they are still mobile as judged by averaging and variance analysis (data not shown).
Enzymatic activity of linker deletion mutants
As expected from the results of Crans-Mileva et al. (2008), our linker deletion mutants were found to interact well with ClpP and to promote ClpP peptidase activity similarly to wild-type ClpA (data not shown). Enzymatic properties with different protein substrates were quite variable, consistent with previous findings that the influence of the N-domains on binding and degradation is substrate-dependent (Xia et al., 2004). Table 1 (see also Fig. S4) shows basic enzymatic parameters (Km, Vmax, and Ka) for activation of ATPase activity and for degradation of a natively unfolded protein (casein), a folded protein that is directly recognized by ClpA (GFP-SsrA), and a protein whose interaction is dependent on the adaptor protein, ClpS (LR-GFPVenus). With respect to the ClpS-independent substrates (casein, GFP-SsrA), the linker-deletion mutants behaved similarly to the N-domain-deleted form of ClpA, suggesting that the N-domain orientations adopted on Δ10 and Δ15 place their substrate-interacting regions in unfavorable positions for interaction with unfolded proteins. The observed increase in Km for casein and lack of change in Km for GFP-SsrA, along with the increase in Vmax for both substrates, are consistent with the previous conclusion that the N-domains bind and recruit unfolded proteins but they also interact with initially folded proteins after they are unfolded on ClpA, retarding their translocation (Xia et al., 2004). For the ClpS-dependent substrate, neither parameter was affected appreciably in the deletion mutants. Since maximum degradation of LR-GFPVenus requires only a single molecule of ClpS per ClpA hexamer (De Donatis, G.M., Singh, S.K., Viswanathan, S., and Maurizi, M.R. - unpublished results), the data suggest that at least one N-domain in the Δ10 and Δ15 proteins can adopt an orientation that favors delivery of the protein into the axial channel. Apparently, the presence of ClpS prevents other N-domains from interacting with the substrate as it is unfolded, thereby impeding translocation. These results point to adaptive roles of the N-domains in regulating access to the interior of ClpA (Cranz-Mileva et al., 2008).
Table 1.
Enzymatic activity of linker deletion mutants of ClpA
| ATP hydrolysis1 |
Protein degradation1 |
||||
|---|---|---|---|---|---|
| ClpA form |
Activity |
Km(ATP)/Ka (casein)2 |
α-casein |
GFP-SsrA |
LR-GFPVenus |
| Vmax/Km | Vmax/Km | Vmax/Km | |||
| Wild type | 990±25 | 0.26±0.1/0.21±0.04 | 9.1±1.5/0.2±0.3 | 10±1/1.7±0.2 | 5.0±0.3/0.95±0.1 |
| Link-Δ10 | 900±35 | 0.86±0.4/0.11±0.03 | ND | 15±2/1.7±0.3 | 5.0±0.2/0.8±0.1 |
| Link-Δ15 | 1050±40 | 1.1±0.3/0.45±0.1 | 13±1/0.35±0.02 | 17±2/1.5±0.5 | 5.1±0.4/0.7±0.2 |
| ClpA-ΔN | 1020±50 | 0.25±0.2/6.0±1 | 27±2/6.0±13 | 19±1/1.0±0.3 | NA4 |
Units of ATPase activity are μmol ATP hydrolyzed/min/μmol ClpA6. Units of protein degradation (Vmax) are μmol protein degraded/min/μmol ClpA6. Km or Ka values are expressed as μM protein monomer. Values shown are the averages of three replicates and the error range of the measurements. LR-GFPVenus degradation was carried out in the presence of saturating ClpS (0.4 μmol).
Casein concentration required for half-maximal activation of ClpA6 ATPase activity.
Data from DeDonatis et al (2009 J. Biol. Chem. under revision).
NA, not applicable. ClpA-ΔN does not bind ClpS and therefore has no ClpS-dependent activity against LR-GFPVenus.
Overall ClpAP architecture and functional implications
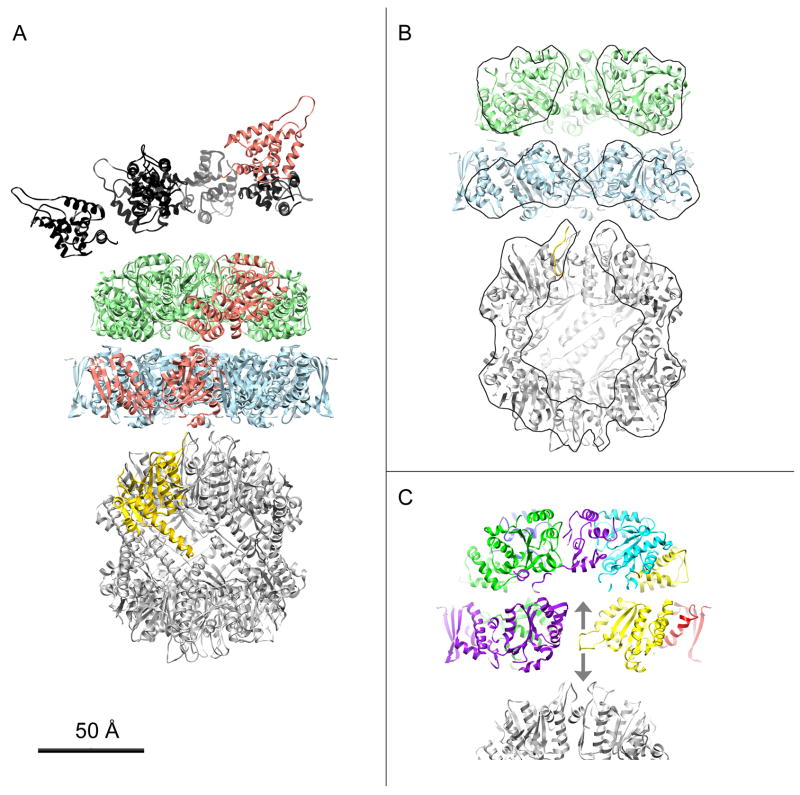
Based on the work presented here and in previous studies, we constructed a pseudo-atomic model of the fully assembled 1:1 complex (Fig. 5A). In it, the ClpA N-domains were allowed to assume random non-overlapping positions in the hemisphere overlying the apical surface of the D1 tier, as demarcated by the variance and difference maps (Fig. 4 and Ishikawa et al. (2004)). The D1 and D2 tiers of ClpA are as specified in our pseudo-atomic model. We used the crystal structure of Bewley et al. (2006) for the ClpP barrel, and adapted the N-loops (Effantin G., Maurizi, M.R., and Steven, A.C., unpublished data). The axial placement of the barrel relative to the D2 tier of ClpA was defined by our reconstruction and its azimuthal position relative to ClpA, as determined by Beuron et al. (1998) - (Fig. S5). The ClpA and ClpP rings were taken to be co-axial.
Figure 5.
Model for the ClpAP holoenzyme. A, Ribbon representation of a side-view of a 1:1 complex with ClpA at top. The pseudo-atomic model for D1 (green) and D2 (light blue) are from Figure 2. The N-domains (black) were randomly placed at the apical surface of D1 in accordance with Figure 3 and the conclusions of Ishikawa et al. (2004). The three domains of one ClpA monomer are colored salmon. The ClpP atomic model is after Bewley et al. (2006) (PDB: 1YG6). One monomer is colored yellow. The azimuthal setting of ClpA relative to ClpP is according to Beuron et al. (1998) - see Methods. B, Cut-away view of the 1:1 complex from which N-domains (black in A) were removed, for clarity. To show the interior structure of the complex and in particular the axial channel, the front half was removed computationally. Regions lying within the sectioning plane are contoured. C, Side-view section of the ClpA – ClpP interface centered on the D2 diaphragm loop of one ClpA monomer (yellow). The two arrows represent the possible directions of the movements this loop could undergo, in response to changes in the nucleotide state of the subunit.
The D2 tier has two distinct interactions with ClpP: one at the periphery via the IGL motif, and one closer to the axis, although the precise nature of the latter interaction is not yet clear. As the IGL-containing motif was not resolved in the crystal structure of ClpA (Guo et al., 2002), this interaction could not be modeled in detail. It has been proposed that a second interaction occurs between ClpA and the N-terminal loops of ClpP (Bewley et al., 2006; Gribun et al., 2005). In our pseudo-atomic model, the only element in the central channel of ClpA and close to the N-terminal loops of ClpP is the D2 diaphragm pore loop (Fig. 5C; yellow loop). The conformation shown in Fig. 5C is likely to be inexact in the context of the working enzyme as the present results attribute mobility to this motif; nevertheless, its position suggests that the loop can reach far enough to engage the N-terminal loops of ClpP (Fig. 5C; bottom arrow). Such a “down” conformation would create a physical link between ClpA and ClpP (Fig. 5B) and could prevent back-slippage of translocating substrates. It would also expand the range of movement this loop may undergo, as it has been proposed that the D2 pore loop can also adopt an “up” conformation (Fig. 5C; op arrow) (Bohon et al., 2008; Farbman et al., 2008; Hinnerwisch et al., 2005).
Discussion
Our cryo-EM reconstruction has led to a pseudo-atomic model of the entire ClpAP complex. Multiple lines of evidence indicate that this complex is highly dynamic, as manifested in several ways. The substrate-binding N-domains undergo large fluctuations relative to the main body of the complex, as observed previously and further characterized in the present study. We find that peri-axial motifs that have been implicated in substrate translocation are also mobile, as registered in our density map by local blurring. It is also likely that, as in other AAA+ ATPases, transitions between different nucleotide-binding states of ClpA generate domain movements that are coupled mechanically to the unfolding and transport of substrates; we have obtained the first clues about what such changes may entail by comparing our cryo-EM analysis of the ATPγS hexamer with a crystal-derived model for the ADP state (Fig. S3).
Mobility of the N-terminal domains
One discrepancy between the “solid state” picture conveyed by the ClpA crystal structure (Guo et al., 2002) and the “solution state” representation from cryo-EM concerns the N-domains. In the former model, they occupy fixed positions around the periphery of the D1 tier, whereas cryo-EM indicates that they are free to fluctuate, mainly in the region distal to the apical surface (Ishikawa et al., 2004). The peripheral positions are far from the central axis and thus hard to reconcile with an involvement in axial translocation of substrates. We conclude that the fixed positions observed in the crystal structure result from crystal contacts and represent sites that are accessible on molecules in solution but seldom visited, to judge from the averaged images and variance maps (Ishikawa et al., 2004).
The N-domain is connected to D1 by a linker of ~ 25 residues (Guo et al., 2002). We studied mutants with deletions in the linker, anticipating that longer deletions should progressively reduce the mobility. This was indeed what was observed for the Δ10 and Δ15 mutants, although even the longest deletion tried, Δ20, failed to completely immobilize the N-domains.
Functional consequences of extending, replacing, or shortening the linker were examined in a previous study (Cranz-Mileva et al., 2008). Our results differ in some details from those reported, but concur with the major finding that restricting the N-domain mobility has relatively modest effects on the enzymatic parameters for degradation of a major class of physiological substrates presented by the adaptor protein, ClpS. For other substrates, a more significant difference was observed. In earlier studies (Singh et al., 2001), we proposed that binding of unfolded proteins by the N-domains has two somewhat conflicting consequences - improved recruitment of unfolded protein substrates but impaired translocation of unfolded proteins bound to D1. Here we found that restricted N-domain mobility confers enzymatic properties approximating those seen when the N-domains are removed (Table 1). Lower N-domain mobility appears to disfavor orientations that promote binding of unfolded proteins and thus increases the overall maximum rates of degradation. As increasing the range of N-domain motion has little effect on protein degradation (Cranz-Mileva et al., 2008), we conclude that high mobility of the N-domains ensures that an optimal number of N-domains will be oriented to interact productively with unfolded substrates, position them for controlled entry into the axial channel, and allow timely release for translocation to ClpP.
Mobility is also manifested by the N-terminal domains of other related ATPases, such as ClpB of T. Thermophilus (Lee et al., 2007) and of E. Coli (Ishikawa, T., Maurizi, M.R., and Steven, A.C., unpublished data). A similar role in substrate engagement has been imputed to the distally extending insertion domain of ClpY/HslU, and indeed, this domain is partially or completely disordered in crystal structures (Sousa et al., 2000; Trame and McKay, 2001; Wang et al., 2001a), although we note that this mobility is on a markedly smaller scale than that of ClpA N-domains, as the I-domains are clearly visible in cryo-EM (Ishikawa et al., 2000). There is, therefore, an emerging trend that mobility of substrate-binding domains is a general feature of AAA+ enzymes (Djuranovic et al., 2009).
Comparison of the ClpA hexamer with other AAA+ ATPases
ClpA is a Type II AAA+ ATPase, with two ATPase domains that have different roles in the working enzyme. D1 is primarily responsible for hexamer assembly, while D2 has higher ATPase activity (Singh and Maurizi, 1994) which probably contributes to rapid substrate translocation, a correlation that has been demonstrated in ClpX (Kenniston et al., 2003). The placement of the D2 domains in our model is very similar to that seen in ClpY/HslU, a Type I enzyme (Bochtler et al., 2000; Sousa et al., 2000; cf. Figs. S2B and S2C)). On the other hand, the D1 model is more similar to the D2 tier of NSF, another Type II ATPase, whose D2 domain has been shown – like ClpA D1 - to be important for hexamerization (Lenzen et al., 1998). In ClpA-D2, the β-sheet of the large RecA subdomain is oriented roughly parallel to the hexamer axis, as in ClpY/HslU, whereas the β-sheet in D1 is oriented more orthogonally to the axis. According to the proposal that the direction of the force vector generated by domain movements in response to changes in nucleotide state is parallel to the strands of the β-sheet (Fig. 2 of Wang, 2004), these orientations are consistent with D2 generating an axial translocating force and D1 generating a radial force that may contribute to substrate unfolding.
Implications for nucleotide-dependent conformational changes
Crystal structures of p97 in various nucleotide states show its D1 domain to be basically fixed while its D2 domains occupy positions related to each other by simple rigid body rotations (Davies et al., 2008;DeLaBarre and Brunger, 2005). In ClpY/HslU, the main structural difference between nucleotide states is a rigid body rotation of the small α domain relative to the large α/β one (Wang et al., 2001b).
In the presence of ADP, ClpA does not form a stable hexamer. Nevertheless, when the crystal structure of the ClpA-ADP monomer was solved, this information was used to construct a hexamer model (Guo et al., 2002), using NSF D2 in the AMP.PNP state (PDB 1D2N - Lenzen et al., 1998) and HslU in the ATP state (PDB 1DO0 - Bochtler et al., 2000) as templates for ClpA D1 and D2, respectively. Its D1 tier fits well into our density map and is in close agreement with the model that we have built. On the other hand, the D2 tier fits poorly, a disagreement that can be resolved by subjecting the D2 domain to a rigid-body rotation of ~ 20° about the axis marked on Fig. S3 and by rotating the small domain relative to the large one as shown in Fig. S2. Pending experimental confirmation of the ClpA.ADP model and with the caveat that apparent changes could be affected by ClpP binding, we nevertheless hypothesize that the differences observed in the D2 ring between these two models reflect nucleotide-dependent conformational changes in ClpA. Thus our results suggest that the D1 domains act as a relatively static platform, with conformational changes mainly affecting the D2 domains. The inferred transition is not incompatible with ClpA and ClpP maintaining contact either via the IGL loop(s) and/or the protruding N-loops of ClpP (Bewley et al., 2006). In vivo, the ADP state would be expected to be shortlived, reverting rapidly to the ATP state before it has the chance to disassemble (as it would under in vitro conditions), thereby enhancing processivity. One translocation-relevant feature of the two models is that the D2 diaphragm loop is displaced ~ 12 Å further down the axial channel towards ClpP between the two putative nucleotide states (Fig. S3E).
Pore loop conformation in the active enzyme
ClpA has several motifs lining its axial channel that have been implicated in binding and translocation of substrates (Hinnerwisch et al., 2005). In the D1 tier, two short loops 1 (res. 247–262) and 2 (res. 291–299) were identified as important for binding of SsrA-tagged proteins. While loop 1 fits well into the cryo-EM map, loop 2 is only partially accounted for, implying that there is some local mobility in this region.
At one end of the D2 diaphragm loop is a GYVG motif (res. 539–542), conserved in other Clp family members, that has also been implicated in substrate binding and translocation (Hinnerwisch et al., 2005). In particular, for ClpX, the aromatic ring of the tyrosine residue was proposed to grip the substrate and drive unfolding and translocation through cyclic nucleotide-dependent movements (Martin et al., 2008). This loop is often disordered in crystal structures of related ATPases; its imputed pliability is consistent with the idea that, upon ATP hydrolysis, it could change conformation in a way that would move the substrate down towards ClpP.
Recently, based on differences in solvent accessibility between ClpA monomers and hexamers as measured by synchrotron X-ray footprinting, it was proposed that the D2 diaphragm loop in what would be its “up” conformation, makes contact with the sensor-1 region in D1 (Bohon et al., 2008) (Fig. 5C – top arrow). Our cryo-EM analysis supports this mechanism or other similarly dynamic movements within the channel. The blurred densities corresponding to the diaphragm loops are located mainly at the D2 level but appear to extend to the bottom of the D1 ring (see Fig. 2B at the 56Å mark and Fig. 2D). Our model of D1 attributes some degree of mobility to an α-helix connected to the sensor-1 region (Fig. 3A – arrow) that corresponds to the motif that has been proposed to contact the D2 diaphragm loop in the ATP state (Bohon et al., 2008).
Axial interactions in the ClpAP complex
In our model, the diaphragm loop lies roughly midway through the D2 tier (Fig. 5C – yellow loop). We posit that the conformation shown here, which is taken from the crystal structure, is only one of several that this loop can adopt. Considering its position and its length, we hypothesize that with a movement as simple as a rigid body rotation, this loop could reach the vicinity of the N-terminal loops of ClpP in what would be a “down” conformation, as opposed to the “up” conformation discussed above. Such loop movements are consistent with mechanisms proposed for ClpA and other related ATPases and would effectively position the translocating substrate at the ClpP pore entrance.
Materials and Methods
Plasmid clones and protein expression
ClpA, ClpP, and ClpS were expressed and purified as described (Guo et al., 2002; Maurizi et al., 1994; Singh et al., 2001). as were ClpA-Δ153 (Singh et al., 2001), the phage P1 RepA protein (DasGupta et al., 1993), and GFP-SsrA (Singh et al., 2000). ClpA-Δ10 and -Δ15 were obtained by deleting 10 (from 145 to 154) and 15 (from 145 to 159) codons from the ClpA coding region, using overlapping PCR. The fragments carrying the desired deletions were inserted to replace the equivalent region in the plasmid encoding ClpA. The sequences were verified by DNA sequencing. Mutant proteins were expressed and purified as described for ClpA (Maurizi et al., 1994). Construction of the expression system for SUMO-LR-GFPVenus and purification of LR-GFPVenus has been described (Heckman and Pease, 2007; Karin et al., 2007). Protein concentration measurements and enzymatic assays are described elsewhere (De Donatis et al., Ms in submission). All assays were conducted at 37 °C.
Cryo-electron microscopy
To obtain wild-type ClpAP complexes, ClpA monomers were incubated in Buffer A (50 mM Tris HCl (pH 7.5), 150 mM KCl, 10 mM MgCl2) with 1mM ATPγS. After 20 min at room temperature, the resulting ClpA hexamers were mixed with ClpP tetradecamers in Buffer A to give final concentrations of ~ 70 μg/mL (ClpP) and ~ 120 μg/mL (ClpA). 3 μl drops of sample were applied to glow-discharged, holey carbon film-mounted grids, blotted, and quenched in liquid ethane, using a KF80 (Leica) or Vitrobot (FEI) cryo-station. Vitrified specimens were then transferred into a Philips CM200 FEG electron microscope and focal pairs were recorded under low-dose conditions at 120 kV and 38,000× magnification. For ClpA-Δ10 and -Δ15, hexamer assembly was performed as above except that ATPγS was increased to 2 and 5 mM, respectively. To increase the number of side-views obtained, experiments were also performed with continuous carbon film substrates, with ClpA hexamers mixed with ClpP tetradecamers to give final concentrations of ~ 15 μg/mL (ClpP) and ~ 30 μg/mL (ClpA). Vitrification was done as described above and grids were transferred to the CM200-FEG (Δ15) and a CM120 (Δ10). Cryo-micrographs were recorded at 50k magnification (Δ15) or 60k (Δ10), using Kodak SO-163 film.
3D reconstruction of wild-type ClpA
Recorded data were screened for drift-free micrographs, showing fields with a good proportion (>50%) of side-views and a minimum of aggregation. Films were digitized on a Zeiss scanner at 7 μm step, giving 1.84 Å/pixel at the specimen. Preprocessing was carried out as follows with the Bsoft package (Heymann and Belnap, 2007). The reconstruction procedure is described fully in the Supplemental material. In brief, micrographs were screened by evaluating their Fourier transforms for the absence of drift or astigmatism and for whose signal extend to at least 10 Å. The CTF parameters were determined for each micrograph and CTF effects corrected by phase-flipping for determination of orientations and alignment and with additional baseline correction for reconstruction. A total of 8260 ClpA focal pairs was compiled and used for 3D reconstruction in SPIDER (Frank et al., 1996). Two quite different initial references were used in the projection-matching scheme and converged to the same solution. The final resolution determined by FSC at 0.5 and 0.3 cutoff was 12.5 Å and 10.5 Å respectively.
2D analysis of ClpA-Δ10 and -Δ15/ClpP
Micrographs were scanned on a Zeiss scanner at 14 μm step, giving final samplings of 2.3 and 2.8 A/pixel for Δ10 and Δ15 respectively. Images were preprocessed as explained above in Bsoft. 968 and 1222 particles were picked for Δ10 and Δ15 respectively. An initial 2D global average was generated by reference-free alignment as implemented in SPIDER. Alignment of the images was refined by running few iterations of template matching using the global average as reference, alignment of the particles, and calculation of the average and variance images.
Building pseudo-atomic models of ClpA wild type and ClpAP
First, HslU (PDB: 1G3I) and NSF D2 (PDB: 1D2N) were fitted semi-automatically into the cryo-EM map with Chimera (Pettersen et al., 2004). Then the monomer crystal structure of ClpA D1 and D2 was structurally aligned to one monomer copy of the docked hexameric HslU and NSF for ClpA D2 and D1 respectively. From this initial position, the ClpA D1 and D2 monomer were moved manually to improve the fit in Chimera. The symmetry related monomers were generated with the bmolsym command in Bsoft. Built hexamers were checked for any steric clashes between subunits and their positions were automatically locally refined in Chimera. For the ClpAP model, the azimuthal position of ClpA relative to ClpP was set according to Beuron et al (1998). In Chimera, we used the ClpA portions of the two cryo-EM models of ClpAP to align the ClpA hexamers consistently (see Supp. Fig. 6). Both models used crystal structures of ClpP. In our case, we used that of Bewley et al (2006) (PDB: 1YG6). The N-terminal domains of ClpA (residues 1–153) are from the crystal structure (Guo et al., 2002). In theory, a linker peptide of 25 amino acids could be as long as ~ 90 Å in a fully extended conformation. From our results and previous work, the N-domains of ClpA were found to be on average closer to D1 than the theoretical limit. They were therefore placed randomly within a half sphere (radius in the order of the ClpA D1 radius i.e. ~ 60 Å) at the apical surface of ClpA and their positions checked to avoid clashes.
Supplementary Material
Acknowledgments
We thank Drs N. Cheng and D. Winkler for their help with electron microscopes and Dr. B. Heymann for his support in computational matters. This work was supported by the Intramural Research Programs of NIAMS and NCI at the National Institutes of Health.
Footnotes
Accession code.
The cryo-EM map of ClpA has been deposited in the 3D-EM database under the accession number EMD-1673.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J Struct Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuron F, Maurizi MR, Belnap DM, Kocsis E, Booy FP, Kessel M, Steven AC. At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J Struct Biol. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- Bewley MC, Graziano V, Griffin K, Flanagan JM. The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. J Struct Biol. 2006;153:113–128. doi: 10.1016/j.jsb.2005.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- Bohon J, Jennings LD, Phillips CM, Licht S, Chance MR. Synchrotron protein footprinting supports substrate translocation by ClpA via ATP-induced movements of the D2 loop. Structure. 2008;16:1157–1165. doi: 10.1016/j.str.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Licht S. Control of peptide product sizes by the energy-dependent protease ClpAP. Biochemistry. 2005;44:13921–13931. doi: 10.1021/bi0505060. [DOI] [PubMed] [Google Scholar]
- Cranz-Mileva S, Imkamp F, Kolygo K, Maglica Z, Kress W, Weber-Ban E. The flexible attachment of the N-domains to the ClpA ring body allows their use on demand. J Mol Biol. 2008;378:412–424. doi: 10.1016/j.jmb.2008.02.047. [DOI] [PubMed] [Google Scholar]
- DasGupta S, Mukhopadhyay G, Papp PP, Lewis MS, Chattoraj DK. Activation of DNA binding by the monomeric form of the P1 replication initiator RepA by heat shock proteins DnaJ and DnaK. J Mol Biol. 1993;232:23–34. doi: 10.1006/jmbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- De Donatis GM, Singh SK, Viswanathan S, Maurizi MR. A single ClpS monomer is sufficient to direct the activity of the ClpA hexamer. J Biol Chem. 2010 doi: 10.1074/jbc.M109.053736. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Farbman ME, Gershenson A, Licht S. Role of a conserved pore residue in the formation of a prehydrolytic high substrate affinity state in the AAA+ chaperone ClpA. Biochemistry. 2008;47:13497–13505. doi: 10.1021/bi801140y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Gribun A, Kimber MS, Ching R, Sprangers R, Fiebig KM, Houry WA. The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J Biol Chem. 2005;280:16185–16196. doi: 10.1074/jbc.M414124200. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Heckman KL, Pease LR. Gene splicing and mutagenesis by PCR-driven overlap extension. Nat Protoc. 2007;2:924–932. doi: 10.1038/nprot.2007.132. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Beuron F, Kessel M, Wickner S, Maurizi MR, Steven AC. Translocation pathway of protein substrates in ClpAP protease. Proc Nat’l Acad Sci USA. 2001;98:4328–4333. doi: 10.1073/pnas.081543698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Maurizi MR, Belnap D, Steven AC. Docking of components in a bacterial complex. Nature. 2000;408:667–668. doi: 10.1038/35047165. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Maurizi MR, Steven AC. The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease. J Struct Biol. 2004;146:180–188. doi: 10.1016/j.jsb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Katayama Y, Gottesman S, Pumphrey J, Rudikoff S, Clark WP, Maurizi MR. The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988;263:15226–15236. [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kessel M, Maurizi MR, Kim B, Kocsis E, Trus BL, Singh SK, Steven AC. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK. Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem. 2003;278:50664–50670. doi: 10.1074/jbc.M305882200. [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Lenzen CU, Steinmann D, Whiteheart SW, Weis WI. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell. 1998;94:525–536. doi: 10.1016/s0092-8674(00)81593-7. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi MR, Thompson MW, Singh SK, Kim SH. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- Ortega J, Heymann JB, Kajava AV, Ustrell V, Rechsteiner M, Steven AC. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J Mol Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Reid BG, Fenton WA, Horwich AL, Weber-Ban EU. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci USA. 2001;98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseman AM, Chen S, White H, Braig K, Saibil HR. The chaperonin ATPase cycle: mechanism of allosteric switching and movements of substrate-binding domains in GroEL. Cell. 1996;87:241–251. doi: 10.1016/s0092-8674(00)81342-2. [DOI] [PubMed] [Google Scholar]
- Rotanova TV, Botos I, Melnikov EE, Rasulova F, Gustchina A, Maurizi MR, Wlodawer A. Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 2006;15:1815–1828. doi: 10.1110/ps.052069306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci USA. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Maurizi MR. Mutational analysis demonstrates different functional roles for the two ATP-binding sites in ClpAP protease from Escherichia coli. J Biol Chem. 1994;269:29537–29545. [PubMed] [Google Scholar]
- Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem. 2001;276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- Snider J, Houry WA. AAA+ proteins: diversity in function, similarity in structure. Biochem Soc Trans. 2008;36:72–77. doi: 10.1042/BST0360072. [DOI] [PubMed] [Google Scholar]
- Sousa MC, Trame CB, Tsuruta H, Wilbanks SM, Reddy VS, McKay DB. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Szyk A, Maurizi MR. Crystal structure at 1.9Å of E. coli ClpP with a peptide covalently bound at the active site. J Struct Biol. 2006;156:165–174. doi: 10.1016/j.jsb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Trame CB, McKay DB. Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning. Acta Crystallogr D Biol Crystallogr. 2001;57:1079–1090. doi: 10.1107/s0907444901007673. [DOI] [PubMed] [Google Scholar]
- Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J Struct Biol. 2004;148:259–267. doi: 10.1016/j.jsb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Franklin MC, Kamtekar S, Im YJ, Rho SH, Seong IS, Lee CS, Chung CH, Eom SH. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001a;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. structure. 2001b;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Xia D, Esser L, Singh SK, Guo F, Maurizi MR. Crystallographic investigation of peptide binding sites in the N-domain of the ClpA chaperone. J Struct Biol. 2004;146:166–179. doi: 10.1016/j.jsb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Yu AY, Houry WA. ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 2007;581:3749–3757. doi: 10.1016/j.febslet.2007.04.076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.