Abstract
Background and Objective
Cells with osteoprogenitor potential are present within periodontal tissues during development and in postnatal life. To identify an osteoprogenitor population, this study utilized a transgenic model in which an α-smooth muscle actin (αSMA) promoter directed green fluorescent protein (GFP) expression.
Material and Methods
Observation of GFP expression was complemented with analysis of osteogenic differentiation by determining the expression of RNA of bone markers, by histochemical staining for alkaline phosphatase and by the detection of mineralized nodules using xylenol orange. Flow cytometry was utilized to determine the proliferative potential and cell-surface phenotype of cultured αSMA-positive cells.
Results
αSMA–GFP expression was detected within the dental follicle and in the apical region of the root (i.e. areas rich in vascularization) but not in mature bone. αSMA–GFP expression was observed during the early stages of primary cultures derived from the dental follicle and periodontal ligament and was diminished in areas undergoing mineralization. Intense alkaline phosphatase activity and the presence of mineralized nodules was observed 2 wk after osteogenic induction. Consequently, the expression of bone sialoprotein, osteocalcin and dentin matrix protein-1 was increased. Flow cytometry revealed that in vitro expansion enriched for an αSMA–GFP-positive population in which 55–65% of cells expressed the cell-surface markers Thy1+ and Sca1+. The αSMA–GFP-positive population exhibited high proliferative and osteogenic potentials when compared with an αSMA–GFP-negative population.
Conclusion
Our data indicate that the αSMA promoter can be used to identify a population of osteoprogenitor cells residing within the dental follicle and periodontal ligament that can differentiate into mature osteoblasts.
Keywords: smooth muscle alpha actin, periodontal ligament, dental follicle, osteoprogenitor cell
Previous studies have indicated that tissues associated with the periodontium contain a cell population with properties characteristic of a progenitor cell (1–3). The characterization of these cell types within the dental follicle (DF) and the periodontal ligament(PDL) has been difficult because of a lack of specific markers with which to identify cells with distinctive progenitor properties.
The supportive tissues of the tooth, including PDL, cementum and alveolar bone, are developmentally derived from the cells of the DF through a mechanism of migration and differentiation of mesenchymal cells. In addition to this classical mesenchymal origin of periodontal tissues, it has also been proposed that epithelial–mesenchymal transformation can be, at least in part, responsible for producing cells of the cementoblast lineage (4–9). Thus, the DF, a major component of the tooth germ, may undergo differentiation into cell lineages of biologically functional and competent tooth-supporting tissues (10,11).
The PDL is positioned between the thin mineralized outer layer of the cementum and the inner wall of the alveolar bone socket (12). It has been well documented that the PDL contains a population of progenitor cells that can differentiate into osteoblasts and cementoblasts. The ability of PDL-derived stem cells to differentiate and to mineralize has already been documented in different species, including rats (13,14), humans (15,16), mice (17), sheep (18) and cows (19). In addition, the process of periodontal regeneration is thought to involve the recruitment of locally derived uncommitted cell populations with the capacity to develop into osteoblasts, cementoblasts or PDL cells (12).
The characterization and evaluation of progenitor cell populations in primary cultures of DF and PDL is critical for understanding remodeling and regeneration of the adjacent alveolar bone and cementum. Previous studies have utilized cell-surface markers to identify and characterize cell populations with osteoprogenitor potential within the periodontal tissues and the dental pulptissues. Thesestrategies used markers such as STRO-1, CD106 (vascular cell adhesion molecule-1), CD44, CD105 and CD146 that are known to be expressed by bone marrow-derived mesenchymal stem cells (1,20,21). It has been shown that cell populations obtained from dental pulp, deciduous teeth, bone marrow or PDL express cell-surface markers associated with the perivascular compartment [α-smooth muscle actin (αSMA), 3G5, CD146], as well as markers of bone/dentin/cementum [Col1a1, alkaline phosphatase (ALP), osteocalcin and osteonectin)] and fibroblasts (Coll3a1 and scleraxis) (12). It has also been demonstrated that αSMA-expressing cells are localized to the DF and to the highly vascular areas in the apical regions of the root of adult rats (22). These data are indicative for the presence of stem cells within those tissues and underline the heterogeneity of the populations within tissues and in the in vitro assays.
We have previously shown that the αSMA promoter – green fluorescent protein (GFP) transgene construct can be used to identify a population of cells within bone marrow stromal cells and adipose-derived vascular stromal cells that exhibit osteogenic and adipogenic potential (23). The goal of our study was to characterize a cell population residing within the periodontium that expresses GFP under the control of a smooth muscle type α-actin promoter (αSMA–GFP)(24). Development and characterization of the visual marker for identification of the DF and PDL-derived osteoprogenitor cell population would greatly enhance the ability to selectively isolate and utilize these cells in tissue-regeneration studies.
Material and methods
Transgenic mice
αSMA-directed GFP expression
To identify the perivascular population of cells, we utilized an αSMA promoter (termed SMP8) to direct the expression of enhanced GFP, a transgenic model developed by Dr Jen-Yue Tsai (24,25).
Dentin matrix protein-1 promoter-directed GFP expression
We utilized previously developed transgenic mice in which a dentin matrix protein-1 (DMP-1) promoter directs the expression of the GFP transgene. Expression of GFP was localized primarily to pre-osteocytes, osteocytes and odontoblasts (26). The Institutional Animal Care Committee approved the protocol and procedure for use of the αSMA–GFP and DMP-1–GFP transgenic mice.
Histological evaluation of GFP expression
Mandibles from 3–5-d-old mice and from 4-wk-old mice were fixed in 10% buffered formalin at 4°C for 1–3 d. Following fixation, teeth were decalcified in 15% EDTA (pH 7.1) for 1 and 3 wk respectively, placed in 30% sucrose overnight and embedded in tissue-embedding medium (Cryomatrix; Thermo Shandon, Pittsburg, PA, USA). Teeth were cut into 5-μm thin sections using a CryoJane tape transfer system (Instrumedics, St Louis, MO, USA). Green fluorescent protein expression was detected using a fluorescein isothiocyanate/Texas Red dual filter cube on a Zeiss Axiovert 200 M microscope (Carl Zeiss, MicroImaging Inc., Thornwood, NY, USA) and photographed using an Axiocam digital camera (Carl Zeiss, MicroImaging Inc.).
Immunohistochemistry
We utilized immunohistochemistry to detect the presence of CD31 as a marker of the endothelial cell population. Mandibles were processed and sectioned as indicated above. Briefly, slides were immersed in phosphate-buffered saline (PBS) for 30 min and the cover glasses were removed. Thereafter, sections were treated with 3% H2O2 in PBS for 30 min to inactivate endogenous peroxidases and were then incubated in Power Blocking reagent for 20 min at 20°C. After washing with PBS, the sections were incubated with primary CD31 antibody (1:100 dilution; BD Biosciences, San Jose, CA, USA) at 4°C overnight. Sections were washed and exposed to secondary biotinylated antibody (1:200 dilution) for 1 h at 20°C and reaction complexes were visualized using the 3′-diaminobenzidime reaction (Vector Laboratories, Burlingame, CA, USA).
Preparation of murine PDL and murine DF cultures
Dental follicle cells and PDL cells were isolated from 3–5-d-old neonatal mice, and from 4–6-wk-old αSMA–GFP transgenic mice, using a procedure described by D’Errico et al. (27). Briefly, after removal of DF or molars, together with adherent PDL, samples were initially placed in 15 mL of digestion buffer containing PBS, 1.5 U/mL of collagenase P and 0.05% trypsin, and rotated for 90 min at 37°C in a shaker. Cell suspensions were collected and enzyme activity was terminated by the addition of Dulbecco’s modified Eagle’s minimal essential medium/20% fetal bovine serum (DMEM/20% FBS). Cells were pelleted by centrifugation at 500 g for 5 min. The supernatant was carefully aspirated and the cells were resuspended in DMEM/20% FBS containing antibiotics and nonessential amino acids then filtered through a 70-μm pore size cell strainer. Cells were plated at a density of 105 cells/cm2 in 24-well plates containing DMEM/20% FBS. At 1 wk of culture when the cells reached confluence, the medium was changed to differentiation medium (alpha-minimal essential medium/10% FBS, 50 μg/mL of ascorbic acid and 4 mM β glycerophosphate). The medium was replaced every 2 d until mineralized nodules were formed. The expression of GFP was visualized using an Olympus IX50 inverted reflected light fluorescence microscope (Olympus America, Inc., Melville, NY, USA). A specific excitation wavelength was obtained using filters for GFPtopaz (exciter, D500/20; dichoroic longpass, 525DCLP; emitter, D550/40) and GFPemerald (exciter, D470/40; dichroic, 495LP; emitter, D525/50) and recorded using a SPOT Camera (Diagnostic Instruments, Sterling Heights, MI, USA).
In vitro determination of mineralized nodules in murine dental follicle cultures and in murine periodontal ligament cultures
Cell cultures were monitored for mineralization at three different time-points: week 1 (without osteogenic medium); and at weeks 2 and 3 (with osteogenic medium). Consequently, to visualize mineralized nodules at the end of each time-point, culturedcellswereincubated with 20 μM xylenol orange (XO) for a period of 4 h before scanning. Prior to imaging, XO-containing medium was replaced with fresh medium to remove the fluorescent background. Xylenol orange produced a red color when visualized using a Tetramethyl rhodamine iso-thiocyanate (TRITC) filter (excitation 545/30, emission 620/60).
Histochemical analysis of cell cultures
Histochemical staining for ALP activity was performed using a commercially available kit (86-R ALP; Sigma Aldrich, Inc., St Louis, MO, USA) according to the manufacturer’s instructions. Mineralization was assessed using the von Kossa silver nitrate staining method. The results of staining procedures were recorded using a scanner (UMax Astra 4000U; UMax Technologies, Inc., Dallas, TX, USA) and Adobe Photoshop (Adobe Systems).
RNA extraction and real-time quantitative RT-PCR
Total RNA was extracted from DFs and PDL cultures using TRIzol reagent (InvitrogenCorp., Carlsbad, CA, USA). RNA samples were used for complementary DNA synthesis, and gene-expression analysis wascompleted using semiquantitative and real-time PCR. Three micrograms of total RNA was reverse transcribed in a 30 μL reaction mixture containing 50 ng of random hexamers mixed together with 200 U of Superscript III reverse transcriptase, according to the manufacturer’s instructions. Real-time PCR was carried out using the TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) for bone sialoprotein (Mm00492555_m1) and DMP (Mm01208365_m1). The data were analyzed using the 7500 fast system SDS software (Applied Biosystems). Semi-quantitative PCR was used to detect the expression of osteocalcin and dentin sialophosphoprotein using the following primers: osteocalcin, forward, TCCAAGCAGGA GGGCAATAAG, and reverse, GCGTTTGTAGGCGGTCTTCAAG; dentin sialophosphoprotein, forward, GGCATAATCAAAACACCGCTGC, and reverse, GGGGAAATAGGGAAATGACAAAGG).
Flow cytometric analysis and cell sorting
The freshly digested murine DF (mDF) cells and murinePDL(mPDL)cells were prepared as described above. The process was terminated by the addition of medium containing serum followed by centrifugation. Cells were resuspended in PBS/FBS 12% and filtered through a 70-μm cell strainer. In order to obtain a higher proportion of αSMA–GFP-positive cells, mDF and mPDL were expanded by culture for 5–7 d. Cells were grown in DMEM/20% FBS and themediumwaschangedevery2 d. Cells were prepared by washing in PBS and by digesting the matrix with 0.25% trypsin/1 mM EDTA. Samples were resuspended in PBS/2% FBS and filtered. Phenotypic characterization using fluorescence-activated cell sorter (FACS) analysis was carried out on a FACS Calibur flow cytometer (Becton-Dickinson, San Jose, CA, USA) and analysis was performed using CELLQUEST software (BD Biosciences, San Jose, CA, USA). Our analysis utilized anti-mouse CD45-allophycocyanin (APC) (clone RA3-6B2), CD11b–phycoerythrin (clone M1/70), CD117 (clone 2B8), anti-mouse Sca1–phycoerythrin or CD90–phycoerythrin. All antibodies were purchased from Pharmingen or e-Bioscience (San Diego, CA, USA).
To analyze the cell cycle of the mDF and the mPDL, 3-d-old cultures were trypsinized and pelleted by centrifugation. A nuclear dye (Hoechst 33342; Molecular Probes, Carlsbad, CA, USA) was added to samples from wild-type and transgenic mice at a concentration of 4 mg/mL. As negative controls we utilized the same cell sources, but without addition of nuclear dye. The labeled cells were analyzed in a BD LSRII flow cytometer (Becton-Dickinson). Data analysis was performed using MODFIT LT software (Verity Software House, Inc., Topshame, ME, USA). Both wild-type and αSMA–GFP-positive samples were analyzed to determine the distribution of cells present in G0 + G1, G2 + M and S phases.
For cell sorting, cells were prepared as previously described (28). Briefly, cell cultures obtained from 4-d-old mDF primary cultures were digested in 0.25% trypsin/1 mM EDTA for 5 min. Following centrifugation, the cells were resuspended in PBS, washed and filtered. Cell sorting was carried out using a FACS Vantage (BD Biosciences) with a 488-nm excitation filter and a 530/30 emission filter. Cells were separated using a 130-μm nozzle and collected into DMEM/20% FBS. Cells were counted and replated at a density of 20,000 cells/cm2, and osteogenic differentiation was induced after the cells reached confluence.
Results
Defining the expression of αSMA–GFP in DF and PDL
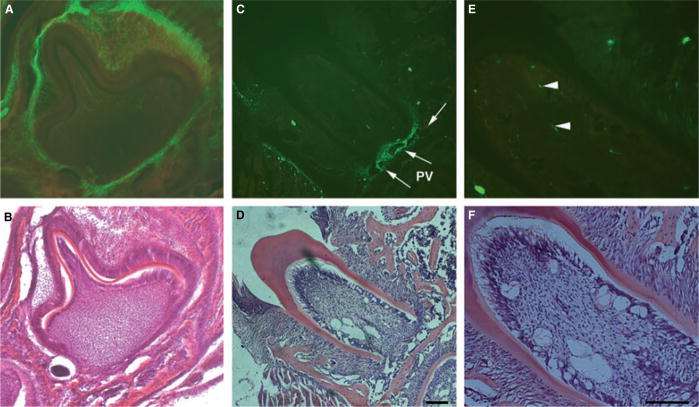
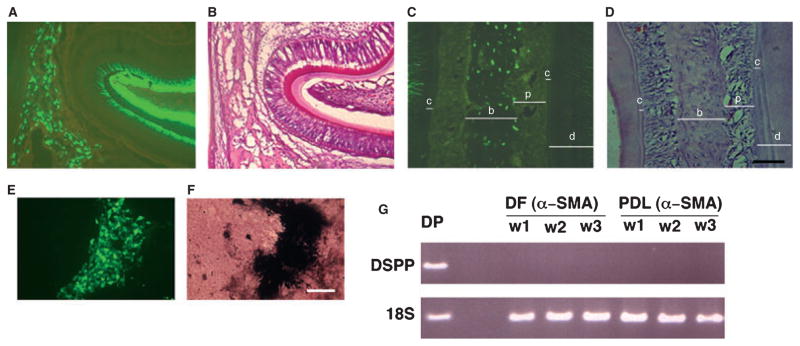
Decalcified frozen mandibular sections of 3–5-d-old neonatal mice and 4–6-wk-old adult αSMA–GFP transgenic mice were prepared for histological examination. Strong expression of αSMA–GFP was observed in the DF area and weaker expression was observed within the stellate reticulum of the enamel organ (Fig. 1A). In sections of the periodontium of a 4-wk-old mouse, cells within the perivascular areas in the apical part of the PDL showed intense expression of αSMA–GFP (Fig. 1C). The expression of αSMA–GFP in different areas of the PDL was also examined. This analysis revealed the presence of αSMA–GFP+ cells associated with the microvasculature within the alveolar crest, the horizontal and oblique fibers, and the rich vascular area of the apical fibers of the PDL (data not shown). A similar pattern of expression was also found in the dental pulp (Fig. 1E).
Fig. 1.
Expression of the α-smooth muscle actin–green fluorescent protein (αSMA–GFP) transgene in dental follicle and periodontal ligament. αSMA–GFP expression was evaluated in decalcified frozen sections of mandibles derived from 3–5-d-old neonatal mice (A, B) and from 4-wk-old αSMA–GFP transgenic mice (C–F). Epifluorescence was detected in cells within the dental follicular area of the developing tooth and in the outer dental epithelium (A). The perivascular (PV) area in the apical part of the root showed intense expression of GFP (C, E, see arrows). Green fluorescent protein was also detected in a few cells within the dental pulp (E, arrowheads). Hematoxylin & eosin staining of adjacent sections are shown in panels B, D and F. Images were taken at 10× (A–D) and 20× (E, F) magnifications (bar = 100 μm).
αSMA–GFP-expressing cells in developing tooth and periodontium are associated with the microvasculature
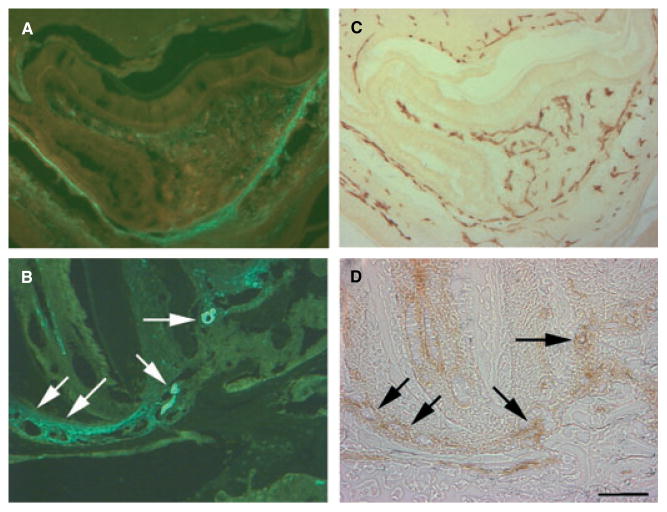
To localize the expression of αSMA–GFP to cells associated within the microvasculature, sections obtained from the developing tooth organ and the periodontium were analyzed immunohistochemically for CD31 expression. The endothelial cells associated with the microvasculature within a DF, with developing layers of the enamel organ and with the apical region of the PDL were positive for CD31 (Fig. 2C,D). The expression of αSMA–GFP was localized to the cells in close proximity to CD31-positive cells (Fig. 2A,B). These results indicated that perivascular cells are localized in association with the microvascular network within the developing layers and the PDL fibers.
Fig. 2.
Localizing α-smooth muscle actin–green fluorescent protein (αSMA–GFP) to the perivascular area. Images were taken to evaluate epifluorescence (A, B) and as brightfield for immunohistochemical staining (C, D). The expression of GFP was localized in proximity to blood vessels or capillaries that were lined with CD31 endothelial cells (B, D, see arrows). The cells within the dental follicle area and the developing layers of the enamel organ, including the stellate reticulum, were positive for αSMA–GFP (A). The expression of αSMA–GFP was localized proximal to the endothelial cells, as identified by CD31 expression (C, D). Images were taken using 20× magnification (bar = 100 μm).
Differentiation potential of primary mDF cells and mPDL cells
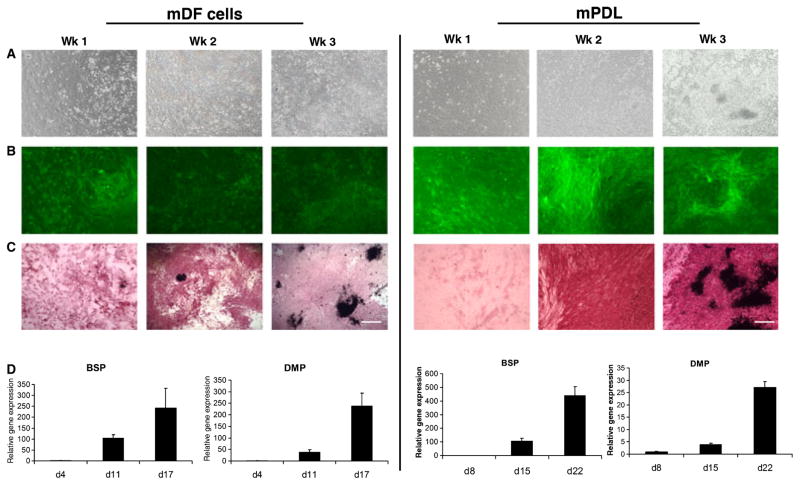
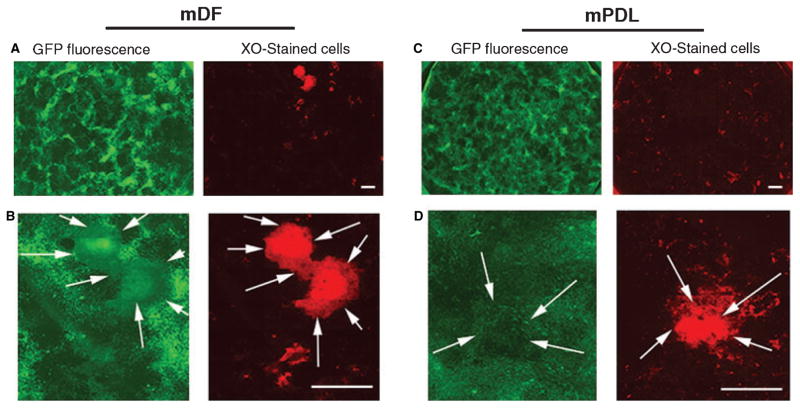
The patterns of αSMA–GFP expression in both mDF cells and mPDL cells were analyzed. The expression of αSMA–GFP was monitored at the following time-points during differentiation: week 1 (confluent cell stage); week 2 (nodule-formation stage); and week 3 (mineralized nodule stage). Phase-contrast microscopy images of cultured cells showed the degree of confluence before and after osteogenic induction (Fig. 3A). Both mDF (Fig. 3B left panel) and mPDL (Fig. 3B right panel) expressed αSMA–GFP from weeks 1 to 3 of differentiation. After 1 wk in culture, but before the addition of osteogenic-inductive media, cells were analyzed for ALP activity. Alkaline phosphatase activity was detected at week 1 and was maximal at week 2. von Kossa staining revealed the presence of mineralized nodules between weeks 2 and 3 in culture (Fig. 3C). Within the nodule-containing areas, a decrease was observed in the expression of αSMA–GFP. This coincided with the progression of mineralization in both cultures derived from mDF and mPDL. To confirm these observations, we utilized a supravital staining method to detect mineralized nodules. The presence of mineralized nodules was detected using XO staining procedures. Both mDF and mPDL cells were monitored at different time-points from weeks 1 to 3. A number of XO-stained nodules were observed at week 3. Higher magnification showed the XO-stained nodules in red (indicated by white arrows), with their corresponding GFP expression indicating that areas undergoing mineralization exhibit lower levels of GFP expression (Fig. 4A–D).
Fig. 3.
In vitro analysis of the osteogenic potential of primary dental follicle and periodontal ligament (PDL)-derived cells. Murine dental follicle (mDF) cells (left panel) derived from 3–5-d-old mice, and murine PDL (mPDL) cells (right panel) from 4–6-wk-old α-smooth muscle actin–green fluorescent protein (αSMA–GFP) transgenic mice were imaged using phase-contrast microscopy (A). The GFP expression of both mDF and mPDL cells was monitored at different time points and the corresponding stage of differentiation: week 1 (confluent cell stage), week 2 (multilayer formation stage) and week 3 (mineralized nodule stage) (B). The cells were also analyzed for alkaline phosphatase (ALP) activity at these three time points. ALP activity was detected at week 1 and was maximal at week 2 (C). von Kossa staining revealed the presence of mineralized nodules at week 3. The mDF and mPDL cells were analyzed for the expression of bone sialoprotein (BSP) and dentin matrix protein (DMP) by real-time PCR. Maximum expression of these genes was observed after 3 wk in culture along with an increase in mineralization (D). Images were taken under 10× magnification (bar = 200 μm).
Fig. 4.
Observation of α-smooth muscle actin–green fluorescent protein (αSMA–GFP) expression during mineralization. The expression of αSMA–GFP was evaluated in primary murine dental follicle (mDF) cultures (A, B) and in murine periodontal ligament (mPDL) cultures (C, D) that were induced to mineralize. Mineralization was detected by xylenol orange (XO) deposition that was added to culture 4 h before imaging. Cultures grown in 35-mm-diameter wells were imaged for GFP expression (fluorescein isothiocyanate filter) and for XO deposition (TRITC filter) using a motorized stage, and images were assembled (A, C). High-power images of the same positions show areas of mineral deposition that exhibited weaker expression of the αSMA–GFP transgene (indicated by the arrows). Original images were taken at 5× magnification, bar = 2 mm.
Real-time PCR analysis of mDF cells and mPDL cells subjected to differentiation conditions showed a time-dependent up-regulation of bone-related genes such as bone sialoprotein, DMP-1 and osteocalcin (Fig. S1). Maximum expression of these genes was observed at the later stage of mineralization, namely week 3 (Fig. 3D).
mDF and mPDL do not differentiate into odontoblast-lineage cells
Because primary cultures of mDF and mPDL may be contaminated with dental pulp, it is important to clearly distinguish osteogenic differentiation from odontogenic differentiation. A high level of expression of DMP-1–GFP was detected in odontoblasts and osteocytes in the 5-d-old mandible (Fig. 5A–D). No expression of DMP-1–GFP was detected in the dental pulp or in the DF (Fig. 5A,B). The primary mDF cultures, derived from DMP-1–GFP transgenic mice and grown in osteogenic media, showed a correlation of the GFP signal with mineral deposition (Fig. 5E,F). The presence of DMP-1–GFP in the cultures confirmed that progenitors achieve terminal differentiation into mineralized tissues. We utilized RT-PCR analysis to identify dentin sialophosphoprotein expression in samples derived from primary cultures. Dentin sialophosphoprotein expression was detected only in samples of dental pulp cultures that were utilized as a positive control (Fig. 5G, dental pulp); no detectable levels of dentin sialophosphoprotein were present in mDF or in mPDL. Real-time PCR also demonstrated that cell cultures of mDF and of mPDL do not express dentin sialophosphoprotein (data not shown). This observation indicated that both mDF and mPDL did not exhibit the ability to differentiate into odontoblast-like cells and that the method of primary culture preparation did not result in any significant contamination with dental pulp cells.
Fig. 5.
Observation of dentin matrix protein-1–green fluorescent protein (DMP-1–GFP) expression as a marker of mature osteoblast and odontoblast lineage cells. The expression of DMP-1–GFP was evaluated in 5-d-old mandibles (A, B) and in the periodontal ligament area (C, D). The expression of DMP-1–GFP was localized to odontoblasts (A, B) and to osteocytes within the alveolar bone (C, D; c, cementum; b, bone; p, periodontal ligament; d, dentin). The expression of DMP-1–GFP was observed at a late stage and was localized to the mineralized areas of differentiated murine dental follicle (mDF) cultures (E, F). The absence of dentin sialophosphoprotein expression as an indicator of odontoblastic differentiation and the contamination of mDF culture with pulp cells was confirmed by real-time PCR (panel G: DSPP, dentin sialophosphoprotein; DP, dental pulp; DF, DF culture; PDL, periodontal ligament culture; 18S, ribosomal RNA; w, week) (A, D: images were taken under 20× magnification, bar = 100 μm; E, F: images were taken under 10× magnification, bar = 200 μm). 18S.
Phenotyping of the αSMA–GFP+ population derived from mDF and mPDL cultures
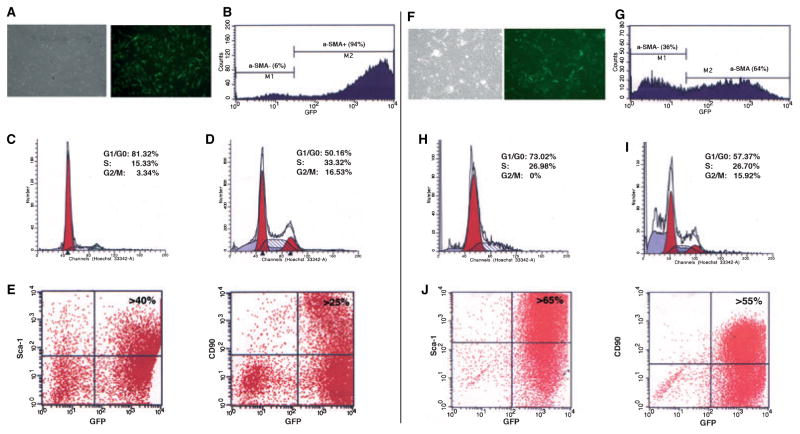
The profiling of mDF and mPDL was performed using flow cytometry for cell cycle analysis and detection of cell-surface markers. Flow cytometric analysis of 6-d-old cultures revealed the presence of a high proportion of αSMA–GFP+ cells (Fig. 6A,B,F,G). Detection of the GFP transgene was combined with cell cycle analysis using a Hoechst 33342 nuclear stain. The αSMA–GFP+ cells derived from mDF showed a high proportion of cells undergoing division, with 50% of cells in the G1/G0 phase, 33% in the S phase and 17% in the G2M phase (Fig. 6D). Similarly, in mPDL cell populations, 57% of cells were in the G1/G0 phase, 27% were in the S phase and 16% were in the G2M phase (Fig. 6I). αSMA–GFP+ cells derived from both mDF and mPDL showed a significantly higher cell-division rate compared with αSMA–GFP− cells (Fig. 6).
Fig. 6.
Phenotype of α-smooth muscle actin–green fluorescent protein (αSMA–GFP)-positive cells derived from primary murine dental follicle (mDF) and murine periodontal ligament (mPDL) cultures. (A) Transmitted and epifluorescence images of primary mDF cultures on day 7, and (B) fluorescence-activated cell sorter analysis revealed that a high proportion of cells express αSMA–GFP (> 90%). The histogram of cell cycle analysis was performed for αSMA–GFP− cells and αSMA–GFP+ cells, indicated in Fig. 5B as M1 and M2 gated populations. (D) Three-day-old αSMA–GFP+ mDF cells underwent a high division rate, with > 30% of cells in the S phase of the cell cycle and > 15% of cells in the G2M phase of the cell cycle, while the αSMA–GFP− population (C) showed a very limited potential for cell division. (E) A high proportion of αSMA–GFP+ cells expressed Sca-1 (> 40%) and CD90 (> 25%). (F, G) The αSMA–GFP+ population represents about 60% of the primary mPDL culture on day 3. Similarly to mDF, αSMA–GFP+ cells derived from mPDL showed a high cell division rate, with > 25% of cells in the S phase of the cell cycle and > 15% of cells in the G2M phase of the cell cycle (I), while the αSMA–GFP− population (H) showed very few cells undergoing cell division. (J) More than 65% of mPDL cells expressed Sca-1 and > 55% were positive for CD90. Representative plots are shown after the CD45− population was excluded from analysis using CD45–APC staining.
Immunolabeling for CD45, Sca-1, CD90, CD117 and CD11b was performed on cells after 5–7 d in culture. A high proportion (94%) of mDF cells were αSMA–GFP+ (Fig. 6B) and did not express CD117 or CD11b (data not shown). Large proportions of αSMA–GFP+ cells were Sca1+ (40%) and CD90+ (25%), indicative of the presence of a mesenchymal progenitor population within the αSMA–GFP+ cells (Fig. 6E,J). Fluorescence-activated cell sorter analysis revealed that 55–65% of αSMA–GFP+ cells obtained after short-term culture of mPDL cells expressed Sca-1 and CD90. Our results indicated that αSMA–GFP+ cells express markers characteristic of mesenchymal progenitor cells and exhibit a high proliferation when grown in primary cultures.
Evaluating osteoprogenitor ability of sorted αSMA–GFP+ cells
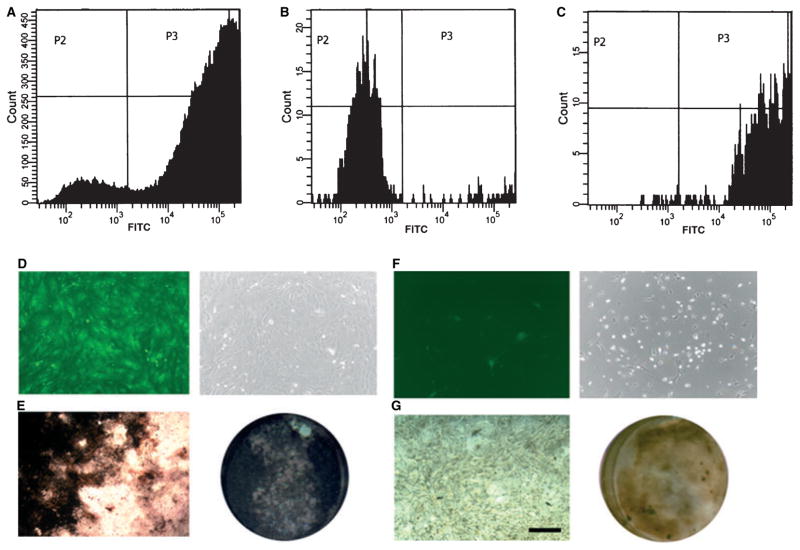
Cell populations obtained from 5-d-old primary cultures of mDF cells were used for isolating αSMA–GFP+ cells by cell sorting (Fig. 7A–C). Isolated cells were replated and, upon reaching confluence, cell populations were evaluated for their capacity to differentiate into the osteoblastic lineage (Fig. 7D,F). After 3 wk of culture in an osteogenic induction medium, αSMA–GFP+ cells derived from primary mDF showed intense ALP activity and large areas of mineralization, as detected by von Kossa staining (Fig. 7E). By contrast, the αSMA–GFP− cells failed to undergo mineralization (Fig. 7G). These data strongly support our initial observations that αSMA–GFP+ cells contain a population of osteoprogenitor cells with the ability to generate mature osteoblast-lineage cells.
Fig. 7.
Osteoprogenitor potential of an isolated population of α-smooth muscle actin–green fluorescent protein-positive (αSMA–GFP+) cells. Fluorescence-activated cell sorter (FACS) analysis showing an unsorted population of primary murine dental follicle (mDF) cells (A). Following cell sorting, the purity of sorted cell populations was reconfirmed by FACS analysis of αSMA–GFP− cells (B) and αSMA–GFP+ cells (C). GFP imaging of replated cells was carried out to assess for αSMA–GFP+ (D) and αSMA–GFP− (F) populations. Sorted cell populations were replated and grown in osteogenic differentiation media for 3 wk. (E) The αSMA–GFP+ sorted cells showed intense staining for alkaline phosphatase (ALP) activity, with almost the whole culture area covered by von Kossa-positive mineralized nodules. By contrast, the αSMA–GFP− cells (G) showed very weak expression of alkaline phosphatase and only a few mineralized areas were evident after 3 wk in culture. Images were obtained using 10× magnification (bar = 200 μm). FITC, fluorescein isothiocyanate.
Discussion
Previous studies have revealed the existence of mesenchymal progenitors within the PDL cell population that exhibits the ability to differentiate into an osteoprogenitor lineage (3). These cells, termed PDL stem cells, can be isolated based on their expression of the cell-surface markers STRO-1 and CD146 (29). Although these markers can enrich for the stem cell population, the STRO-1+ CD146+ population still remains heterogeneous. In addition, the STRO-1 marker is restricted to studies on human PDL stem cells, with data on its expression in the murine model being very limited (30). Recent publications indicate that the CD146 population is localized to the perivascular niche, and that these cells derived from bone marrow (31) and from human PDL have the ability to differentiate into an osteoprogenitor lineage (1,2,29). Most in vitro studies using the DF cells obtained from mouse, bovine, porcine, rat and human sources exhibited various levels of ALP activity upon stimulation with growth factors or differentiating medium, or with 1.25(OH)2 vitamin D3. These variations can be attributed to the heterogeneity of DF cells (12,28,30,32,33).
In this study, we characterized an αSMA–GFP-expressing cell population as a potential source for osteoprogenitors. These cells were localized to perivascular areas and in close proximity to CD31+ endothelial cells. There have been numerous reports indicating that pericytes residing within the retina and the bone marrow represent a cell population with mesenchymal progenitor properties (23,31–33). Similarly, a recent study by Zannettino et al. (34) described the presence of multipotential cells within adult human adipose tissue, which appears to be intimately associated with perivascular cells. The pericytes were characterized by their expression of the cell-surface marker, 3G5, and by the expression of cytoskeletal proteins such as SM22α and smooth muscle α-actin (33). We utilized the expression of GFP under the control of an αSMA promoter to identify, visually, an αSMA-expressing population of cells within periodontal tissue. In addition, we evaluated the expression of the αSMA–GFP transgene in primary cultures derived from DF or PDL cells. The mDF exhibited distinctive features of cells undergoing mineralization in vitro as shown by their ALP activity and by von Kossa staining. Our study demonstrated that ALP activity in cultured mDF cells was detected when the cells became confluent, markedly increasing during the multilayer-formation stage. Similar results were shown in a study that utilized cloned DF cell lines (11). The primary cultures of mDF and mPDL become confluent within 1 wk and, following osteogenic induction, mineralization and expression of the osteogenic markers bone sialoprotein, osteocalcin and DMP occured. As differentiation progressed, the expression of αSMA–GFP decreased in the areas undergoing mineralization. The definite answer regarding a decrease in the expression of αSMA–GFP in individual cells within the mineralized areas could be obtained by combining the αSMA–GFP with a visual marker of mature osteoblasts. Following utilization of a two-color approach, and by using flow cytometry, a better insight into transition of the cellular phenotype could be obtained. However, observation of a decreased expression of αSMA–GFP is supported by in vivo findings (Figs 1 and 2) and by our previous observations that αSMA–GFP is not expressed in mature cells of the osteoblast lineage (23).
Preparation of a primary culture of DF cells using enzymatic digestion could potentially result in significant contamination with dental pulp stem cells. Numerous studies have shown that dental pulp contains mesenchymal stem cells, termed dental pulp stem cells (35). The dental pulp stem cell population exhibits the ability to differentiate into odontoblast-lineage cells, as described by the expression of dentin sialophosphoprotein, a marker with high specificity for odontoblasts (36–38). We evaluated the expression of dentin sialophosphoprotein in mDF and in mPDL after the addition of differentiation media (containing 50 μg/mL of ascorbic acid and 4 mM β-glycerolphosphate). While dental pulp-derived cells (i.e. the positive control) showed a high level of dentin sialophosphoprotein expression, dentin sialophosphoprotein was not detected in primary mDF and mPDL using conventional and real-time PCR. These results support the conclusion that mature cells with mineralization potential are derived from DF and not from contaminating dental pulp.
The αSMA–GFP+ cells are a rare population within the periodontal tissues of adult mice, as detected by histological evaluation. The expression of αSMA–GFP in mDF is also present in a defined population that accounts for 18% of cells of freshly digested DF tissue (data obtained by flow cytometry analysis). An intriguing observation was the potential of the αSMA–GFP+ cells to divide and multiply rapidly under in vitro conditions. Cell cycle analysis showed that a high proportion of αSMA–GFP+ mDF and mPDL cells undergoe cell division (S, G2 and M). This finding is very promising for tissue-regeneration purposes, as any procedure involving transplantation of progenitor cells would require their in vitro expansion. To characterize the progenitor characteristics of αSMA–GFP+ cells in greater detail, flow cytometry was utilized to evaluate the expression of cell-surface markers. Large proportions of αSMA–GFP+ cells were positive for the stem cell markers, Sca-1+ and Thy-1 (CD90+), while expression of CD117 (c-kit) or CD11b (Mac1) (data not shown) was not detected. Sca1+ and Thy1+ are indicative of the presence of a progenitor population within the αSMA–GFP+ cells (39–41). Putative stem cells in healthy and diseased PDL were largely detected within the perivascular region and hardly any were present in the extravascular region (29). Our previous study using an αSMA–GFP reporter revealed the presence of a mesenchymal progenitor population in the perivascular niches of adipose tissues, and culturing these cells highly enriched for αSMA+/CD45−/Sca1+ progenitors (23).
In this study, we evaluated the osteogenic potential of the αSMA–GFP+ cells using a primary tissue culture system. Following expansion and separation of cell populations by fluorescence-activated cell sorter analysis, yielding a purity of > 99%, cells were induced to osteogenesis. Sorted αSMA–GFP+ cells continued to proliferate and differentiate into mineralized nodules with strong expression of ALP. The αSMA–GFP− population contained only a few ALP-positive cells. Previous studies showed that progenitors derived from PDL and DF can differentiate into osteoblast/cementoblast cells and PDL fibroblasts (3). As osteoblast lineage markers are expressed by cementoblasts, a clear distinction of these lineages in a murine model has not yet been established (5). Therefore, the availability and development of a specific marker for the cementoblast has remained very elusive in the field. The cementum attachment protein (CAP) and cementum protein 23 (CP23) have been previously been shown to be relatively nonspecific for cementoblasts and are also expressed by PDL cells and osteoblasts (42). Defining differentiation of progenitors into periodontium-related lineages with mineralization potential (osteoblasts/cementoblasts) and nonmineralizing cells (PDL fibroblasts) will require the use of adequate lineage markers. Currently, several strains of transgenic mice expressing GFP under the control of different promoters can be used to identify cellular components of the periodontium. The scleraxis promoter directs GFP expression (43) to PDL fibroblasts with only a few odontoblasts expressing a weak GFP signal (data not shown). Scleraxis is a basic helix–loop–helix transcription factor, which is expressed in tendon progenitor populations and mature tendons (44). Utilization of the DMP-1 promoter GFP transgene (Fig. 5) allows identification of pre-osteocytes/osteocytes in vivo and in vitro. Dentin matrix protein-1 is expressed in mineralized nodules and shows a strong signal in osteocytes and odontoblasts in vivo (26). Primary cultures of DF and PDL do not show evidence of differentiation into mature odontoblasts and therefore in these cultures DMP-1–GFP expression can serve as a marker for mature osteoblast lineage cells. Identifying markers expressed selectively in cementoblasts would allow the generation of a cementoblast-specific visual marker, using gene promoter sequences.
In summary, we have characterized a population of cells expressing αSMA–GFP with the ability to generate cells with a mature osteoblast lineage phenotype. Identification of adult progenitor cells using a combination of various markers will permit the development of better protocols for purification of these cells. In combination with markers of mature lineages of periodontium (alveolar bone, cementum, PDL) isolation, expansion and transplantation procedures have the potential to lead to the full restoration of damaged periodontal tissues.
Supplementary Material
Osteocalcin expression in differentiating mDF and mPDL cultures.
Acknowledgments
This work was supported by grants from the National Institutes of Health: 5U24-DE016495-02 providing institutional support and R03-AR053275 to IK. The authors would like to thank Dr Sanai Sato and Dr Jen-Yue Tsai for providing us with the αSMA–GFP transgenic mice. We are thankful to Ms Diane Gran for assistance with flow cytometry analysis, and to Dr Ana Maria Balic and Dr Mina Mina for providing us with the RNA sample of cultured dental pulp cells.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.Gay IC, Chen S, MacDougall M. Isolation and characterization of multipotent human periodontal ligament stem cells. Orthod Craniofac Res. 2007;10:149–160. doi: 10.1111/j.1601-6343.2007.00399.x. [DOI] [PubMed] [Google Scholar]
- 2.Ivanovski S, Gronthos S, Shi S, Bartold PM. Stem cells in the periodontal ligament. Oral Dis. 2006;12:358–363. doi: 10.1111/j.1601-0825.2006.01253.x. [DOI] [PubMed] [Google Scholar]
- 3.Seo BM, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 4.MacNeil RL, Somerman MJ. Development and regeneration of the periodontium: parallels and contrasts. Periodontol 2000. 1999;19:8–20. doi: 10.1111/j.1600-0757.1999.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 5.Bosshardt DD. Are cementoblasts a sub-population of osteoblasts or a unique phenotype? J Dent Res. 2005;84:390–406. doi: 10.1177/154405910508400501. [DOI] [PubMed] [Google Scholar]
- 6.Zeichner-David M. Regeneration of periodontal tissues: cementogenesis revisited. Periodontol 2000. 2006;41:196–217. doi: 10.1111/j.1600-0757.2006.00162.x. [DOI] [PubMed] [Google Scholar]
- 7.Thesleff I, Nieminen P. Tooth morphogenesis and cell differentiation. Curr Opin Cell Biol. 1996;8:844–850. doi: 10.1016/s0955-0674(96)80086-x. [DOI] [PubMed] [Google Scholar]
- 8.Melcher AH. Cells of periodontium: their role in the healing of wounds. Ann R Coll Surg Engl. 1985;67:130–131. [PMC free article] [PubMed] [Google Scholar]
- 9.Ten Cate AR. The development of the periodontium – a largely ectomesenchymally derived unit. Periodontol 2000. 1997;13:9–19. doi: 10.1111/j.1600-0757.1997.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 10.Hou LT, Liu CM, Chen YJ, et al. Characterization of dental follicle cells in developing mouse molar. Arch Oral Biol. 1999;44:759–770. doi: 10.1016/s0003-9969(99)00033-3. [DOI] [PubMed] [Google Scholar]
- 11.Luan X, Ito Y, Dangaria S, Diekwisch TG. Dental follicle progenitor cell heterogeneity in the developing mouse periodontium. Stem Cells Dev. 2006;15:595–608. doi: 10.1089/scd.2006.15.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod Craniofac Res. 2005;8:191–199. doi: 10.1111/j.1601-6343.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 13.Nohutcu RM, McCauley LK, Koh AJ, Somerman MJ. Expression of extracellular matrix proteins in human periodontal ligament cells during mineralization in vitro. J Periodontol. 1997;68:320–327. doi: 10.1902/jop.1997.68.4.320. [DOI] [PubMed] [Google Scholar]
- 14.Lekic P, Rojas J, Birek C, Tenenbaum H, McCulloch CA. Phenotypic comparison of periodontal ligament cells in vivo and in vitro. J Periodontal Res. 2001;36:71–79. doi: 10.1034/j.1600-0765.2001.360202.x. [DOI] [PubMed] [Google Scholar]
- 15.Hou LT, Liu CM, Lei JY, Wong MY, Chen JK. Biological effects of cementum and bone extracts on human periodontal fibroblasts. J Periodontol. 2000;71:1100–1109. doi: 10.1902/jop.2000.71.7.1100. [DOI] [PubMed] [Google Scholar]
- 16.Lin DG, Kenny DJ, Barrett EJ, Lekic P, McCulloch CA. Storage conditions of avulsed teeth affect the phenotype of cultured human periodontal ligament cells. J Periodontal Res. 2000;35:42–50. doi: 10.1034/j.1600-0765.2000.035001042.x. [DOI] [PubMed] [Google Scholar]
- 17.Saito Y, Yoshizawa T, Takizawa F, et al. A cell line with characteristics of the periodontal ligament fibroblasts is negatively regulated for mineralization and Runx2/Cbfa1/Osf2 activity, part of which can be overcome by bone morphogenetic protein-2. J Cell Sci. 2002;115:4191–4200. doi: 10.1242/jcs.00098. [DOI] [PubMed] [Google Scholar]
- 18.Gronthos S, Mrozik K, Shi S, Bartold PM. Ovine periodontal ligament stem cells: isolation, characterization, and differentiation potential. Calcif Tissue Int. 2006;79:310–317. doi: 10.1007/s00223-006-0040-4. [DOI] [PubMed] [Google Scholar]
- 19.Duarte WR, Iimura T, Takenaga K, Ohya K, Ishikawa I, Kasugai S. Extracellular role of S100A4 calcium-binding protein in the periodontal ligament. Biochem Biophys Res Commun. 1999;255:416–420. doi: 10.1006/bbrc.1999.0214. [DOI] [PubMed] [Google Scholar]
- 20.Lin NH, Menicanin D, Mrozik K, Gronthos S, Bartold PM. Putative stem cells in regenerating human periodontium. J Periodontal Res. 2008;43:514–523. doi: 10.1111/j.1600-0765.2007.01061.x. [DOI] [PubMed] [Google Scholar]
- 21.Miura M, Gronthos S, Zhao M, et al. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100:5807–5812. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosoya A, Nakamura H, Ninomiya T, et al. Immunohistochemical localization of alpha-smooth muscle actin during rat molar tooth development. J Histochem Cytochem. 2006;54:1371–1378. doi: 10.1369/jhc.6A6980.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalajzic Z, Li H, Wang LP, et al. Use of an alpha-smooth muscle actin GFP reporter to identify an osteoprogenitor population. Bone. 2008;43:501–510. doi: 10.1016/j.bone.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokota T, Kawakami Y, Nagai Y, et al. Bone marrow lacks a transplantable progenitor for smooth muscle type alpha-actin-expressing cells. Stem cells (Dayton, OH) 2006;24:13–22. doi: 10.1634/stemcells.2004-0346. [DOI] [PubMed] [Google Scholar]
- 25.Maeda S, Sutliff RL, Qian J, et al. Targeted overexpression of parathyroid hormone-related protein (PTHrP) to vascular smooth muscle in transgenic mice lowers blood pressure and alters vascular contractility. Endocrinology. 1999;140:1815–1825. doi: 10.1210/endo.140.4.6646. [DOI] [PubMed] [Google Scholar]
- 26.Kalajzic I, Braut A, Guo D, et al. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 27.D’Errico JA, Ouyang H, Berry JE, et al. Immortalized cementoblasts and periodontal ligament cells in culture. Bone. 1999;25:39–47. doi: 10.1016/s8756-3282(99)00096-4. [DOI] [PubMed] [Google Scholar]
- 28.Kalajzic I, Staal A, Yang WP, et al. Expression profile of osteoblast lineage at defined stages of differentiation. J Biol Chem. 2005;280:24618–24626. doi: 10.1074/jbc.M413834200. [DOI] [PubMed] [Google Scholar]
- 29.Chen SC, Marino V, Gronthos S, Bartold PM. Location of putative stem cells in human periodontal ligament. J Periodontal Res. 2006;41:547–553. doi: 10.1111/j.1600-0765.2006.00904.x. [DOI] [PubMed] [Google Scholar]
- 30.Kemoun P, Laurencin-Dalicieux S, Rue J, et al. Localization of STRO-1, BMP-2/-3/-7, BMP receptors and phosphorylated Smad-1 during the formation of mouse periodontium. Tissue Cell. 2007;39:257–266. doi: 10.1016/j.tice.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 31.Sacchetti B, Funari A, Michienzi S, et al. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Doherty MJ, Ashton BA, Walsh S, Beresford JN, Grant ME, Canfield AE. Vascular pericytes express osteogenic potential in vitro and in vivo. J Bone Miner Res. 1998;13:828–838. doi: 10.1359/jbmr.1998.13.5.828. [DOI] [PubMed] [Google Scholar]
- 33.Farrington-Rock C, Crofts NJ, Doherty MJ, Ashton BA, Griffin-Jones C, Canfield AE. Chondrogenic and adipogenic potential of microvascular pericytes. Circulation. 2004;110:2226–2232. doi: 10.1161/01.CIR.0000144457.55518.E5. [DOI] [PubMed] [Google Scholar]
- 34.Zannettino AC, Paton S, Arthur A, et al. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cell Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 35.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci USA. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukamoto Y, Fukutani S, Shin-Ike T, et al. Mineralized nodule formation by cultures of human dental pulp-derived fibroblasts. Arch Oral Biol. 1992;37:1045–1055. doi: 10.1016/0003-9969(92)90037-9. [DOI] [PubMed] [Google Scholar]
- 37.Takeda T, Tezuka Y, Horiuchi M, et al. Characterization of dental pulp stem cells of human tooth germs. J Dent Res. 2008;87:676–681. doi: 10.1177/154405910808700716. [DOI] [PubMed] [Google Scholar]
- 38.Zheng L, Iohara K, Ishikawa M, et al. Runx3 negatively regulates Osterix expression in dental pulp cells. Biochem J. 2007;405:69–75. doi: 10.1042/BJ20070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Vlasselaer P, Falla N, Snoeck H, Mathieu E. Characterization and purification of osteogenic cells from murine bone marrow by two-color cell sorting using anti-Sca-1 monoclonal antibody and wheat germ agglutinin. Blood. 1994;84:753–763. [PubMed] [Google Scholar]
- 40.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science (New York, NY) 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 41.Welm BE, Tepera SB, Venezia T, Graubert TA, Rosen JM, Goodell MA. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Dev Biol. 2002;245:42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 42.Foster BL, Popowics TE, Fong HK, Somerman MJ. Advances in defining regulators of cementum development and periodontal regeneration. Curr Top Dev Biol. 2007;78:47–126. doi: 10.1016/S0070-2153(06)78003-6. [DOI] [PubMed] [Google Scholar]
- 43.Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 44.Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Osteocalcin expression in differentiating mDF and mPDL cultures.