Summary
NLRX1 is the only member of the Nod-like receptor (NLR) family that is targeted to the mitochondria, and its overexpression induces the generation of reactive oxygen species (ROS), thus impacting on NFκB- and JNK-dependent signaling cascades. In addition, NLRX1 has been shown to interact with MAVS (also known as IPS-1, VISA and Cardif) at the mitochondrial outer membrane and to modulate antiviral responses. Here we report that NLRX1 has a functional leader sequence and fully translocates to the mitochondrial matrix via a mechanism requiring the mitochondrial inner-membrane potential, ΔΨm. Importantly, we failed to detect NLRX1 at the mitochondrial outer membrane. We also show that the leader sequence of NLRX1 is removed, which generates a mature protein lacking the first 39 amino acids through a maturation process that is common for mitochondrial-matrix proteins. Finally, we identified UQCRC2, a matrix-facing protein of the respiratory chain complex III, as an NLRX1-interacting molecule, thus providing a molecular basis for the role of NLRX1 in ROS generation. These results provide the first identification of a protein belonging to the NLR family that is targeted to the mitochondrial matrix.
Keywords: MAVS, Nod-like receptors, Antiviral signaling, Innate immunity, Mitochondria
Introduction
Innate immunity relies on the detection of conserved microbial- or danger-associated molecular patterns (MAMPs or DAMPs, respectively) by pattern-recognition molecules (PRMs). In mammals, several families of PRMs have been recently identified, including the transmembrane Toll-like receptors (TLRs), cytosolic Nod-like receptors (NLRs) and RIG-I-like receptors (RLRs) (Akira et al., 2006). NLR proteins include Nod1 and Nod2, which trigger pro-inflammatory pathways such as NFκB and MAP kinases in response to bacterial peptidoglycan, and NLRPs (also known as Nalps), such as NLRP1 and NLRP3, which induce the activation of the caspase-1 inflammasome in response to various MAMPs and DAMPs (Fritz et al., 2006). NLR proteins are grouped in subfamilies, in part on the basis of their N-terminal domain. Whereas Nod1, Nod2 and NLRC4 have an N-terminal caspase-activation and -recruitment domain (CARD), NLRP proteins display a PYRIN domain, initially identified in the eponymous protein (Fritz et al., 2006). NLRX1 has an unconventional N-terminal domain that shares no apparent homology to any other protein. Recently, NLRX1 has been shown to represent the first NLR protein to localize to a specific organelle: this protein targets the mitochondria (Moore et al., 2008; Tattoli et al., 2008). We observed that mitochondrial localization of NLRX1 resulted in the generation of reactive oxygen species (ROS), leading to the potentiation of NFκB and JNK signaling (Tattoli et al., 2008). Concomitant with our studies, Moore et al. demonstrated that NLRX1 localized to the mitochondrial outer membrane, where it modulated the antiviral function of MAVS (also known as IPS-1, VISA or Cardif) through direct interaction (Moore et al., 2008). MAVS is a crucial adaptor protein for RIG-I- and Mda-5-dependent antiviral NFκB and type-I-interferon signaling (Meylan et al., 2006). Interestingly, MAVS is anchored in the mitochondrial outer-membrane protein via a single C-terminal transmembrane (TM) domain and, consequently, MAVS is entirely exposed to the cytosolic side of mitochondria (Seth et al., 2005).
Mitochondria are composed of an outer and an inner membrane (MOM and MIM, respectively), and the large majority of mitochondrial proteins are encoded in the nucleus, synthesized in the cytosol and transported to specific mitochondrial compartments via mechanisms that have been extensively studied (Neupert and Herrmann, 2007). Importantly, import of most proteins to the intermembrane space (IMS), the MIM or the matrix commonly depends on the presence of an N-terminal addressing sequence, which targets the translocase of the outer membrane (TOM) complex. Subsequent translocation of proteins to the MIM or the matrix requires the energy provided by the MIM potential, ΔΨm. Once in the matrix or the IMS, a number of mitochondrial proteins are matured by mitochondrial processing peptidases (MPPs), thus resulting in the removal of their N-terminal leader sequence (Neupert and Herrmann, 2007).
Here we investigated in detail the mitochondrial subcellular localization of NLRX1 and provide evidence that the protein has a functional N-terminal mitochondrial-addressing sequence allowing targeting to the mitochondrial matrix. Importantly, NLRX1 was undetectable at the MOM, which does not support a direct role for NLRX1 as a regulator of MAVS signaling. We also demonstrate that this translocation required a functional ΔΨm, which is a common feature of proteins that get access to the mitochondrial matrix. Moreover, we used mass spectrometry to show that NLRX1 was matured by MPPs, resulting in a protein lacking its first 39 amino acids. Finally, using pulldown assays to identify NLRX1-binding partners, we identified UQCRC2, a matrix-facing protein of the respiratory chain complex III (bc1 complex) as an NLRX1-interacting protein. Because the bc1 complex plays a key role in the generation of mitochondrial ROS, these results provide a molecular basis for the role of NLRX1 in ROS generation. Together, our observations point to a role for NLRX1 in the mitochondrial matrix and provide the first identification of a PRM family member being localized to the mitochondrial matrix.
Results
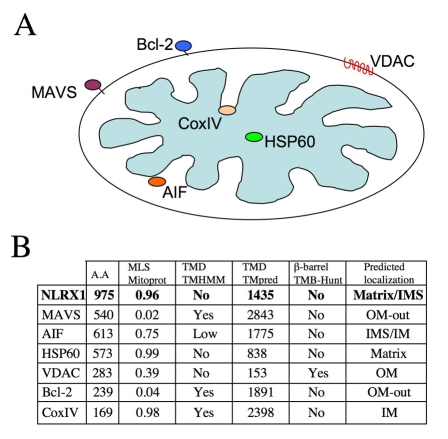
We first performed an analysis of the NLRX1 sequence, using several protein-analysis algorithms. Similar to most proteins internalized through the TOM machinery, including CoxIV, AIF and HSP60 (Fig. 1A), NLRX1 was predicted to have a canonical N-terminal mitochondrial-addressing sequence (using MitoProt) (Fig. 1B). MAVS and Bcl-2, which are anchored into the MOM, do not contain a predicted leader sequence (Fig. 1B). Using two algorithms designed to predict transmembrane domains (TMHMM and TMpred), we found no strong prediction for a TM domain in NLRX1, whereas MAVS and Bcl-2 C-terminal TM domains, as well as Cox IV, obtained a very high score with both algorithms (Fig. 1B). Moreover, NLRX1 was not predicted to display a β-barrel organization (using TMB-Hunt), which is a very specific secondary structure observed in some MOM proteins, such as VDAC. Finally, we analyzed the N-terminal domain of NLRX1 in all animal species for which the sequence is available in public databases. We observed that, despite poor primary-sequence conservation between some species, a strong N-terminal mitochondrial-addressing sequence was always identified (supplementary material Fig. S1), suggesting that this characteristic was under selective pressure in evolution. Together, this initial in silico analyses predicted that NLRX1 was most probably targeted to the matrix, or possibly to the IMS.
Fig. 1.
In silico analysis of the NLRX1 protein sequence. (A) Mitochondrial subcellular localization of MAVS, VDAC, Bcl-2, AIF, Cox IV and HSP60. (B) Algorithms were used to evaluate the presence of an N-terminal mitochondrial-addressing sequence (Mitoprot), transmembrane domains (TMHMM and TMpred) and β-barrels (TMB-Hunt). Numerical values computed by Mitoprot predict N-terminal mitochondrial-addressing sequences, ranging from 0 (lowest probability) to 1 (highest probability), and TMpred usually predicts a transmembrane domain for values >1750. Mitochondrial proteins of known subcellular localization (MAVS, AIF, HSP60, VDAC, Bcl-2, Cox IV) were also analyzed, to evaluate the strength of the algorithms. The resulting predicted subcellular localization was deduced from these analyses.
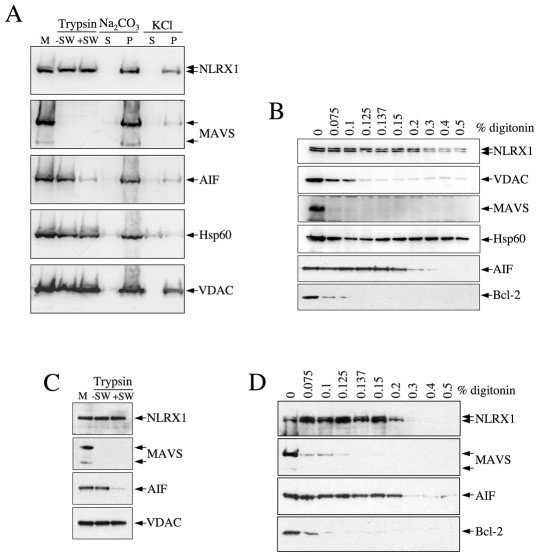
Next, we overexpressed C-terminal FLAG-tagged NLRX1 in HEK293T cells and purified mitochondria, through a standard procedure using sucrose density-gradient centrifugation. Resuspended mitochondria were then incubated on ice in either isotonic or swelling buffers, the latter being specifically designed to rupture the MOM without altering the MIM by taking advantage of the superior expanding capacity of the MIM over the MOM. Trypsin digested only cytosol-exposed MOM proteins in an isotonic buffer and also degraded IMS proteins if added to a swelling buffer, but could not digest matrix proteins. Interestingly, we observed that NLRX1 remained resistant to trypsin action in regular and swelling buffers, suggesting that the protein was either in the matrix or fully embedded into a mitochondrial membrane (Fig. 2A). As controls, we observed that MAVS was fully digested by trypsin in both buffers, in agreement with its reported localization to the cytosolic side of the MOM (Seth et al., 2005). HSP60 and VDAC were, as expected, trypsin-resistant owing to different reasons: HSP60 is a matrix protein, whereas VDAC escapes the action of the enzyme because it is fully inserted into the MOM. Finally, AIF, an IMS mitochondrial protein, was resistant to trypsin action in an isotonic buffer, but was partially degraded in a swelling buffer (Fig. 2A). Next, we treated isolated mitochondria with sodium carbonate, which results, after centrifugation, in the partial separation of soluble (in the supernatant) and membrane-anchored (pellet) proteins. We failed to detect NLRX1 in the supernatant of sodium-carbonate-treated mitochondria, suggesting that, following translocation to the matrix, the protein is possibly anchored to the MIM (Fig. 2A, lane 4). By contrast, a fraction of HSP60 was found in the supernatant, in agreement with the fact that this protein is soluble in the matrix. It must be noted, however, that the majority of HSP60 remained associated with the pellet of the sodium-carbonate-treated mitochondria, which is explained by the fact that this treatment poorly targets matrix proteins in general. To get more definitive information, we sonicated isolated mitochondria, treated them with a high concentration of KCl (to disrupt electrostatic protein-protein interactions) and separated soluble (in the supernatant) from membrane-bound (pellet) proteins by centrifugation. Most HSP60 was found in the supernatant, whereas the membrane-bound proteins MAVS and VDAC were found in the pellet. AIF displayed an intermediate profile, in agreement with the fact that the protein is partially membrane-bound and is also found soluble in the IMS (Otera et al., 2005). Interestingly, in these conditions, NLRX1 was exclusively found in the pellet fraction (Fig. 2A). On the basis of these results, we concluded that NLRX1 most probably localized to the mitochondrial matrix, where it associated strongly with the inner membrane. However, another possibility (although unlikely on the basis of our in silico analysis) remained that NLRX1 could be, similar to VDAC, a protein of the MOM that would be fully inserted in the membrane. Indeed, with the experiments described above, the profiles of NLRX1 and VDAC were indistinguishable (Fig. 2A). Therefore, in order to differentiate these two possible mitochondrial subcellular localizations, we treated isolated mitochondria with increasing concentrations of the membrane-permeabilizing toxin digitonin and analyzed proteins present in the centrifugation pellet. As previously reported (Schmidt et al., 1984), low doses of digitonin specifically disrupt the MOM, resulting in the loss of MOM proteins from the mitochondrial pellet, and, at higher digitonin concentrations, similar disruption of the MIM starts to occur. We observed that VDAC, Bcl-2 and MAVS, three MOM proteins, were lost from the mitochondrial pellet at digitonin concentrations below 0.1%, whereas much higher concentrations (0.3% and more) were required to decrease the presence of AIF, HSP60 or NLRX1 in the mitochondrial pellet (Fig. 2B), thus showing that NLRX1 was not an integral MOM protein. Taken together, these results strongly suggest that NLRX1 is a matrix protein.
Fig. 2.
NLRX1 is a mitochondrial-matrix protein. (A) Purified mitochondria (M) from HEK293T cells overexpressing NLRX1-FLAG were treated with trypsin, in regular buffer (–SW) or in swelling conditions (+SW), or treated with Na2CO3 or KCl prior to centrifugation. S, supernatant; P, pellet. The resulting protein samples were analyzed by western blot with antibodies against FLAG, or against several mitochondrial markers, as indicated. (B) Purified mitochondria from HEK293T cells overexpressing NLRX1-FLAG were treated with increasing concentrations of digitonin, as indicated, and the pellet after centrifugation was analyzed by western blot for NLRX1, using an anti-FLAG antibody. Other mitochondrial markers were also analyzed, as indicated. (C) Purified mitochondria (M) from HEK293T cells were treated with trypsin in regular buffer (–SW) or in swelling conditions (+SW) and the presence of endogenous NLRX1 was determined by western blot using an anti-NLRX1 antibody, together with other mitochondrial markers, as indicated. (D) As in B but without transfection, and the indicated endogenous proteins were analyzed.
In order to ascertain that the results presented with overexpressed NLRX1 were also true for the endogenous protein, we studied the mitochondrial targeting of endogenous NLRX1 using a polyclonal anti-NLRX1 antibody (described in supplementary material Fig. S2). In agreement with the results presented above, we observed that endogenous NLRX1 was resistant to trypsin treatment of purified mitochondria in isotonic or swelling buffers (Fig. 2C), thus showing that NLRX1 is neither a cytosolic-exposed MOM nor an IMS protein. Moreover, digitonin treatment of purified mitochondria demonstrated a similar profile for endogenous NLRX1 (Fig. 2D) as the one described above for the overexpressed protein.
The results that we have presented so far clearly demonstrate that NLRX1 localizes to the mitochondrial matrix, and most likely associates tightly with the MIM, but we cannot formally exclude that a minor pool of NLRX1 could localize to another mitochondrial compartment. Indeed, in such a hypothesis, a putative minor fraction of trypsin-sensitive NLRX1 would have probably been missed in the assays described above if the large majority of NLRX1 was in the matrix, and therefore trypsin-resistant. To circumvent this limitation, we applied a protocol of MOM purification on swollen mitochondria, using mechanical disruption and ultracentrifugation, thereby allowing the separation of mitoplasts (enriched in MIM, but still containing residual MOM) from the pure MOM fraction (Arnoult, 2008; Riezman et al., 1983). Using this technique, we observed that, whereas MAVS and VDAC were found in the MOM fraction, both endogenous NLRX1 and the MIM marker Cox IV were undetectable (Fig. 3). These findings therefore suggest that, if a minor pool of MOM-localized NLRX1 existed, it would remain under the detection limit of our experimental assays.
Fig. 3.
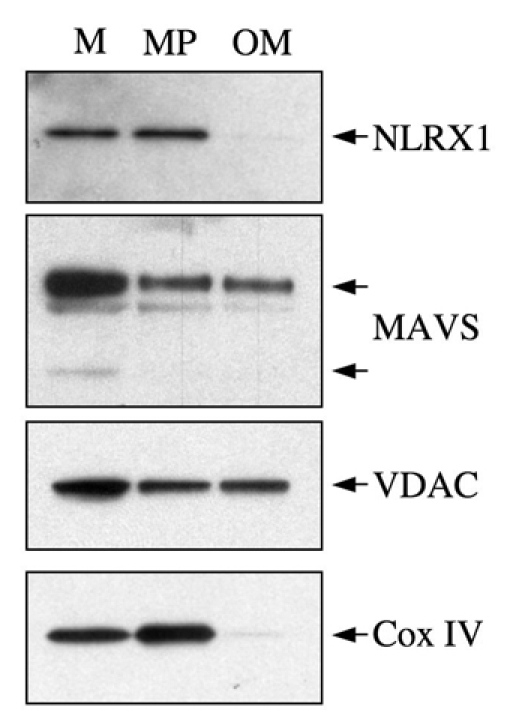
NLRX1 is undetectable at the mitochondrial outer membrane. Purified mitochondria (M) were fractioned by ultracentrifugation into mitoplasts (MP), which are enriched in inner-membrane proteins but still contain outer-membrane proteins, and pure outer membrane (OM). The presence of the endogenous NLRX1, as well as known mitochondrial markers, was analyzed by western blotting.
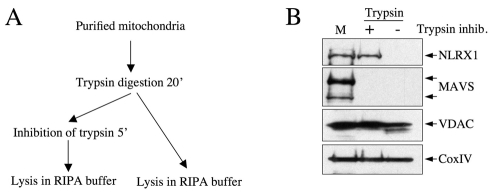
Having demonstrated that NLRX1 is a matrix protein that is tightly associated with the inner membrane, we aimed to determine whether NLRX1 was fully inserted into the MIM or whether it remained exposed to the matrix. To do so, we designed a modification of our trypsin assay described above, and omitted the step in which the trypsin inhibitor is added, thereby allowing continuous action of the enzyme once mitochondria are fully degraded in lysis buffer (Fig. 4A). Using these conditions, NLRX1 became sensitive to trypsin action, whereas fully membrane-embedded proteins, such as VDAC (in the MOM) and CoxIV (in the MIM), remained trypsin-resistant (Fig. 4B). Together, these results demonstrate that NLRX1 is a mitochondrial protein that is tightly associated with the MIM, but not fully inserted into this membrane.
Fig. 4.
NLRX1 is not an integral MIM protein. (A) Schematic representation of the procedure applied. (B) Purified mitochondria (M) were treated with trypsin for 20 minutes and then lysed in RIPA buffer in the presence (+) or absence (–) of a trypsin inhibitor. The presence of endogenous NLRX1, as well as of known mitochondrial markers, was analyzed by western blotting.
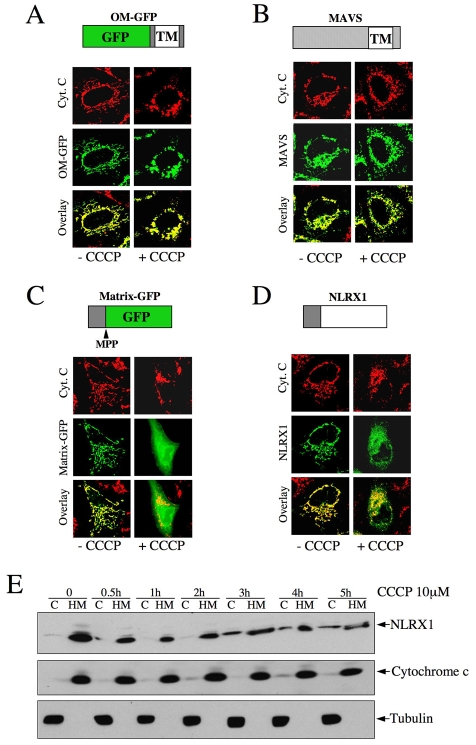
The import of mitochondrial proteins to the matrix via the TIM23 complex, as well as via some MIM proteins inserted via the TIM22 complex, requires energy provided by the MIM potential, ΔΨm, whereas proteins targeted to other mitochondrial compartments do not (Neupert and Herrmann, 2007). We aimed to characterize whether the transport of NLRX1 to the mitochondrial matrix was ΔΨm-dependent. To do so, we overexpressed NLRX1 in HeLa cells and dissipated the ΔΨm for 8 hours by adding the H+ ionophore CCCP. As controls, we also overexpressed OM-GFP and MAVS, two constructs targeting the MOM, and therefore not requiring the ΔΨm for mitochondrial localization. We also overexpressed Matrix-GFP, an engineered GFP construct with an N-terminal mitochondrial-addressing sequence, which requires the ΔΨm for targeting into the mitochondrial matrix. The addition of CCCP did not interfere with the mitochondrial localization of OM-GFP (Fig. 5A) and MAVS (Fig. 5B), as observed by the unaltered overlay between these overexpressed proteins and the mitochondrial marker, cytochrome c, in the presence or absence of CCCP. By contrast, the mitochondrial localization of Matrix-GFP was highly sensitive to the addition of CCCP (Fig. 5C), and so was that of NLRX1 (Fig. 5D; supplementary material Fig. S3). We noted, however, that the cytosolic retention of NLRX1 was not as complete as the one of Matrix-GFP, which might be due to a longer half-life of NLRX1 than Matrix-GFP once in the mitochondrial matrix. In agreement with the immunofluorescence studies, time-course experiments revealed that CCCP treatment resulted in cytosolic retention of NLRX1 as early as 3 hours post-stimulation (Fig. 5E). The presence of NLRX1 in the cytosol was indeed due to cytosolic retention and was unlikely to be caused by release of the matrix-localized NLRX1 from damaged mitochondria, because our treatments did not result in cytochrome-c release, as observed by immunofluorescence (Fig. 5A-D) or western blotting (Fig. 5E). Together, these results demonstrate that the import of NLRX1 into the mitochondrial matrix requires energy from the ΔΨm.
Fig. 5.
The import of NLRX1 into mitochondria requires the ΔΨm. (A-D) HeLa cells were grown on coverslips, transfected overnight with OM-GFP (A), FLAG-MAVS (B), Matrix-GFP (C) or NLRX1-FLAG (D) and incubated or not with 10 μM CCCP for 8 hours before fixation and analysis by immunofluorescence. Mitochondria were visualized using an anti-cytochrome-c antibody. MAVS and NLRX1 were visualized using an anti-FLAG antibody. (E) HeLa cells transfected with NLRX1-FLAG were incubated for various times with 10 μM CCCP, and cytosol (C) and heavy membrane (HM) that contained mitochondria were analyzed by western blotting, using antibodies against FLAG (for NLRX1), cytochrome c and tubulin.
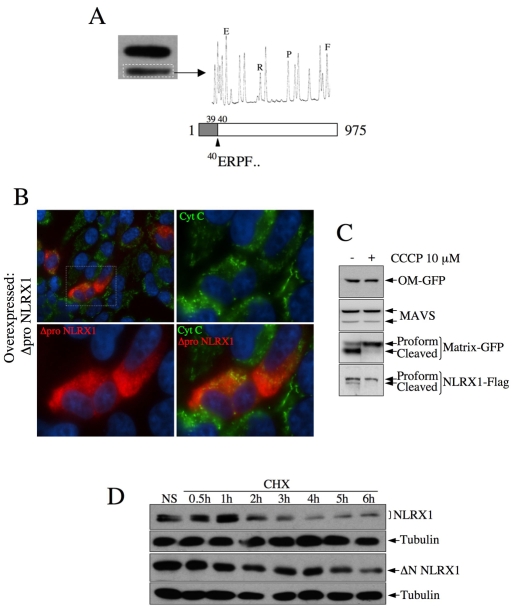
In our western blot assays against either the overexpressed NLRX1-FLAG or endogenous NLRX1, we repeatedly noticed that NLRX1 could be found as a doublet (see Fig. 2), which prompted us to hypothesize that the protein might be matured by MPPs, a process common for many proteins of the matrix and some of the IMS (Neupert and Herrmann, 2007). We overexpressed NLRX1-FLAG in HeLa cells, isolated mitochondria, immunoprecipitated proteins from lysates with anti-FLAG antibody and resolved the bound proteins on SDS-PAGE. We cut the band corresponding to the faster migrating entity and performed protein identification using Edman degradation procedure. We identified that the faster migrating entity displayed `ERPF' at the N-terminal end, which is found starting at position 40 in the NLRX1 amino acid sequence (Fig. 6A), therefore showing that the faster migrating band of NLRX1 represents a mature form processed between amino acids 39 and 40. Next, we generated an N-terminal-truncated form of NLRX1 lacking the first 39 amino acids (Δpro NLRX1) and observed by immunofluorescence that this truncated protein was unable to localize to the mitochondria (Fig. 6B), thus showing that the N-terminal prodomain of NLRX1 serves as a mitochondrial-addressing sequence. In order to demonstrate that the maturation of NLRX1 occurs in the mitochondrial matrix, where MPPs reside, we performed an experiment similar to the one described in Fig. 5, which uses CCCP to dissipate the ΔΨm, but followed the processing of proteins by western blotting. Whereas CCCP treatment did not affect the profile of proteins targeted to the MOM (MAVS and OM-GFP), it abolished the maturation of the two matrix proteins tested, Matrix-GFP and NLRX1 (Fig. 6C), therefore demonstrating that NLRX1 maturation requires the ΔΨm and must occur once the protein has reached the mitochondrial matrix. The presence of a doublet in a number of our experiments (see Fig. 2) suggested that, following translocation of the protein to the matrix, NLRX1 was not fully matured. In order to determine whether this was representative of the existence of two distinct pools of NLRX1 in the matrix, or simply reflected an overload of proteins on a saturated MPP complex, we treated cells overexpressing NLRX1 with cycloheximide, in order to stop the synthesis of new NLRX1 proteins. Two hours of cycloheximide treatment were sufficient to induce the disappearance of the proform of NLRX1 (Fig. 6D), thus suggesting that the doublet commonly observed is due to an incomplete maturation by MPPs. Because we also commonly observed this doublet with the endogenous protein (see Fig. 2D), this suggested that the pool of matrix NLRX1 must be actively replaced in resting conditions. This assumption is also supported by the fact that, in our cycloheximide experiments, the stability of matrix-targeting NLRX1 seemed lower compared with a truncated form that was unable to target the mitochondria (Fig. 6D), but comparable to the stability of the endogenous protein (supplementary material Fig. S4).
Fig. 6.
The N-terminal leader sequence of NLRX1 is cleaved following transport. (A) The identity of the faster migrating band of NLRX1 was determined, after membrane excision, by Edman degradation, revealing maturation between amino acids 39 and 40 of the NLRX1 prosequence. (B) A truncated form of NLRX1, lacking the (1-39) prosequence, is unable to localize to mitochondria, as determined by immunofluorescence. Mitochondria and Δpro-NLRX1–FLAG were visualized using anti-cytochrome-c and anti-FLAG antibodies, respectively. (C) HeLa cells transfected and stimulated with 10 μM CCCP, as in Fig. 2, were analyzed by western blot with the indicated antibodies. (D) HeLa cells were transfected with either NLRX1-FLAG or ΔN-NLRX1–FLAG, treated with 100 μM cycloheximide (CHX) for the indicated times, and cell lysates analyzed by western blot using an anti-FLAG antibody. Membranes were stripped and reblotted with an anti-tubulin antibody for normalization.
Because we identified that NLRX1 mitochondrial targeting relies on an N-terminal leader sequence, and because these addressing mechanisms are extremely conserved in evolution, we overexpressed human NLRX1 in Drosophila S2 cells and followed the subcellular localization of the protein by immunofluorescence. Using an antibody against the complex V of the respiratory chain in Drosophila (CV-α) as a mitochondrial marker, we observed that NLRX1 localized completely to mitochondria (supplementary material Fig. S5), therefore strongly implying that the NLRX1 leader sequence directs the protein to the highly conserved TOM-TIM complex of mitochondrial import.
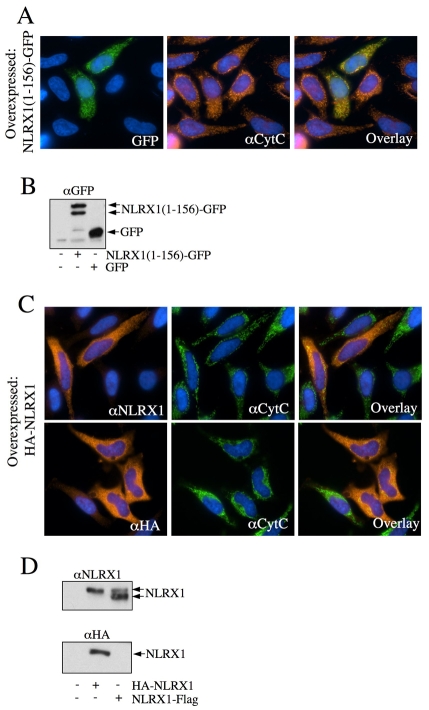
In order to determine whether the N-terminal domain of NLRX1 was sufficient to target the mitochondrial matrix, or whether other domains such as the NACHT or LRR were also participating in internalization, we constructed a chimeric protein containing the first 156 amino acids of NLRX1 fused to GFP [NLRX1(1-156)-GFP]. We observed by immunofluorescence that, whereas overexpressed GFP was found in the nucleus and the cytosol (data not shown), NLRX1(1-156)-GFP colocalized fully with the mitochondrial marker cytochrome c (Fig. 7A), thus demonstrating that the N-terminal domain of NLRX1 is sufficient for mitochondrial targeting. In addition, NLRX1(1-156)-GFP, but not GFP, was found as a doublet by western blotting (Fig. 7B), thus suggesting that the fusion construct reached the mitochondrial matrix, where it was matured by an MPP. This observation also implied that MPP-dependent processing of NLRX1 in the mitochondrial matrix only required amino acid sequences from 1 to 156, but neither the NACHT nor the LRR domains.
Fig. 7.
A free N-terminal NLRX1 addressing sequence is necessary and sufficient for mitochondrial targeting. (A,C) HeLa cells were grown on coverslips, transfected with NLRX1(1-156)-GFP (A) or HA-NLRX1 (C), fixed and analyzed by immunofluorescence using anti-cytochrome-c (A,C), anti-NLRX1 and anti-HA (C), as indicated. (B,D) HEK293T cells were transfected with NLRX1(1-156)-GFP (B) or HA-NLRX1 (D) and analyzed by western blotting using anti-GFP (B), anti-NLRX1 and anti-HA (D), as indicated.
A conserved feature of mitochondrial-addressing sequences is the requirement for the leader sequence to be at the N-terminal end of the protein, rather than internally. This dogma has been challenged by a recent study involving NLRX1, in which the authors suggested that mitochondrial targeting required amino acids 7-12 of the protein, whereas addition of an N-terminal HA tag did not alter targeting of the ectopically expressed HA-NLRX1 protein to mitochondria (Moore et al., 2008). Contrary to these observations, we showed by immunofluorescence that overexpressed HA-NLRX1 was found exclusively in the cytosol and did not colocalize with cytochrome c (Fig. 7C). In addition, the overexpression of HA-NLRX1 resulted in a single band by western blotting, migrating approximately at the level of the unprocessed form of overexpressed NLRX1-FLAG (Fig. 7D), showing that HA-NLRX1 was not matured by a matrix MPP, in agreement with the fact that the protein failed to target mitochondria.
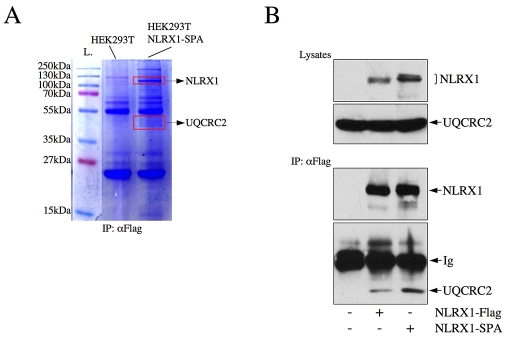
Finally, we aimed to identify the target(s) of NLRX1 in the mitochondria. To do so, we cloned NLRX1 into a sequential peptide affinity (SPA)-tag expression vector and generated HEK293T cell lines stably expressing NLRX1-SPA. Total cellular lysates were immunoprecipitated using a monoclonal anti-FLAG antibody (SPA-tag contains three repeats of FLAG epitope), separated by SDS-PAGE and stained with Coomassie Blue (Fig. 8A). Bands found only in HEK293T cells expressing NLRX1-SPA were excised and their sequences were determined by mass spectrometry. As a control for the quality of the immunoprecipitation, we observed that NLRX1 was the most-abundant protein identified in the pulldown (see Fig. 8A, upper rectangle; and data not shown). Besides NLRX1, we identified a single mitochondrial protein, UQCRC2, which was found in the lysates of HEK293T cells expressing NLRX1-SPA but not of control cells. The interaction between NLRX1-SPA or NLRX1-FLAG and the endogenous form of UQCRC2 was confirmed by co-immunoprecipitation, using a monoclonal antibody against FLAG (Fig. 8B). UQCRC2 is an integral member of the complex III (also known as the bc1 complex) of the oxidative phosphorylation respiratory chain, and is fully exposed to the matrix, thus supporting our biochemical fractionation assays presented above, which showed localization of NLRX1 to the matrix side of the MIM. Moreover, because the bc1 complex is central to the induction of ROS by mitochondria (Nohl et al., 2005), the identification of an interaction between NLRX1 and UQCRC2 provides a molecular basis for the modulation of ROS-dependent pathways by NLRX1.
Fig. 8.
Pulldown assays identify UQCRC2 as a binding partner of NLRX1. (A) Lysates from HEK293T and HEK293T NLRX1-SPA cell lines were immunoprecipitated using a monoclonal anti-FLAG antibody and bound proteins were resolved by SDS-PAGE. Bands found only in immunoprecipitates from HEK293T NLRX1-SPA cells were identified by mass spectrometry. L, ladder. (B) HEK293T cells were transfected with NLRX1-FLAG or NLRX1-SPA. Cell lysates were analyzed using anti-FLAG and anti-UQCRC2 antibodies. Lysates were immunoprecipitated using anti-UQCRC2 antibody and analyzed by western blot using anti-FLAG and anti-UQCRC2 antibodies. Ig, immunoglobulins.
Discussion
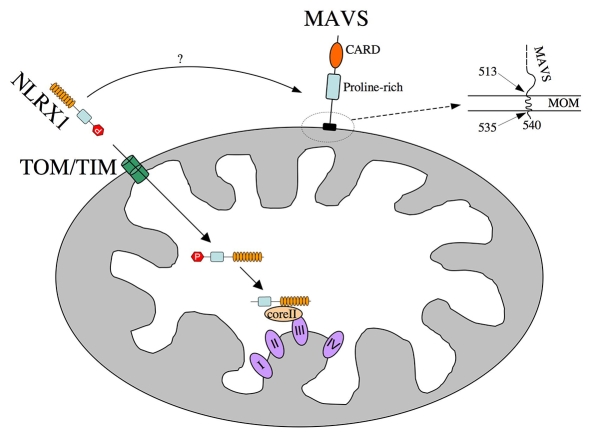
We demonstrated here that NLRX1 is a mitochondrial protein that is targeted to the matrix, and that the protein uses an N-terminal addressing sequence for import, which most probably occurs through the TOM-TIM complex (Fig. 9). Once in the mitochondrial matrix, we showed that NLRX1 is matured, probably by MPPs, which cleave the first 39 amino acids of the prosequence. Our fractionation assays strongly suggest that the protein then associates very tightly with the MIM, because it cannot be solubilized by sodium carbonate or KCl treatments. However, the protein remains accessible to trypsin-mediated digestion (in lysis buffer and without trypsin inhibitor), thus showing that it is not fully inserted into the MIM. However, our biochemical fractionation assays could not distinguish whether NLRX1 was anchored to the MIM through one or multiple TM domains, or whether the protein associated very strongly with MIM proteins, through association that were not removed by our sodium carbonate or KCl treatments. In this regard, the identification of UQCRC2 as a binding partner of NLRX1 in the mitochondrial matrix supports the latter hypothesis.
Fig. 9.
Schematic model for the internalization of NLRX1 into the mitochondrial matrix. Schematic representation of the proposed steps for the import of NLRX1 to the matrix face of the inner membrane. MAVS is also shown in this representation, and the precise insertion of the MAVS C-terminal sequence into the MOM is illustrated, and is derived from the study by Seth et al. (Seth et al., 2005). The four complexes (I-IV) of the mitochondrial respiratory chain are shown, as well as the core II protein (UQCRC2) of the complex III (bc1 complex). P, prosequence (aa 1-39); CARD, caspase-activation and -recruitment domain.
UQCRC2 is an integral component of the bc1 complex. This complex is composed of 11 subunits and is responsible for the transfer of electrons from the donor ubihydroquinone to the acceptor cytochrome c, with generation of a proton gradient across the inner membrane (Iwata et al., 1998). Moreover, a large amount of literature has shown that the bc1 complex is crucial for the generation of mitochondrial ROS. At present, it remains unclear how the interaction between UQCRC2 and NLRX1 could modulate ROS generation, and understanding this issue will warrant further investigation. One interesting possibility is that NLRX1 might interfere with UQCRC2-mediated processing and maturation of the rieske iron-sulfur protein, another integral member of the bc1 complex. Indeed, the maturation of this protein by UQCRC2 is required for its incorporation into the bc1 complex, and the rieske protein is essential for bc1-complex-mediated ROS generation (Bell et al., 2007). Interestingly, UQCRC2 displays strong homology with MPPs (Iwata et al., 1998) and it is possible that NLRX1 is a substrate for UQCRC2-mediated signal-peptidase activity.
Our results contrast with those reported recently by Moore et al., which showed that NLRX1 localized to the cytosolic side of the MOM, where it interacted with MAVS (Moore et al., 2008). Although we do not have definitive explanations for these discrepancies, we demonstrated that NLRX1 localized to the matrix and we failed to detect the protein at the MOM, arguing against the possibility that NLRX1 could have a dual localization in mitochondria. Our results with Δpro NLRX1 and HA-NLRX1 also argue against the existence of a strong interaction between NLRX1 and MAVS on the cytosolic side of the MOM, because this would result in the localization of these NLRX1 constructs to mitochondria by immunofluorescence. It must also be noted that the interaction studies between MAVS and NLRX1 were mostly performed using N-terminally HA-tagged NLRX1 constructs (Moore et al., 2008), which, according to our observations, are mislocalized to the cytosol. Finally, our results using Drosophila cells, in which MAVS is not present, demonstrate that the localization of NLRX1 to mitochondria does not require MAVS, but necessitates the presence of an N-terminal addressing sequence.
Despite these differences, however, it might be argued that, while being targeted to the mitochondrial matrix, NLRX1 could still interact with MAVS at contact points between the MIM and the MOM, where the distance between the matrix and the MOM is minimal. However, this hypothesis is extremely unlikely given the following observations: (1) MAVS is fully exposed to the cytosolic side of the MOM, with a single transmembrane domain at the very end of the C-terminal region, leaving at best five amino acids to face the IMS compartment of the mitochondria (Seth et al., 2005) (see Fig. 9); (2) the co-immunoprecipitation data reported by Moore et al. identified an interaction between NLRX1 and MAVS that occurred through the CARD domain of MAVS (Moore et al., 2008). Therefore, the localization of NLRX1 to the mitochondrial matrix is incompatible with a modulation of MAVS-dependent signaling through direct interaction, as previously proposed (Moore et al., 2008). However, it remains possible that, in conditions in which the import of NLRX1 into the mitochondrial matrix would be impaired (such as loss of ΔΨm), a cytosol-retained fraction of NLRX1 could interact with MAVS and modulate its function.
To our knowledge, our results are the first that identify the targeting of a PRM family member to the mitochondrial matrix. These observations are consistent with a role for NLRX1 in the modulation of ROS (Tattoli et al., 2008), because the generation of these is intimately linked to the activity of the inner-membrane respiratory chain. However, in the current view of the role of NLRs as sensors of MAMPs, it seems unclear how such microbial molecular patterns could actually access the mitochondrial matrix. Therefore, we speculate that NLRX1 might, as has been recently shown for NLRP3 (Benko et al., 2008), represent a sensor of danger signals (or DAMPs) and would in turn modulate some crucial mitochondrial functions. The identification of such danger signal(s) would undoubtedly represent a significant advance for the understanding of the role of NLRX1 in innate immunity.
Materials and Methods
Cell culture and reagents
HeLa cells and HEK293 cells were cultured under standard conditions in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mM L-glutamine, 50 IU of penicillin and 50 μg/ml streptomycin.
Transient transfection of HeLa cells was performed with FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. Transient transfection of HEK293 cells was performed using a standard calcium-phosphate method. Cycloheximide (Sigma) was at 100 μM; CCCP (Sigma) was at 10 μM.
Treatment of isolated mitochondria with trypsin, carbonate and KCl
Mitochondria were isolated intact from HeLa cells or transfected HEK293 cells by sucrose density-gradient centrifugation, as previously described (Nemoto and De Camilli, 1999). Isolated mitochondria were incubated for 20 minutes on ice with 50 μg/ml trypsin in 500 μl of Mitochondrial Isolation Buffer (MIB: 210 mM mannitol, 70 mM sucrose, 1 mM EDTA and 10 mM HEPES, pH 7.5) supplemented with the protease inhibitor mixture Complete (Roche Molecular Biochemicals) or 500 μl of 20 mM HEPES-KOH buffer for mitochondrial swelling. Next, trypsin was neutralized with trypsin inhibitor (120 μg/ml; Sigma) for 5 minutes at 4°C. The pellet was recovered by centrifugation (13,000 g, 10 minutes, 4°C). For carbonate extraction, isolated mitochondria were resuspended in a 0.1 M Na2CO3 buffer (pH 11.0) and incubated on ice for 30 minutes; the pellet was then recovered by centrifugation (130,000 g, 30 minutes, 4°C). For salt-wash experiments, mitochondria were diluted tenfold in buffers consisting of either 30 mM KCl or 500 mM KCl in 30 mM Tris-HCl (pH 7.4) and sonicated at the highest level ten times for 10 seconds per cycle. The pellet was recovered by centrifugation (130,000 g, 30 minutes, 4°C). Proteins from the resulting supernatants were concentrated by precipitation with 12% (w/v) trichloroacetic acid.
Digitonin treatment of isolated mitochondria
Isolated mitochondria were incubated for 15 minutes on ice with an increasing concentration of digitonin in 100 μl MIB. The pellet was recovered by centrifugation (13,000 g, 10 minutes, 4°C).
Mitochondria sub-fractionation
Isolated mitochondria were resuspended at a protein concentration of 4-8 mg/ml in a hypotonic buffer (5 mM potassium phosphate, pH 7.2, 5 mM EDTA and 1 mM PMSF) to allow swelling on ice for 10 minutes. Separation of the outer membrane from the remaining mitoplasts was achieved by 20 strokes in a glass-Teflon homogenizer. A total of 15 ml homogenate was loaded onto a sucrose gradient consisting of 9 ml of 0.25 M sucrose and 12 ml of 0.9-M sucrose steps in EMP buffer (2.5 mM EDTA, 10 mM MOPS-KOH, pH 7.2, and 1 mM PMSF). After ultracentrifugation for 1 hour (SW28 rotor at 141,000 g), the outer-membrane fraction was harvested from the 0.25 and 0.9 M sucrose interface. Both outer membrane and the pellet containing mitoplasts were next resuspended in MIB for immunoblotting analyses.
Heavy-membrane and cytosolic fractionation
Cells were collected in phosphate-buffered saline (PBS, pH 7.4) containing 1 mM EDTA, followed by digitonin-permeabilization on ice for 5 minutes using cytosolic extraction buffer (CEB: 250 mM sucrose, 70 mM KCl, 137 mM NaCl, 4.3 mM Na2PO4, 1.4 mM KH2PO4, 300 μM digitonin and Protease Inhibitor Cocktail, Sigma P8340). Following centrifugation at 1000 g for 5 minutes, the supernatant containing the cytosolic fraction was collected and the pellet resuspended in mitochondrial lysis buffer (MLB: 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.2% Triton-X-100, 0.3% NP40 and Protease Inhibitor Cocktail). Centrifugation at 10,000 g for 10 minutes resulted in a supernatant-enriched heavy-membrane fraction.
Immunofluorescence microscopy
Cells grown in LabTek chambers were transfected then 8 hours later were treated or not with 10 μM CCCP to neutralize mitochondrial import. Twelve hours later, cells were fixed for 15 minutes with PFA 4% followed by permeabilization with 0.15% Triton X-100 in PBS for 15 minutes. The cells were then incubated for 1 hour in blocking buffer (2% bovine serum albumin in PBS) followed by incubation overnight at 4°C with a mouse monoclonal anti-cytochrome-c (BD Biosciences Pharmingen, clone 6H2.B4) (1:800 dilution in blocking buffer) antibody to label mitochondria. Cells were washed three times for 10 minutes each in blocking buffer and incubated for 2 hours with Alexa-Fluor-568 secondary anti-mouse antibody (Molecular Probes). Images were acquired using a Leica confocal microscope through a 63× oil fluorescence objective.
Antibodies
Antibodies used in immunoblotting were: mouse mAb against FLAG (clone M2, Sigma; 1/2000 dilution), MAVS (clone Adri-1, Axxora; 1/2000), AIF (clone E1, Santa Cruz; 1/1000), Hsp60 (clone 24, BD Bioscience; 1/4000), VDAC (clone 31HL, Calbiochem; 1/8000), Bcl-2 (clone 100, Santa Cruz; 1/1000), Cox IV (clone 10G8, Molecular Probes; 1/2000), complex III subunit core 2 (UQCRC2) monoclonal (clone 13G12AF12BB11, Mitosciences; 1/1000), anti-HA rabbit polyclonal (Y-ll, Santa Cruz; 1/1000), rabbit polyclonal anti-GFP (ab290, Abcam) and rabbit polyclonal anti-NLRX1, designed against CQLVAQRYTPLKEV (214-226aa) of NLRX1 (Cederlane Laboratories, Hornby, Canada).
NH2-terminal sequence determination
Mitochondrial proteins immunoprecipitated with a monoclonal anti-FLAG antibody were separated by SDS-PAGE, electroblotted onto PVDF membrane and stained with Coomassie Blue. The lower band from the stained blot was excised and submitted to Edman sequencing. N-terminal NLRX1 sequencing was achieved by sequential Edman degradation (Advanced Protein Technology Center, SickKids, Toronto, Canada).
Expression vectors
The NLRX1-FLAG and ΔN-NLRX1–FLAG vectors were described previously (Tattoli et al., 2008). The C-terminally FLAG-tagged Δpro-NLRX1 expression construct was generated by PCR, between the nucleotides 115 and 2925 of NLRX1, using standard techniques and the construct was fully sequenced. NLRX1-FLAG was subcloned by PCR (using BamHI and KpnI restriction enzymes) into a pCMA vector to allow expression in Drosophila cells. Matrix-GFP was from Clontech Laboratories, and OM-GFP was previously described (Nemoto and De Camilli, 1999). A low-level ubiquitous promoter (Rbp1) on pMZ3F vector containing a C-terminal SPA-tag [calmodulin-binding peptide (CBP) and 3× FLAG epitope separated by Tobacco Etch Virus Protease) was used to generate NLRX1-SPA using standard PCR-based cloning (EcoRI and NotI restriction enzymes). Both HA-NLRX1 (phCMV2, KpnI and XhoI) and NLRX1(1-156)-GFP (pEGFP N1, EcoRI and BamHI) were generated using standard PCR-based cloning and the constructs were fully sequenced.
Generation of NLRX1-SPA stable cell lines
HEK293T cells were transfected with NLRX1-SPA expression vector using polyethylenimine (PEI, 3 μl:1 μg DNA). At 48 hours post-transfection, cells were passed and selected using 1 mg/ml Geneticin (Sigma). Single colonies were isolated and screened for SPA-tagged protein expression through western blot using the anti-M2-FLAG antibody.
Identification of NLRX1-binding partners by mass spectrometry
Lysates from NLRX1-SPA and control cell lines were prepared as described above using RIPA (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid and Protease Inhibitor Cocktail) buffer. A total of 50 μl of Ezview Red anti-FLAG M2 affinity gel (Sigma) washed with RIPA was added to the lysate and incubated overnight at 4°C. The resin was washed three times, each for 60 minutes with RIPA buffer followed by the addition of SDS-PAGE loading buffer. SDS-PAGE analysis followed by Coomassie Blue staining was performed and bands of interest were excised and submitted for mass spectrometry (Advanced Protein Technology Center, SickKids, Toronto, Canada).
Co-immunoprecipitation
Lysates were prepared from HEK293T cells transfected with pcDNA3, NLRX1 or NLRX1-SPA. Cells were lysed in cold RIPA buffer for 10 minutes on ice followed by centrifugation at 14,000 g for 15 minutes. Lysates were incubated overnight at 4°C with protein-G beads and UQCRC2 monoclonal antibody (1/200). Beads were washed three times, each for 45 minutes with RIPA and resuspended in 6× SDS-PAGE loading buffer. Lysates and immunoprecipitates were analyzed using standard western blot techniques.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/17/3161/DC1
This work was supported by grants from the Canadian Institutes of Health Research (MOP81360 to S.E.G.; MOP480142 to D.J.P.), Burroughs Wellcome Fund (to S.E.G.), and ANRS (Agence Nationale de La Recherche sur le SIDA), FRM (Fondation pour La Recherche Médicale), La Ligue contre le Cancer and Université Paris Sud (to D.A). We thank Angus McQuibban (University of Toronto) for the kind gift of reagents for Drosophila studies. Deposited in PMC for release after 6 months.
References
- Akira, S., Uematsu, S. and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783-801. [DOI] [PubMed] [Google Scholar]
- Arnoult, D. (2008). Apoptosis-associated mitochondrial outer membrane permeabilization assays. Methods 44, 229-234. [DOI] [PubMed] [Google Scholar]
- Bell, E. L., Klimova, T. A., Eisenbart, J., Moraes, C. T., Murphy, M. P., Budinger, G. R. and Chandel, N. S. (2007). The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J. Cell Biol. 177, 1029-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko, S., Philpott, D. J. and Girardin, S. E. (2008). The microbial and danger signals that activate Nod-like receptors. Cytokine 43, 368-373. [DOI] [PubMed] [Google Scholar]
- Fritz, J. H., Ferrero, R. L., Philpott, D. J. and Girardin, S. E. (2006). Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 7, 1250-1257. [DOI] [PubMed] [Google Scholar]
- Iwata, S., Lee, J. W., Okada, K., Lee, J. K., Iwata, M., Rasmussen, B., Link, T. A., Ramaswamy, S. and Jap, B. K. (1998). Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281, 64-71. [DOI] [PubMed] [Google Scholar]
- Meylan, E., Tschopp, J. and Karin, M. (2006). Intracellular pattern recognition receptors in the host response. Nature 442, 39-44. [DOI] [PubMed] [Google Scholar]
- Moore, C. B., Bergstralh, D. T., Duncan, J. A., Lei, Y., Morrison, T. E., Zimmermann, A. G., Accavitti-Loper, M. A., Madden, V. J., Sun, L., Ye, Z. et al. (2008). NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451, 573-577. [DOI] [PubMed] [Google Scholar]
- Nemoto, Y. and De Camilli, P. (1999). Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. EMBO J. 18, 2991-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert, W. and Herrmann, J. M. (2007). Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723-749. [DOI] [PubMed] [Google Scholar]
- Nohl, H., Gille, L. and Staniek, K. (2005). Intracellular generation of reactive oxygen species by mitochondria. Biochem. Pharmacol. 69, 719-723. [DOI] [PubMed] [Google Scholar]
- Otera, H., Ohsakaya, S., Nagaura, Z., Ishihara, N. and Mihara, K. (2005). Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 24, 1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman, H., Hay, R., Gasser, S., Daum, G., Schneider, G., Witte, C. and Schatz, G. (1983). The outer membrane of yeast mitochondria: isolation of outside-out sealed vesicles. EMBO J. 2, 1105-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, B., Wachter, E., Sebald, W. and Neupert, W. (1984). Processing peptidase of Neurospora mitochondria: two-step cleavage of imported ATPase subunit 9. Eur. J. Biochem. 144, 581-588. [DOI] [PubMed] [Google Scholar]
- Seth, R. B., Sun, L., Ea, C. K. and Chen, Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122, 669-682. [DOI] [PubMed] [Google Scholar]
- Tattoli, I., Carneiro, L. A., Jehanno, M., Magalhaes, J. G., Shu, Y., Philpott, D. J., Arnoult, D. and Girardin, S. E. (2008). NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 9, 293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.