Abstract
We review recent structural and biophysical studies of the mechanism of action of formins, proteins that direct the assembly of unbranched actin filaments for cytokinetic contractile rings and other cellular structures. Formins use free actin monomers to nucleate filaments and then remain bound to the barbed ends of these filaments as they elongate. In addition to variable regulatory domains, formins typically have formin homology 1 (FH1) and formin homology 2 (FH2) domains. FH1 domains have multiple binding sites for profilin, an abundant actin monomer binding protein. FH2 homodimers encircle the barbed end of a filament. Most FH2 domains inhibit actin filament elongation, but FH1 domains concentrate multiple profilin-actin complexes near the end of the filament. Actin transfers very rapidly from the FH1 domains onto the barbed end of the filament, allowing elongation at rates that exceed elongation by the addition of free actin monomers diffusing in solution. Binding of actin to the end of the filament provides the energy for the highly processive movement of the FH2 as a filament adds thousands of actin subunits. These biophysical insights provide the context to understand how formins contribute to actin assembly in cells.
Keywords: actin, cytokinesis, formin, profilin, polymerization
Eukaryotic cells use a diverse array of proteins to control the polymerization of actin filaments for various processes. Purified actin monomers can self-assemble into filaments, but such spontaneous polymerization process is initiated slowly, because formation of actin filament nuclei is kinetically unfavorable (Cooper et al. 1983; Frieden 1983; Sept and McCammon 2001). Furthermore cells contain proteins such as profilin and thymosin-β4 that suppress spontaneous nucleation. Thus, cells use several families of proteins to initiate actin filaments at specific times and sites (Chhabra and Higgs, 2007). The best-studied actin filament nucleating proteins are Arp2/3 complex (Pollard 2007), Spire (Quinlan et al. 2005), cofilin (Andrianantoandro and Pollard 2006), leiomodin (Chereau et al. 2008) and formins (Goode and Eck 2007). Arp2/3 complex initiates filaments as branches on the sides of pre-existing filaments, building networks similar to the twigs on a bush for cellular motility and endocytosis (Pollard and Borisy 2003). Spire and leiomodin use multiple WH2 (WASp homology 2) domains to bring together actin monomers to initiate unbranched filaments. Cofilin binds actin monomers and stabilizes nuclei in addition to severing filaments.
Formins assemble diverse cellular structures composed of unbranched actin filaments including the cytokinetic contractile ring, polarized actin cables, stress fibers and filopodia [reviewed in (Faix and Grosse 2006; Goode and Eck 2007)]. Single formin molecules have the remarkable ability to remain bound to the fast-growing barbed end of an actin filament through hundreds of rounds of actin subunit addition – a property known as processive association (Higashida et al. 2004; Kovar and Pollard 2004). Formins protect barbed ends from capping by proteins that block elongation (Harris et al. 2004; Kovar et al. 2005; Moseley et al. 2004; Zigmond et al. 2003). The ability of formins to track faithfully on growing barbed ends provides a means for continuous elongation of actin filaments.
The cellular functions of formin proteins are best understood in yeast where the limited number of formin genes makes experimental analysis much easier than in animals having more than a dozen formin genes encoding proteins with overlapping functions. Neither of the two formin genes of budding yeast S. cerevisiae is essential, because the proteins have overlapping functions, but at least one of these genes is required for viability (Pruyne et al. 2004; Sagot et al. 2002a). Both formins assemble polarized actin cables emanating from the bud towards the mother cell and around the site of cleavage (Evangelista et al. 2002; Sagot et al. 2002a). Each of the three formins in fission yeast S. pombe has a unique, non-redundant function in making filaments for either cytokinesis (Chang et al. 1997), interphase actin cables (Feierbach and Chang 2001) or mating (Petersen et al. 1998). Metazoan cells depend on formins to assemble actin filaments for cytokinesis (Watanabe et al. 2008), filopodia (Schirenbeck et al. 2005), and lamellipodia (Yang et al. 2007), although overlapping contributions from more than one formin complicate analysis (Faix and Grosse 2006). The contributions of formins and Arp2/3 complex are generally distinct. For example, both yeasts depend entirely on Arp2/3 complex to assemble actin filaments at sites of clathrin-mediated endocytosis called actin patches (Li et al. 1995; Morrell et al. 1999) and on formins to polymerize actin filaments for the cytokinetic contractile ring (Chang et al. 1997; Evangelista et al. 2002). On the other hand, in animals both Arp2/3 complex and formins appear to contribute to actin filament assembly at the leading edge of motile cells and in the formation of filopodia (Korobova and Svitkina 2008; Yang et al. 2007).
To understand the diverse functions of formins in eukaryotic biology, it is essential to understand the mechanisms by which formins nucleate actin filaments and cooperate with profilin to promote rapid elongation while remaining processively associated with the barbed end of a filament. Here we review how crystal structures and biophysical studies of single formin molecules have advanced our understanding of the contributions of formins to these processes.
Domain organization of formin proteins
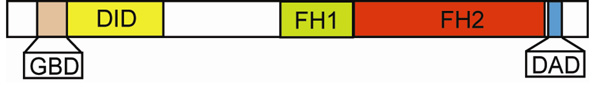
In pioneering work on the diaphanous gene from D. melanogaster, Castrillon and Wasserman (Castrillon and Wasserman 1994) discovered genes with related sequences from other eukaryotes (mouse formin IV, budding yeast Bni1p and a rice EST). They identified two distinct conserved regions in these formin sequences, which they called the FH1 and FH2 domains. Each FH1 domain contained short tracks of proline residues (Figure 1). The FH2 domain was a more strongly conserved region among these formins. These researchers identified it as a region of ~130 amino acids containing a GNXMN motif. Subsequent comparisons with more formins showed that sequence homology of FH2 domains spans ~500 amino acids (Higgs and Peterson 2005; Pruyne et al. 2002; Xu et al. 2004). FH1 and FH2 domains are the focus of this review.
Figure 1. Domain map of the formin mDia1.
The arrangement of the GTPase-binding domain (GBD), the Diaphanous-Inhibitory Domain (DID), the Formin-homology (FH1) Domain, the Formin-homology (FH2) Domain, and Diaphanous Autoregulatory Domain (DAD), are delineated at their approximate, relative scales according to primary sequence of the full-length mDia1 formin molecule (Higgs 2005).
When tested experimentally, FH1 domains are found to be essential for the physiological functions of formins. Less is known about the few formins that lack FH1 domains (Higgs and Peterson 2005; Rivero et al. 2005). Inclusion of a FH1 domain was required for formin Bni1p constructs to rescue the defects of S. cerevisiae caused by deletion of the formin bni1 gene (Evangelista et al. 2002; Sagot et al. 2002b). Moreover, inclusion of an FH1 domain was required for over expression of Bni1p to induce polymerization of excessive and aberrant polarized actin cables (Evangelista et al. 2002; Sagot et al. 2002b).
The signature FH2 domain is the most conserved part of formins. A FH2 domain is essential for a formin to induce actin assembly in cells (Evangelista et al. 2002; Sagot et al. 2002b). Biochemical studies detailed below subsequently showed that recombinant FH2 domains suffice for actin filament nucleation (Pruyne et al. 2002; Sagot et al. 2002b) and processive association with growing barbed ends (Kovar et al. 2006; Kovar and Pollard 2004).
In addition to FH1 and FH2 domains, gene sequences revealed that many formins consist of other domains linked together from the N-termini as follows: GBD-DID-FH1-FH2-DAD (Figure 1). The DAD “Diaphanous Autoregulatory Domain” interacts intramolecularly with DID, the “DAD Interacting Domain,” to inhibit actin assembly by the FH2 domain (Alberts 2001). Rho-family GTPases bind the GBD and partially overcome this autoinhibition (Li and Higgs 2005; Otomo et al. 2005a; Rose et al. 2005; Wallar et al. 2006).
It is important to appreciate that some proteins with FH2 domains are highly divergent from the GBD-DID-FH1-FH2-DAD formins (Grunt et al. 2008; Higgs 2005; Higgs and Peterson 2005). For example, delphilin, an FH2-containing protein expressed in Purkinje neurons, lacks GBD, DID and DAD, but one of its alternatively spliced isoforms bears a palmitoylation tag and a PDZ domain near its N-terminus. The palmitoylation tag localizes the protein to synaptic spines and the PDZ domain is essential for interaction with the GluRδ2 subunit of AMPA receptors (Matsuda et al. 2006; Miyagi et al. 2002). In plant genomes, none of the open reading frames encoding FH2 domains contains GBD, DID or DAD domains (Higgs and Peterson 2005). Instead, these formin sequences cluster into three distinct classes that suggest various modes of membrane localization: class I formins contain transmembrane sequences; class II formins bear PTEN domains; and class III formins contain catalytically-inactive RhoGAP-like domains (Grunt et al. 2008). These and many other examples show that evolution has crafted genes for formin proteins that couple basic actin assembly mechanisms by FH1FH2 domains with a wide range of regulatory mechanisms.
FH1 domain structure
All FH1 domains contain discrete tracks of contiguous proline residues and are typically located just N-terminal to the FH2 domain (Higgs and Peterson 2005; Rivero et al. 2005) (Figure 1). The number of polyproline tracks a FH1 domain may contain varies widely – Fus1p from S. pombe contains a single polyproline track, while mouse mDia1 contains 14 such tracks. Non-proline residues often interrupt an otherwise continuous run of proline residues. Though the significance of these non-proline residues has not been widely studied, a leucine residue at the penultimate position of one of the two Cdc12p FH1 polyproline tracks is critical for profilin binding (Yonetani et al. 2008). The polyproline tracks are expected to form rigid type-II polyproline helices. The sequences between polyproline tracks in FH1 domains are not conserved and are predicted to be flexible (Higgs 2005).
Polyproline oligomers were known to bind profilin (Perelroizen et al. 1994; Petrella et al. 1996; Tanaka and Shibata 1985), but no physiologically relevant polyproline receptors were identified until co-immunoprecipitation studies in fission yeast (Chang et al. 1997) and mammalian cells (Watanabe et al. 1997) showed that profilin binds FH1 domains. Profilin is abundant and binds most actin monomers in the cellular milieu (Kaiser et al. 1999).
FH1 polyproline tracks (Kursula et al. 2008) and polyproline oligomers (Archer et al. 1994; Kovar et al. 2006; Mahoney et al. 1997; Mahoney et al. 1999) bind profilin in a groove consisting of a patch of highly conserved aromatic residues on the face opposite the actin binding site (Schutt et al. 1993). These crystal structures explain how profilin can bind actin and polyproline simultaneously without mutual interference (Perelroizen et al. 1994; Tanaka and Shibata 1985).
FH2 domain structure
The FH2 domain from budding yeast Bni1p is the best-characterized FH2 domain in terms of its biophysical properties and the only one with atomic structures with and without actin (Otomo et al. 2005b; Xu et al. 2004). In agreement with hydrodynamic studies showing that the FH2 domain dimerizes (Moseley et al. 2004), a crystal structure of the whole Bni1p FH2 domain revealed a donut-like structure in which the two FH2 polypeptides associate such that the head of each subunit contacts the tail of the other subunit (Xu et al. 2004) (Figure 2A). This FH2 domain is composed largely of bundles of alpha-helices. The “lasso” region at the N-terminus of each subunit binds the “post” site near the C-terminus of the other subunit. A peptide of 17 amino acids links the lasso to the “knob” region of the main body of the domain. This linker is mostly unstructured but contains a short alpha-helix. In a different crystal form, this linker is fully extended (Xu et al. 2004).
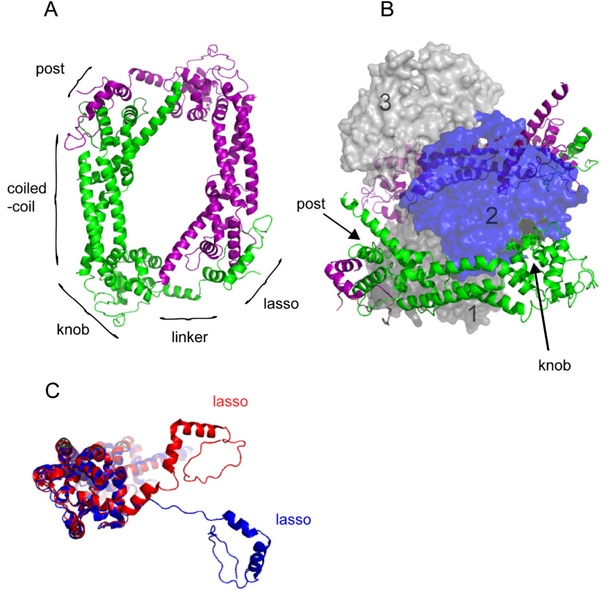
Figure 2. The structure of the Bni1p-FH2 domain.
(A) Ribbon diagram of the crystal structure of the head to tail homodimer of FH2 domains of Bni1p (residues 1350–1760) (Xu et al. 2004) (PDB Accession Code 1UX5). The two subunits are shown in green and purple. Labels indicate the approximate positions of the lasso, flexible linker, knob, coiled-coil, and post regions of the green subunit. (B) Crystal structure of the complex of Bni1p-FH2 (residues 1350–1760) with muscle actin (Otomo et al. 2005b) (PDB Accession Code 1Y64). Three contiguous actin subunits along the filament-like polymer are shown as space-filling representation in shades of gray and blue and numbered 1 to 3 from the barbed end. Ribbon diagrams of two FH2 subunits are colored as in (A). A continuous chain of FH2 domains wraps around the actin polymer with the lasso of one FH2 subunit joined to the post of the next. For clarity the density for the linker (residues 1401–1417) is omitted. This view shows the knob of the green FH2 subunit bound in the groove between subdomains 1 and 3 of actin subunit 2, as well as the post site of the green FH2 subunit bound to subdomain 1 of actin subunit 2. The partial transparency of the actin subunits reveals the symmetrical attachments of the purple FH2 subunit to actin subunits 2 and 3. (C) Comparison of the FH2 subunits from (A) and (B). When the knob and post regions of Bni1p-FH2 (residues 1418–1760) from the homodimer (Xu et al. 2004) (red) and the cocrystal with actin (Otomo et al. 2005b) (blue) were overlaid with PYMOL using the “align” function, the positions of the lasso-linkers diverge substantially with the linker more extended in complex with actin. All images were rendered with PYMOL (Delano Scientific).
A co-crystal of the FH2 domain of Bni1p with actin (Otomo et al. 2005b) provided important clues about interactions of FH2 domains with actin filaments (Figure 2B), particularly because the arrangement of actin subunits in the crystal differs from that observed in actin filaments. The head to tail (pointed end to barbed end) arrangement of subunits in two strands is similar in the crystal and filaments. However, the polymer in the crystal is flat with a two-fold axis of symmetry between these two parallel strands of subunits. Thus successive subunits are rotated by 180° relative to their nearest neighbors in the other strand. On the other hand, successive subunits in actin filaments are offset by 167° about the long axis (Holmes et al. 1990; Huxley 1963) forming twisted short pitch helix.
The most striking feature of the co-crystal structure is that the FH2 domains encircle the flat actin polymer with the FH2 domains linked into a continuous chain rather than in head to tail dimers (Holmes et al. 1990; Huxley 1963). The structures and relative orientations of the knob, coiled-coil and post regions of the FH2 domain are similar in the crystals of the free homodimer and co-crystal with actin, but the flexible linkers are dramatically different (Figure 2C). First, domain swapping during crystallization rearranged the linkers between FH2 domains into a continuous helix (post-1 to lasso-2 to post-2 to lasso-3, etc.) around the flat actin polymer. Second the linkers are fully extended to form contacts around the circumference of the filament.
Manually rearranging the connections between FH2 domains into head to tail dimers around a planar actin filament (Otomo et al. 2005b) (Figure 2C) suggests how an FH2 dimer binds to the three terminal subunits at the barbed end of a filament with all knob and post sites engaged. The FH2 knob binds in a hydrophobic groove between subdomains 1 and 3 of actin, and the post makes principally electrostatic contacts along subdomain 1 of actin (Otomo et al. 2005b; Xu et al. 2004). A key insight is that an FH2 dimer can bind with all of its strong contacts to the actin subunits in the 180° conformation, but steric constraints allow only a subset of these contacts around an actin filament with a 167° twist (Otomo et al. 2005b).
Elongation by FH2 domains
Pioneering studies suggested that formins interact with the barbed end: (i) electron micrographs showed gold-labeled formin molecules near the barbed ends of actin filaments (Pruyne et al. 2002); (ii) formins prevent annealing of the barbed end of one filament to the pointed end of another (Kovar et al. 2003); and (iii) formins inhibit the ability of capping protein to block growth from barbed ends (Harris et al. 2004; Kovar et al. 2005; Moseley et al. 2004; Zigmond et al. 2003). Observations (ii) and (iii) were particularly informative, because formins prevented association of other proteins (actin monomers and capping protein) with high specificity for barbed ends. Kinetic assays of assembly of actin in bulk solution (Pruyne et al. 2002; Zigmond et al. 2003) also suggested processive association of formins with growing barbed ends.
The strongest support for processive association of formin FH2 dimers with the barbed ends of growing actin filaments came from fluorescence microscopy. Fluorescent spots translocate at rates of 2.0 µm/sec (740 subunits/sec) through the cytoplasm of live Xenopus fibroblasts transfected with a plasmid encoding a GFP tag fused to the N-terminus of an active form of mouse formin mDia1 or mDia1 FH2 domains (Higashida et al. 2004). These spots were interpreted as single formin molecules associated with the ends of growing actin filaments. Total-internal-reflection-fluorescence (TIRF) microscopy showed that the barbed ends of single actin filaments grow from fixed points of attachment on glass microscope slides coated with four types of purified formins (Kovar et al. 2006; Kovar and Pollard 2004). Growth of barbed ends immobilized on slides was interpreted to arise from interaction of a barbed end with a single formin dimer bound to the slide and evidence for processive association. Formin dimers have now been labeled with quantum dots and observed directly to translocate on the barbed ends of growing filaments (Paul and Pollard 2009).
Barbed ends associated with most FH2 dimers grow slower than free barbed ends (Kovar et al. 2006; Kovar et al. 2003; Kovar and Pollard 2004), so it was proposed that a formin dimer on a barbed end “gates” subunit addition by equilibrating rapidly between an open state permissive for subunit addition and a closed state that that prevents it (Kovar et al. 2006;Otomo et al. 2005b; Vavylonis et al. 2006). The degrees to which FH2 domains from different formins inhibit actin filament elongation vary widely: S. pombe Cdc12p-FH2 inhibits elongation by 99% (Kovar et al. 2003; Paul and Pollard 2009), while mouse mDia1-FH2 inhibits elongation by only 10% (Harris et al. 2004; Kovar et al. 2006). The interpretation according to the gating hypothesis is that these formins differ in their equilibrium between open and closed states. The physiological significance, if any, of the variation in this equilibrium constant is not known.
Processive association of formins with growing barbed ends requires that FH2 domains (i) have a higher affinity for the barbed end than interior subunits of the actin filament and (ii) translocate onto the barbed end with the addition of each new actin subunit. The crystal structure of Bni1p FH2 homodimer suggested that multiple sites on the dimer (two knobs and two post sites) interact with the barbed end (Otomo et al. 2005b; Xu et al. 2004). One model for translocation postulated that both subunits of the FH2 dimer bind actin in the closed state and block the barbed end. This model postulates that a fraction of the strong FH2-actin contacts dissociate from actin in the open state (Figure 3). This partially dissociated state was proposed to allow an actin subunit from solution to bind both the exposed FH2 domain and the terminal actin subunit on the end of the filament, thereby reestablishing completely the attached formin in the closed state. This and related ideas are called stair stepping models, because the FH2 dimer must “step” off the end of the filament prior the addition of each new actin subunit.
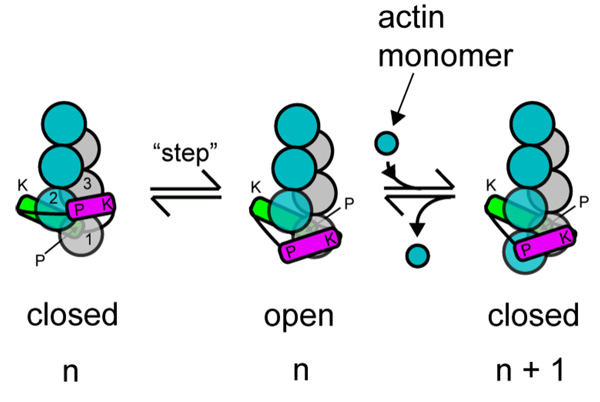
Figure 3. Two-state “stair-stepping” model for processive association of the formin-FH2 dimer with a growing barbed end.
The schematic depicts addition of one subunit (from “n” to “n+1”) onto barbed end associated with a FH2 dimer, where the formin equilibrates between a closed state that prevents subunit addition and an open state that allows addition (Otomo et al. 2005b; Xu et al. 2004). The subunits of the two long-pitch strands of the actin filament are shown as blue or silver spheres numbered 1, 2 and 3 from the barbed end. The leading subunit of the FH2 dimer is green and the trailing subunit purple. In the closed state, the FH2 dimer is engaged at both of its knob (K) and post (P) sites to the three terminal subunits. The leading FH2 subunit binds to actin subunits 1 and 2 and the trailing subunit binds to actin subunits 2 and 3 in the closed state. To enter the open state, the trailing subunit disengages both its knob and post sites, translocates or “steps” in the barbed end direction, and reattaches only its knob to the terminal barbed end subunit. In this open state, this FH2 subunit’s post site is exposed to solution. An actin monomer in solution binds to this post site and the two terminal actin subunits to complete the cycle of subunit addition and reestablish the closed state.
The structure of the Bni1p FH2 domain complexed with actin provided important clues for an alternative hypothesis regarding the interplay between the formin and the filament in the open and closed states (Otomo et al. 2005b). Specifically, the structure showed that the formin fully engages the filament only with the actin subunits in a planar structure with an 180° rotation between consecutive subunits along the short pitch helix. This planar conformation was proposed to be the closed state, because barbed end does not present favorable contacts for the incoming actin subunit. On the other hand, the 167° helical twist found in the interior of filaments might compromise FH2 binding in two ways. First, both subunits in the FH2 cannot engage fully with the actin, and, second, the two linkers between the FH2 domains must be distorted from the 180° conformation.
These structural features suggested that full engagement with a formin dimer traps the end of the actin filament in a high energy 180° state unfavorable for subunit addition, while the geometry of the 167° open state allows subunit addition but compromises formin binding to actin and strains the formin linkers (Otomo et al. 2005b). A new structure of the actin filament based on fiber diffraction shows that actin subunit is flattened upon incorporation into the polymer (Oda et al. 2009). This conformation differs from free actin monomers and actin subunits in the cocrystal with Bni1p FH2 dimers (Otomo et al. 2005b). Therefore an FH2 dimer bound to the barbed end might influence not only local the helical twist but also the conformations of the actin subunits and vice versa.
The ends are the only places in filaments where the 180° twist is likely, given that surrounding subunits trap that rest of the filament in the 167° conformation. We suggested that after each round of actin subunit addition in the open state, the formin subunit is transiently bound to interior subunits of the filament. The strained FH2 will then move onto the new end where it may lower its free energy by entering the closed state (Paul and Pollard 2008) (Figure 4). We call this a “stepping second” mechanism. All models for FH2 translocation involve one or more steps where the FH2 dimer is not fully engaged with all four strong contacts on actin. The stepping second model has the leading FH2 domain step only after the incorporation of the new subunit, so steps depend on elongation. Stair stepping models assume that the leading FH2 domain dissociates before actin subunit addition. Such a mechanism implies that an FH2 domain on a barbed end should be in a rapid equilibrium on and off the actin independent of subunit addition.
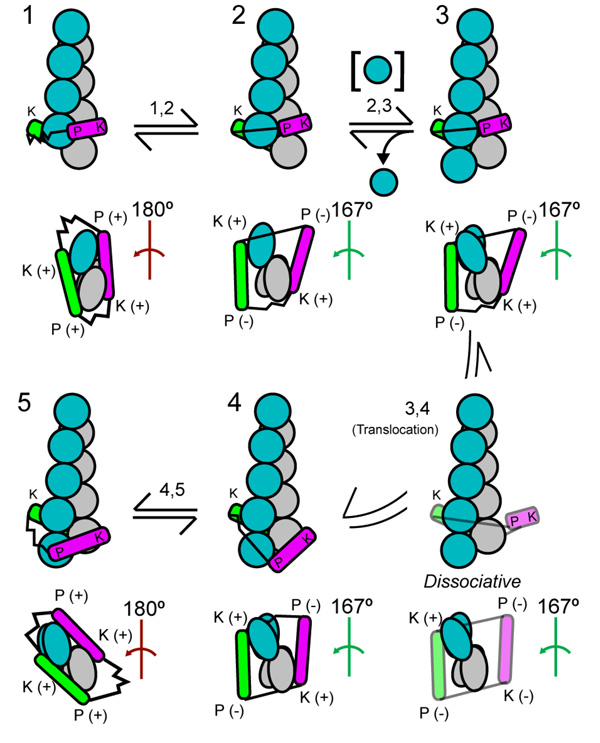
Figure 4. The “stepping second” hypothesis for actin subunit addition to a barbed end associated with a formin FH2 domain.
The drawing gives five steps (1–5) and transitions in a hypothetical mechanical cycle of actin subunit addition coupled to translocation of a formin FH2 dimer (green and magenta). States 5 and 4 are equivalent to states 1 and 2 but the filament is one subunit longer. The upper images for each state show a side view with the barbed end down. The lower images are views of the barbed end. The actin subunits are grey along one long pitch strand and blue along the other strand. The short-pitch helical twist of the 3 terminal barbed end subunits is either 180° as found in cocrystals of Bni1pFH2 with actin or the 167° as in the core of actin filaments. The red angle symbol indicates closed 180° conformations that do not accept subunit addition. The green angle symbol indicates open 167° conformations that permit subunit addition. Each FH2 subunit has two sites that can interact with actin: the knob (K) and post (P). Sites engaged with the filament are labeled (+). Sites dissociated from the filament are labeled (−). The flexible linkers between the two FH2 subunits are depicted as either stretched or relaxed springs. States 1 and 2 (as 5 and 4) are rapid equilibria between the open and closes states. A new actin subunit adds to open state 2 to form intermediate state 3. The leading FH2 subunit steps onto the new terminal subunit to complete the cycle. (From Paul and Pollard, Paul and Pollard 2008)
Assuming that the formin dimer is most likely to dissociate from the barbed end during a step when its contacts with actin are minimal, these mechanisms can be distinguished by determining the dependence of the formin dissociation rate on the rate of elongation. Direct microscopic observations showed that FH2 domains track with high fidelity on growing barbed ends, but that the rate of formin dissociation from growing ends is proportional to the rate of actin subunit addition (Paul and Pollard 2008). This finding is consistent with the stepping second hypothesis where dissociation is expected to be proportional to the number of cycles of subunit addition and inconsistent with stair stepping mechanisms, which predict that dissociation is a first order reaction.
Figure 4 shows a structure-based model for formin-mediated elongation based on the “stepping second” mechanism in which actin subunit addition onto the barbed end precedes translocation of the formin dimer. The key concept is that the translocation step is the point in the elongation cycle when the formin dimer is most tenuously associated with the barbed end and thus most prone to dissociation from the end.
FH2 domains from different formins dissociate from growing ends at substantially different rates (Table). For example the C. elegans cytokinesis formin CYK-1 dissociates ~50-fold faster than the S. pombe cytokinesis formin Cdc12p (Neidt et al. 2008). Experiments on chimeras constructed from a Bni1p FH2 dimer with different linkers showed that the linkers strongly influence processivity, with the rate of dissociation roughly proportional to the length of the linker. To explain this finding, we suggested that the lifetime of the translocating intermediate is proportional to the length of the linker (Paul and Pollard 2009).
In all models of processive translocation of formin FH2 domains on the barbed ends result in the FH2 domain rotating as the helical actin polymer grows. On the other hand, when a formin FH2 dimer is attached to a microscope slide the associated polymer can grow without rotating (Kovar and Pollard 2004). Shemesh et al. called this the “rotation paradox” and proposed a reasonable mechanism that allows the FH2 dimer to slip backwards around the filament to relieve strain accumulated during subunit addition (Shemesh et al. 2005). The details of this mechanism have yet to be investigated.
Contributions of FH1 domains and profilin to elongation of actin filaments by FH2 domains
Given that the bulk of unpolymerized actin in cells is bound to profilin (Kaiser et al. 1999), profilin must have a strong influence on actin polymerization with formins. Profilin binds to the barbed end of actin monomers and strongly inhibits nucleation and actin addition to filament pointed ends (Pollard and Cooper 1984). However, profilin bound to actin monomers does not inhibit elongation of barbed ends (Pollard and Cooper 1984), because profilin dissociates rapidly from the end after the complex of actin and profilin adds to the barbed end (Kang et al. 1999). Profilin binds weakly to actin filament barbed ends, so high concentrations of profilin can inhibit elongation of barbed ends (Gutsche-Perelroizen et al. 1999; Kaiser et al. 1999).
Low concentrations of profilin stimulate polymerization of bulk samples of actin and Bni1(FH1FH2)p (Sagot et al. 2002b), a finding inconsistent with the well-established ability of profilin to inhibit nucleation. This finding was subsequently explained by the finding that profilin speeds elongation of individual barbed ends associated with FH1FH2-formins (Kovar et al. 2006; Kovar et al. 2003; Kovar and Pollard 2004; Romero et al. 2004). This stimulatory effect of profilin requires the presence of the FH1 domain. This ability of profilin to speed elongation with FH1FH2-formin is so substantial that barbed ends associated with FH1FH2 domains of mDia1 grow substantially faster than free barbed ends in the same conditions (Romero et al. 2004). Because the addition of actin monomers onto barbed ends is diffusion-limited (Drenckhahn and Pollard 1986), this finding showed that addition of profilin-actin onto ends associated with an FH1FH2-formin is more complex than the simple bimolecular association of barbed ends with profilin-actin dimers diffusing freely in solution.
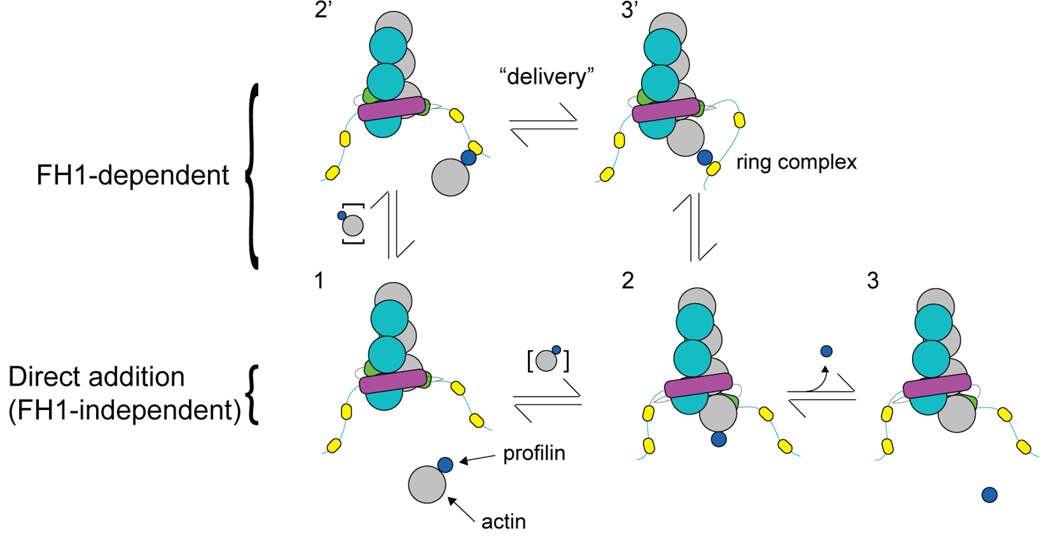
Systematic experimental and theoretical studies (Kovar et al. 2006; Paul and Pollard 2008; Vavylonis et al. 2006) revealed how profilin mediates rapid elongation of actin filament barbed ends associated with FH1FH2 domains from four different formins (Figure 5). Bulk phase profilin-actin binds to multiple sites on the two FH1 domains, concentrating profilin-actin near the barbed end. The flexible FH1 domains allow for rapid collisions between actin tethered to the FH1 by profilin and the barbed end. When the FH2/actin complex is in the open conformation, FH1-bound profilin-actin may bind the barbed end, forming the so-called ring complex (FH2-FH1-profilin-actin-barbed end). Then FH1 and profilin dissociate from the newly incorporated actin subunit to disassemble the ring complex and complete the elongation cycle (Vavylonis et al. 2006). Simulations of these reactions were consistent with the elongation rates with four different formins at profilin concentrations up to about 10 µM. To account for the observed rates of elongation through this FH1-dependent pathway, the rate of profilin-actin delivery to the filament by a FH1 polyproline track is on the order of 104 s−1.
Figure 5. Comparison of FH1-dependent and independent pathways of actin subunit addition.
Through the process of subunit addition, each state of the formin-FH1FH2-associated end is denoted by a different number. The actin filament and formin dimer subunits are colored as in Figure 3. Each FH1 domain has multiple polyproline tracks (yellow ovals). Profilin (small blue circle) binds to an actin monomer in solution (large gray circle). To complete one cycle of subunit addition, the profilin-actin complex may add onto the formin-FH1FH2-associated end via the FH1-independent pathway (1-2-3) or the FH1-dependent pathway (1-2’-3’-2-3) (Vavylonis et al. 2006). The rapid delivery step by which FH1-bound-profilin-actin is transferred directly to the FH2-associated barbed end and the resultant “ring complex” (Vavylonis et al. 2006) are labeled.
In the presence of profilin, subunit addition mediated by this FH1-dependent pathway vastly exceeds FH1-independent addition whereby actin monomers or dimers of profilin-actin in solution add directly onto formin-associated ends (Paul and Pollard 2008; Vavylonis et al. 2006). High concentrations of profilin inhibit elongation because free profilin competes with profilin-actin for binding FH1 domains (Vavylonis et al. 2006). Further experiments and modeling established that individual FH1 polyproline tracks bind profilin-actin and deliver actin onto the formin-associated barbed end and that under most conditions the rate-limiting step in elongation is binding of profilin-actin to the FH1 domain (Paul and Pollard 2008).
Two new studies show that formin FH1 domains may be very selective for profilin isoforms. Consequently only specific profilins are capable of supporting actin filament elongation by certain formins both in vitro and in cells (Ezezika et al. 2009; Neidt et al. 2009).
Energy for processive elongation by FH2 domains
Romero et al. (Romero et al. 2004; Romero et al. 2007) presented evidence that processive elongation by FH1FH2 formins in the presence of profilin is coupled to ATP hydrolysis and/or release of the γ-phosphate from actin. However, formins can use ADP-actin monomers for processive elongation (Kovar et al. 2006) and phosphate release occurs well after incorporation of ATP-actin at barbed ends (Paul and Pollard 2009). Furthermore, Bni1(FH1FH2)p with profilin does not stimulate the release of inorganic phosphate from polymerizing actin in the presence (Paul and Pollard 2009) or absence (Blanchoin and Pollard 2002) of formins. A careful thermodynamic analysis of the known interactions between the FH1 domain, profilin, actin monomers and the barbed end showed that subunit addition alone can provide the energy for processive elongation (Paul and Pollard 2009). These simulations also support the hypothesis that a common mechanism describes actin subunit addition mediated by diverse FH1FH2-formins and profilin (Kovar et al. 2006; Vavylonis et al. 2006).
Nucleation by FH2 domains
Formins strongly promote nucleation of filaments from free actin monomers but only weakly from profilin-actin (Pruyne et al. 2002; Sagot et al. 2002b; Zigmond et al. 2003). Now that the mechanism of formin-mediated elongation is understood reasonably well, it is possible to interpret how formins influence actin filament nucleation in these bulk samples. The structure of the cocrystal of Bni1p FH2 with actin suggested how a formin dimer might promote nucleation of new filament ends (Otomo et al. 2005b) by stabilizing an actin dimer or trimer, normally unstable intermediates on the nucleation pathway (Cooper et al. 1983; Frieden 1983; Sept and McCammon 2001). Simulations of a model where a FH2 dimer stabilizes an actin dimer, which serves as nucleus for elongation, fit the time course of polymerization in bulk samples with Bni1p (Pring et al. 2003).
In cells the concentration of profilin-actin vastly exceeds that of free actin monomers (Kaiser et al. 1999) and profilin strongly inhibits nucleation of actin (Pollard and Cooper 1984; Tobacman et al. 1983). Comparison of the time course of bulk actin polymerization with the rates of elongation of individual filaments in a range of profilin concentrations showed formins are likely to nucleate ends principally from free actin monomers and not profilin-actin (Paul and Pollard 2008). Thus, formins must rely on the small pool of free actin monomers to initiate new filaments in cells. Once these filaments start growing profilin strongly stimulates their elongation in association with FH1FH2-formins to rates exceeding the diffusion-limited elongation rate of free barbed ends.
Implications for cellular physiology
All well-characterized formins elongate actin filaments processively and dissociate rarely, so they generate long unbranched filaments. In the lamella of fibroblasts a recombinant fragment of the formin mDia1 seems to move freely with filaments as they grow in the cytoplasm (Higashida et al. 2004). By contrast, if a formin is anchored, the filaments can grow rapidly from the anchoring site. For example, formin Bni1p is localized in the bud of S. cerevisiae and extends cables of actin filaments at 100 subunits per second (Yang and Pon 2002) into the mother cell (Pruyne et al. 2004). Similarly S. pombe formin For3p associates with the cortex at the two poles of the cell and extends polarized actin cables along the length of the cell (Martin and Chang 2006). In both yeasts, these cables serve as tracks for Type V myosins to move particulate cargo (Huckaba et al. 2004; Pruyne et al. 1998). In preparation for cytokinesis, another fission yeast formin Cdc12p associates with clusters of other proteins (called nodes) in the cortex around the equator of the cell and extends filaments (Chang et al. 1997). Myosin-II Myo2p in other protein nodes captures filaments from other nodes, pulls on the actin filaments and condenses the nodes into the contractile ring (Vavylonis et al. 2008; Wu et al. 2006).
Owing to their processive association with a barbed end, formins prevent capping protein from terminating the growth of these filaments (Harris et al. 2004; Kovar et al. 2005; Moseley et al. 2004; Zigmond et al. 2003) and enable sustained elongation from the barbed of single filaments. In contrast, free barbed ends are capped in a second or less, accounting for the short branches at the leading edge of motile cells (Schafer et al. 1996).
The wide range of formin processivities is reminiscent of the variations in processivity among members of the myosin superfamily of motor proteins. The rates of the steps in the common ATPase cycle of all myosins are tuned to allow myosins to take just one or a large number of contiguous steps before dissociating from the actin filament [reviewed in (De La Cruz and Ostap 2004)]. These processivities are related to biological functions. For example, fast contraction of muscles depends on the myosins pulling on an actin filament for only a few milliseconds during each round of ATP hydrolysis. The asynchronous activities of multiple myosin molecules on aligned bundles of actin filaments produce sustained contraction over longer periods. By stark contrast to these muscle myosins, single molecules of myosin V transport vesicles over micrometer distances with single runs on actin filaments lasting up to a few seconds (Baker et al. 2004). Its ATPase cycle allows the two motor domains to interact processively with an actin filament as the myosin walks hand over hand and spends a large fraction of its time strongly bound to actin.
Compared to myosins, understanding of the relation of formin biophysics to their biological functions is immature, but it seems likely that formin processivities are also tuned for specific tasks. The broad range in the processive capabilities of FH2 domains suggests that sequence variation between formin homologs is strongly related to this key parameter of formin function. Further work is also required to test the mechanism proposed to explain how FH2 dimers translocate on actin filament barbed ends.
Acknowledgement
The research work from our laboratory on formins is supported by NIH research grants GM-026132 and GM-026338.
References
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276(4):2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24(1):13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Archer SJ, Vinson VK, Pollard TD, Torchia DA. Elucidation of the poly-L-proline binding site in Acanthamoeba profilin-I by NMR spectroscopy. FEBS Lett. 1994;337:145–151. doi: 10.1016/0014-5793(94)80262-9. [DOI] [PubMed] [Google Scholar]
- Baker JE, Krementsova EB, Kennedy GG, Armstrong A, Trybus KM, Warshaw DM. Myosin V processivity: multiple kinetic pathways for head-to-head coordination. Proc Natl Acad Sci USA. 2004;101:5542–5546. doi: 10.1073/pnas.0307247101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41(2):597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- Castrillon D, Wasserman S. diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 1997;137(1):169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320(5873):239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Buhle EL, Jr, Walker SB, Tsong TY, Pollard TD. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry. 1983;22:2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM. Relating biochemistry and function in the myosin superfamily. Curr Opin Cell Biol. 2004;16:61–67. doi: 10.1016/j.ceb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Drenckhahn D, Pollard TD. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J Biol Chem. 1986;261(27):12754–12758. [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4(1):32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- Ezezika OC, Younger NS, Lu J, Kaiser DA, Corbin ZA, Nolen BJ, Kovar DR, Pollard TD. Incompatibility with Formin Cdc12p Prevents Human Profilin from Substituting for Fission Yeast Profilin: Insights from crystal structures of fission yeast profilin. J Biol Chem. 2009;284:2088–2097. doi: 10.1074/jbc.M807073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Grosse R. Staying in shape with formins. Dev Cell. 2006;10(6):693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11(21):1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Frieden C. Polymerization of actin: mechanism of the Mg2+-induced process at pH8 and 20C. Proc. Natl. Acad. Sci. USA. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Grunt M, Zárský V, Cvrcková F. Roots of angiosperm formins: the evolutionary history of plant FH2 domain-containing proteins. BMC Evol Biol. 2008;8:115. doi: 10.1186/1471-2148-8-115. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutsche-Perelroizen I, Lepault J, Ott A, Carlier MF. Filament assembly from profilin-actin. J Biol Chem. 1999;274(10):6234–6243. doi: 10.1074/jbc.274.10.6234. [DOI] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279(19):20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303(5666):2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30(6):342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16(1):1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Huckaba TM, Gay AC, Pantalena LF, Yang HC, Pon LA. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 2004;167(3):519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. The structure of F-actin and of actin filaments isolated from muscle. J. Mol. Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- Kaiser DA, Vinson VK, Murphy DB, Pollard TD. Profilin is predominantly associated with monomeric actin in Acanthamoeba. J Cell Sci. 1999;112(Pt 21):3779–3790. doi: 10.1242/jcs.112.21.3779. [DOI] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J. Biol. Chem. 1999;274(52):36963–36972. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124(2):423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161(5):875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101(41):14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Wu JQ, Pollard TD. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol Biol Cell. 2005;16(5):2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursula P, Kursula I, Massimi M, Song YH, Downer J, Stanley WA, Witke W, Wilmanns M. High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDia1 and the proline-rich domain of VASP. J Mol Biol. 2008;375:270–290. doi: 10.1016/j.jmb.2007.10.050. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280(8):6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- Li R, Zheng Y, Drubin DG. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J. Cell Biol. 1995;128(4):599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4(11):953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Mahoney NM, Rozwarski DA, Fedorov E, Fedorov AA, Almo SC. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat Struct Biol. 1999;6(7):666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16(12):1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Matsuda S, Gladding CM, Yuzaki M. Characterization of the delta2 glutamate receptor-binding protein delphilin: Splicing variants with differential palmitoylation and an additional PDZ domain. J Biol Chem. 2006;281:25577–25587. doi: 10.1074/jbc.M602044200. [DOI] [PubMed] [Google Scholar]
- Miyagi Y, Yamashita T, Fukaya M, Sonoda T, Okuno T, Yamada K, Watanabe M, Nagashima Y, Aoki I, Okuda K, et al. Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci. 2002;22:803–814. doi: 10.1523/JNEUROSCI.22-03-00803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell JL, Morphew M, Gould KL. A mutant of Arp2p causes partial disassembly of the Arp2/3 complex and loss of cortical actin function in fission yeast. Mol. Biol. Cell. 1999;10(12):4201–4215. doi: 10.1091/mbc.10.12.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15(2):896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidt EM, Scott BJ, Kovar DR. Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J Biol Chem. 2009;284:673–684. doi: 10.1074/jbc.M804201200. [DOI] [PubMed] [Google Scholar]
- Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–23883. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005a;18(3):273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005b;433(7025):488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Paul A, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2 formins. J Biol Chem. 2009;284 doi: 10.1074/jbc.M808587200. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18(1):9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33(28):8472–8478. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141(5):1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella EC, Machesky LM, Kaiser DA, Pollard TD. Structural requirements and thermodynamics of the interaction of proline peptides with profilin. Biochemistry. 1996;35(51):16535–16543. doi: 10.1021/bi961498d. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry. 1984;23(26):6631–6641. doi: 10.1021/bi00321a054. [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 2003;42(2):486–496. doi: 10.1021/bi026520j. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297(5581):612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Gao L, Bi E, Bretscher A. Stable and dynamic axes of polarity use distinct formin isoforms in budding yeast. Mol Biol Cell. 2004;15:4971–4989. doi: 10.1091/mbc.E04-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433(7024):382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Rivero F, Muramoto T, Meyer AK, Urushihara H, Uyeda TQ, Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6(1):28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Didry D, Larquet E, Boisset N, Pantaloni D, Carlier MF. How ATP hydrolysis controls filament assembly from profilin-actin: implication for formin processivity. J Biol Chem. 2007;282:8435–8445. doi: 10.1074/jbc.M609886200. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119(3):419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Rose R, Weyand M, Lammers M, Ishizaki T, Ahmadian MR, Wittinghofer A. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435(7041):513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002a;4(1):42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002b;4(8):626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 1996;135(1):169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7(6):619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365(6449):810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys. J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemesh T, Otomo T, Rosen MK, Bershadsky AD, Kozlov MM. A novel mechanism of actin filament processive capping by formin: solution of the rotation paradox. J Cell Biol. 2005;170(6):889–893. doi: 10.1083/jcb.200504156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Shibata H. Poly (L-proline)-binding proteins from chick embryos are a profilin and profilactin. European J. Biochem. 1985;151:291–297. doi: 10.1111/j.1432-1033.1985.tb09099.x. [DOI] [PubMed] [Google Scholar]
- Tobacman LS, Brenner SL, Korn ED. Effect of Acanthamoeba profilin on the pre-steady state kinetics of actin polymerization and on the concentration of F-actin at steady state. J. Biol. Chem. 1983;258(14):8806–8812. [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21(4):455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319(5859):97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wallar BJ, Stropich BN, Schoenherr JA, Holman HA, Kitchen SM, Alberts AS. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281(7):4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Madaule P, Reid T, Ishizaki T, Watanabe G, Kakizuka A, Saito Y, Nakao K, Jockusch B, Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16(11):3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ando Y, Yasuda S, Hosoya H, Watanabe N, Ishizaki T, Narumiya S. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116(5):711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Pon LA. Actin cable dynamics in budding yeast. Proc Natl Acad Sci U S A. 2002;99(2):751–756. doi: 10.1073/pnas.022462899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, Chang F. Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol Biol Cell. 2008;19:2208–2219. doi: 10.1091/mbc.E07-07-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13(20):1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]