Summary
A number of small-molecule inhibitors have been developed that target the catalytic domains of protein kinases that are not in an active conformation. An inactive form that has been observed in several kinases is the DFG-out conformation. This conformation is characterized by an almost 180° rotation of the con served Asp-Phe-Gly (DFG) motif in the ATP-binding cleft relative to the active form. However, the sequence and structural determinants that allow a kinase to stably adopt the DFG-out conformation are not known. Here, we characterize a series of inhibitors based on a general pharmacophore for this inactive form. We demonstrate that modified versions of these inhibitors can be used to study the thermodynamics and kinetics of ligand binding to DFG-out-adopting kinases and for enriching these kinases from complex protein mixtures.
INTRODUCTION
Comprised of over 500 members, protein kinases are the largest enzyme family encoded by the human genome (Manning et al., 2002). These enzymes are the major mediators of intracellular protein phosphorylation cascades and are involved in complex phenotypic responses such as growth, differentiation, adhesion and motility (Cohen, 2000). The proper spatial and temporal control of intracellular phosphorylation events by protein kinases is essential for normal cellular function. Indeed, aberrant kinase activity has been implicated in numerous diseases including cancer, diabetes and chronic inflammation (Cohen, 2002). For this reason, there has been considerable effort towards the development of pharmacological agents that target this enzyme family.
The overall structure of the catalytic domain of protein kinases is highly conserved (Figure 1A) (Hanks and Hunter, 1995). It consists of 250-300 residues that form a bi-lobal structure containing a smaller, mostly β-stranded, N-terminal domain and a larger, mostly α-helical, C-terminal domain (Taylor et al., 1988). Adenosine 5’-triphosphate (ATP) sits in a narrow hydrophobic cleft between the N- and C-terminal lobes, with the purine moiety forming a pair of hydrogen bonds with the hinge region (Engh and Bossemeyer, 2001). Most protein kinases possess at least two distinct catalytic states: an active state where all of the catalytic residues are in an optimal position for phosphate transfer and an inactive state (or states) with reduced catalytic activity (Huse and Kuriyan, 2002). The conformation of the ATP-binding site in these two states can be quite distinct, characterized by large conformational shifts in several conserved catalytic residues. A number of phosphorylation events and protein-protein interactions regulate the conformational state of these enzymes (Jeffrey et al., 1995; Pellicena and Kuriyan, 2006; Wong et al., 2004).
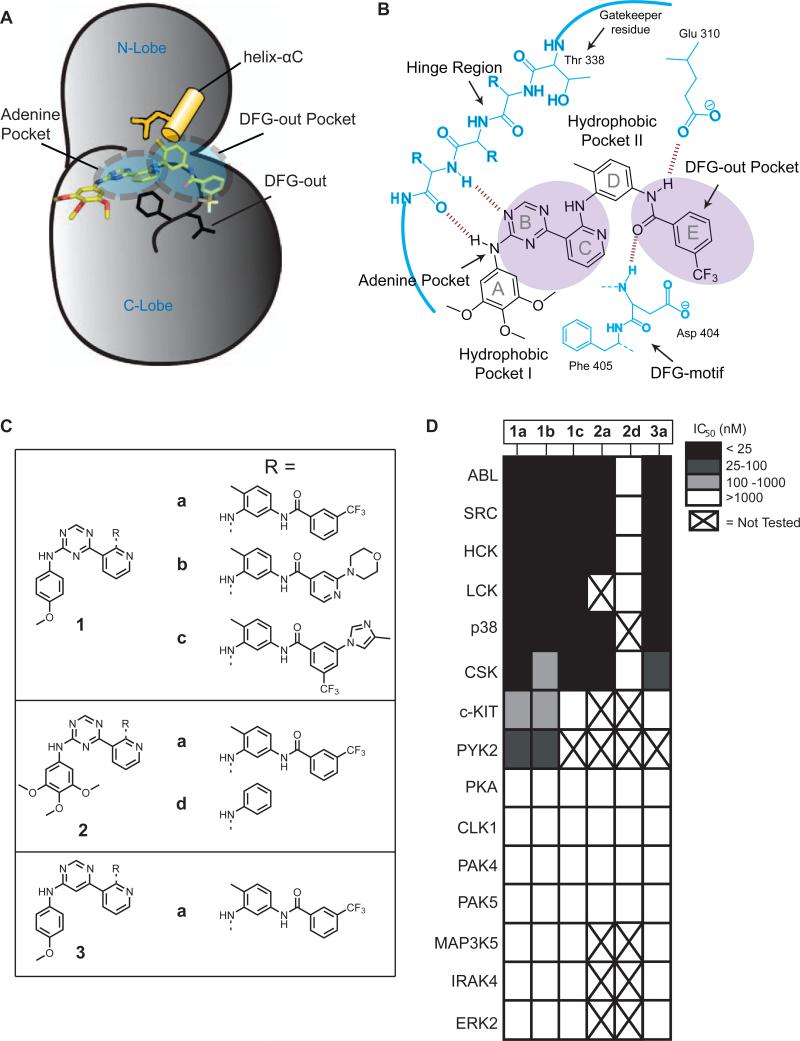
Figure 1.
The DFG-out conformation of protein kinases and type II inhibitors (A) The catalytic domain of the tyrosine kinase SRC bound to the type II inhibitor 2a. The ATP-binding cleft is in the DFG-out conformation. The conserved glutamate in helix-αC (shown in yellow) and the conserved aspartate and phenylalanine in the DFG-motif (shown in black) are in conformations observed for kinases bound to type II inhibitors. Kinase sub-pockets occupied by type II inhibitors are indicated by dashed circles. (B) A schematic representation of 2a bound to the tyrosine kinase SRC. This inhibitor forms two hydrogen bonds with the hinge region of the kinase. The characteristic set of hydrogen bonds that type II inhibitors form with the conserved glutamate residue in the helix-αC and the amide backbone of the aspartate of the DFG-motif are shown. The DFG-out Pocket that is generated by the movement of the phenylalanine residue of the DFG motif is shaded. (C) Chemical structures of inhibitors 1a-1c, 2a, 2d and 3a. (D) IC50s of the inhibitors shown in Figure 1C against a diverse panel of kinases. Potencies are represented in gray-scale. Note: We have previously reported the IC50s of inhibitors 1a and 2a for SRC, ABL and HCK (Seeliger et al., 2009). In this publication these compounds are referred to as DSA7 and DSA1, respectively. X = not tested. All assays were run in triplicate or quadruplicate.
The ATP-binding sites of protein kinases in the active conformation are very similar, while the structures of inactive kinases are much more varied (Engh and Bossemeyer, 2001; Kornev et al., 2006). A specific inactive form of the ATP-binding cleft that has been observed in a number of kinases is the DFG-out conformation (Liu and Gray, 2006; Okram et al., 2006). A number of potent kinase inhibitors have been found to selectively bind to the DFG-out conformation of their kinase targets (type II inhibitors). In many cases, these type II inhibitors exhibit a high degree of selectivity. Despite the selectivity demonstrated by some type II inhibitors, a set of conserved binding interactions in the ATP-binding pocket has been observed (Jacobs et al., 2007; Liu and Gray, 2006; Nagar et al., 2002; Okram et al., 2006; Simard et al., 2009a). While the DFG-out conformation appears to be energetically accessible to a number of kinases, the sequence and structural determinants that allow this transition are still poorly understood. Furthermore, it is still not possible to predict which members of the kinome can be effectively targeted with type II inhibitors. Identifying the full complement of kinases that are able to stably adopt this inactive conformation will shed light on the catalytic regulation of protein kinases and may yield new targets for drug discovery.
Here we report the development of a set of affinity probes that are derived from a general type II ligand for the DFG-out conformation of protein kinases. We show that fluorophore-conjugated analogs of these ligands are useful probes for measuring the thermodynamics and kinetics of type II inhibitor binding to protein kinases. In addition, we demonstrate that these reagents are capable of selectively enriching protein kinases that can adopt this inactive form from complex protein mixtures. Using these probes we provide evidence that the STE20 kinase LOK behaves like a kinase that can adopt the DFG-out conformation.
RESULTS
A General Type II Inhibitor Scaffold
In order to develop a set of reagents for investigating inactive forms of protein kinases we wished to identify a general ligand for the DFG-out conformation. Recently, we have characterized a series of triazolo-pyridines that are equipotent inhibitors of the tyrosine kinases SRC and ABL (Seeliger et al., 2009). Through structural and kinetic studies we demonstrated that this class of inhibitors binds to the DFG-out conformation of these kinases. The general binding mode of the triazolo-pyridine ligands is similar to other type II inhibitors that have been described (Figure 1B. (pdbs: 3G6G, 3G6H)). The aniline and triazine groups (rings A and B) form a pair of hydrogen bonds with the hinge region and occupy the hydrophobic cleft utilized by adenine. Rings C and D serve as a linker connecting the adenine-binding region to a hydrophobic pocket (DFG-out Pocket) that is created by the nearly 180° rotation of the Phe in the DFG-motif to a more solvent exposed position. Upon entering the DFG-out Pocket, the amide group connecting rings D and E forms a pair of hydrogen bonds with a conserved glutamate in the helix-αC and the backbone of the DFG-motif. Finally, ring E occupies the DFG-out Pocket. A small panel of inhibitors based on the triazolo-pyridine scaffold was generated to determine the generality of this ligand class (Figure 1C). For inhibitor series 1 and 2, the substituents that occupy the DFG-out Pocket and exit the adenine-binding site were varied (rings A and E), while the core scaffold (rings B, C and D) was held constant. Compounds 1a and 2a contain a 3-trifluoromethylbenzamide group which is a common binding element in many type II inhibitors (Liu and Gray, 2006; Okram et al., 2006). Compounds 1b and 1c contain functional groups that are components of selective type II inhibitors: 1b contains a 2-morphilinopyridyl group, which is the pharmacophore that occupies the DFG-out Pocket in the selective type II p38 MAPK inhibitor MPAQ and compound 1c contains the functional groups that are present in the selective type II BCR-ABL inhibitor nilotinib (Cumming et al., 2004; Sullivan et al., 2005; Weisberg et al., 2005). Finally, compound 2d was generated as a control compound due to its lack of a substituent that occupies the DFG-out Pocket. In addition to the triazolo-pyridine inhibitors from series 1, a pyrimidine-pyridine compound (3a) was also generated to determine the role of scaffold structure.
To determine whether the inhibitors generated are potent ligands for kinases that adopt the DFG-out conformation, they were tested against a panel of kinases in which high resolution crystal structures of the catalytic domain are available. Crystal structures of the tyrosine kinases SRC, LCK, ABL, PYK2, CSK and c-KIT and the serine/threonine kinase p38α in the DFG-out conformation have been obtained and these kinases have been found to be sensitive to type II inhibitors (Han et al., 2009; Mol et al., 2004; Seeliger et al., 2009; Simard et al., 2009b). In contrast, the serine/threonine kinases PKA, PAK4, PAK5, CLK1, MAP3K5, IRAK4, and ERK2 have not been observed in this inactive form and inhibitor selectivity screens have shown them to only be sensitive to type I inhibitors (Fedorov et al, 2007). The results of the inhibition assays are shown in Figure 1D. SRC, LCK, ABL, PYK2, CSK, HCK, p38 and c-KIT were potently inhibited by most of the type II inhibitors (1a-1c, 2a and 3a), while kinases that have not previously been observed in the DFG-out conformation show little or no inhibition at the highest concentration tested (1 μM). Consistent with these inhibitors binding to the DFG-out conformation, control compound 2d shows minimal inhibition of kinases that are sensitive to type II inhibitors. Surprisingly, the identity of the aryl group that projects into the DFG-out Pocket has only a small impact on the selectivity profile amongst DFG-out adopting kinases.
BODIPY-Labeled Compounds as Binding Probes for Protein Kinases
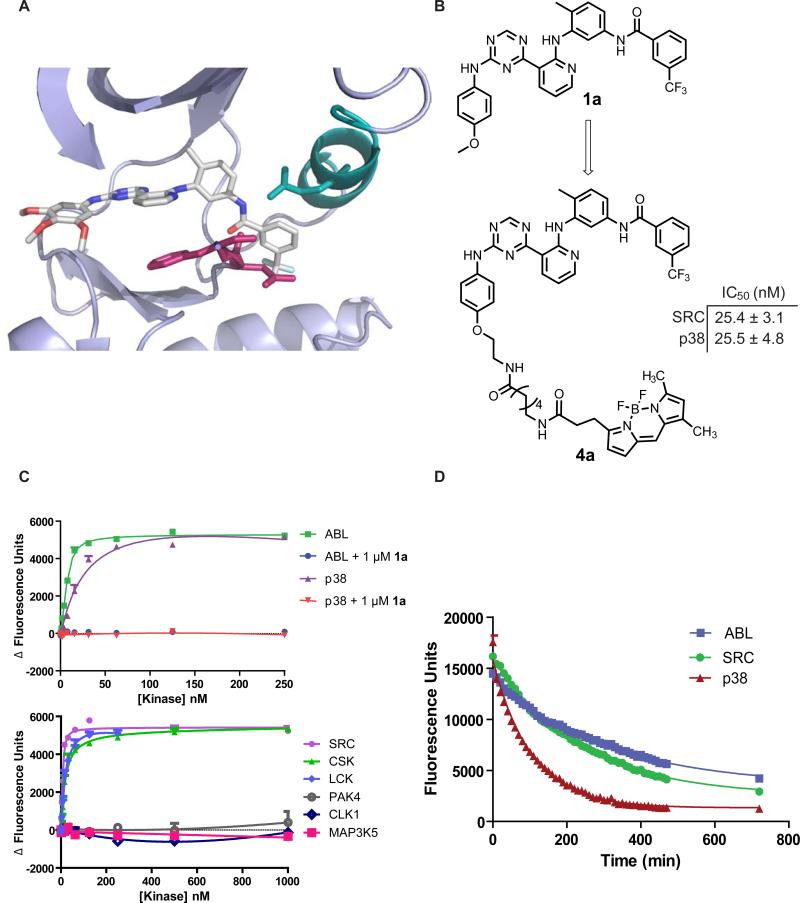
In most cases, the potencies of compounds that bind the DFG-out inactive conformation have been determined by their ability to inhibit activated kinases. However, this method of analysis does not allow kinase forms with low catalytic activity to be probed and it is often difficult to correlate the observed inhibition to the binding of a specific kinase conformation. A direct binding assay has the ability to measure ligand affinity for any activation state of a protein kinase and allows binding kinetics to be determined. For this reason, we determined the feasibility of converting our general type II inhibitors into fluorescently-labeled binding probes. Based on a previously reported crystal structure of inhibitor 2a bound to SRC, the solvent exposed 4-position of ring A was selected as the site of fluorophore attachment (Figure 2A). Because inhibitors 1a and 2a have similar potencies against most kinases, a BODIPY-FL fluorophore was conjugated to the monoalkoxy derivative 1a. To determine whether fluorophore attachment significantly affects the potency of this inhibitor against protein kinases, in vitro activity assays were performed with p38 and SRC. Gratifyingly, the IC50 of probe 4a for SRC and p38 is similar to inhibitor 1a (Figure 2B).
Figure 2.
Fluorophore-labeled ligands for studying the kinetics and thermodynamics of type II inhibitor binding (A) Crystal structure of 2a bound to the catalytic domain of SRC T338I (PDB: 3g6h); magenta; DFG-motif, teal; helix-αC. (B) Conversion of compound 1a into a BODIPY-labeled binding probe (4a). IC50 values of 4a against SRC and p38 in activity assays. (C) Top graph: change in fluorescence observed with increasing concentrations of kinases (green, ABL: purple, p38) in the presence of fluorescent probe 4a (10 nM) and in the presence of both 4a (10 nM) and competitor 1a (1000 nM). Bottom graph: change in fluorescence observed with increasing concentrations of the kinases SRC, CSK, LCK, PAK4, CLK1 and MAP3K5 in the presence of 4a (10 nM). (D) Determination of the SRC-4a, ABL-4a and p38-4a dissociative half lives (t1/2). The catalytic domains of these kinases were incubated under conditions in which >95% of 4a (500 nM) is bound. The complexes were then diluted 30-fold into a solution containing an excess of competitor 1a (5 μM) to prevent reformation of the kinase-probe complex. The rate of dissociation is measured by the decrease in fluorescence over time. Fluorescence was read every 10 minutes for 8 h, then at 12 h. Data was fit to a mono-exponential decay curve. Assays were run in triplicate.
We hypothesized that probe 4a would show an increase in fluorescence when bound to the ATP-binding cleft of a protein kinase. To test this, 10 nM of probe 4a was incubated with increasing concentrations of the catalytic domains of ABL or p38 (1-250 nM). In the presence of these kinases, probe 4a demonstrated a concentration-dependent increase in fluorescence (Figure 2C, top graph). To confirm that the observed increase in fluorescence is due to a specific interaction in the ATP-binding pocket, this experiment was repeated in the presence of an excess of a competitor that does not contain a fluorophore (1 μM of 1a). As expected, no increase in fluorescence was observed for either kinase in the presence of excess competitor (Figure 2C). Having validated our binding assay with p38 and ABL, the Kd values of probe 4a for a panel of kinases was determined (Figure 2C, bottom graph). Consistent with the in vitro activity assays described above, kinases that have previously been demonstrated to be able to adopt the DFG-out conformation (ABL, SRC, HCK, LCK, p38, EPHA3 and CSK) have a high affinity for probe 4a, while other kinases (CLK1, PAK4, PAK5 and MAP3K5) do not show a detectable level of binding (Table 1).
Table 1.
Kd values of kinases determined with 5 nM (ABL, SRC, HCK) or 10 nM (LCK, p38, CSK, EPHA3, CLK1, PAK4, PAK5, MAP3K5) 4a. Values shown are the average of three assays ± SEM.
| Table 1 | Kd (nM) |
|---|---|
| ABL | 9.9 ± 1.6 |
| SRC | 13 ± 2 |
| HCK | 8.7 ± 0.6 |
| LCK | 20 ± 2 |
| p38 | 35 ± 11 |
| CSK | 17 ± 3 |
| EPHA3 | 27 ± 5 |
| CLK1 | >1000 |
| PAK4 | >1000 |
| PAK5 | >1000 |
| MAP3K5 | >1000 |
Because a large conformational change in the ATP-binding pocket of kinases is required to accommodate inhibitor binding, most type II inhibitors demonstrate slow binding kinetics. To determine if 4a can be used to obtain type II inhibitor binding kinetics, an assay that allows the measurement of the rate of kinase-4a complex dissociation was developed. For this assay, the catalytic domains of SRC, ABL and p38 were incubated with probe 4a under conditions in which >95% of the fluorescent probe was bound. The reaction mixture was then diluted 30-fold into a solution containing a large excess of competitor 1a, to ensure that dissociated 4a cannot re-bind to the kinase. By measuring the time-dependent loss in fluorescence, the rate of dissociation of the SRC-4a, ABL-4a and p38-4a complexes were determined (Figure 2D). The data obtained under these conditions fit well to a mono-exponential decay curve, with t1/2s for the SRC, ABL and p38 complexes of 162, 220 and 72 minutes, respectively (Table 2). These data correlate well with kinetic data that has been obtained for this class of compounds and other type II inhibitors (Pargellis et al., 2002; Seeliger et al., 2007; Seeliger et al., 2009; Sullivan et al., 2005).
Table 2.
Kd, t1/2 and koff values for imatinib-resistant mutants of ABL and the corresponding SRC mutants. Kd values for ABL and SRC were determined with 10 nM 4a. Values shown are the average of three assays ± SEM.
| Table 2 | Kd (nM) | t1/2 (min) | koff (sec−1) |
| ABL wt | 9.9 ± 1.6 | 220 ± 3 | 5.2 × 10−5 |
| ABL E255V | 45 ± 4 | 234 ± 5 | 4.9 × 10−5 |
| ABL Y253H | 9.4 ± 1.2 | 195 ± 7 | 5.9 × 10−5 |
| SRC wt | 13 ± 2 | 162 ± 4 | 7.3 × 10−5 |
| SRC E280V | 15 ± 2 | 133 ± 18 | 9.0 × 10−5 |
| SRC F278H | 12 ± 3 | 167 ± 4 | 6.9 × 10−5 |
| SRC T338I | 11 ± 1 | 67 ± 9 | 1.8 × 10−4 |
| p38 | 35 ± 11 | 72 ± 1 | 1.6 × 10−4 |
We next investigated how mutations in the catalytic domains of protein kinases affect the conformational dynamics of the ATP-binding cleft. Specifically, probe 4a was used to study how imatinib-resistance mutations in the P-loop affect the interaction of ABL kinase with type II inhibitors (Table 2). The Kd of this probe for two resistance mutants that are frequently observed in patients with chronic myelogenous leukemia (CML) that are undergoing imatinib treatment, Y253H and E255V, was determined (Quintas-Cardama et al., 2007). Consistent with inhibitors of this class not making significant interactions with this flexible region of the kinase, the Y253H mutant of ABL was found to have an identical binding affinity for 4a as the wild type enzyme. However, the E255V mutant demonstrated a 4-to-5-fold weaker interaction. The analogous mutants of SRC, F278H and E280V, were also characterized. The less flexible nature of SRC's P-loop is reflected in the identical kinetics and thermodynamics observed for both mutants and wild type. Finally, a gatekeeper mutant of SRC, SRC T338I, was characterized. Similar to the underivatized inhibitor, 4a has an equal affinity for wild type and SRC T338I. Despite this similarity in binding thermodynamics, the on and off rates for 4a are 2-to-3 times faster for the gatekeeper mutant.
Generation of Affinity Matrices 14a and 15a
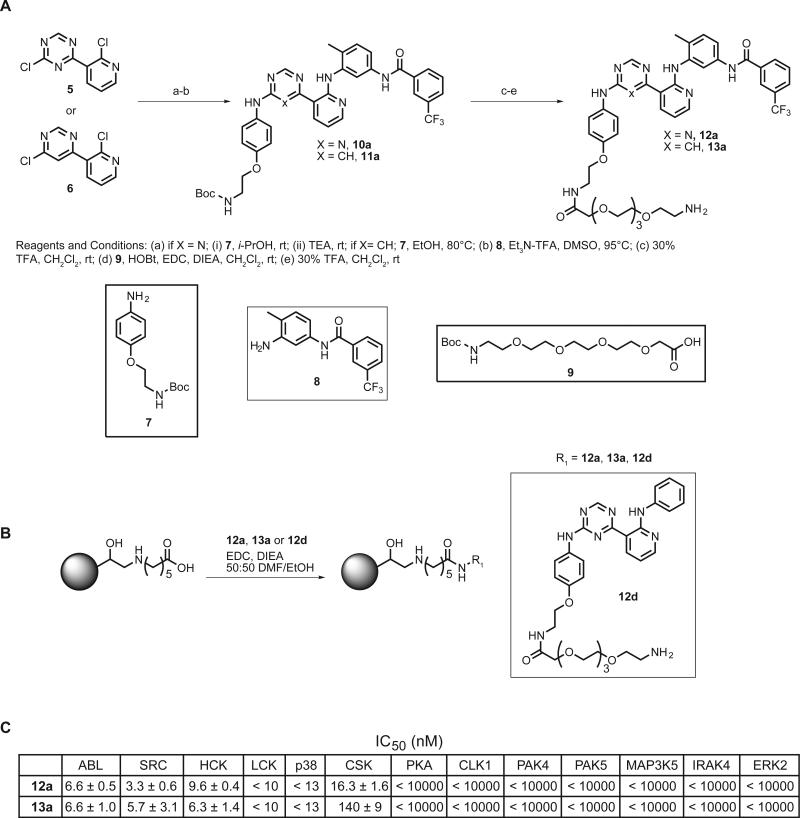
We determined the feasibility of converting compounds 1a and 3a into affinity pull-down reagents. These proteomic tools should allow unbiased analyses of the kinases that are able to adopt the DFG-out conformation in diverse cellular lysates. The 4-position of ring A was modified with a flexible polyethylene glycol (PEG) linker that contains a primary amino group to allow selective immobilization to a solid support (Compounds 12a and 13a, Figure 3A). Triazolo-pyridine 12d, which is an analog of control compound 2d, was also generated as a negative control for enrichment studies. To determine whether linker attachment affects the potency of this scaffold for kinases that adopt the DFG-out conformation, in vitro activity assays were performed (Figure 3C). Gratifyingly, attachment of a flexible PEG linker does not alter the selectivity or potency of these inhibitors. Inhibitor-tethered resins containing 12a and 13a were generated by the procedure shown in Figure 3B. In each coupling reaction, the corresponding non-linkable analogs (1a, 3a or 2d) were included as controls. Addition of this internal standard allowed the progress of the coupling reaction to be monitored by observing the selective loss of the amine-containing compounds 12a, 13a and 12d by HPLC (Figures S1 and S2).
Figure 3.
Synthesis and characterization of immobilized type II inhibitors (A) Synthetic scheme for generating type II inhibitors that can be immobilized. (B) Procedure for coupling 12a, 13a and control 12d to ECH Sepharose 4B. (C) IC50s of inhibitors 12a and 13a against a panel of kinases.
Affinity Matrices 14a and 15a Enrich Kinases that Adopt the DFG-Out Conformation
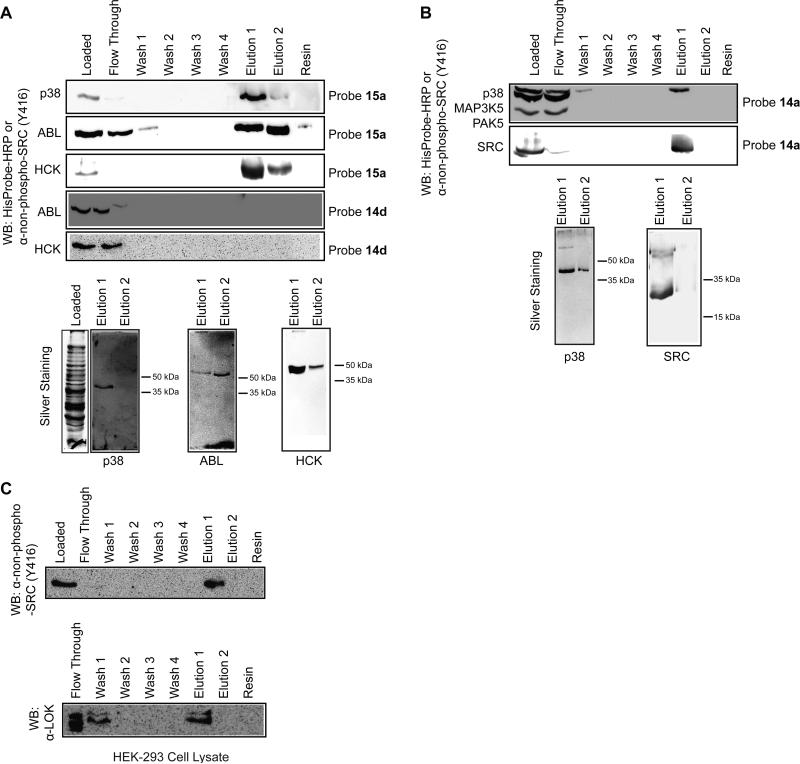
A model system was developed to determine the ability of affinity matrices 14a and 15a to enrich kinases that are able to adopt the DFG-out conformation. Optimal conditions for affinity capture, resin washing and protein elution were determined by adding purified protein kinases to cellular lysates from E. coli (Figures 4A and 4B). Because E. coli lack eukaryotic and eukaryotic-like protein kinases only exogenously added kinases should be retained by the affinity matrix (Kannan et al., 2007). We first tested the ability of affinity matrix 15a to enrich p38 kinase from lysates. A majority of His6-p38 was retained by affinity matrix 15a after 2 h of incubation as demonstrated by the lack of detectable levels of His6-tagged kinase in the soluble fraction (Figures 4A). Furthermore, no immobilized p38 was found to dissociate from the resin after multiple washes. Repeated efforts to elute p38 from the affinity matrix with high concentrations (>mM) of soluble, ATP-competitive inhibitors were unsuccessful. This is most likely due to the high affinity of the immobilized ligand for kinases that adopt the DFG-out conformation and the slow rate of dissociation of these inhibitor-kinase complexes. However, a mild denaturant (0.5% SDS) allowed quantitative elution of this kinase. Furthermore, these conditions appear to be selective for disruption of the kinase-inhibitor complex as demonstrated by the lack of other proteins in the eluted fractions detected via silver staining. The same conditions were found to be effective for enriching ABL and the SRC-family kinase HCK (Figure 4A). As expected, none of these kinases were retained by control matrix 14d. To ensure that the affinity matrix selectively enriches kinases that adopt the DFG-out conformation, p38 was subjected to the standard affinity enrichment protocol in an E. coli protein lysate containing the kinases PAK5 and MAP3K5 (Figure 4A). As expected, only p38 was bound and retained by affinity matrix 14a. Furthermore, p38 was the only protein that could be detected in the eluted fractions. Similar results were observed for SRC.
Figure 4.
Characterization of immobilized type II inhibitors 14a and 15a (A) Enrichment of protein kinases from E. coli lysates with affinity resins 14a, 15a or control matrix 14d. Purified His6-p38 (4.6 μg), His6-ABL (5 μg) or HCK (18 μg) were added to E. coli lysate (550-700 μg) and subjected to standard enrichment conditions with affinity matrix 15a or control matrix 14d. Top panels: All fractions were subjected to SDS-PAGE. ABL and p38 were detected with HisProbe-HRP (Pierce); SRC and HCK were detected with an antibody that specifically recognizes SRC-family kinases that are dephosphorylated at Tyr416 in the activation loop (α-non-phospho-SRC(Y416)) (Cell Signaling). Bottom gels: Elutions 1 and 2 from the enrichment experiments described above were subjected to SDS-PAGE and silver stained. (B) Enrichment of kinases that are sensitive to type II inhibitors in the presence of kinases that have not been characterized in the DFG-out conformation. His6-p38 (4 μg), His6-MAP3K5 (10 μg) and His6-PAK5 (10 μg) were added to E. coli lysate (500 μg) and subjected to standard enrichment conditions with affinity matrix 14a. The catalytic domain of SRC (4 μg), His6-MAP3K5 (4.5 μg) and His6-STK16 (4.5 μg) were added to E. coli lysate (600 μg) and subjected to standard enrichment conditions with affinity matrix 14a. Top panels: All fractions were subjected to SDS-PAGE and probed with HisProbe-HRP or α-non-phospho-SRC(Y416). Bottom gels: Elutions 1 and 2 from the enrichment experiments described above were subjected to SDS-PAGE and silver stained. Note: Procedures for protein kinase expression and purification are in the Supplemental Data. (C) Enrichment of endogenous kinases from HEK-293 cell lysate. 600 μg of HEK-293 lysate was subjected to standard enrichment conditions with affinity matrix 14a. All fractions were subjected to SDS-PAGE and probed with α-non-phospho-SRC(Y416) or α-LOK.
The ability of immobilized inhibitor 14a to enrich endogenous protein kinases from mammalian lysates was tested. Western blot analysis demonstrated that >90% of total SRC was retained by the beads when 650 μg of HEK-293 protein lysate was incubated with affinity resin 14a (Figure 4C). SRC remained bound to the affinity matrix during repeated washes and was eluted with 0.5% SDS. Comparison of the proteins eluted from control matrix 14d and affinity matrix 14a by SDS-PAGE followed by silver staining demonstrated that several unique proteins were enriched from HEK-293 and HeLa cell lysates by immobilized inhibitor 14a (Figure S3). However, a number of prominent bands were present in the eluted fractions for both support-bound ligands. These appear to be proteins that bind non-specifically to the matrix, as pre-incubation with control matrix 14d prior to enrichment with 14a did not deplete a majority of these bands. Mass spectrometric analysis of the proteins that were enriched from HEK-293 and HeLa cells is ongoing and will be reported elsewhere. In parallel, a candidate approach for identifying kinases that can adopt the DFG-out conformation was pursued. Previous inhibitor screens have shown that the serine/threonine kinase LOK is sensitive to several type II inhibitors (Angell et al., 2008; Fabian et al., 2005; Karaman et al., 2008). However, biochemical studies have not been performed to establish whether LOK can adopt the DFG-out conformation. To determine whether endogenous LOK binds to the triazolo-pyridine scaffold, HEK-293 cell lysate was incubated with affinity matrix 14a. Immuno-blot analysis of the bound and unbound fractions with an α-LOK antibody demonstrated that a percentage of this kinase was retained by the beads and selectively eluted.
The Catalytic Domain of LOK Behaves Similarly to Other DFG-Out-Adopting Protein Kinases
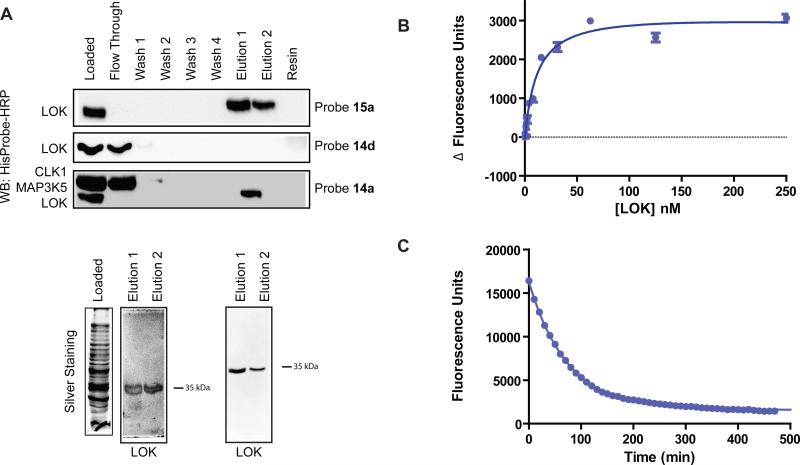
To further characterize LOK, the His6-tagged catalytic domain of this kinase was expressed and purified using a previously described protocol (Fedorov et al., 2007). An in vitro activity assay was developed for LOK and compounds 1a-c, 2a and 3a were tested for their ability to inhibit this kinase. LOK was found to be equally sensitive to inhibition by type II inhibitors as other kinases that adopt the DFG-out conformation (Table S1). Furthermore, resins 14a and 15a were able to enrich the catalytic domain of LOK from E. coli lysate over other protein kinases (Figure 5A). In addition, LOK was not retained by control matrix 14d. To further confirm the affinity of LOK for the type II inhibitor class, binding studies with probe 4a were performed. Similar to other kinases that adopt the DFG-out conformation, LOK has a high affinity for this fluorescently-labeled probe (Kd = 12 nM, Figure 5B). Finally, the t1/2 for the LOK-4a complex was determined (Figure 5C). Consistent with LOK demonstrating slow binding kinetics for type II inhibitors, the t1/2 for this complex was found to be 51 minutes. While the off rate for probe 4a is 3-4-fold faster than for SRC and ABL, the kinetics observed are consistent with a conformational change being necessary for ligand binding.
Figure 5.
The catalytic domain of LOK behaves like a kinase that can adopt the DFG-out conformation. (A) Enrichment of the catalytic domain of LOK from E. coli lysates with affinity resins 14a, 15a or control matrix 14d. Top panel: Purified His6-LOK (4.5 μg) was added to E. coli lysate (600 μg) and subjected to standard enrichment conditions with affinity matrix 15a. LOK was detected with HisProbe-HRP (Pierce). Middle panel: Purified His6-LOK (4.5 μg) was added to E. coli lysate (600 μg) and subjected to standard enrichment conditions with control matrix 14d. LOK was detected with HisProbe-HRP. Bottom panel: Purified His6-LOK (4 μg), His6-CLK1 (10 μg) and His6-MAP3K5 (10 μg) were added to E. coli lysate (500 μg) and subjected to standard enrichment conditions with affinity matrix 14a. All fractions were subjected to SDS-PAGE and kinases were detected with HisProbe-HRP (Pierce). Bottom gels: Elutions 1 and 2 from the enrichment experiments performed in the top and bottom panels above. Samples were subjected to SDS-PAGE and silver stained. (B) Change in fluorescence observed with an increasing concentration of kinase LOK in the presence of 5 nM of fluorescent probe 4a. Kd = 12 ± 1 nM. Value shown is the average of three assays ± SEM. (C) Determination of the LOK-4a complex dissociative half life (t1/2). For LOK-4a t1/2 = 51 minutes and koff = 2.3 × 10-5 s-1. Assays were run in triplicate.
DISCUSSION
After it was discovered that the inhibitors imatinib and BIRB796 bind to the inactive forms of their kinase targets, there has been widespread interest in the identification of small molecules that selectively recognize alternate active site conformations (Liu and Gray, 2006; Nagar et al., 2002; Okram et al., 2006; Schindler et al., 2000). High selectivity can be achieved with inhibitors that bind to inactive kinase conformations and the characteristically long target residence times of these protein-small molecule complexes may be beneficial from a therapeutic standpoint. The DFG-out conformation is a specific inactive form that has been successfully targeted by a number of type II inhibitors. While it was once thought that only a few kinases are able to adopt this conformation, recent studies have demonstrated that it is accessible to many members of the human kinome (Choi et al., 2009; Dar et al., 2008; DiMauro et al., 2006; Han et al., 2009; Hodous et al., 2007; Mol et al., 2004; Schroeder et al., 2009; Seeliger et al., 2009; Wan et al., 2004). However, the sequence and structural determinants that allow a kinase to adopt the DFG-out conformation are still not known and it is not possible to predict which kinases will be sensitive to type II inhibitors. To address this issue, our goal was to identify a general ligand for kinases that adopt the DFG-out conformation. We have found that compounds based on the triazolo-pyridine scaffold, which we have previously demonstrated through kinetic and structural studies to bind the DFG-flipped forms of the kinases ABL and SRC, potently inhibit a broad spectrum of kinases that are sensitive to type II inhibitors. Furthermore, these compounds do not inhibit other well-characterized kinases that have not previously been observed in this specific inactive form.
A small panel of analogs based on the triazolo-pyridine scaffold was generated to determine how substituents that occupy the pocket created by the DFG-motif flipping to a more solvent exposed position affect kinase selectivity (Figure 1). As expected, a control compound that lacks a hydrophobic moiety that occupies the DFG-out Pocket and an amide group that makes a set of characteristic hydrogen bonds with the backbone amide of the DFG-motif and a conserved glutamic acid in the helix-αC does not inhibit any of the DFG-out-adopting kinases. Furthermore, an analog that contains a 3-trifluormethylbenzamide (1a), which is a general pharmacophore that occupies the DFG-out Pocket in many type II inhibitors, appears to be a non-selective ligand for kinases that have previously been characterized to adopt the DFG-out conformation. Unexpectedly, replacing this pharmacophore with functional groups from more selective type II inhibitors does not greatly enhance the selectivity profile for compounds based on the triazolo-pyridine scaffold. For example, compound 1b, which contains a 2-morphilinopyridyl group, is a nearly equipotent inhibitor of p38, ABL, SRC, HCK and CSK; however, inhibitors that display this moiety from a 4-anilinoquinazoline scaffold are >500-fold selective for p38 over these kinases (Perera and Maly, 2008). Similarly, 1c, which contains the functional groups present in the highly selective BCR-ABL inhibitor nilotinib, is a fairly non-selective inhibitor of DFG-out-adopting kinases. These data highlight that the ability of some type II inhibitors to discriminate between closely-related kinases is most likely due to several unique protein-ligand interactions rather than a single selectivity filter. Indeed, we have demonstrated that a unique interaction between the P-loop of ABL and imatinib makes a major contribution to the ability of this compound to inhibit this kinase over closely-related SRC (Seeliger et al., 2009). Several recent reports that describe type II inhibitors with limited selectivity profiles highlight that ligands of this class have the potential to inhibit a much broader spectrum of kinases than was previously appreciated (Angell et al., 2008; Choi et al., 2009; Okram et al., 2006).
To gain a better understanding of how inhibitors based on the triazolo-pyridine scaffold interact with protein kinases, we converted compound 1a into a fluorescently-labeled ligand that can be used in binding assays (Figure 2). The use of a binding assay allows the affinity of this probe to be determined for kinases in any activation state and facilitates the direct measurement of ligand binding kinetics. As many type II inhibitors have long residence times and slow on rates, determination of slow binding kinetics with a general probe for the DFG-out conformation can be used to verify the ability of a kinase to adopt this inactive form. In the presence of DFG-out-adopting kinases, fluorescent probe 4a demonstrated a concentration-dependent increase in fluorescence. An assay was developed to measure the residence time of this probe when bound to p38, SRC and ABL. The slow binding kinetics demonstrated by this probe are similar to those observed for other type II inhibitors of these kinases (Pargellis et al., 2002; Seeliger et al., 2007; Seeliger et al., 2009; Sullivan et al., 2005). This assay was also used to study how drug-resistance mutations affect the interactions of kinases with type II inhibitors. A mutation to the flexible P-loop region of ABL, E255V, was found to reduce the affinity of this kinase for the probe. Interestingly, the observed loss in affinity is due to a slower on rate rather than a slower rate of dissociation. None of the analogous P-loop mutations in SRC were found to affect the interaction of this kinase with type II inhibitors, which is consistent with this region not interacting with inhibitors. Finally, mutation of the gatekeeper residue of SRC to a bulkier residue was found to affect both the on and off rates of ligand binding. This result is consistent with recent findings that mutation of the gatekeeper residue in ABL causes increased conformational disturbances in a specific catalytic domain region of this kinase (Iacob et al., 2009).
While structural studies and kinetic assays can be used to determine if a kinase can adopt the DFG-out conformation, these methods can only be performed on purified proteins. Because it is difficult to know whether the conformational preferences of a recombinantly-expressed purified kinase are representative of those in a cell, probes that allow interrogation of endogenous kinases are highly desirable. For this reason, we developed a support-bound affinity matrix based on inhibitor 1a that allows an unbiased sampling of mammalian lysates. We first verified that our affinity matrix was capable of enriching purified kinases that are known to be sensitive to type II inhibitors from complex mixtures (Figure 4). Indeed, affinity matrices 14a and 15a selectively enriched SRC, HCK and p38 from an E. coli lysate but not kinases that are insensitive to type II inhibitors. Importantly, control matrix 14d, which does not contain a pharmacophore that occupies the specificity pocket, does not enrich any of the kinases tested. We next tested this reagent in mammalian cell lysates. First, it was verified that affinity matrix 14a was able to enrich endogenous SRC from HEK-293 cells. Analysis of the proteins that were selectively enriched with affinity resin 14a over control matrix 14d from HeLa and HEK-293 lysates demonstrated that there are several unique proteins. Mass spectrometric identification of these proteins is currently being performed.
A candidate approach using Western blot analysis was adopted to discover novel DFG-out-adopting kinases. We found that the STE20 kinase LOK could be selectively enriched from HEK-293 lysates with affinity matrix 14a. Furthermore, the purified catalytic domain could be enriched from E. coli lysates and was sensitive (IC50 < 20 nM) to type II inhibitors 1a-1c, 2a and3a in activity assays (Table S1). Finally, LOK demonstrated similarly slow binding kinetics with type II inhibitors as ABL, SRC and p38. Besides p38 and RAF, no other serine/threonine kinases have been characterized in the DFG-out conformation. Furthermore, no members of the STE20 kinase family have been found to be sensitive to type II inhibitors. Indeed, MAP3K5, PAK4 and PAK5, which are STE20 kinases and much more closely related to LOK than SRC or ABL, were not sensitive to any of the affinity reagents that we have developed. LOK is also of interest because it contains a residue with a bulky side-chain (isoleucine) at the gatekeeper position. To date, a vast majority of the kinases that have been characterized to adopt the DFG-out conformation (except Tie-2, PYK2 and c-MET) contain either a threonine or valine at this position. However, the presence of a larger gatekeeper residue does not preclude a kinase from adopting this form. While conversion of the threonione gatekeeper of ABL to an isoleucine results in this kinase being resistant to the type II inhibitors imatinib and nilotinib, several type II inhibitors that are extremely potent against this mutant have been described (Huang et al., 2009; O'Hare et al., 2009; Seeliger et al., 2009). Expanding the number and diversity of kinases that are able to stably adopt the DFG-out conformation will allow the identification of the sequence determinants that allow this transition.
SIGNIFICANCE
A number of type II inhibitors have been developed that target an inactive form of the ATP-binding site of kinases called the DFG-out conformation. Once thought to be energetically accessible to only a few kinases, the DFG-out conformation has been characterized with an increasing number of enzymes from this family. However, it is still not possible to predict which members of the kinome are able to stably adopt this inactive conformation. In addition, the structural and sequence determinants of this transition remain poorly understood. Here, we have identified a scaffold that appears to be a general pharmacophore for the DFG-out conformation. Derivatives of this scaffold are potent inhibitors of kinases that have previously been characterized to be sensitive to type II inhibitors. Furthermore, fluorophore-conjugated versions of these molecules allow the determination of the thermodynamics and kinetics of ligand binding to any activation state of the kinase catalytic domain. In addition, immobilized analogs of the general scaffold were found to be effective reagents for enriching DFG-out-adopting kinases from complex protein mixtures. Affinity reagents 14a and 15a were used to demonstrate that the STE20 kinase LOK behaves like other DFG-out adopting kinases. LOK is distantly related to other members of the kinase family that have been characterized in the DFG-out conformation. Use of these reagents to discover the full complement of kinases that are able to stably adopt the DFG-out conformation will allow a greater understanding of the thermodynamics and kinetics of the catalytic domains of this enzyme family.
EXPERIMENTAL PROCEDURES
Synthetic Methods
Detailed synthetic procedures are in the Supplemental Data.
Affinity Measurements with Probe 4a
Purified kinase (initial concentration = 250 nM, 2 fold serial dilution down to 0.2 nM) was incubated with 5 or 10 nM 4a in 120 μL of buffer containing 20 mM Tris (pH = 7.5), 2% DMSO, 100 mM KCl, 10% glycerol, 1 mM DTT, 0.05% Pluronic F-68 and 1 mM MgCl2. Fluorescence was read after 30 min of incubation at rt (excitation = 485 nm; emission = 535 nm). The Kd was determined by fitting the data to non-linear regression analysis (one site total binding) with Prism Graphpad software.
Determination of t1/2 with Probe 4a
Purified kinase (700 nM) was incubated with 4a (500 nM) in 120 μL of buffer containing 20 mM Tris (pH = 7.5), 2% DMSO, 100 mM KCl, 10% glycerol, 1 mM DTT, 0.05% Pluronic F-68 and 1 mM MgCl2. After 4 h, the mixture was diluted 30-fold ([4a] = 17 nM) into the same buffer containing 5 μM of 1a. Fluorescence was read every 10 min for 8 h and at 12 h (excitation = 485 nm; emission = 535 nm). Off rates (koff) and dissociative half lives (t1/2) were determined by fitting the data to a one phase exponential decay equation with Prism Graphpad software.
Generation of Affinity Matrix 14a
A modification of a previously published procedure was used. (Wissing et al., 2007). 1.5 mL of ECH Sepharose resin was washed with 10 bed volumes of H2O (pH = 4.5, 0.5 M NaCl), and 1:1 DMF/EtOH. 3 mL of 0.75 mM 12a and 0.46 mM 1a in 1:1 DMF/EtOH was added to the resin followed by 30 μL of DIEA and 450 μL of 1 M EDCI in 1:1 DMF/EtOH. The reaction mixture was rotated for 48 h and monitored by analytical HPLC (Figures S2 and S3). After 48 h, the reaction mixture was drained and washed with 2 bed volumes of 1:1 DMF/EtOH. The resin was then incubated with 56 μL ethanolamine, 933 μL of 20 mM acetic acid and 450 μL of EDCI in 1:1 DMF/EtOH for 12 h, followed by washing with 6 bed volumes of 1:1 DMF/EtOH (3x), 6 bed volumes of 0.5 M NaCl (2x) and 10 bed volumes of 20% EtOH. The resin was stored at 4°C in 20% EtOH until further use. Affinity matrices 14d and 15a were generated by the same method.
General Method for Affinity Enrichment from E. coli Lysate
50 μL of affinity matrix 14a, 14d or 15a was washed with 8 bed volumes of a buffer containing 50 mM HEPES (pH = 7.5), 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA and 100 mM NaCl. Purified kinase (4-18 μg) and E. coli lysate protein (500-800 μg) in 250 μL of binding buffer (50 mM HEPES (pH = 7.5), 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA, 20 mM MgCl2, 1 M NaCl and 100 μg/mL PMSF) was added to the resin and incubated for 2 h at 4°C. The resin was then drained, incubated with 8 bed volumes of binding buffer for 30 min at 4°C and drained again (4 washes). After the final wash, 1.5 bed volumes of 0.5% SDS in binding buffer was added to the resin and incubated for 10 min at rt. The eluted protein was collected and this process was repeated. All collected fractions were subjected to SDS-PAGE, transferred to nitrocellulose and subjected to Western blot analysis. Eluted fractions were also subjected to SDS-PAGE and stained with Silver Stain Xpress (Invitrogen).
General Method for Affinity Enrichment from Mammalian Cell Lysate
50-150 μL of affinity matrix 14a, 14d or 15a was washed with 8 bed volumes of a buffer containing 50 mM HEPES (pH = 7.5), 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA and 100 mM NaCl. HEK-293 or HeLa lysate proteins (0.6-8 mg) in 0.25-1.0 mL of buffer containing 50 mM HEPES (pH = 7.5), 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium orthovanadate, 1 mM PMSF, Roche protease inhibitor cocktail and 20 mM MgCl2 was added to the resin and incubated for 2 h at 4°C. The resin was then drained, incubated with 8 bed volumes of binding buffer for 30 min at 4°C and dra ined again (4 washes). After the final wash, 3-4 bed volumes of 0.5% SDS in binding buffer was added to the resin and incubated for 10 min at rt. The eluted protein was collected and this process was repeated. All collected fractions were lyophilized, subjected to SDS-PAGE, transferred to nitrocellulose and subjected to Western blot analysis with an α-non-phospho-SRC(Y416) or α-LOK antibody. Eluted fractions were subjected to SDS-PAGE and stained with Silver Stain Xpress (Invitrogen).
Supplementary Material
ACKNOWLEDGEMENTS
We thank John Kuriyan (UC, Berkeley) and Markus Seeliger (SUNY, Stony Brook) for providing expression plasmids for ABL (cat), SRC(cat), HCK(cat), HCK(3D), LCK(3D), SRC(3D) and ABL(3D). We thank Stefan Knapp (Structural Genomics Consortium (SGC)) for providing expression plasmids for CLK1, STK16, EPHA3, PAK4, PAK5, MAP3K5 and LOK. This work was supported by the National Institute of General Medical Science (R01GM086858 to D. J. M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, three figures and one table can be found with this article online at:
REFERENCES
- Angell RM, Angell TD, Bamborough P, Bamford MJ, Chung CW, Cockerill SG, Flack SS, Jones KL, Laine DI, Longstaff, et al. Biphenyl amide p38 kinase inhibitors 4: DFG-in and DFG-out binding modes. Bioorg. Med. Chem. Lett. 2008;18:4433–4437. doi: 10.1016/j.bmcl.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Choi Y, Syeda F, Walker JR, Finerty PJ, Jr., Cuerrier D, Wojciechowski A, Liu Q, Dhe-Paganon S, Gray NS. Discovery and structural analysis of Eph receptor tyrosine kinase inhibitors. Bioorg. Med. Chem. Lett. 2009;19:4467–4470. doi: 10.1016/j.bmcl.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem. Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat. Rev. Drug Discov. 2002;1:309–315. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- Cumming JG, McKenzie CL, Bowden SG, Campbell D, Masters DJ, Breed J, Jewsbury PJ. Novel, potent and selective anilinoquinazoline and anilinopyrimidine inhibitors of p38 MAP kinase. Bioorg. Med. Chem. Lett. 2004;14:5389–5394. doi: 10.1016/j.bmcl.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Dar AC, Lopez MS, Shokat KM. Small molecule recognition of c-Src via the Imatinib-binding conformation. Chem. Biol. 2008;15:1015–1022. doi: 10.1016/j.chembiol.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro EF, Newcomb J, Nunes JJ, Bemis JE, Boucher C, Buchanan JL, Buckner WH, Cee VJ, Chai L, Deak HL, et al. Discovery of aminoquinazolines as potent, orally bioavailable inhibitors of Lck: synthesis, SAR, and in vivo anti-inflammatory activity. J. Med. Chem. 2006;49:5671–5686. doi: 10.1021/jm0605482. [DOI] [PubMed] [Google Scholar]
- Engh RA, Bossemeyer D. The protein kinase activity modulation sites: mechanisms for cellular regulation - targets for therapeutic intervention. Adv. Enzyme Regul. 2001;41:121–149. doi: 10.1016/s0065-2571(00)00010-8. [DOI] [PubMed] [Google Scholar]
- Fabian MA, Biggs WH, III, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, Schwaller J, Sundstrom M, Knapp S. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. USA. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Mistry A, Chang JS, Cunningham D, Griffor M, Bonnette PC, Wang H, Chrunyk BA, Aspnes GE, Walker DP, et al. Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design. J. Biol. Chem. 2009;284:13193–13201. doi: 10.1074/jbc.M809038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Hodous BL, Geuns-Meyer SD, Hughes PE, Albrecht BK, Bellon S, Bready J, Caenepeel S, Cee VJ, Chaffee SC, Coxon A, et al. Evolution of a highly selective and potent 2-(pyridin-2-yl)-1,3,5-triazine Tie-2 kinase inhibitor. J. Med. Chem. 2007;50:611–626. doi: 10.1021/jm061107l. [DOI] [PubMed] [Google Scholar]
- Huang WS, Zhu X, Wang Y, Azam M, Wen D, Sundaramoorthi R, Thomas RM, Liu S, Banda G, Lentini SP, et al. 9-(Arenethenyl)purines as dual Src/Abl kinase inhibitors targeting the inactive conformation: design, synthesis, and biological evaluation. J. Med. Chem. 2009;52:4743–4756. doi: 10.1021/jm900166t. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc. Natl. Acad. Sci. USA. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MD, Caron PR, Hare BJ. Classifying protein kinase structures guides use of ligand-selectivity profiles to predict inactive conformations: Structure of lck/imatinib complex. Proteins. 2007;70:1451–1460. doi: 10.1002/prot.21633. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006;2:358–364. doi: 10.1038/nchembio799. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, Snell GP, Zou H, Sang BC, Wilson KP. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J. Biol. Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- O'Hare T, Eide CA, Deininger MW. Bcr-Abl kinase domain mutations and the unsettled problem of Bcr-AblT315I: looking into the future of controlling drug resistance in chronic myeloid leukemia. Clin. Lymphoma Myeloma. 2007;7(Suppl 3):S120–130. doi: 10.3816/clm.2007.s.012. [DOI] [PubMed] [Google Scholar]
- O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, Metcalf CA, 3rd, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16:401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okram B, Nagle A, Adrian FJ, Lee C, Ren P, Wang X, Sim T, Xie Y, Wang X, Xia G, et al. A general strategy for creating “inactive-conformation” abl inhibitors. Chem. Biol. 2006;13:779–786. doi: 10.1016/j.chembiol.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, et al. Inhibition of p38 MAP kinase by utilizing a novel allosteric binding site. Nat. Struct. Biol. 2002;9:268–272. doi: 10.1038/nsb770. [DOI] [PubMed] [Google Scholar]
- Pellicena P, Kuriyan J. Protein-protein interactions in the allosteric regulation of protein kinases. Curr. Opin. Struct. Biol. 2006;16:702–709. doi: 10.1016/j.sbi.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Perera BG, Maly DJ. Design, synthesis and characterization of “clickable” 4-anilinoquinazoline kinase inhibitors. Mol. BioSyst. 2008;4:542–550. doi: 10.1039/b720014e. [DOI] [PubMed] [Google Scholar]
- Quintas-Cardama A, Kantarjian H, Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat. Rev. Drug Discov. 2007;6:834–848. doi: 10.1038/nrd2324. [DOI] [PubMed] [Google Scholar]
- Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- Schroeder GM, An YM, Cai ZW, Chen XT, Clark C, Cornelius LAM, Dai J, Gullo-Brown J, Gupta A, Henley B, et al. Discovery of N-(4-(2-Amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a Selective and Orally Efficacious Inhibitor of the Met Kinase Superfamily. J. Med. Chem. 2009;52:1251–1254. doi: 10.1021/jm801586s. [DOI] [PubMed] [Google Scholar]
- Seeliger MA, Nagar B, Frank F, Cao X, Henderson MN, Kuriyan J. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Seeliger MA, Ranjitkar P, Kasap C, Shan Y, Shaw DE, Shah NP, Kuriyan J, Maly DJ. Equally potent inhibition of c-Src and Abl by compounds that recognize inactive kinase conformations. Cancer Res. 2009;69:2384–2392. doi: 10.1158/0008-5472.CAN-08-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard JR, Getlik M, Grütter C, Pawar V, Wulfert S, Rabiller M, Rauh D. Development of a fluorescent-tagged kinase assay system for the detection and characterization of allosteric kinase inhibitors. J. Am. Chem. Soc. 2009a;131:13286–13296. doi: 10.1021/ja902010p. [DOI] [PubMed] [Google Scholar]
- Simard JR, Kluter S, Grütter C, Getlik M, Rabiller M, Rode HB, Rauh D. A new screening assay for allosteric inhibitors of cSrc. Nat. Chem. Biol. 2009b;5:394–396. doi: 10.1038/nchembio.162. [DOI] [PubMed] [Google Scholar]
- Sullivan JE, Holdgate GA, Campbell D, Timms D, Gerhardt S, Breed J, Breeze AL, Bermingham A, Pauptit RA, Norman RA, et al. Prevention of MKK6-dependent activation by binding to p38alpha MAP kinase. Biochemistry. 2005;44:16475–16490. doi: 10.1021/bi051714v. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Bubis J, Toner-Webb J, Saraswat LD, First EA, Buechler JA, Knighton DR, Sowadski J. CAMP-dependent protein kinase: prototype for a family of enzymes. FASEB J. 1988;2:2677–2685. doi: 10.1096/fasebj.2.11.3294077. [DOI] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Wissing J, Jansch L, Nimtz M, Dieterich G, Hornberger R, Keri G, Wehland J, Daub H. Proteomics analysis of protein kinases by target class-selective prefractionation and tandem mass spectrometry. Mol. Cell. Proteomics. 2007;6:537–547. doi: 10.1074/mcp.T600062-MCP200. [DOI] [PubMed] [Google Scholar]
- Wong L, Jennings PA, Adams JA. Communication pathways between the nucleotide pocket and distal regulatory sites in protein kinases. Acc. Chem. Res. 2004;37:304–311. doi: 10.1021/ar020128g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.