Abstract
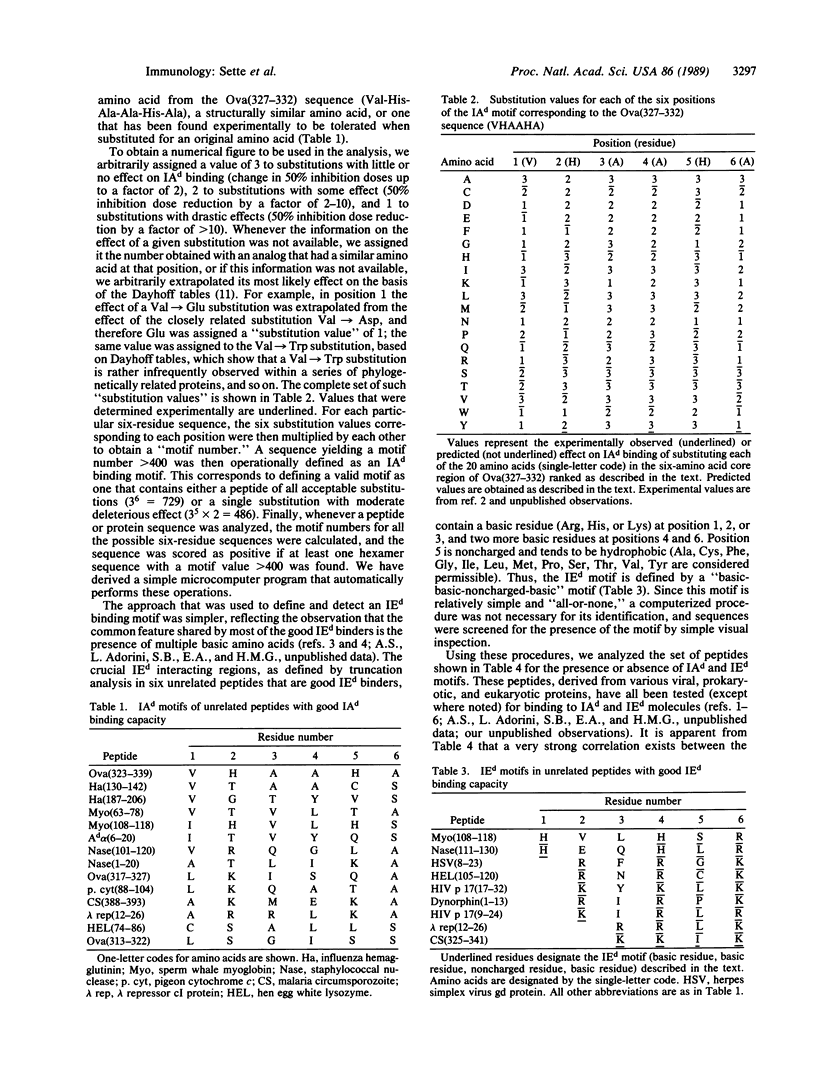
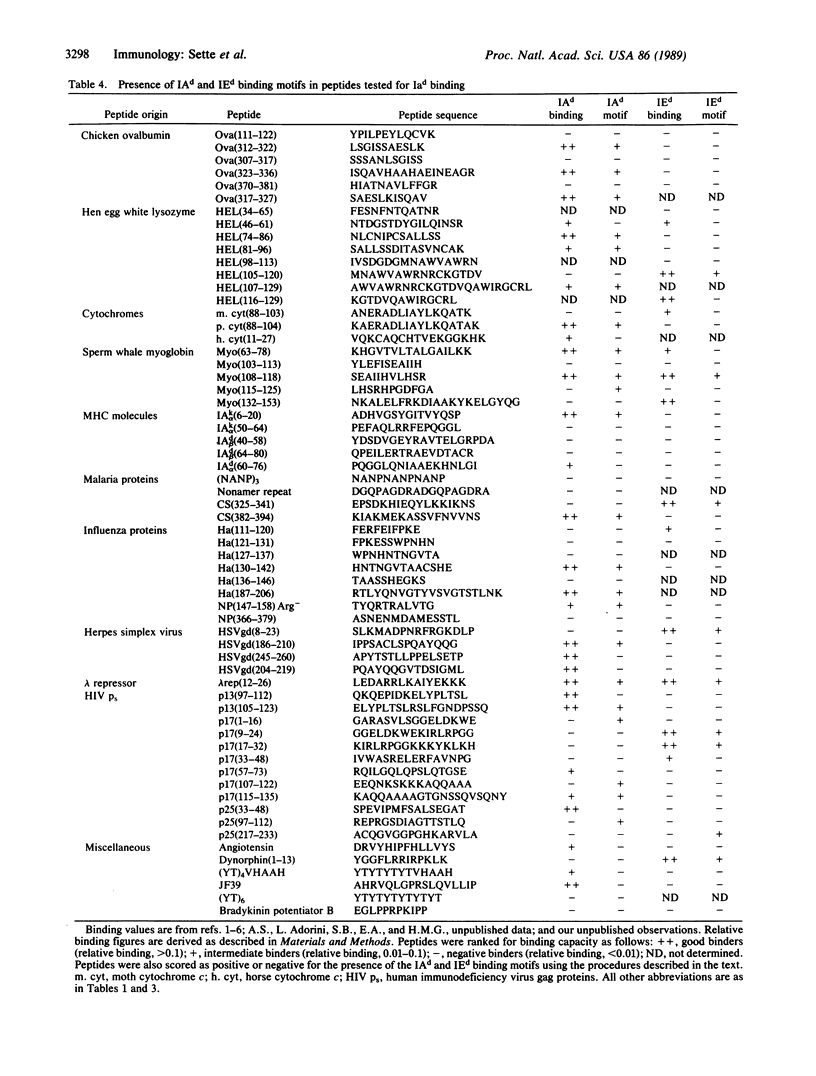
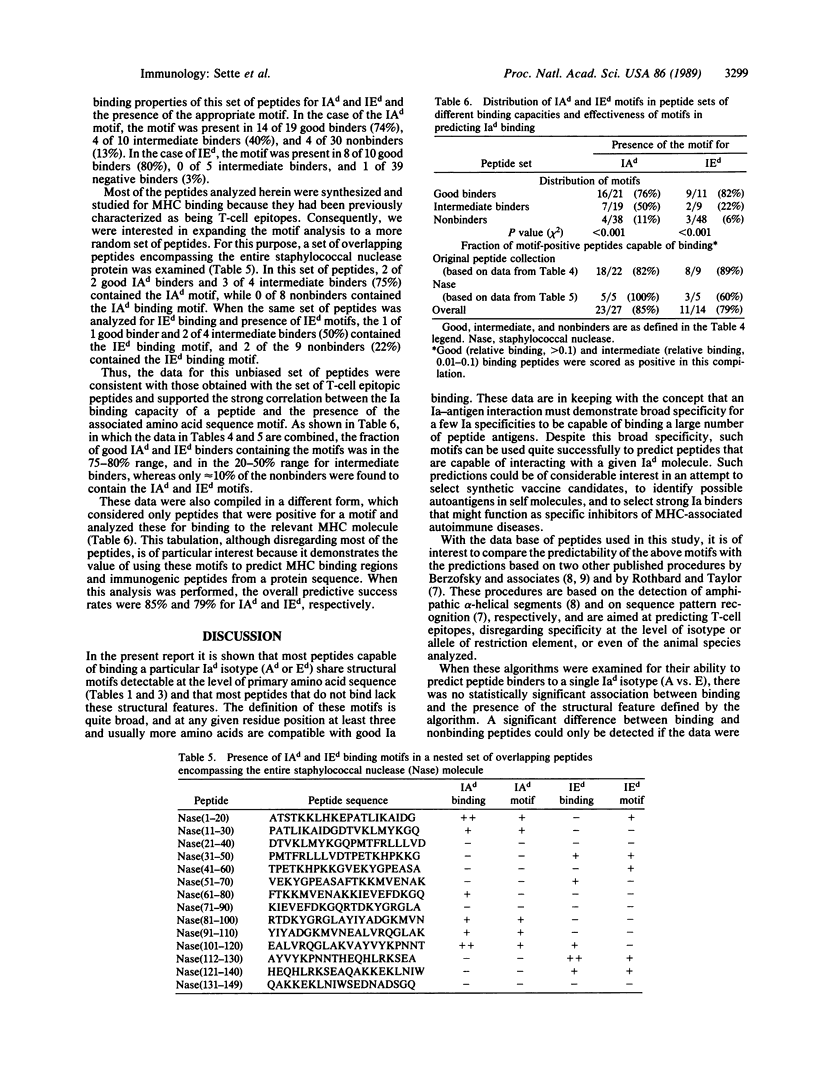
We have previously experimentally analyzed the structural requirements for interaction between peptide antigens and mouse major histocompatibility complex (MHC) molecules of the d haplotype. We describe here two procedures devised to predict specifically the capacity of peptide molecules to interact with these MHC class II molecules (IAd and IEd). The accuracy of these procedures has been tested on a large panel of synthetic peptides of eukaryotic, prokaryotic, and viral origin, and also on a set of overlapping peptides encompassing the entire staphylococcal nuclease molecule. For both sets of peptides, IAd and IEd binding was successfully predicted in approximately 75% of the cases. This suggests that definition of such sequence "motifs" could be of general use in predicting potentially immunogenic peptide regions within proteins.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adorini L., Sette A., Buus S., Grey H. M., Darsley M., Lehmann P. V., Doria G., Nagy Z. A., Appella E. Interaction of an immunodominant epitope with Ia molecules in T-cell activation. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5181–5185. doi: 10.1073/pnas.85.14.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buus S., Sette A., Colon S. M., Jenis D. M., Grey H. M. Isolation and characterization of antigen-Ia complexes involved in T cell recognition. Cell. 1986 Dec 26;47(6):1071–1077. doi: 10.1016/0092-8674(86)90822-6. [DOI] [PubMed] [Google Scholar]
- DeLisi C., Berzofsky J. A. T-cell antigenic sites tend to be amphipathic structures. Proc Natl Acad Sci U S A. 1985 Oct;82(20):7048–7052. doi: 10.1073/pnas.82.20.7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammon G., Shastri N., Cogswell J., Wilbur S., Sadegh-Nasseri S., Krzych U., Miller A., Sercarz E. The choice of T-cell epitopes utilized on a protein antigen depends on multiple factors distant from, as well as at the determinant site. Immunol Rev. 1987 Aug;98:53–73. doi: 10.1111/j.1600-065x.1987.tb00519.x. [DOI] [PubMed] [Google Scholar]
- Guillet J. G., Lai M. Z., Briner T. J., Buus S., Sette A., Grey H. M., Smith J. A., Gefter M. L. Immunological self, nonself discrimination. Science. 1987 Feb 20;235(4791):865–870. doi: 10.1126/science.2433769. [DOI] [PubMed] [Google Scholar]
- Margalit H., Spouge J. L., Cornette J. L., Cease K. B., Delisi C., Berzofsky J. A. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J Immunol. 1987 Apr 1;138(7):2213–2229. [PubMed] [Google Scholar]
- Rothbard J. B., Taylor W. R. A sequence pattern common to T cell epitopes. EMBO J. 1988 Jan;7(1):93–100. doi: 10.1002/j.1460-2075.1988.tb02787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Buus S., Colon S., Miles C., Grey H. M. I-Ad-binding peptides derived from unrelated protein antigens share a common structural motif. J Immunol. 1988 Jul 1;141(1):45–48. [PubMed] [Google Scholar]
- Sette A., Buus S., Colon S., Miles C., Grey H. M. Structural analysis of peptides capable of binding to more than one Ia antigen. J Immunol. 1989 Jan 1;142(1):35–40. [PubMed] [Google Scholar]
- Sette A., Buus S., Colon S., Smith J. A., Miles C., Grey H. M. Structural characteristics of an antigen required for its interaction with Ia and recognition by T cells. 1987 Jul 30-Aug 5Nature. 328(6129):395–399. doi: 10.1038/328395a0. [DOI] [PubMed] [Google Scholar]


