Abstract
We present a novel single-molecule method for rapidly evaluating small-molecule binding to individual DNA molecules using nanopores fabricated in ultrathin silicon membranes. A measurable shift in the residual ion current through a ~3.5 nm pore results from threading of a dye-intercalated DNA molecule, as compared to the typical residual current of native DNA. The average level of the residual current can be used to directly quantify the fraction of bound molecules to DNA, providing a new way to determine binding isotherms. Spatial sensitivity is also demonstrated by designing a two-segment DNA molecule that contains small-molecule binding sites in one of its two segments. Translocations of such molecules exhibit two current levels upon incubation with a DNA-binding dye, caused by selectively bound dye in one of the DNA segments. Our results, as shown here with four different dyes, coincide well with bulk fluorescence measurements performed under identical conditions. The nanopore approach for “reading-out” molecular binding along a DNA molecule, combined with the miniscule amounts of DNA required and the potential for scalability using nanopore arrays, provide a novel platform for future applications in analytical drug screening.
Selection of small molecules that bind genomic DNA or other nucleic acids with high specificity is a central requirement for drug development, necessitating new in vitro methods for rapid and low-cost assessment of the binding affinity and location of drugs along DNA molecules.1,2 While conventional, bulk spectroscopic tools (e.g., NMR, crystallography, mass spectrometry) have yielded a wealth of information on small-molecule binding to macromolecules,3 membrane-embedded nanopores have enjoyed remarkable success as nucleic acid analyzers. For example, membrane embedded alpha-hemolysin channels have been used to thread and detect variations in nucleic acid sequences,4 to investigate DNA–protein interactions,5 and to follow enzyme processivity.6 In addition, chiral nanopores of subnanometer dimensions have been used to discriminate enantiomeric drugs in solution,7 as well as to discriminate among the different mononucleotides in solution.8 Solid-state nanopores have only recently taken the stage as more versatile, synthetic analogs of protein channels, owing to their mechanical robustness and size tunability. Several applications of solid-state nanopores in biophysics have already been demonstrated, such as characterization of DNA duplexes by electromechanical unzipping,9 detection of DNA/protein complexes,10 and characterization of proteins11 and DNA.12–14
In this paper, we show that sub-5 nm solid-state nanopores can be used to quantitatively profile the binding of small molecules to both double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) molecules, potentially facilitating the detection of DNA/drug complexes with spatial sensitivity along the biopolymer. We present the first demonstration that the residual ion current signal through a nanopore during dsDNA translocation is highly affected by the amount of intercalated small molecules, which give the DNA/intercalator complex a bulkier structure than that of native DNA. Furthermore, we demonstrate the capability of using nanopores to discriminate within a DNA molecule among native and small molecule-bound regions, achieved using a ssDNA molecule containing two regions with different affinities toward a cyanine dye. Nucleic acid-binding fluorophores were used here to validate our nanopore data with bulk fluorescence measurements, illustrating the analytical capabilities of the nanopore method. Moreover, as we show here, our method offers a set of unique advantages over conventional techniques, such as not requiring chemical labeling with fluorescent reporters (which can often interact with DNA), detecting small molecule/DNA binding with high spatial resolution, and offering single-molecule sensitivity and rapid analysis.
Our setup, shown in Figure 1, is based on a Si-chip device containing a 20-nm-thick silicon nitride window, through which a nanoscale pore is drilled using the electron beam of a transmission electron microscope (TEM).15,16 A TEM image of a 3.5 nm pore used here is also shown. The cis and trans chambers, separated by the membrane, are filled with 1 M KCl electrolyte buffered to pH 8, such that an electrolyte junction between the chambers forms at the nanopore. When a voltage is applied across the membrane, the baseline ion current is measured (io). This current is reduced when a DNA molecule threads into the pore and linearly translocates through it, driven by the electric field. Reduction of the current signal during translocation is caused by the steric/electrostatic effects of the DNA segment inside the broad region of the nanopore, which due to its hourglass shape has an approximate length of 7 nm.16 We have previously shown that the average translocation speed is a strong function of the pore diameter, such that a short DNA fragment (100–1000 bp) translocates through a 3.5 nm pore with an average speed of ~0.3 μs/bp, yielding a current signal well within our measurement time resolution (~12 μs).14 Each recorded ion current transient corresponds to the translocation (or collision, see ref 14 for details) of a single-molecule and is characterized by its average current amplitude Δib = ῑo − ῑb, where ῑb and ῑo are average blocked-state and open-state currents for each translocation event, respectively (see Figure 1). Unless otherwise indicated, for each experiment we have collected thousands of single-molecule translocations to obtain Δib distributions, the average threading rate, and translocation time distributions.
Figure 1.
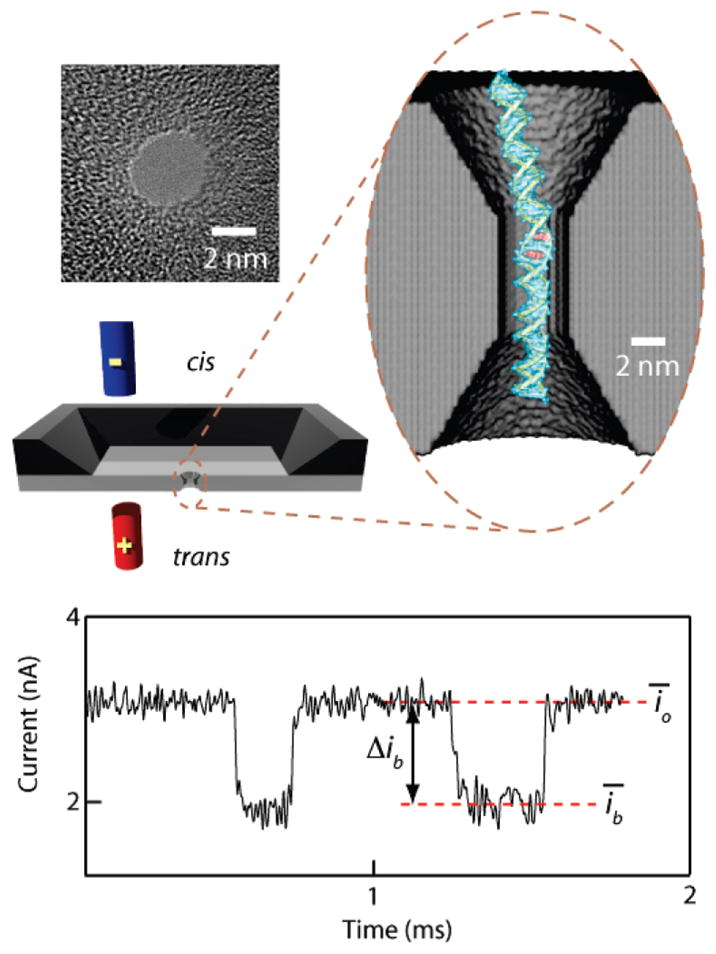
Solid-state nanopore setup for detecting the binding of small molecules to DNA. A pair of electrodes in solution is used to thread individual DNA molecules through a nanopore while recording the current of an electrolyte. Left: a TEM image of a 3.5 nm pore fabricated in an ultrathin silicon nitride membrane. Magnified oval: 3D rendering of a 3.5 nm nanopore with ethidium-intercalated double-stranded DNA. Bottom: ion current signal from DNA translocating through a 3.5 nm pore, in which the downward transients correspond to translocation of double-stranded DNA molecules (the time between events was reduced for clarity). For explanation of the definitions, see text.
Small Molecule Intercalation into Double-Stranded DNA
Intercalation is a common binding mode of many molecules to DNA, in which the molecule inserts itself between two adjacent basepairs.17 We first studied current signatures for a 400 bp DNA fragment translocating through a 3.5 nm pore, exposed to three intercalating dyes with different affinities to DNA (see Figure 2a). Typical single-molecule traces for a free 400 bp DNA fragment exhibit a single characteristic blocked current level of amplitude ~1 nA (Figure 2b), whereas for increasing ethidium bromide (EtBr) concentrations, we observe a deeper blockade level of amplitude ~1.5 nA (red level, Figure 2b). Interestingly, for increasing EtBr concentrations, a greater fraction of the event duration corresponds to the deeper blockage level, suggesting a correlation of the mean event amplitude, Δib, with EtBr loading. The larger blockade amplitude may be related to the EtBr loading fraction, since EtBr is known to widen the normal B-DNA cross section by about 15%.18 In addition, partial charge neutralization of the DNA from the positively charged EtBr may result in release of counterions from the DNA backbone, further reducing the effective ion concentration in the pore. The open pore current did not change by more than 0.2% for all EtBr concentrations.
Figure 2.
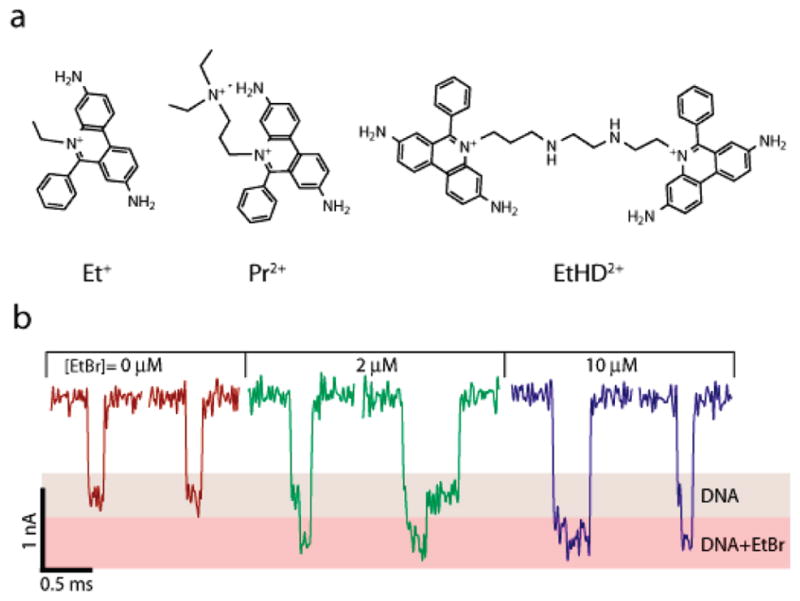
(a) Chemical structures of ethidium (Et+), propidium (Pr2+) and ethidium homodimer (EtHD2+). (b) Representative single-molecule 400 bp DNA translocation events through a 3.5 nm pore at different EtBr concentrations, showing a deeper blocking current amplitude for the DNA/Et+ complex (V = 300 mV for all experiments).
A more detailed correlation of Δib with EtBr loading is shown by nanopore titration experiments. As shown in Figure 3a, we have added aliquots of EtBr to the cis sample chamber and recorded >2000 translocations at each EtBr concentration. The surface plot in Figure 3a shows histograms of the event blockade amplitude from thousands of molecules per EtBr concentration, as a function of EtBr concentration. As we have previously reported, the major and minor Gaussian populations in each histogram correspond to translocations and collisions (brief current blockades with smaller event amplitudes of ~0.75 nA, attributed to DNA molecules that are not fully threaded into the pore), respectively.14 As the EtBr concentration increases, a noticeable shift in the position of the major peak of the current amplitude (red, defined here as ΔIb, see Figure 3a) is seen with negligible change in the collision peak (purple). The effect of EtBr on ΔIb suggests that the translocation signal is extremely sensitive to the additive fraction on the DNA. The dependence of ΔIb on EtBr concentration is shown in Figure 3b, displaying >25% increase in the average event amplitude in the EtBr concentration range used. Analogous binding fraction curves obtained from fluorescence experiments performed in bulk are superimposed on the nanopore data set (see Supporting Information for fluorescence measurements) and show excellent agreement with a measured dissociation constant Kd = 14.7 μM.
Figure 3.
Nanopore titration of a DNA/EtBr complex. (a) Current amplitude histograms for a 400 bp DNA fragment as a function of EtBr concentration. A shift to increasing amplitudes is seen for the translocation peak (major population, red), whereas the collision peak (minor population, purple) remains unchanged. (b) The peak current amplitude value (ΔIb) for ethidium/DNA as a function of ethidium concentration. Each point in the graph is determined from >2000 translocations events, and the error bars are smaller than the marker size. The overlaid curve is a best fit to the fraction bound dye obtained from fluorescence measurements (right axis, see Supporting Information), exhibiting good agreement with the nanopore data.
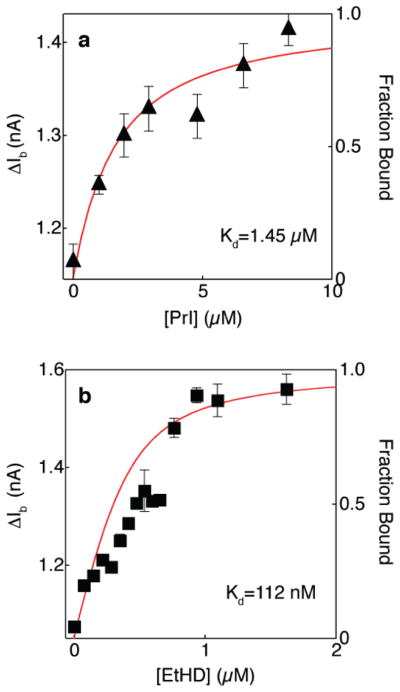
In order to evaluate the generality of the nanopore method, we repeated the experiments performed with EtBr using two other DNA intercalator dyes, propidium (Pr2+) and ethidium homodimer (EtHD2+). These dyes display stronger binding affinities to B-DNA as compared to EtBr, mainly due to their double charge. As described above for EtBr, the peak event amplitudes ΔIb were measured for each of these dyes. Figure 4 displays the dependence of ΔIb on dye concentrations for propidium (a) and for ethidium homodimer (b). Bulk fluorescence measurements of binding to the same DNA molecules are overlaid, displaying good agreement with the nanopore measurements. We also note that as expected, the EtHD2+ displayed a stronger shift (>50%) in the event amplitude, due to its bulkier structure. As with the EtBr experiments, the addition of either PR2+ or EtHD2+ did not appreciably change the open pore current.
Figure 4.
Nanopore titration of 400 bp DNA using propidium (a) and ethidium homodimer (b). The peak current amplitude value (ΔIb) is plotted as a function of dye concentrations (symbols). The overlaid curves are best fit to the fraction bound dye obtained from fluorescence measurements (right axis, see Supporting Information).
Charged DNA intercalators are also known to affect the mobility of DNA in an electric field. For example, it is well known that a DNA stained with EtBr displays retarded mobility in gel electrophoresis as compared to unstained DNA, due to the decreased effective charge and increased stiffness of the polymer.19 These molecular changes to DNA structure upon binding of cationic intercalators have resulted in similar trends with our nanopores. In Table 1, average capture rates (normalized to concentration) and translocation times are given, both before and after the addition of intercalator to saturation. First, we note that average capture rates decreased by a factor of 3–5 upon intercalation of Et+, Pr2+, or EtHD2+. Furthermore, a closer look at average translocation times reveals a slightly higher degree of retardation for the divalent intercalators EtHD and PrI (factor of 5–6) than for EtBr (factor of 4), expected for a DNA with a greater charge reduction. These findings are consistent with the reduced gel electrophoretic mobility of the intercalator/DNA complex (see Supporting Information), manifested in the case of nanopores by suppression of DNA capture and slowed transport through the pore. Apart from the reduced effective charge of the intercalator/DNA complex, other factors that may contribute to slowing down may be (1) increased contour length (and stiffness) of the intercalator/DNA complex,20 (2) increased interactions of the complex with the pore walls, already shown to reduce the DNA velocity in small pores,14 (3) increased stall force of the intercalator/DNA complex, which has a larger cross-section than free DNA, arising from hydrodynamic drag inside the pore.21–23
Table 1.
Average Capture Rates and Translocation Times for Free and Intercalator-Bound DNA
| capture rate (s−1nM−1) |
translocation time (ms) |
|||
|---|---|---|---|---|
| molecule | free DNA | drug-bound | free DNA | drug-bound |
| Et+ | 0.57 ± 0.02 | 0.13 ± 0.01 | 0.11 ± 0.01 | 0.44 ± 0.04 |
| Pr2+ | 0.65 ± 0.02 | 0.12 ± 0.01 | 0.09 ± 0.02 | 0.55 ± 0.06 |
| EtHD2+ | 0.52 ± 0.02 | 0.15 ± 0.01 | 0.10 ± 0.01 | 0.52 ± 0.04 |
Interaction with Single-Stranded DNA
In this part of our study we have used a 2.2 nm pore (see Figure 5a) to probe the interaction of single-stranded DNA molecules with the cyanine dye SYBR Green II (SGII), an RNA-selective stain that preferentially intercalates into single-stranded DNA over double-stranded DNA (Invitrogen Corp., Carlsbad, CA). SGII has particularly low affinity toward a deoxyadenine homopolymer (see Supporting Information), presumably because the purine bases of the homopolymer undergo extensive stacking interactions, resulting in a helical secondary structure.24,25 Using fluorescence measurements, we have determined the binding affinity of SGII to poly(dA) to be >50 times smaller than for a random sequence (see Supporting Information). Since SGII binds with high affinity to a random single-stranded DNA sequence, for our nanopore experiments we have designed a two-segment dA50dN50 molecule (see Supporting Information for sequence) for which SGII provides spatial contrast along the molecule by binding only to the “random sequence” dN region (see Figure 5a). As a negative control, the response of the dA50dN50 to SGII was compared to a dA60 homopolymer molecule.
Figure 5.

Nanopore detection of small-molecule binding to single-stranded DNA. (a) TEM image of a 2.2 nm pore, and the molecules used in this study. Since SYBR Green II (SGII) binds to poly(dA) with very low affinity, dA60 was used as a negative control and dA50dN50 as a positive control. (b) Typical translocation events for dA60, before and after the addition of SGII to the cis chamber (side histogram is an all-point histogram of the current data). Typical translocation events for dA50dN50, before and after the addition of SGII, are shown in (c) and (d), respectively. The two-step events prevalent after SGII binding were characterized by amplitudes of 0.58 and 0.96 nA, corresponding to dA50 and SGII-bound dN50, respectively (see text).
In Figure 5b, a typical concatenated set of translocation events for dA60 through a 2.2 nm pore is shown, both before and after the addition of SGII to the cis chamber. All experiments here were carried out at 5 °C, in order to reduce translocation speeds and to stabilize the poly(dA) secondary structure. As expected, these experiments do not reveal any differences in the poly(dA) translocation properties before and after the addition of SGII with both exhibiting mean current amplitudes of ~0.58 nA and a peak translocation time of ~80 μs. In contrast, typical translocation events for the dA50dN50 sample with SGII (Figure 5d) display strikingly long translocation times and exhibit a second, deeper blocked current level. The all-point histograms for these events clearly show that the deeper blockade is a discrete level with an amplitude of 0.96 nA, much greater than the blockade amplitude for the DNA alone (0.60–0.62 nA).
The translocation time distributions for the dA60 and dA50dN50 samples, shown in Figure 6a,b, respectively, show that SGII binding considerably slows the translocation process (histograms shown in log time units). While the mean translocation time scales for dA60 both before and after the addition of SGII were 0.11 ± 0.03 ms, mean translocation times for dA50dN50 increased from 0.25 ± 0.04 ms to 7.1 ± 1.0 ms upon SGII binding. A closer inspection of the relative time scales of the two blocking levels reveals that >80% of a typical event is spent at the shallow blockade amplitude (i = 0.74 nA). It is also interesting to note that for the vast majority of events (>95%), the shallow blockade level precedes the deeper blockade level. These findings suggest that entry of the dA50 portion of the molecule is favored over entry of the bulkier SGII-bound dN50 portion. This favored entry can be also rationalized by considering the positive charge of SGII, which may decrease the overall charge of the dN50 region upon binding SGII, and/or condense the DNA structure to a coil. Although we do not have an exact mechanism to account for the long dwell times, we note two main possibilities: (1) SGII-binding results in formation of secondary structure by condensing the DNA, thereby stalling the translocation process, or (2) the SGII-bound DNA is bulky enough to interact with the SiN membrane and the pore. However, since the translocation events suggest that the poly(dA) portion is in the pore for most of the translocation process, we favor the explanation that SGII-binding induces secondary structure which stalls entry of the dN50 portion of the molecule into the pore. This conclusion is consistent with the literature, as certain DNA-binding molecules, including clinical drugs, have been shown to induce secondary structure in single-stranded DNA molecules.26
Figure 6.

Translocation-time distributions for dA60 (a) and dA50dN50 (b), both before and after the addition of excess SGII (see text for discussion).
In summary, we have developed a novel, high-throughput single-molecule method for evaluating small molecule binding to DNA. Our method was validated using a set of DNA intercalating fluorophores for which we obtained a complementary set of fluorescence titration curves. For double-stranded DNA, we used three intercalating dyes with different binding affinities to DNA in the range Kd = 10−7−10−5 M. Single-molecule current traces for partially intercalated DNA reveal bilevel shapes with deeper blockades correlated to the fraction of intercalator molecules bound to the DNA and with deeper blockades for larger dyes. The mean fraction of deeper blockades, which corresponds very well to the fraction of bound dye as measured by fluorescence, allows binding curves for DNA-binding drugs to be obtained at high sensitivity. Furthermore, we have shown that binding of small molecules to single-stranded DNA can be analyzed using 2 nm pores, revealing clear differences between the region of the ssDNA molecule that binds the fluorophores and a poly(dA) region that does not bind the dye. Nanopore profiling of DNA interactions with small molecules is extremely rapid, uses only ~103–104 DNA copies, and can be performed using short or long DNAs. Since the method is essentially “label-free”, that is, it does not require fluorogenic or radioactive probes, it is well-suited for future discovery of high affinity, sequence-specific nucleic acid-targeting drugs for epigenetic profiling and applications in environmental and molecular toxicity.
Supplementary Material
Acknowledgments
We acknowledge useful discussion with M. Frank-Kamenetskii, help in sample preparation from B. McNally and J. Larkin, and financial support from NIH award HG-004128, and NSF award PHY-0646637. We thank A. Squires for comments on the manuscript.
Footnotes
Supporting Information Available: (1) Details on the experimental setup and (2) fluorescence titration data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Thurston DE. Br J Cancer. 1999;80:65–85. [PubMed] [Google Scholar]
- 2.Palchaudhuri R, Hergenrother PJ. Curr Opin Biotechnol. 2007;18(6):497–503. doi: 10.1016/j.copbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Krugh TR. Curr Opin Struct Biol. 1994;4(3):351–364. [Google Scholar]
- 4.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Biophys J. 1999;77(6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Nat Methods. 2007;4(4):315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 6.Cockroft SL, Chu J, Amorin M, Ghadiri MR. J Am Chem Soc. 2008;130(3):818–20. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang XF, Cheley S, Guan X, Bayley H. J Am Chem Soc. 2006;128(33):10684–5. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- 8.Astier Y, Braha O, Bayley H. J Am Chem Soc. 2006;128:1705–10. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- 9.McNally B, Wanunu M, Meller A. Nano Lett. 2008;8:3418–3422. doi: 10.1021/nl802218f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, Sligar S, Aksimentiev A, Timp G. Nano Lett. 2007;7(6):1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fologea D, Ledden B, McNabb DS, Li JL. Appl Phys Lett. 2007;91(5):053901. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Phys Rev E. 2005;71(5):051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 13.Fologea D, Brandin E, Uplinger J, Branton D, Li J. Electrophoresis. 2007;28(18):3186–3192. doi: 10.1002/elps.200700047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wanunu M, Sutin J, McNally B, Chow A, Meller A. Biophys J. 2008;95(10):4716–25. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Nat Mater. 2003;2(8):537–40. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 16.Kim MJ, Wanunu M, Bell DC, Meller A. Adv Mater. 2006;18(23):3149–3155. [Google Scholar]
- 17.Waring MJ, Chaires JB. Topics in Current Chemistry. Vol. 253. Springer: Berlin; 2005. DNA Binders and Related Subjects. [Google Scholar]
- 18.Sobell HM, Tsai C, Jain SC, Gilbert SG. J Mol Biol. 1977;114(3):333–365. doi: 10.1016/0022-2836(77)90254-6. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen PE, Zhen WP, Henriksen U, Buchardt O. Biochemistry. 1988;27(1):67–73. doi: 10.1021/bi00401a012. [DOI] [PubMed] [Google Scholar]
- 20.Lepecq JB, Paoletti C. J Mol Biol. 1967;27(1):87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 21.Wong CTA, Muthukumar M. J Chem Phys. 2007;126:164903. doi: 10.1063/1.2723088. [DOI] [PubMed] [Google Scholar]
- 22.Luan B, Aksimentiev A. Phys Rev E. 2008;78(2):021912. doi: 10.1103/PhysRevE.78.021912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorp SV, Keyser UF, Dekker NH, Dekker C, Lemay SG. Nat Phys. 2009;5:347–51. [Google Scholar]
- 24.Holcomb DN, Tinoco I. Biopolymers. 1965;3(2):121–133. [Google Scholar]
- 25.Saenger W, Riecke J, Suck D. J Mol Biol. 1975;93(4):529–534. doi: 10.1016/0022-2836(75)90244-2. [DOI] [PubMed] [Google Scholar]
- 26.Adamcik J, Valle F, Witz G, Rechendorff K, Dietler G. Nanotechnology. 2008;19(38):384016. doi: 10.1088/0957-4484/19/38/384016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.