Abstract
Hair follicles undergo continuous cycles of growth, involution and rest. This process, referred to as the hair growth cycle, has a periodicity of weeks to months. At the same time, skin and hair follicles harbor a functional circadian clock that regulates gene expression with a periodicity of approximately twenty four hours. In our recent study we found that circadian clock genes play a role in regulation of the hair growth cycle during synchronized hair follicle cycling, uncovering an unexpected connection between these two timing systems within skin. This work, therefore, indicates a role for circadian clock genes in a cyclical process of much longer periodicity than twenty four hours.
Keywords: Circadian clock, cell cycle, hair growth cycle, aging, hair loss
The hair growth cycle
Beginning after completion of hair morphogenesis (postnatal day 14 in the mouse), hair growth cycles commence with catagen, an involution process of the hair follicle during which the majority of its epithelial compartments undergo apoptosis [1]. This stage is followed by telogen during which the hair follicle remains in relative quiescence. Telogen is interrupted by activation of epithelial stem and progenitor cells located in specialized stem cell compartments in the bulge and secondary hair germ, followed by rapid proliferation and differentiation of progeny keratinocytes; this growth phase is referred to as anagen [2]. In mice, the two initial hair growth cycles are synchronized such that the majority of hair follicles are in a similar stage of the hair growth cycle at a given time. But as the mouse ages, the hair growth cycles become progressively less synchronized [3].
Circadian gene expression is hair growth cycle dependent
In order to systematically discover transcriptional activity associated with the hair growth cycle, we profiled mRNA expression at a genome-wide level over multiple time points corresponding to morphogenesis and two synchronized hair growth cycles. Interestingly, a large fraction of the genome, more than six thousand genes, exhibits changes in expression that correlates with the progression of the hair growth cycle, thus underscoring the complexity of this process [4,5]. One of the surprises that came from this study was the finding that genes regulated by the core circadian clock mechanism showed expression changes that correlated with the hair growth cycle, with highest expression during the telogen-anagen transition.
On a molecular level, the circadian clock consists of positive and negative feedback loops. At its core are the bHLH-PAS transcriptional activators CLOCK and BMAL1 (ARNTL), which form a heterodimer and activate target genes containing E-boxes in their enhancer regions, including Periods (Per1, 2 and 3) and Cryptochromes (Cry1 and 2). PERs and CRYs form heterodimeric complexes that translocate into the nucleus where they inhibit BMAL1-CLOCK transcriptional activity, thus constituting the negative feedback loop [6]. In other words, the PER/CRY complex inhibits its own expression, allowing for reactivation of BMAL1/CLOCK leading to rhythmic expression with a periodicity of 24 hours. Several other proteins, including kinases, play an important role in generation of rhythmic expression [7]. The CLOCK-BMAL1 heterodimer activates other genes as well, including Dbp, Tef, Hlf, and Rev-Erbα, which codes for an orphan nuclear receptor. REV-ERBα regulates transcription of Bmal1 and other target genes by binding to retinoic acid-related orphan receptor response elements (ROREs) [8]. The clock genes that we identified as upregulated in telogen/early anagen were all CLOCK/BMAL1 target genes, including Pers, Dbp and Rev-Erbα. While these genes show a clear circadian pattern of expression in skin, as was previously demonstrated [9-11], their amplitude was higher during telogen and early anagen (Figure 1), indicating that in skin, the expression of clock controlled genes is dependent both on circadian mechanism and the hair growth cycle.
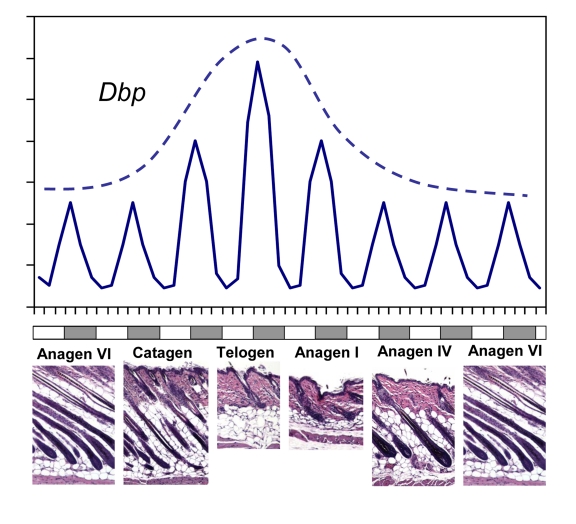
Figure 1. CLOCK-controlled gene expression in skin has a circadian pattern and correlates with synchronized hair growth cycles.
A schematic diagram showing rhythmic circadian expression of clock controlled gene Dbp over different phases of the hair growth cycle (solid line). The circadian amplitude of Dbp expression correlates with progression of the hair follicle cycle with highest expression during telogen (broken line). Skin histology for representative hair growth cycle stages is shown below. Note that this schematic does not show the actual length of each phase of the hair growth cycle.
Since our mRNA expression studies were performed using whole skin, we asked which compartments of the skin and hair follicles contribute to the robust rhythmic circadian gene expression in telogen. The hair follicle contains several functionally and structurally distinct compartments, including the bulge region, which harbors slow-cycling hair follicle stem cells [12]; the secondary hair germ, which contains actively cycling stem and progenitor cells [13-15]; and the dermal papilla, a source of signals for activating the stem cells at the beginning of anagen [16] (Figure 2). While in situ hybridization studies revealed that all cell types of the skin express circadian clock genes, the site of most prominent rhythmic circadian gene expression during telogen and early anagen was the secondary hair germ. This compartment, strategically positioned between the dermal papilla and the bulge, contains proliferative Lrg5-positive stem cells thought to have migrated from the bulge during late catagen and early telogen [13,17]. The secondary hair germ cells are the first to be activated during anagen initiation, giving rise to transient amplifying cells of the hair matrix and eventually differentiating into the hair shaft [15,17,18]. Additionally, our data shows that as anagen progresses, the circadian amplitude within the hair follicle proper becomes dampened, while the circadian amplitude in the dermis and interfollicular epidermis continues to be robust. Interestingly, suspension of circadian rhythm has been previously noted in other highly proliferative and differentiating tissues, including testis and thymus [19-21].
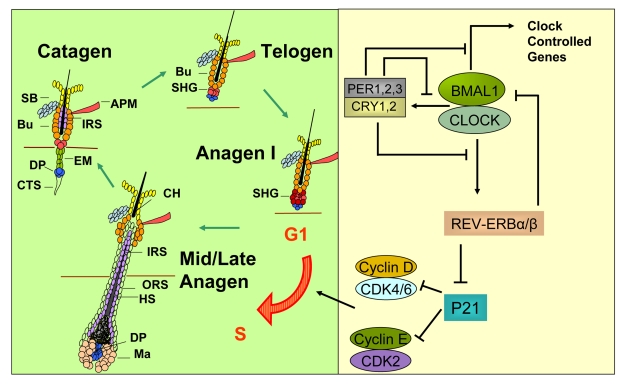
Figure 2. A model for how circadian clock genes participate in regulation of the synchronized hair growth cycle through regulation of cell cycle progression in the secondary hair germ.
The hair growth cycle (left panel) is a continuous process consisting of the quiescent telogen phase followed by the growth phase (anagen) where signals, presumably originating in the dermal papilla, activate stem and progenitor cell proliferation leading to growth and differentiation of the hair shaft. Anagen is followed by catagen where the lower two-thirds of the follicle undergo apoptosis, sparing the stem cell compartments and the dermal papilla. The CLOCK/BMAL1 complex is at the core of the mammalian circadian clock mechanism (right panel). It activates multiple genes, collectively referred to as clock controlled genes. Among these genes are Per1,2,3 and Cry1,2 whose protein products translocate into the nucleus to inhibit the transcriptional activity of the CLOCK/BMAL1 complex. Rev-erbα is another clock controlled gene whose protein product negatively regulates expression of Bmal1. Additionally, REV-ERBα directly inhibits expression of the G1-S cell cycle inhibitor p21WAF1/CIP. In the absence of BMAL1, downregulation of Rev-erbα leads to high P21 expression and G1 arrest in the hair germ cells during anagen I of the hair follicle cycle, thus delaying anagen progression. APM - arrector pili muscle, Bu - bulge, CH - club hair, CTS - connective tissue sheath, DP - dermal papilla, EM - epithelial membrane, HS - hair shaft, IRS - inner root sheath, Ma - matrix, ORS - outer root sheath, SB - sebaceous gland, SHG - secondary hair germ.
Role for circadian clock genes in hair growth cycling
The robust circadian clock gene expression within the secondary hair germ led us to test the possibility that circadian clock genes might play a role in the hair growth cycle. For these studies, we turned to Clock and Bmal1 mutant mouse models. We found a significant delay in anagen progression in both mutants and this delay was more pronounced in Bmal1 deficient mice, possibly due to partial functional redundancy between Clock and its homologue Npas2. Clock and Bmal1 mutant mice have no visible defects in hair follicle morphogenesis and enter the first stage of anagen, characterized by the expansion of the secondary hair germ, at approximately the same time (post-natal day 22). Yet by day 28, when the majority of hair follicles in control littermates have developed hair matrix and hair shaft with the hair bulb growing into the subcutis, the Bmal1 mutant mice remained in the first anagen phase [5]. After experiencing a nearly week-long delay, the Bmal1 deficient hair follicles resumed normal progression of the hair cycle. There were no abnormalities in the structure of the mature anagen follicles in the Bmal1 or Clock mutant mice, supporting the idea that circadian clock genes are primarily involved in timing mechanisms during the telogen-anagen transition.
Further analysis revealed absence of mitotic cells in the early anagen secondary hair germ in Bmal1 mutant hair follicles, while wild-type secondary hair germs at the same stage contained mitotic cells. Importantly, epidermis and dermis of Bmal1 mutant mice contained mitotic cells, indicating that the proliferation defect was hair follicle specific. Phosphorylated Retinoblastoma Protein (Rb), a marker of cell cycle progression through the G1-S cell cycle checkpoint [22], was absent in the secondary hair germ of Bmal1 mutant hair follicles while it was abundant in control mice. These results indicate that in Bmal1 mutant hair follicles, progenitor cells of the early anagen secondary hair germ are arrested at the G1-S cell cycle checkpoint.
To gain insights into the molecular mechanisms underlying the G1 arrest of progenitor cells in the secondary hair germ, we profiled gene expression in the skin of Bmal1 deficient mice during telogen. As expected, the expression of multiple known CLOCK-BMAL1 target genes was affected, including that of Rev-Erbα, which was downregulated approximately fifteen fold. Studies in hepatocytes have demonstrated that REV-ERBα directly represses expression of the gene encoding the G1 cell cycle inhibitor p21WAF1/CIP [23], and consistently p21 is upregulated approximately 2.5 fold in Bmal1 mutant skin. These findings led us to propose that hair growth cycling in Bmal1 mutant mice is delayed due to upregulation of p21, leading to slowed G1-S cell cycle progression in progenitor cells of the secondary hair germ (Figure 2). These results are consistent with the known extensive crosstalk between the circadian clock and the cell division cycle [24]. We have also considered the possibility that circadian gene regulation of the hair growth might involve a mechanism that "counts" the number of circadian peaks to regulate timing in the hair growth cycle. However, results from our preliminary experiments in mice entrained to 22 and 26 hour days argue against this possibility.
In the mouse, the first two hair growth cycles are synchronized. After completion of the second telogen, which can last up to thirty days, the coat begins to grow asynchronously in complex domains created by waves of anagen moving through the domain until a wave reaches "refractory" telogen, an area of skin unresponsive to the propagating anagen stimulus. As the mouse ages, this process creates increasingly complex patterns of hair growth with each domain consisting of a telogen competent to be activated, a propagating anagen wave, a catagen, and a refractive telogen [3,25]. In preliminary experiments, we did not observe differential expression of clock controlled genes in skin corresponding to different hair growth phases in asynchronously cycling skin, suggesting the possibility that hair cycle related regulation of clock gene expression may be particularly important in synchronized hair follicle cycling.
One plausible role for circadian mechanisms in the hair growth cycle is in animals with seasonal hair growth, commonly found in mammals living in the wild [26-30]. In these animals, the circadian clock-regulated hormones melatonin and prolactin are thought to be key regulators of seasonal changes in hair growth [31-34]. Intriguingly, seasonal hair growth has been found to be regulated at the telogen-anagen transition; in several breeds of sheep, the winter coat is in telogen and in the spring when duration of daylight increases, hair follicles enter anagen and the winter hair fibers are shed [35]. Therefore telogen, and specifically the secondary hair germ, could serve as an important interpreter of photic hair growth cycle timing in animals bearing seasonal fur.
The aging hair follicle
Hair loss and hair graying are commonly recognized symptoms of aging in mammals. In addition to the visible location of hair, the highly regenerative nature of hair follicles may explain why hair loss is a prominent feature of aging syndromes. Several mouse models with premature aging phenotypes show progressive hair loss or graying [36-38], and human conditions with progeria-like symptoms, such as Werner Syndrome and Hutchinson-Gilford Progeria, present with premature hair loss or hair graying [39]. The common form of human hair loss, androgenetic alopecia, shows a clear age-related progression [40]. In addition, some authorities have argued for a distinct age-related entity, referred to as senescent alopecia [40-43]. This syndrome as well as androgenetic alopecia are characterized by a reduction in the large diameter pigmented (terminal) hair and an increased prevalence of thin (vellus-like) hair [40]. Thus, both syndromes are thought to represent hair growth cycle defects characterized by increased telogen to anagen hair follicle ratio due to a shortened anagen phase and persistent telogen follicles [42,44].
Age-associated hair graying has been linked to ultraviolet light and reactive oxygen species (ROS)-induced cell damage. The hair follicle bulge harbors melanocyte stem cells that give rise to mature melanocytes which synthesize and secrete hair pigments during anagen. Graying human hair follicles have been shown to contain melanocytes with accumulated oxidative stress [45], and consistent with this finding, genotoxic stress induced by ionizing radiation in mice leads to premature differentiation of melanocyte stem cells, followed by stem cell depletion and hair graying [46]. Furthermore, deficiency in the ATM gene enhanced ectopic differentiation of the melanocyte stem cells [46]. Additionally, both Werner Syndrome and Hutchinson-Gilford Progeria are associated with accumulation of DNA damage [47]. Together, this data suggests that hair graying may be due to melanocyte stem cell depletion caused by UV radiation and genotoxic ROS.
The mechanisms underlying senescent alopecia have not been extensively studied. However, a plausible hypothesis is that analogous to hair graying, alopecia is related to loss of hair follicle epithelial stem cells either through decreased renewal, premature differentiation, apoptosis or cellular senescence. While the link between genotoxic stress and hair graying has been established using mouse models, only correlative data from human patients are available in regards to life-long UV exposure and hair loss [40,45]. Skin and hair follicles are heavily bombarded by UV radiation and also contain actively dividing keratinocytes, a likely source of mito-chondrial ROS.
Circadian clock and the aging hair follicle
Recent studies indicate that circadian clock proteins may be involved in DNA repair and in regulating accumulation of cellular ROS, thus making them plausible actors in the aging processes [48]. Fu et. al. demonstrated that a mutation in the Per2 gene leads to an increase in tumor development as well as hair graying and hair loss after gamma irradiation [49]. PER1 is also known to interact with ATM and CHK2, thus affecting the proper initiation of double strand break repair [50]. In addition, recent work from several groups has revealed an important link between the circadian clock and cellular metabolism [51-54]. This work suggests that cell division and DNA synthesis are temporally segregated from the oxidative phase of the metabolic cycle [55], raising the possibility that the circadian clock has evolved to coordinate cell division with cellular metabolism, thus minimizing DNA damage. Furthermore, the NADH/NAD+ ratio and heme, indicators of the redox state, have been shown to directly modulate activity of circadian clock proteins, suggesting that the circadian clock can read and interpret the cellular metabolic state [56]. In addition, SIRT1, the mammalian orthologue of yeast SIR2, is a conserved NAD+-dependent protein deacetylase that deacetylates BMAL1 and PER2, and functions as a histone deacetylase at clock-regulated promoters [51,52,57]. SIR2 and its orthologues are important regulators of longevity in yeast, worms and flies [58-60], and in mice, several studies demonstrate SIRT1 contribution to genome stability and DNA repair [61,62]. Together, these data suggest the possibility that sirtuins could regulate longevity in part through circadian clock mechanisms [56].
Direct evidence for clock involvement in the aging process comes from the study of Kontdravov et. al. [38], showing that the lifespan of Bmal1 mutant mice is decreased by half and that the mice exhibit a range of premature aging phenotypes. Among the phenotypes reported by the authors of this study were age-related lens and cornea defects, reduced subcutaneous fat, and hair regeneration defects, pointing to strong effect of the Bmal1 deletion in exposed cutaneous tissues, including hair follicles. The authors demonstrate that by thirty weeks of age, Bmal1 deficient mice accumulate significantly more ROS than control animals, thus potentially explaining the progeria-like phenotype. While our work focused on younger Bmal1 mutated mice [5], prior to the development of aging symptoms, the finding of circadian clock involvement in cell cycle progression within the secondary hair germ may provide a partial explanation for the hair regeneration defect. Alternatively, in the absence of BMAL1, deregulation of DNA damage and oxidative stress responses could cause depletion of stem populations necessary for hair regeneration. Furthermore, in several rodent models there is deregulated suprachiasmatic nucleus electric activity and photic entrainment as well as abnormal periodicity and amplitude of circadian gene expression during the normal aging process [63-66]. Thus, a decline in the robustness of circadian rhythms may contribute to the aging process.
In conclusion, our study demonstrated that circadian clock genes can regulate the non-circadian cyclical hair growth cycle, presumably via an effect on the progression of the cell cycle in a progenitor cell compartment of the hair follicle, the secondary hair germ [5]. We speculate that circadian genes may play a role in aging-related alopecia which is characterized by aberrations in the hair growth cycle.
Acknowledgments
This work was supported by NIH grant AR44882 (to B.A.) and California Breast Cancer Research Fellowship 14GB-0163 (to M.G.). We thank Ambica Bhandari and Amelia Soto for reading the manuscript.
Footnotes
The authors of this manuscript have no conflict of interest to declare.
References
- 1.Paus R, Foitzik K. In search of the "hair cycle clock": a guided tour. Differentiation. 2004;72:489–511. doi: 10.1111/j.1432-0436.2004.07209004.x. [DOI] [PubMed] [Google Scholar]
- 2.Schneider MR, Schmidt-Ullrich R, Paus R. The hair follicle as a dynamic miniorgan. Curr Biol. 2009;19:R132–142. doi: 10.1016/j.cub.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Plikus MV, Mayer JA, de la Cruz D. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin KK, Chudova D, Hatfield GW, Smyth P, Andersen B. Identification of hair cycle-associated genes from time-course gene expression profile data by using replicate variance. Proc Natl Acad Sci U S A. 2004;101:15955–15960. doi: 10.1073/pnas.0407114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin KK, Kumar V, Geyfman M. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009;5:e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 Spec No 2:R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 8.Ukai-Tadenuma M, Kasukawa T, Ueda HR. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–1163. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- 9.Bjarnason GA, Jordan RC, Wood PA. Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol. 2001;158:1793–1801. doi: 10.1016/S0002-9440(10)64135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanello SB, Jackson DM, Holick MF. Expression of the circadian clock genes clock and period1 in human skin. J Invest Dermatol. 2000;115:757–760. doi: 10.1046/j.1523-1747.2000.00121.x. [DOI] [PubMed] [Google Scholar]
- 11.Tanioka M, Yamada H, Doi M. Molecular clocks in mouse skin. J Invest Dermatol. 2009;129:1225–1231. doi: 10.1038/jid.2008.345. [DOI] [PubMed] [Google Scholar]
- 12.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 13.Ito M, Kizawa K, Hamada K, Cotsarelis G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation. 2004;72:548–557. doi: 10.1111/j.1432-0436.2004.07209008.x. [DOI] [PubMed] [Google Scholar]
- 14.Panteleyev AA, Jahoda CA, Christiano AM. Hair follicle predetermination. J Cell Sci. 2001;114:3419–3431. doi: 10.1242/jcs.114.19.3419. [DOI] [PubMed] [Google Scholar]
- 15.Jaks V, Barker N, Kasper M. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 16.Botchkarev VA, Botchkareva NV, Sharov AA, Funa K, Huber O, Gilchrest BA. Modulation of BMP signaling by noggin is required for induction of the secondary (nontylotrich) hair follicles. J Invest Dermatol. 2002;118:3–10. doi: 10.1046/j.1523-1747.2002.01645.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greco V, Chen T, Rendl M. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–69. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez JD, Sehgal A. The thymus is similar to the testis in its pattern of circadian clock gene expression. J Biol Rhythms. 2005;20:111–121. doi: 10.1177/0748730404274078. [DOI] [PubMed] [Google Scholar]
- 20.Morse D, Cermakian N, Brancorsini S, Parvinen M, Sassone-Corsi P. No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol Endocrinol. 2003;17:141–151. doi: 10.1210/me.2002-0184. [DOI] [PubMed] [Google Scholar]
- 21.Okamura H. Clock genes in cell clocks: roles, actions, and mysteries. J Biol Rhythms. 2004;19:388–399. doi: 10.1177/0748730404269169. [DOI] [PubMed] [Google Scholar]
- 22.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 23.Grechez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–4342. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 24.Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 25.Plikus MV, Widelitz RB, Maxson R, Chuong CM. Analyses of regenerative wave patterns in adult hair follicle populations reveal macro-environmental regulation of stem cell activity. Int J Dev Biol. 2009;53:857–868. doi: 10.1387/ijdb.072564mp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler MP, Zucker I. Seasonal pelage changes are synchronized by simulated natural photoperiods in Siberian hamsters (Phodopus sungorus) J Exp Zool A Ecol Genet Physiol. 2009;311:475–482. doi: 10.1002/jez.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebling FJ, Alexander IH, Urbanski HF, Hastings MH. Effects of N-methyl-D-aspartate (NMDA) on seasonal cycles of reproduction, body weight and pelage colour in the male Siberian hamster. J Neuroendocrinol. 1995;7:555–566. doi: 10.1111/j.1365-2826.1995.tb00792.x. [DOI] [PubMed] [Google Scholar]
- 28.al-Khateeb A, Johnson E. Action of hormones on seasonal changes of pelage in the vole. J Endocrinol. 1969;43:43–44. [PubMed] [Google Scholar]
- 29.Rust CC, Shackelford RM, Meyer RK. Hormonal control of pelage cycles in the mink. J Mammal. 1965;46:549–565. [PubMed] [Google Scholar]
- 30.Randall VA, Thornton MJ, Messenger AG, Hibberts NA, Loudon AS, Brinklow BR. Hormones and hair growth: variations in androgen receptor content of dermal papilla cells cultured from human and red deer (Cervus elaphus) hair follicles. J Invest Dermatol. 1993;101:114S–120S. doi: 10.1111/1523-1747.ep12363039. [DOI] [PubMed] [Google Scholar]
- 31.Kauffman AS, Cabrera A, Zucker I. Energy intake and fur in summer- and winter-acclimated Siberian hamsters (Phodopus sungorus) Am J Physiol Regul Integr Comp Physiol. 2001;281:R519–527. doi: 10.1152/ajpregu.2001.281.2.R519. [DOI] [PubMed] [Google Scholar]
- 32.Rose J, Oldfield J, Stormshak F. Apparent role of melatonin and prolactin in initiating winter fur growth in mink. Gen Comp Endocrinol. 1987;65:212–215. doi: 10.1016/0016-6480(87)90168-7. [DOI] [PubMed] [Google Scholar]
- 33.Rose J, Stormshak F, Oldfield J, Adair J. Induction of winter fur growth in mink (Mustela vison) with melatonin. J Anim Sci. 1984;58:57–61. doi: 10.2527/jas1984.58157x. [DOI] [PubMed] [Google Scholar]
- 34.Sarkar M, Prakash BS. Circadian variations in plasma concentrations of melatonin and prolactin during breeding and non-breeding seasons in yak (Poephagus grunniens L.) Anim Reprod Sci. 2005;90:149–162. doi: 10.1016/j.anireprosci.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Rogers GE. Biology of the wool follicle: an excursion into a unique tissue interaction system waiting to be re-discovered. Exp Dermatol. 2006;15:931–949. doi: 10.1111/j.1600-0625.2006.00512.x. [DOI] [PubMed] [Google Scholar]
- 36.Tyner SD, Venkatachalam S, Choi J. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 37.Sun LQ, Lee DW, Zhang Q. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamenisch Y, Berneburg M. Progeroid syndromes and UV-induced oxidative DNA damage. J Investig Dermatol Symp Proc. 2009;14:8–14. doi: 10.1038/jidsymp.2009.6. [DOI] [PubMed] [Google Scholar]
- 40.Trueb RM. Aging of hair. J Cosmet Dermatol. 2005;4:60–72. doi: 10.1111/j.1473-2165.2005.40203.x. [DOI] [PubMed] [Google Scholar]
- 41.Kligman AM. The comparative histopathology of male-pattern baldness and senescent baldness. Clin Dermatol. 1988;6:108–118. doi: 10.1016/0738-081x(88)90074-0. [DOI] [PubMed] [Google Scholar]
- 42.Courtois M, Loussouarn G, Hourseau C, Grollier JF. Ageing and hair cycles. Br J Dermatol. 1995;132:86–93. doi: 10.1111/j.1365-2133.1995.tb08630.x. [DOI] [PubMed] [Google Scholar]
- 43.Courtois M, Loussouarn G, Hourseau C, Grollier JF. Hair cycle and alopecia. Skin Pharmacol. 1994;7:84–89. doi: 10.1159/000211279. [DOI] [PubMed] [Google Scholar]
- 44.Ellis JA, Sinclair RD. Male pattern baldness: current treatments, future prospects. Drug Discov Today. 2008;13:791–797. doi: 10.1016/j.drudis.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 45.Arck PC, Overall R, Spatz K. Towards a "free radical theory of graying": melanocyte apoptosis in the aging human hair follicle is an indicator of oxidative stress induced tissue damage. FASEB J. 2006;20:1567–1569. doi: 10.1096/fj.05-4039fje. [DOI] [PubMed] [Google Scholar]
- 46.Inomata K, Aoto T, Binh NT. Genotoxic stress abrogates renewal of melanocyte stem cells by triggering their differentiation. Cell. 2009;137:1088–1099. doi: 10.1016/j.cell.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 47.Cox LS, Faragher RG. From old organisms to new molecules: integrative biology and therapeutic targets in accelerated human ageing. Cell Mol Life Sci. 2007;64:2620–2641. doi: 10.1007/s00018-007-7123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antoch MP, Kondratov RV. Circadian proteins and genotoxic stress response. Circ Res. 2010;106:68–78. doi: 10.1161/CIRCRESAHA.109.207076. [DOI] [PubMed] [Google Scholar]
- 49.Fu L, Pelicano H, Liu J, Huang P, Lee C. The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell. 2002;111:41–50. doi: 10.1016/s0092-8674(02)00961-3. [DOI] [PubMed] [Google Scholar]
- 50.Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 51.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324:654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakahata Y, Kaluzova M, Grimaldi B. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yin L, Wu N, Curtin JC. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 54.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 55.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 56.Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Asher G, Gatfield D, Stratmann M. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 58.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–6003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 60.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang RH, Sengupta K, Li C. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oberdoerffer P, Michan S, McVay M. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Effects of aging on central and peripheral mammalian clocks. Proc Natl Acad Sci U S A. 2002;99:10801–10806. doi: 10.1073/pnas.152318499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sutin EL, Kilduff TS. Circadian and light-induced expression of immediate early gene mRNAs in the rat suprachiasmatic nucleus. Brain Res Mol Brain Res. 1992;15:281–290. doi: 10.1016/0169-328x(92)90119-v. [DOI] [PubMed] [Google Scholar]
- 65.Van Reeth O, Zhang Y, Reddy A, Zee P, Turek FW. Aging alters the entraining effects of an activity-inducing stimulus on the circadian clock. Brain Res. 1993;607:286–292. doi: 10.1016/0006-8993(93)91518-w. [DOI] [PubMed] [Google Scholar]
- 66.Kolker DE, Fukuyama H, Huang DS, Takahashi JS, Horton TH, Turek FW. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J Biol Rhythms. 2003;18:159–169. doi: 10.1177/0748730403251802. [DOI] [PubMed] [Google Scholar]