Abstract
Background
MFN1032 is a clinical Pseudomonas fluorescens strain able to grow at 37°C. MFN1032 cells induce necrosis and apoptosis in rat glial cells at this temperature. This strain displays secretion-mediated hemolytic activity involving phospholipase C and cyclolipopeptides. Under laboratory conditions, this activity is not expressed at 37°C. This activity is tightly regulated and is subject to phase variation.
Results
We found that MFN1032 displays a cell-associated hemolytic activity distinct from the secreted hemolytic activity. Cell-associated hemolysis was expressed at 37°C and was only detected in vitro in mid log growth phase in the presence of erythrocytes. We studied the regulation of this activity in the wild-type strain and in a mutant defective in the Gac two-component pathway. GacS/GacA is a negative regulator of this activity. In contrast to the Pseudomonas fluorescens strains PfO-1 and Pf5, whose genomes have been sequenced, the MFN1032 strain has the type III secretion-like genes hrcRST belonging to the hrpU operon. We showed that disruption of this operon abolished cell-associated hemolytic activity. This activity was not detected in P.fluorescens strains carrying similar hrc genes, as for the P. fluorescens psychrotrophic strain MF37.
Conclusions
To our knowledge this the first demonstration of cell-associated hemolytic activity of a clinical strain of Pseudomonas fluorescens. Moreover, this activity seems to be related to a functional hrpU operon and is independent of biosurfactant production. Precise link between a functional hrpU operon and cell-associated hemolytic activity remains to be elucidated.
Background
Pseudomonas fluorescens is a highly heterogeneous species, as shown the extensive literature on the taxonomy and phylogeny of this species [1-4]. These studies include saprophytic, rhizopheric and phytopathogenic strains of P. fluorescens, illustrating the ubiquity of this species. Most studies describe P. fluorescens as a psychrotrophic bacterium unable to grow at temperatures greater than 32°C and therefore as an avirulent bacterium in humans. Nevertheless, previous studies of the infectious potential of P. fluorescens have demonstrated that the rifampicin spontaneous mutant MF37 [5] derived from the environmental psychrotrophic strain MF0 [6] can bind specifically to the surface of neurons and glial cells [7]. This adhesion to the host cell is associated with the induction of apoptosis and necrosis in glial cells [8]. Lipopolysaccharides (LPS) produced or released by P. fluorescens have a clear role in cytotoxicity, but other factors released at the same time during adhesion also seem to be essential for the virulence of this bacterium [9]. Thus the various enzymes secreted by this species may also be considered as potential high virulence factors [5].
We recently demonstrated that the clinical strain MFN1032 is a Pseudomonas fluorescens sensus stricto Biovar1 strain able to grow at 37°C [10]. This strain has hemolytic activity mediated by secreted factors, similar to the hemolytic activity seen for the opportunistic pathogen Pseudomonas aeruginosa, involving phospholipase C (PlcC) and biosurfactant [11]. Under specific conditions, MFN1032 forms colonies of phenotypic variants, which are defective in secreted hemolysis. Spontaneous mutations of the genes encoding the two-component regulatory system GacS/GacA have been identified as the cause of phenotypic variation in one such group of variants. We hypothesized that phenotypic variation increases the virulence potential of this strain. However these group variants (group 1 variants) do not produce secondary metabolites and have impaired biofilm formation [12]. Then, these results suggested that virulence of MFN1032 is not dependent solely on secreted factors or LPS and thus must involve other factors.
Some bacterial virulence factors are only expressed in the presence of eukaryotic cells. This is the case of the type III secretion system (TTSS), one of the most frequently described contact dependent secretion systems in Pseudomonas. TTSSs are found in many Gram-negative pathogens. They allow the direct translocation of bacterial effector proteins into the cytoplasm of eukaryotic host cells. P. aeruginosa uses the TTSS to translocate four effector proteins (ExoS, ExoT, ExoU, and ExoY) with antihost properties [13]. The P. aeruginosa TTSS consists of nearly 40 genes, regulated in a coordinated manner and encoding structural components of the secretion and translocation machinery, effectors proteins, and regulatory factors [14]. Transcription of the TTSS is induced under calcium-limited growth conditions or following intimate contact of P. aeruginosa with eukaryotic host cells [15]. Pseudomonas syringae pv. tomato DC3000 is a phytopathogenic bacterium that harbors a gene cluster hrp (for hypersensitive reaction and pathogenicity). This hrp cluster is essential for pathogenicity in susceptible plants and for the ability to elicit the hypersensitive reaction in nonhosts or resistant cultivars of host plants [16]. hrp genes are expressed in planta or in media mimicking plant apoptotic conditions [17]. Sequence analyses have uncovered a shared subset of nine hrp genes that were renamed hrc (for hrp and conserved) and that encode proteins homologous to Yersinia ysc gene products [18]. The existence of these genes suggests evolutionary conservation of molecular mechanisms of pathogenicity used by both mammalian and phytopathogenic bacteria [19].
In P. fluorescens, the presence of the hrc genes belonging to hrpU operon depends on the strain. The feature of TTSS and the origin of hrc genes remain to clarify in this species [20-23].
In the present study, we describe the detection of cell-associated hemolytic activity of P. fluorescens MFN1032 in contact with sheep erythrocytes. This hemolytic activity was compared with the hemolytic activity of other P. fluorescens strains: a spontaneous MFN1032 gacA mutant and the opportunistic pathogen Pseudomonas aeruginosa CHA [24]. Cell-associated hemolytic activity and its regulation were compared with the activity and regulation of the previously described secreted hemolytic activity of MFN1032. We then looked for hrc genes in our strain and determined their role in the cell-associated hemolytic activity of MFN1032, using hrpU operon disruption mutant.
Results
MFN1032 displays cell-associated hemolytic activity
Hemolytic activity of Pseudomonas fluorescens biovar I MFN1032 and Pseudomonas aeruginosa CHA (positive control for TTSS-mediated hemolysis) was measured by the technique employed by Dacheux [25], adapted as described in methods.
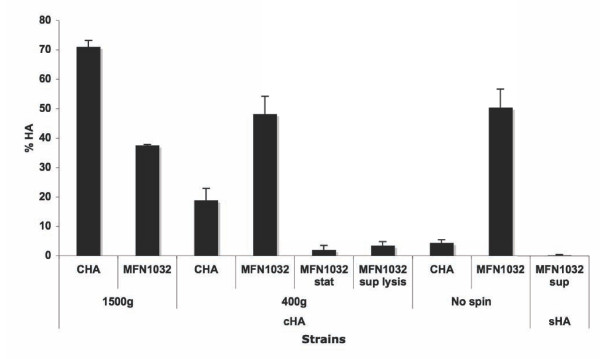
Bacteria were grown at 37°C to mid exponential growth phase and were used at a multiplicity of infection (MOI) of 1, without spin (which enhance contact between bacteria and RBCs). CHA induced lysis of 5% of red blood cells (RBCs) and MFN1032, 50% lysis, within 1 hour at 37°C. Hemolytic activity of CHA was increased by a 10 min centrifugation at 400 g (20% lysis) or 1500 g (70% lysis). By contrast, the hemolytic activity of MFN1032 was unchanged after a 10 min centrifugation at 400 g and reduced by centrifugation at 1500 g (35% lysis) (Figure 1). For further experiments we used a 10 min centrifugation at 400 g since this protocol is allowing close contact between bacterial cells and RBCs and appears compatible with maximum lysis by MFN1032. Supernatants from MFN1032 cells tested in the same conditions had no hemolytic activity. Additionally, we collected supernatants from RBC lysed by MFN1032. Supernatants were filtered and incubated with fresh RBCs for 1 h at 37°C. This supernatant from lysed RBC samples did not induce further RBC lysis. Thus, the factor mediating RBC lysis is not a factor released into the supernatant, but is dependent on the presence of MFN1032 cells.
Figure 1.
Comparison of the hemolytic activity of MFN1032 and CHA and characterization of MFN1032 cell-associated hemolysis. Hemolysis of RBCs (% HA) incubated with MFN1032 and CHA, at 37°C and with a multiplicity of infection (MOI) of 1. Cells were subjected or not to centrifugation at 1500 g or 400 g for 10 min to enhance cell-cell contact. cHA indicates cell-associated hemolytic activity and sHA indicates secreted hemolytic activity. MFN1032 sup indicates MFN1032 cell-free supernatant. MFN1032 stat indicates MFN1032 cells in stationary growth phase. MFN1032 sup lysis indicates supernatants obtained after RBC lysis by MFN1032. Hemolytic activity was measured as described in the materials and methods. Results are means of at least three independent experiments. Standard deviation is shown.
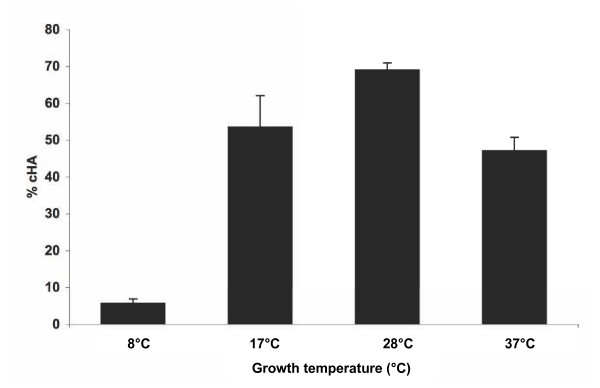
MFN1032 cells from cultures grown to the exponential growth phase at various temperatures were incubated with RBCs for 1 h at 37°C. MFN1032 bacteria grown at 17°C and 37°C showed the same levels of hemolysis (50% of RBCs lysed), whereas bacteria grown at 8°C were almost devoid of hemolytic activity (5% lysis). The maximal hemolytic activity of MFN1032 was observed at 28°C (70% lysis), the optimal growth temperature of this strain (Figure 2).
Figure 2.
Influence of growth temperature on MFN1032 cell-associated hemolytic activity. Cell-associated hemolytic activity (cHA %) was measured for MFN1032 grown at 8°C, 17°C, 28°C (optimum growth temperature) or 37°C, as described in the materials and methods. Results are means of at least three independent experiments. Standard deviation is shown. Contact was enhanced by centrifugation at 400 g for 10 min.
Lysis of RBCs is caused by a pore-forming toxin from MFN1032
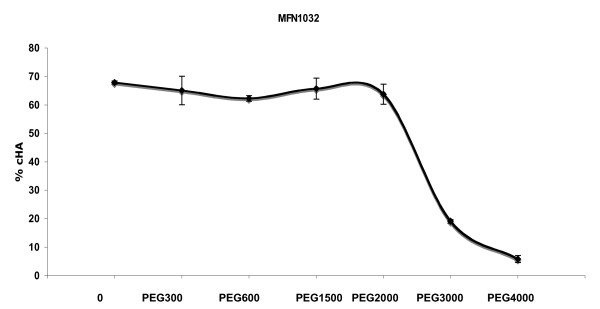
We investigated the nature of the factor involved in RBC lysis by osmoprotection experiments. Osmoprotectants protect RBCs against osmotic shock provoked by bacterial pore-forming toxins. We used different sized molecules in hemolysis experiments to estimate the size of the pore formed in the RBC membrane (Figure 3). We did not observe any effects on hemolysis with PEG300, PEG600, PEG1500 or PEG2000. Molecules larger than PEG2000 protected against MFN1032 cell-associated hemolysis as observed for PEG3000. A maximal level of protection was reached with PEG4000, resulting in the protection of 90% of RBCs against this hemolytic process. Based on these results, we estimated the size of the pore formed in RBC membranes by MFN1032 is between 2.4 nm and 3.2 nm.
Figure 3.
Protection of RBCs from cell-associated hemolysis by osmoprotectants. Omoprotectants were added at a final concentration of 30 mM. All experiments were performed at least three times in triplicate. MFN1032 was grown at 28°C. Standard deviation is shown.
MFN1032 cell-associated hemolytic activity is higher than in other Pseudomonas fluorescens strains
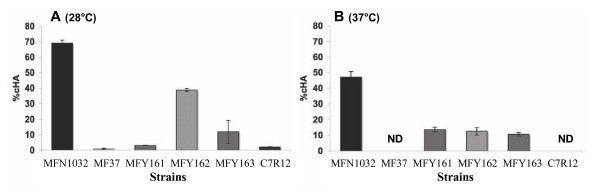
We tested the hemolytic activity of MFN1032, MFY162 (a clinical isolate of Pseudomonas fluorescens Biovar I), MFY161 and MFY163 (clinical isolates of Pseudomonas mosselli) [10], MF37 (Pseudomonas fluorescens biovar V, derived from a psychrotrophic environmental strain) and C7R12 (a psychrotrophic rhizospheric strain) cells at 28 and 37°C.
The hemolytic effect of MFN1032 cells was much higher than the other strains tested, at both growth temperatures (Figure 4). At 28°C, MFY162 was the only other strain showing high levels of hemolytic activity (40% lysis); MFY161 and MFY163 displayed only weak hemolytic activity (5-10% lysis). All clinical isolates showed some hemolytic activity (15% lysis) at 37°C, but at a lower level than that observed for MFN1032 one's. The environmental strains tested were not hemolytic at 28°C and did not grow at 37°C.
Figure 4.
Cell-associated hemolytic activity of different fluorescent Pseudomonas strains. Cell-associated hemolytic activity (cHA %) was measured as described in the materials and methods. Results are means of at least three independent experiments. Standard deviation is shown. A: Hemolysis of RBCs incubated with MFN1032, MF37, C7R12, MFY161, MFY162, MFY163 at 28°C and MOI of 1. Contact was enhanced by centrifugation at 400 g for 10 min. B: Hemolysis of RBCs incubated with MFN1032, MF37, C7R12, MFY161, MFY162, MFY163 at 37°C and MOI of 1. Contact was enhanced by centrifugation at 400 g for 10 min. ND: not determined. MF37 and C7R12 were unable to grow at 37°C.
The hemolytic activities of MFN1032, MFY162 and MFY161, were maximal at their optimal growth temperature (28°C for MFN1032 and MFY162, 37°C for MFY 161). The hemolytic activity of the strain MFY163 was the same at 28°C and 37°C.
Involvement of the Gac two-component system on cell-associated hemolytic activity
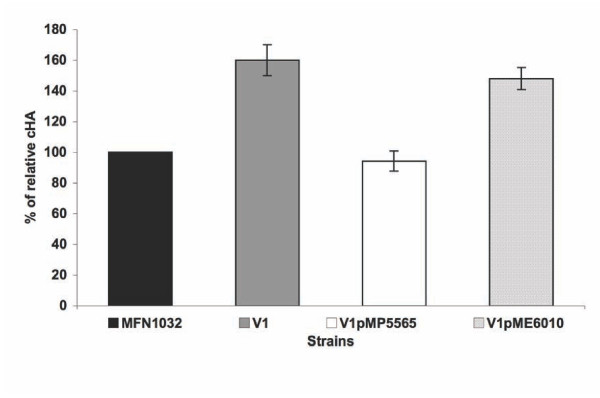
We investigated the possible involvement of the Gac two-component system in the regulation of this cell-associated hemolytic activity using a group1 variant of MFN1032, V1. This variant strain is a gacA mutant and has impaired secreted hemolytic activity [12]. V1 was tested with or without transformation by electroporation with plasmid carrying the gacA gene (pMP5565) or the parental plasmid pME6010, as a control [26] (Figure 5).
Figure 5.
Effect of GacA on MFN1032 cell-associated hemolytic activity. Cell-associated hemolytic activity (cHA) for MFN1032 cells, V1 (gacA mutant) and V1 cells carrying the gacA gene-containing plasmid (pMP5565), or the parental plasmid pME6010 used as a control. The cHA of MFN1032 was taken as the reference value (100%); results are expressed as percent of this value (% relative cHA). The strains were grown at 28°C. Results are means of at least three independent experiments. Standard deviation is shown. Contact was enhanced by centrifugation at 400 g for 10 min.
The non-transformed V1 strain displayed enhanced hemolytic activity (160% lysis), using MFN1032 as a reference value (100%). Introduction of a gacA gene in V1 cells by electroporation with pMP5565 restored wild-type hemolytic levels.
MFN1032 has hrcRST genes
The lysis of RBCs by a number of bacteria requires the formation of a pore in the erythrocyte membrane by TTSS protein. To determine whether TTSS-like genes are present in MFN1032 and MF37, we used PCR primers targeting hrpU operon, (so called U operon of the hrp cluster of type I) encoding the conserved core proteins of fluorescent Pseudomonas TTSS, described by Mazurier et al. [23]. The region amplified by these primers includes the 3'end of hrcR, hrcS and the 5'end of hrcT.
A single fragment of 0.9 kb was obtained for MFN1032 and MF37 and cloned with the pMOS kit. Fragments were sequenced by Genome Express (France). Sequences were registered in the Genbank database (accession number: EU811174 for MFN1032 and FJ694188 for MF37) and named "hrc operon", partial sequence. These sequences predict an 87 residues HrcS protein in these two strains.
A NCBI nucleotide and protein database search showed that the putative HrcS from MFN1032 was very similar to the putative MF37 HrcS (90% identity) and to RscS (94% identity), a type III secretion protein from the Pseudomonas fluorescens strain SBW25 (belonging to the HrcS/YscS/FliQ family), but is more distantly related to the HrcS of C7R12 (73.9% identity) (Table 1). The P. aeruginosa PAO1 FliP partial protein showed 47% identity.
Table 1.
Comparison of the MFN1032 HrcS sequence with other Hrc-like sequences
| Strain | HrcS-like GenBank number |
% Identity to HrcS MFN1032 |
|---|---|---|
| P.fluorescens MFN1032 | Putative HrcS ACE88958 |
- |
| P.fluorescens SBW25 | Putative type III secretion membrane protein RscS CAY46985 |
94% |
| P. fluorescens Pf-5 | Flagellar biosynthesis protein FliQ AAY90949 |
NS |
| P. fluorescens MF37 | Putative HrcS ACO58571 |
90% |
| P. fluorescens Pf0-1 | Flagellar biosynthesis protein FliQ ABA73293 |
NS |
| P. fluorescens C7R12 | Putative HrcS CAC24707 |
74% |
| P.aeruginosa PAO1 | FliP AAG04835 |
47% |
| Pseudomonas syringae pv. tomato str. DC3000 | Type III secretion protein HrcS AAO54916 |
76% |
| Pseudomonas syringae pv. phaseolicola 1448A | Type III secretion component protein HrcS CAE53643 |
74% |
NS: not significant (< at 40%)
Effect of disruption of the hrpU operon
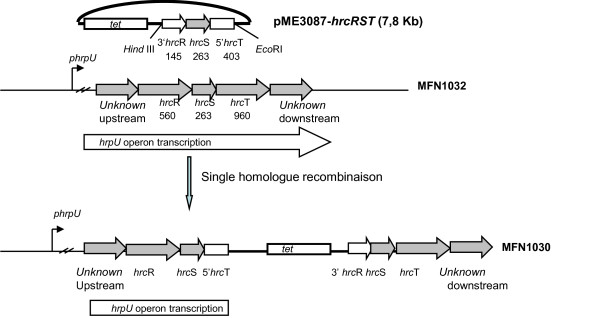
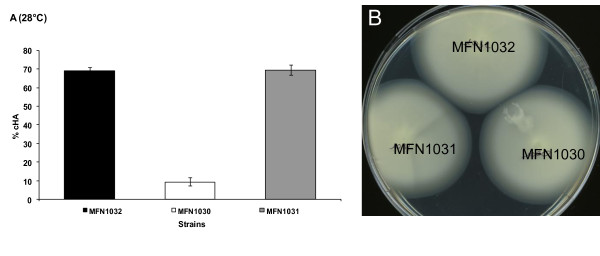
We investigated a possible link between this hrpU operon and the cell-associated hemolytic activity of MFN1032. We used a mutant strain, MFN1030, in which the hrpU operon was disrupted, to determine whether TTSS proteins are required for the hemolytic activity observed in MFN1032. In this construction, the single homologue recombination provokes at least the deletion of the 5'-end of hrcT (58% of HrcT) and of genes situated downstream hrcT in this operon (Figure 6). We observed an almost total loss of cell-associated hemolytic activity (10% lysis) in the mutant strain. Revertant of MFN1030, the strain MFN1031, had a restored hemolytic phenotype, showing activity levels similar to those of MFN1032 (Figure 7A). These results demonstrate a link between the hrpU operon and this cell-associated hemolytic activity in P. fluorescens MFN1032.
Figure 6.
Construction of MFN1030 hrpU operon disrupted mutant. phrpU indicates the promoter of hrpU operon. tet is the tetracycline resistance gene of pME3087. The length of each hrc gene is indicated below its designation (in base pairs). 3'hrcR indicates hrcR gene deleted in 5'-end and 5'hrcT indicates hrcT gene deleted in 3'-end. The arrow above the genes represents the operon transcription. A bold line represents DNA from pME3087. HindIII and EcoRI are enzymes used to clone hrcRST in pME3087. Unknown indicates putatives hrc genes located upstream or downstream hrcRST genes.
Figure 7.
Cell-associated hemolytic activity and swimming motility of MFN1032, MFN1030 (MFN1032 hrcRST-disrupted mutant) and MFN1031 (revertant). A: Hemolysis of RBCs incubated with MFN1032, MFN1030 and MFN1031 at 28°C and a MOI of 1. Results are means of at least three independent experiments. Standard deviation is shown. Contact was enhanced by centrifugation at 400 g for 10 min. B: Swimming motility of MFN1032, MFN1030 and MFN1031. Swimming motility was determined, as described in the methods, on 0.3% LB agar after 16 h of incubation at 28°C. MFN1032, MFN1031 and MFN1030 formed concentric halos corresponding to swimming motility.
Given the homology between hrcS and fliP, we investigated the potential role of hrcRST genes in flagellar synthesis. The effect of disrupting the hrpU operon in MFN1030 was measured in swimming mobility assays, as described in the methods. At 28°C, we observed no differences in swimming ability between MFN1032, MFN1030 and MFN1031 (Figure 7B), suggesting that disruption of this operon has no effect on flagella motility.
Discussion
To our knowledge this is the first study to demonstrate cell-associated hemolytic activity in clinical isolates of a Pseudomonas fluorescens. P.fluorescens MFN1032 cell in exponential growth phase displayed hemolytic activity at 37°C, whereas no hemolytic activity was detected using MFN1032 supernatant. This hemolytic activity was thus dependent on the presence of MFN1032 cells. MFN1032 cells caused hemolysis of RBCs without requiring prior centrifugation to reduce the distance between bacterial and red cell membrane below a critical threshold. Such a centrifugation step has previously been shown to be necessary to induce the "contact-dependent" hemolytic activity displayed by several other bacteria, for example Yersinia [27], Shigella [28] and Pseudomonas aeruginosa [25]. In contrast, "induced" hemolysis does not require close RBC-bacterial contact for enteropathogenic Escherichia coli EPEC, due to the long EspA (TTSS secreted protein) filaments that form a connection between bacteria and host cells for protein translocation [29,30]. MFN1032 hemolytic activity was not strictly contact-dependent but depended on the presence of MFN1032 cells. We therefore propose the term "cell-associated" hemolytic activity.
This activity is independent of the secreted hemolytic activity previously described for this strain. For all tested conditions, we have previously demonstrated that secreted hemolytic activity only occurs at the end of the exponential growth phase [11]. MFN1032 supernatants were not hemolytic at 37°C in vitro and hemolysis was maximal at 17°C. Cell-associated hemolysis measured here was maximal during the exponential growth phase and retrieved at 37°C. Moreover, a gacA mutant of MFN1032 (V1), for which several extracellular activities are impaired (including secreted hemolytic activity), showed increased cell-associated hemolytic activity. In psychrotrophic bacteria, most secreted enzymes are generally found at 17°C (critical temperature), whereas membrane-associated activities are enhanced with decreased generation time [6,31]. Thus, the maximum expression of this new hemolytic activity at 28°C (optimal growth temperature) is consistent with a cell surface associated process.
This hemolytic activity is not common to all Pseudomonas fluorescens species. Indeed, we only observed significant cell-associated hemolysis in the clinical strains MFN1032 and MFY162 and not in the environmental strains tested. Although our panel of studied strains is limited and can not be considered as representative, the presence of this activity seems to be dependent on strain origin, i.e clinical source. Cell-associated hemolytic activity has been rarely observed in environmental strains. Nevertheless, two hemolytic strains showing such phenotype have been described for Plesiomonas shigelloides (former Pseudomonas) [32].
We amplified TTSS-like genes hrcRST from MFN1032 and MF37 cells while P.fluorescens PfO-1 and Pf5 strains [21,33] lack the TTSS genes found in related pathogens or plant-associated bacteria. hrpU operon-like has previously been found in the P. fluorescens strains KD (phytoprotection strain) and SBW25 (biocontrol strain) [22,34]. In one study of a group of fluorescent Pseudomonas, TTSS-like genes were detected in 75% of the phytopathogenic but only in 32% of the saprophytic strains tested [23]. The presence of hrcRST genes is not systematically correlated to hemolytic activity. Indeed, P. fluorescens MF37 and C7R12 have similar hrcRST genes to MFN1032 but are not hemolytic. Thus, the presence of these genes does not strictly imply hemolytic function. Lysis is dependent upon the ability of TTSS translocator proteins to form a pore in the erythrocyte membrane causing hemoglobin leakage. The presence of these hrcRST genes does not necessarily result in the assembly of a functional TTSS. Some TTSS genes are absent from SBW25 and TTSS virulence genes in KD have been suggested to have been recently acquired horizontally from phytopathogenic bacteria and recycled for biocontrol function [22].
TTSS-dependent lysis of erythrocytes has been observed in a number of bacteria. Contact-dependent hemolysis assays have been used to identify the genes required for a functional Salmonella TTSS 1 [35]. MFN1032 cell-associated hemolytic activity shares common features with TTSS-mediated hemolysis. The various mechanisms involved include formation of a pore with an estimated size of 2.4 to 3.2 nm, consistent with TTSS-hemolysis mechanism previously described by Dacheux for Pseudomonas aeruginosa [25]. MFN1032 cells did not show this cell-associated hemolysis during the stationary growth phase. Previous studies have shown a negative effect of high cell density, through a RpoS-mediated mechanisms [36] or by quorum-sensing [37], on TTSS gene expression in Pseudomonas aeruginosa. We found increased hemolytic activity in the MFN1032 gacA mutant (V1). This result suggests that the Gac two-component system is a negative regulator of cell-associated hemolytic activity. Studies on TTSS regulation in Pseudomonas aeruginosa have demonstrated that the GacA response regulator inhibits TTSS function and that, in a gacA mutant, the TTSS effector ExoS is hypersecreted [38]. Opposite, in Pseudomonas syringae, GacA is a positive regulator of the TTSS [39]. The homology between MFN1032 genes and plant-associated TTSS genes is not in favour of a direct negative transcriptional regulation by the system Gac.
To investigate the potential role of TTSS in this hemolytic process, we constructed a mutant with hrpU operon disruption, MFN1030, in which hemolytic activity was severely impaired. Hemolysis was restored in revertant MFN1031 cells, with hemolytic activity levels similar to wild type. Thus, cell-associated hemolytic activity seems to require an intact hrpU operon. In contrast, hrpU operon disruption did not affect swimming motility, suggesting that hrpU operon is not involved in flagella biosynthesis. In MFN1030 the single homologue recombinaison event with PME3087-hrcRST would result in, at least, a lack of HrcT protein. In Pseudomonas cichorri, an insertion of transposon in hrcT was described as sufficient to lost virulence on eggplant [40]. This large insertion in MFN1030 would have a polar effect on genes situated downstream this operon. In Pseudomonas fluorescens, hrcRST genes are highly conserved. Other genes of the hrpU operon, however, seem to vary considerably [22,34]. PCR experiments based on SBW25 and KD sequences did not lead to an amplification of any hrc genes located downstream or upstream hrcRST (data not shown). An experiment of chromosome walking should allow us to identify these genes.
The hrcRST genes from Pseudomonas fluorescens MFN1032 show a high level of homology with hrcRST genes from Pseudomonas syringae, a plant pathogen. TTSS-dependent pore formation is due to the insertion of the translocation pores into host cell membranes. In Pseudomonas syringae, Hrpzpsph forms pores in vitro and is exported by the TTSS. However, when introduced into Yersinia enterocolitica cells, this protein is exported via the Yersinia SSTT but cannot replace YopB functions and do not cause RBC hemolysis [19]. HrpZ is unable to induce pore formation. Moreover, in the two strains of Pseudomonas fluorescens already described no hrpZ homologue was found. We tried to amplify this gene with primers design from hprZ from other pseudomonad, but without success.
The nature of the TTSS component implicates in the cell hemolytic activity of MFN1032 remains to be determined. We will address this issue in future studies.
Conclusion
Pseudomonas fluorescens MFN1032 is a clinical strain isolate that displays two distinct types of hemolytic activity, described here for the first time. The first type is observed in the cell-free supernatant of rich media cultures at 28°C, whereas the second, cell-associated type of hemolysis, is detected at 37°C in the presence of erythrocytes. This strain has hrcRST genes, a feature that is not shared by all Pseudomonas fluorescens strains. Our study establishes an unexpected link between these hrc genes and cell-associated hemolytic activity. These initial findings are consistent, although not sufficient, to demonstrate that this cell-associated hemolysis is due to a functional TTSS. Investigation of type III effector genes in the genome of this strain and the construction of targeted mutants are now needed to confirm these findings. Nevertheless, this study suggests that certain strains of the highly heterogeneous species Pseudomonas fluorescens, which is usually considered to be a saprophytic species, express virulence with characteristic of pathogenic species belonging to the Pseudomonas genus. Nevertheless the principal role of this TTSS homologue to the one of plant-associated bacteria is probably not the pathogenicity against endotherms. The first target of this system would rather be unicellular eukaryotes of the rhizosphere, as mycetes or amoebas.
Methods
Bacterial strains and culture conditions
The MFN1032 strain was collected from a hospital patient suffering from pulmonary tract infection (expectoration) and was considered to be the cause of the infection. MFN1032 was identified as a Pseudomonas fluorescens biovar I strain [10] and was able to grow at 37°C.
CHA is a bronchopulmonary isolate of Pseudomonas aeruginosa from a cystic fibrosis patient [24]. This strain induces TTSS-dependent but ExoU-independent oncosis of neutrophils and macrophages. CHA-induced macrophage death results from a pore forming activity that is dependent on the TTSS. Contact dependent hemolysis provoked by CHA requires the same pore forming activity. CHA has a well inducible and tightly regulated TTSS [41], and is used in our study as a positive control of RBC-TTSS hemolysis.
MF37 is a spontaneous rifampicin-resistant mutant of the MFO strain, a psychrotrophic strain of Pseudomonas fluorescens biovar V, isolated from raw milk and extensively studied in our laboratory [5].
MFY162 is a clinical isolate of Pseudomonas fluorescens Biovar I, MFY161 and MFY163 are clinical isolates of Pseudomonas mosselli [10] and C7R12 a Pseudomonas fluorescens psychrotrophic rhizospheric strain [42].
These bacteria were cultured in Luria Bertani medium (LB), at various temperatures between 8 and 37°C, with shaking at 180 rpm. When necessary, 20 μg/mL tetracycline or 100 μg/mL ampicillin was added. Bacterial density was determined by measuring optical density at 580 nm (Spectronic Unicam spectrophotometer).
Swimming motility
Each strain was incubated on LB agar plates for 24 h at 28°C. Plates of LB medium solidified with 0.3% agar were inoculated by stabbing colonies with a toothpick and inserting the end of the toothpick just below the surface of the agar. Three colonies were picked from three plates and incubated at 28°C until a migration halo appeared.
Hemolysis assay
Hemolysis was performed essentially as described by Dacheux [25]. Sheep red blood cells (RBCs), obtained from Eurobio (France), were washed three times in PBS (pH 7.2, 0.8% NaCl, 0.02% KCl, 0.17% Na2HPO4, 0.8% KH2PO4) and resuspended in RPMI-1640 medium without pH indicator (Sigma) at a density of 5 × 108 RBCs mL-1 at 4°C. Bacteria were grown in LB to an OD580 nm of 0.7 - 1.5, centrifuged and resuspended in RPMI-1640 at 5 × 108 bacteria mL-1. Hemolysis assays were started by mixing 100 μL of RBCs and 100 μL of bacteria, which were than centrifuged at 1500 g or 400 g for 10 minutes and incubated at 37°C for 1 h. The release of hemoglobin was measured at 540 nm, after centrifugation, in 100 μL of cell supernatant.
The percentage (%) of total lysis was calculated as follows: % = [(X -B)/(T-B)] × 100, where B (baseline), a negative control, was corresponding to RBCs incubated with 100 μL of RPMI-1640, and T, a positive control, was corresponding to total RBCs lysis, obtained by incubating cells with 0.1% SDS. X is the OD value of the analysed sample.
When indicated, RBCs were resuspended in 60 mM sterile solutions of osmoprotectant in RPMI-1640, to give a final concentration of 30 mM. For these experiments, a control of hemoglobin precipitation in presence of PEG 4000 and PEG 3000 was realized [43]. PEG 3000 or 4000 were added to a RBCs lysis supernatant obtained after incubation with MFN1032 at a final concentration of 30 mM. No variation of hemoglobin OD value was observed in our conditions during incubation at 37°C for 1 h.
Oligonucleotides and polymerase chain reactions
MFN1032 and MF37 strains were resuspended in 500 μL sterile ultrapure water. The suspension (2 μL) was then used for PCR amplification of DNA from bacterial colonies. PCR was carried out in a 25 μL reaction volume, in a GeneAmp PCR system 2400 (Perkin-Elmer Corporation, USA). Each reaction mixture contained DNA, 0.25 μL Taq polymerase (Q-Biogen, Illkrirch, France), 2.5 μL corresponding buffer, 2.5 μL primers (20 μM) and 2 μL deoxyribonucleoside triphosphate (2.5 mM). After initial denaturation for three minutes at 95°C, the reaction mixture was subjected to 35 cycles of 1 minute at 94°C, 1 minute at 41°C and two minutes at 72°C, followed by a final 3 minutes extension at 72°C. PCR primers, HRCR8092 (5'-CCITT(C/T)ATCGT(C/T)AT(C/T)GA(C/T)(C/T)T-3') and HRCT8986 (5'-CTGTCCCAGATIAICTGIGT-3') (where I indicates inosine), synthesized by Eurogentec (Angers, France), were designed to amplify the "hrc operon", a region of operon U of the hrp cluster of type I (hrpU operon), including the 3' end of hrcR (26%), hrcS (100%), and the 5' end of hrcT (42%) (approximately 897 bp) [23].
Aliquots (5 μL) of the PCR products were analyzed by electrophoresis in 1% agarose gels, stained with ethidium bromide and photographed under UV illumination.
Cloning and sequencing the hrcRST PCR fragment
PCR products were cloned with the pMOSBlue blunt-ended cloning kit (Amersham/Biosciences). MOS cells were transformed and, after blue/white colony screening, clones were picked and plasmid DNA was isolated with the QIAprep Spin Miniprep Kit (Qiagen).
The PCR products were sequenced by Genome Express (France). The predicted sequences of MFN1032 hrcRST and MF37 hrcRST were submitted for BLAST queries http://www.ncbi.nlm.nih.gov/BLAST/.
Construction of MFN1030, an hrcU operon-disrupted mutant of MFN1032 and MF1031, its revertant
The hrcRST-pMOS plasmid from MFN1032 was digested with EcoRI/HindIII and subsequently hrcRST fragment was inserted into the transferable suicide plasmid pME3087 (6,9 Kb) digested by the same enzymes [44]. This construction, pME3087-hrcRST (7,8 kb), was then introduced into Escherichia coli DH5α MCR cells by electroporation. Plasmids were isolated using the QIAprep Spin Miniprep Kit (Qiagen), checked by digestion with HindIII/EcoRI and transferred into the Escherichia coli conjugative strain S17.1 [45]. Colonies were selected for their resistance to tetracycline (20 μg/mL). MFN1032 (naturally ampicillin resistant) cells were conjugated with S17.1 cells carrying the pME3087-hrcRST plasmid and strains were selected for their resistance to tetracycline (20 μg/mL) and ampicillin (100 μg/mL) that corresponds to insertion of the whole plasmid via a single homologue recombinaison. One of the clones was selected and corresponded to an hrpU operon disruption mutant. This disruption mutant was called MFN1030. The reversion of the mutant MFN1030 was obtained after incubating MFN1030 cells on an LB agar plate for 72 hours. Of all the colonies obtained, 100 were subcultured in parallel on LB agar plates with or without tetracycline (20 μg/mL).
Colonies growing on LB agar plates without tetracycline but not on LB agar plates with tetracycline (20 μg/mL) reflect a second recombination event and an excision of the plasmid. One clone was selected and named MFN1031, a revertant of MFN1030 strain.
Authors' contributions
DS carried out most experiments and analyzed most of the data. AM wrote the manuscript, participated in the design of the study and analyzed most of the data. GR participated in the molecular genetic studies, and participated in the design of the study. JG initiated and participated in the design of the study. NC helped set up general laboratory experimental conditions. MF and NO were involved in designing the study. All authors read and approved the final manuscript.
Contributor Information
Daniel Sperandio, Email: daniel.sperandio@etu.univ-rouen.fr.
Gaelle Rossignol, Email: gaelle.rossignol@univ-rouen.fr.
Josette Guerillon, Email: Josette.guerillon@univ-rouen.fr.
Nathalie Connil, Email: nathalie.connil@univ-rouen.fr.
Nicole Orange, Email: nicole.orange@univ-rouen.fr.
Marc GJ Feuilloley, Email: marc.feuilloley@univ-rouen.fr.
Annabelle Merieau, Email: annabelle.merieau@univ-rouen.fr.
Acknowledgements
The Région Haute-Normandie supported this work. We thank Magalie Barreau for technical help.
References
- Couillerot O, Prigent-Combaret C, Caballero-Mellado J, Moenne-Loccoz Y. Pseudomonas fluorescens and closely-related fluorescent pseudomonads as biocontrol agents of soil-borne phytopathogens. Lett Appl Microbiol. 2009;48(5):505–512. doi: 10.1111/j.1472-765X.2009.02566.x. [DOI] [PubMed] [Google Scholar]
- Tourkya B, Boubellouta T, Dufour E, Leriche F. Fluorescence spectroscopy as a promising tool for a polyphasic approach to pseudomonad taxonomy. Curr Microbiol. 2009;58(1):39–46. doi: 10.1007/s00284-008-9263-0. [DOI] [PubMed] [Google Scholar]
- Bodilis J, Calbrix R, Guerillon J, Merieau A, Pawlak B, Orange N, Barray S. Phylogenetic relationships between environmental and clinical isolates of Pseudomonas fluorescens and related species deduced from 16S rRNA gene and OprF protein sequences. Syst Appl Microbiol. 2004;27(1):93–108. doi: 10.1078/0723-2020-00253. [DOI] [PubMed] [Google Scholar]
- Meyer JM, Geoffroy VA, Baida N, Gardan L, Izard D, Lemanceau P, Achouak W, Palleroni NJ. Siderophore typing, a powerful tool for the identification of fluorescent and nonfluorescent pseudomonads. Appl Environ Microbiol. 2002;68(6):2745–2753. doi: 10.1128/AEM.68.6.2745-2753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burini JF, Gugi B, Merieau A, Guespin-Michel JF. Lipase and acidic phosphatase from the psychrotrophic bacterium Pseudomonas fluorescens: two enzymes whose synthesis is regulated by the growth temperature. FEMS Microbiol Lett. 1994;122(1-2):13–18. doi: 10.1111/j.1574-6968.1994.tb07136.x. [DOI] [PubMed] [Google Scholar]
- Gugi B, Orange N, Hellio F, Burini JF, Guillou C, Leriche F, Guespin-Michel JF. Effect of growth temperature on several exported enzyme activities in the psychrotrophic bacterium Pseudomonas fluorescens. J Bacteriol. 1991;173(12):3814–3820. doi: 10.1128/jb.173.12.3814-3820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot L, Abdelmoula SM, Merieau A, Leroux P, Cazin L, Orange N, Feuilloley MG. Pseudomonas fluorescens as a potential pathogen: adherence to nerve cells. Microbes Infect. 2001;3(12):985–995. doi: 10.1016/S1286-4579(01)01462-9. [DOI] [PubMed] [Google Scholar]
- Feuilloley MGJ, Mezghani-Abdelmoula S, Picot L, Lesouhaitier O, Merieau A, Guerillon J, Orange N. Involvement of Pseudomonas and related species in central nervous system infections. Recent Adv Dev Microbiol. 2003;7:55–71. [Google Scholar]
- Picot L, Mezghani-Abdelmoula S, Chevalier S, Merieau A, Lesouhaitier O, Guerillon J, Cazin L, Orange N, Feuilloley MG. Regulation of the cytotoxic effects of Pseudomonas fluorescens by growth temperature. Res Microbiol. 2004;155(1):39–46. doi: 10.1016/j.resmic.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Chapalain A, Rossignol G, Lesouhaitier O, Merieau A, Gruffaz C, Guerillon J, Meyer JM, Orange N, Feuilloley MG. Comparative study of 7 fluorescent pseudomonad clinical isolates. Can J Microbiol. 2008;54(1):19–27. doi: 10.1139/W07-110. [DOI] [PubMed] [Google Scholar]
- Rossignol G, Merieau A, Guerillon J, Veron W, Lesouhaitier O, Feuilloley MG, Orange N. Involvement of a phospholipase C in the hemolytic activity of a clinical strain of Pseudomonas fluorescens. BMC Microbiol. 2008;8:189. doi: 10.1186/1471-2180-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol G, Sperandio D, Guerillon J, Duclairoir Poc C, Soum-Soutera E, Orange N, Feuilloley MG, Merieau A. Phenotypic variation in the Pseudomonas fluorescens clinical strain MFN1032. Res Microbiol. 2009;160(5):337–344. doi: 10.1016/j.resmic.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Barbieri JT, Sun J. Pseudomonas aeruginosa ExoS and ExoT. Rev Physiol Biochem Pharmacol. 2004;152:79–92. doi: 10.1007/s10254-004-0031-7. full_text. [DOI] [PubMed] [Google Scholar]
- Frank DW. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26(4):621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- Vallis AJ, Yahr TL, Barbieri JT, Frank DW. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun. 1999;67(2):914–920. doi: 10.1128/iai.67.2.914-920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284(5418):1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- Huynh TV, Dahlbeck D, Staskawicz BJ. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989;245(4924):1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- Huang HC, He SY, Bauer DW, Collmer A. The Pseudomonas syringae pv. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992;174(21):6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Klusener B, Tsiamis G, Stevens C, Neyt C, Tampakaki AP, Panopoulos NJ, Noller J, Weiler EW, Cornelis GR. HrpZ(Psph) from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc Natl Acad Sci USA. 2001;98(1):289–294. doi: 10.1073/pnas.011265298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Bertrand N, Rainey PB. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol. 2001;41(5):999–1014. doi: 10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Zhai Y, Schneider JC, Ramseier TM, Saier MH Jr. Protein secretion systems of Pseudomonas aeruginosa and P fluorescens. Biochim Biophys Acta. 2003;1611(1-2):223–233. doi: 10.1016/S0005-2736(03)00059-2. [DOI] [PubMed] [Google Scholar]
- Rezzonico F, Binder C, Defago G, Moenne-Loccoz Y. The type III secretion system of biocontrol Pseudomonas fluorescens KD targets the phytopathogenic Chromista Pythium ultimum and promotes cucumber protection. Mol Plant Microbe Interact. 2005;18(9):991–1001. doi: 10.1094/MPMI-18-0991. [DOI] [PubMed] [Google Scholar]
- Mazurier SLM, Siblot S, Mougel C, Lemanceau P. Distribution and diversity of type III secretion system-like genes in saprophytic and phytopathogenic fluorecent Pseudomonas. FEMS Microbiology Ecology. 2004;49:455–467. doi: 10.1016/j.femsec.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Toussaint B, Delic-Attree I, Vignais PM. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochem Biophys Res Commun. 1993;196(1):416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- Dacheux D, Goure J, Chabert J, Usson Y, Attree I. Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol Microbiol. 2001;40(1):76–85. doi: 10.1046/j.1365-2958.2001.02368.x. [DOI] [PubMed] [Google Scholar]
- Broek D van den, Chin AWTF, Bloemberg GV, Lugtenberg BJ. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J Bacteriol. 2005;187(2):593–600. doi: 10.1128/JB.187.2.593-600.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson S, Schesser K, Persson C, Galyov EE, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. Embo J. 1996;15(21):5812–5823. [PMC free article] [PubMed] [Google Scholar]
- Clerc P, Baudry B, Sansonetti PJ. Plasmid-mediated contact haemolytic activity in Shigella species: correlation with penetration into HeLa cells. Ann Inst Pasteur Microbiol. 1986;137A(3):267–278. doi: 10.1016/S0769-2609(86)80033-3. [DOI] [PubMed] [Google Scholar]
- Shaw RK, Daniell S, Ebel F, Frankel G, Knutton S. EspA filament-mediated protein translocation into red blood cells. Cell Microbiol. 2001;3(4):213–222. doi: 10.1046/j.1462-5822.2001.00105.x. [DOI] [PubMed] [Google Scholar]
- Crepin VF, Martinez E, Shaw RK, Frankel G, Daniell SJ. Structural and functional properties of chimeric EspA-FliCi filaments of EPEC. J Mol Biol. 2008;378(1):243–250. doi: 10.1016/j.jmb.2008.02.042. [DOI] [PubMed] [Google Scholar]
- Merieau A, Gügi B, Guespin-Michel JF, Orange N. Temperature regulation of lipase secretion by Pseudomonas fluorescens strain MF0. Appl Microbiol Biotechnol. 1993;39:104–109. [Google Scholar]
- Gonzalez-Rodriguez N, Santos JA, Otero A, Garcia-Lopez ML. Cell-associated hemolytic activity in environmental strains of Plesiomonas shigelloides expressing cell-free, iron-influenced extracellular hemolysin. J Food Prot. 2007;70(4):885–890. doi: 10.4315/0362-028x-70.4.885. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23(7):873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer BA, Jelenska J, Greenberg JT. Bioinformatics correctly identifies many type III secretion substrates in the plant pathogen Pseudomonas syringae and the biocontrol isolate P. fluorescens SBW25. Mol Plant Microbe Interact. 2005;18(8):877–888. doi: 10.1094/MPMI-18-0877. [DOI] [PubMed] [Google Scholar]
- Field TR, Layton AN, Bispham J, Stevens MP, Galyov EE. Identification of novel genes and pathways affecting Salmonella type III secretion system 1 using a contact-dependent hemolysis assay. J Bacteriol. 2008;190(9):3393–3398. doi: 10.1128/JB.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogardt M, Roeder M, Schreff AM, Eberl L, Heesemann J. Expression of Pseudomonas aeruginosa exoS is controlled by quorum sensing and RpoS. Microbiology. 2004;150(Pt 4):843–851. doi: 10.1099/mic.0.26703-0. [DOI] [PubMed] [Google Scholar]
- Bleves S, Soscia C, Nogueira-Orlandi P, Lazdunski A, Filloux A. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J Bacteriol. 2005;187(11):3898–3902. doi: 10.1128/JB.187.11.3898-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soscia C, Hachani A, Bernadac A, Filloux A, Bleves S. Cross talk between type III secretion and flagellar assembly systems in Pseudomonas aeruginosa. J Bacteriol. 2007;189(8):3124–3132. doi: 10.1128/JB.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Cui Y, Yang H, Collmer A, Alfano JR, Chatterjee AK. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol Plant Microbe Interact. 2003;16(12):1106–1117. doi: 10.1094/MPMI.2003.16.12.1106. [DOI] [PubMed] [Google Scholar]
- Hojo H, Koyanagi M, Tanaka M, Kajihara S, Ohnishi K, Kiba A, Hikichi Y. The hrp genes of Pseudomonas cichorii are essential for pathogenicity on eggplant but not on lettuce. Microbiology. 2008;154(Pt 10):2920–2928. doi: 10.1099/mic.0.2008/021097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filopon D, Merieau A, Bernot G, Comet JP, Leberre R, Guery B, Polack B, Guespin-Michel J. Epigenetic acquisition of inducibility of type III cytotoxicity in P. aeruginosa. BMC Bioinformatics. 2006;7:272. doi: 10.1186/1471-2105-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirleau P, Delorme S, Philippot L, Meyer J, Mazurier S, Lemanceau P. Fitness in soil and rhizosphere of Pseudomonas fluorescens C7R12 compared with a C7R12 mutant affected in pyoverdine synthesis and uptake. FEMS Microbiol Ecol. 2000;34(1):35–44. doi: 10.1111/j.1574-6941.2000.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Quijano JC, Lemeshko VV. Hemoglobin precipitation by polyethylene glycols leads to underestimation of membrane pore sizes. Biochim Biophys Acta. 2008;1778(12):2775–2780. doi: 10.1016/j.bbamem.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Schnider U, Keel C, Voisard C, Defago G, Haas D. Tn5-directed cloning of pqq genes from Pseudomonas fluorescens CHA0: mutational inactivation of the genes results in overproduction of the antibiotic pyoluteorin. Appl Environ Microbiol. 1995;61(11):3856–3864. doi: 10.1128/aem.61.11.3856-3864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon RPU, Pehle A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram-negative bacteria. biotechology. 1983;1:784–790. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]