Abstract
Urea is highly concentrated in mammalian kidney to produce the osmotic gradient necessary for water re-absorption. Free diffusion of urea across cell membranes is slow due to its high polarity, and specialized urea transporters have evolved to achieve rapid and selective urea permeation. Here we present the 2.3 Å structure of a functional urea transporter from the bacterium Desulfovibrio vulgaris. The transporter is a homotrimer, and each subunit contains a continuous membrane-spanning pore formed by the two homologous halves of the protein. The pore contains a constricted selectivity filter that can accommodate multiple dehydrated urea molecules in single file. Backbone and side chain oxygen atoms provide continuous coordination of urea as it progresses through the filter, and well-placed α-helix dipoles provide additional compensation for dehydration energy. These results establish that the urea transporter operates by a channel-like mechanism and reveal the physical and chemical basis of urea selectivity.
Urea is ubiquitous in nature. Bacteria take up urea and convert it to ammonia for use as a nitrogen source, and in certain enteric pathogens, a buffer for surviving the extreme acidic conditions in the stomach1, 2. In higher organisms such as mammals, urea is produced as an end product of protein catabolism because it is less toxic than ammonia and more soluble than uric acid. In addition to being a vehicle for nitrogen excretion, urea is utilized as an osmolyte. For example, sharks and rays use urea to maintain isosmotic with sea water3, and mammals are able to concentrate urea hundreds of fold in the kidney to provide the osmotic gradient essential for water re-absorption4, 5.
Urea has a stronger dipole moment (4.6 D) than water (1.8 D) and its un-assisted diffusion across lipid bilayers is slow6. There are at least four families of transporters that facilitate selective permeation of urea: an ATP-dependent ABC type urea transporter7; an ion-motive force-dependent urea transporter8; an acid-activated urea channel that belongs to the urea/amide channel family2; and finally the urea transporter (UT) family, which is the most widely distributed family and the focus of this research. Since the first cloning of a UT from mammalian kidney by expression cloning9, UT members have been found in bacteria, fungi, insects, and many vertebrates including all mammals.
Previous characterizations of both mammalian and bacterial UTs1, 9–12 have demonstrated that UTs facilitate the diffusion of urea and urea analogs along their concentration gradients at rates between 104 to 106/sec10, 13, consistent with a channel-like mechanism. In the absence of structural information, how urea transporters coordinate and stabilize an at least partially dehydrated urea to achieve selectivity and facilitate permeation remains unknown. To address these questions, we focused on the functional and structural characterization of a UT from the bacterium Desulfovibrio vulgaris, dvUT, which is homologous to mammalian UTs (Fig. S1a). All UT sequences have two homologous halves which probably arose from duplication of an ancient gene14, and are predicted to contain ten transmembrane helices. Identity between the transmembrane domains of dvUT and human UTs is around 35%, and clusters of highly conserved residues are distributed throughout the primary sequence. We demonstrate that dvUT is a functional urea transporter and present the atomic resolution structure of dvUT and the molecular basis it reveals for selective permeation of urea.
Functional characterization of dvUT
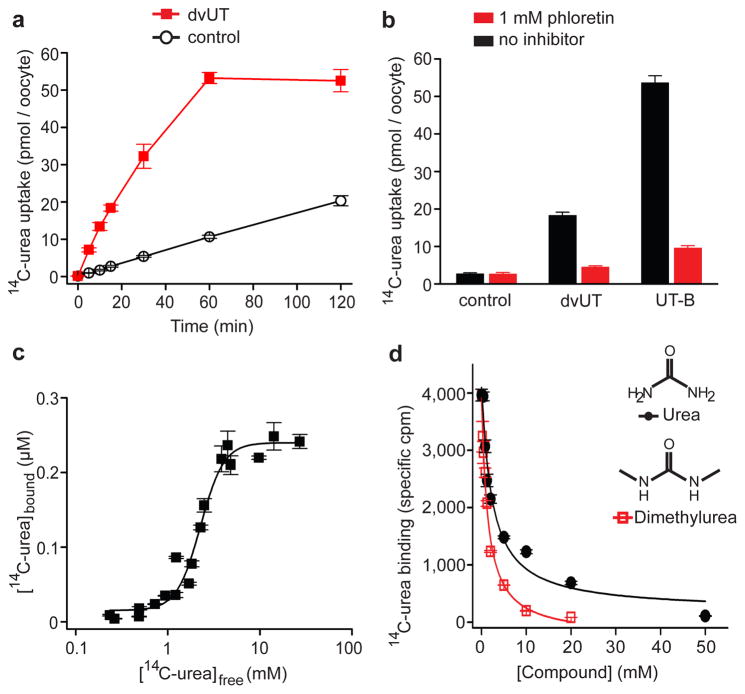
Function of dvUT was examined in two assays. First, urea flux through dvUT was measured in a tracer uptake assay2, 9. dvUT was expressed in Xenopus laevis oocytes by injection of complimentary RNA and uptake of 14C-labelled urea (~181 μM) by individual oocytes was measured. Uptake of labelled urea increased with time, and reached ~50 pmol per oocyte in about 60 minutes (Fig. 1a). Assuming the volume of an oocyte is ~0.25 μl9, the estimated intracellular concentration of labelled urea is ~200 μM, essentially in equilibrium with the extracellular urea. In contrast, urea accumulation in water-injected oocytes showed a much slower time course and had not reached equilibrium even after 120 minutes (Fig 1a).
Figure 1. dvUT mediated urea flux and binding.
a. Timecourse of 14C-urea uptake in oocytes injected with dvUT cRNA (squares) or water (circles). b. Uptake of radio-labelled urea in the presence (red) or absence (black) of 1 mM phloretin. c. Saturation equilibrium urea binding by dvUT. The solid line represents fitting to the Hill equation. d. SPA-based 14C-urea equilibrium binding in the presence of increasing concentrations of urea (circles) and N,N′-dimethylurea (squares). The solid lines correspond to data fit with a single-site binding isotherm. Error bars in all panels are standard errors of the mean of 3–10 measurements.
Since urea flux through mammalian UTs is inhibited by phloretin9, 15, we examined its effect on both human UT-B and dvUT (Fig 1b). Uptake of urea by oocytes expressing UT-B reached equilibrium in ~15 minutes, comparable to a previous report9, and is faster than oocytes expressing dvUT. However, this does not necessary indicate that dvUT has a slower flux rate because the relative amounts of the two functional UT proteins expressed on the membrane are unknown. Phloretin (1 mM) inhibited facilitated urea transport through UT-B and dvUT (Fig. 1b) by 86% and 87% respectively, suggesting a similar mode of interaction of phloretin with both proteins. In fact, phloretin also inhibits a urea transporter from Actinobacillus pleuropneumoniae (apUT)11, indicating that a common architecture likely is shared by both prokaryotic and eukaryotic UTs.
Second, equilibrium binding of urea to dvUT was measured in a scintillation proximity assay (SPA)16, 17. In this assay, purified His-tagged dvUT was immobilized on copper-coated scintillation beads which emit light only when a radioactive ligand stably binds to dvUT. Addition of 14C-labelled urea induced light emission from the scintillant, indicating urea binding to dvUT. Fitting the data to the Hill equation revealed an apparent equilibrium dissociation constant (Kd) of 2.3 ± 0.14 mM (Fig. 1c) with a Hill coefficient of 3.4 ± 0.7, indicative of cooperative binding of about three urea molecules per molecule of dvUT, and suggesting that dvUT has multiple urea binding sites.
Interaction of N, N′-dimethylurea (DMU), a urea analog that was used in the structural studies, with dvUT was measured in an SPA-based competition assay (Fig. 1d). Addition of increasing concentrations of DMU to dvUT incubated with a fixed concentration of labelled urea caused progressive loss of light emission, indicating that the bound urea was displaced. The concentration of DMU necessary to achieve half maximum inhibition (IC50) was 1.4 ± 0.2 mM. As a control experiment, unlabelled urea was used for isotopic dilution of the 14C-labeled urea. A 50 % reduction of 14C-urea binding was obtained at 2.3 ± 0.7 mM urea, consistent with the Kd measured in the previous equilibrium binding assay (Fig. 1c) and slightly higher than the IC50 of DMU (Fig. 1d). These results suggest that DMU and urea may occupy similar sites in dvUT.
Channel architecture
The structure of dvUT was determined by single-wavelength anomalous dispersion using a mercury derivatized crystal, and refined to 2.3 Å using a gold derivatized crystal (Methods and Table S1). The final structure contains one protomer in the asymmetric unit, which has residues 1 to 163 and 168 to 334 of dvUT along with 9 gold atoms and 55 waters. Three residues from the C-terminus and four residues from a long loop connecting the homologous halves of the protein are disordered and omitted from the model.
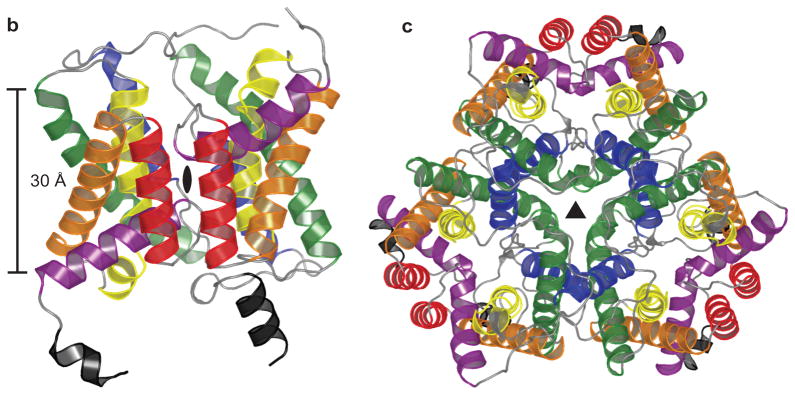
The dvUT protomer contains two hemi-cylindrical domains of six helices each, and the two domains are related by a rotational pseudo-two-fold symmetry axis lying in the plane of the membrane (Fig. 2a and S1b). The first helix of each domain, which we call pore helix a or b (Pa or Pb), is tilted at a roughly 50º angle with respect to the membrane norm and extends ~10 Å into the membrane. The pore helices end in loops that turn sharply to exit on the same side of the membrane. The next four helices from each domain, T1a-T4a and T1b-T4b, span the entire membrane. The remaining helices, T5a and T5b, are perpendicular to the membrane and unwind at the middle of the membrane into an extended coil to cross the membrane. The α-carbon atoms of the first half, Pa and T1a-T5a, can be superposed onto the second half, Pb and T1b-T5b, by a rotation of 179.5° with a root mean square deviation (r.m.s.d.) of 1.13 Å.
Figure 2. Fold and oligomeric structure of dvUT.
a. Cartoon representation of the dvUT protomer. The two-fold pseudo-symmetry axis, marked black as a black oval, is normal to the plane of the figure. Color of helices matches that in the topology diagram (Fig. S1b). b. Cartoon representation of the full dvUT trimer. The crystallographic three-fold symmetry axis is marked as a black triangle.
The N- and C-termini exit the membrane on the same side of the protein and both end in short helical segments (Fig. S1b). Although the orientation of dvUT in the plasma membrane has not been experimentally determined, the excess of positive charge on the termini-containing face suggests that this side orients towards the cytoplasm18, and this orientation is consistent with the known topology of its mammalian homologues19–21.
dvUT crystallizes as a homotrimer (Fig. 2b), with its three-fold rotational axis coincident with a crystallographic symmetry axis. Helices T4a and T5a of one subunit interact with helices T4b and T5b of the neighboring subunit. The interface between the subunits is largely hydrophobic with a total interacting surface area of 5010 Å2. On the three-fold symmetry axis is a large cavity (Fig. S2a). Both ends of the cavity are sealed off from the bulk solvent by two layers of tightly packed hydrophobic sidechains circling the 3-fold rotational axis: L160 and P287 on the periplasmic face, and F120 and W124 on the cytoplasmic face (Fig. S2b and c). The residues lining the walls of the central cavity are largely hydrophobic; tryptophan, phenylalanine and leucine residues predominate. Several large electron density peaks in the cavity most likely correspond to partially ordered detergent or lipid molecules.
Several lines of evidence suggest that the observed trimer is not an artefact of crystal packing. In addition to the large area of the interface, chemical crosslinking experiments support that detergent-solubilized dvUT is a trimer (Fig. S3a); also, the same homotrimer was observed in a lower resolution structure obtained from the native protein, which crystallizes in a lower symmetry space group with different packing (Fig. S3b and c).
Selectivity filter
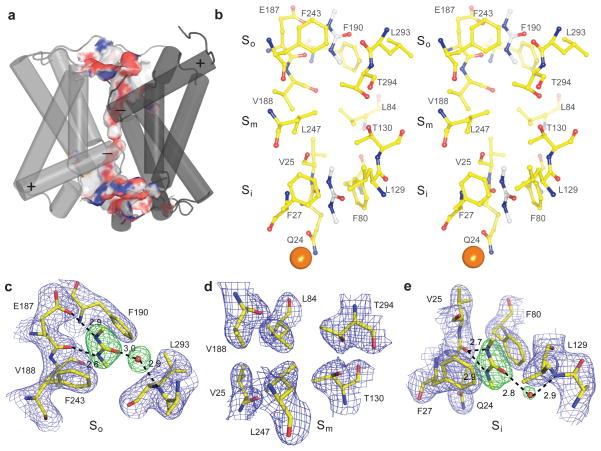
Each dvUT protomer has a continuous solvent accessible permeation pathway that is formed between the two homologous halves of the protein (Fig. 3a). This pore has a constricted region, ~16 Å long, that opens into two wide vestibules on either side. We define the constricted region as the selectivity filter. Highly conserved residues from six different regions of the protein, Pa and Pb, T3a and T3b, and T5a and T5b, are brought together to form the selectivity filter (Fig. S6a). The residues that line the selectivity filter are colored in Fig. S1a and shown in Fig. 3b. One side of the selectivity filter has two linear arrays of three oxygen atoms, which we call the oxygen ladders (Fig. 3b). Each oxygen ladder is flanked by two parallel and closely spaced phenylalanine side chains that compress the filter into a slot-like shape. Hydrophobic phenylalanine and leucine residues complete the lining of the filter opposite to each of the oxygen ladders. Between the two oxygen ladders, there is a gap (~ 6 Å) packed by two valine side chains, V25 and V188. A pair of leucine side chains. L84 and L247, constricts this part of the filter just like the phenylalanines above and below, and on the opposite side of the valines are two threonine side chains, T130 and T294. Despite the relatively weak overall conservation between the two halves of the protein, the nature and positioning of the side chains forming the wall of the selectivity filter are remarkably symmetrical: with the exception of a single Q24/E187 pair, every residue in the filter has an identical symmetry-related partner (Fig. 3b). For each oxygen ladder, the distances between the oxygen atoms are 3.4–3.6 Å, except for the distance between the side chain and backbone oxygens of Q24, which is 4 Å. However, the position of this side chain may be perturbed in the crystal structure by its interaction with a gold atom, added during the crystallization process, sitting at the entrance of the pore (Fig. 3b).
Figure 3. Structure of the dvUT pore and dimethylurea binding sites.
a. A surface representation of the dvUT pore with T1a, T1b and the N- and C-termini removed. Oxygen and nitrogen atoms are colored in red and blue, respectively. b. Stereo view of residues lining the selectivity filter. Two dimethylurea, coordinates taken from the dvUT-dimethylurea complex, are shown in the So and Si sites. A gold atom that co-crystallizes with dvUT is shown in gold. c-e. Views of the So (c), Sm (d) and Si (e) regions of the selectivity filter for dvUT-dimethyl urea complex. The dark blue mesh corresponds to 2Fo-Fc electron density map contoured at 1.5 σ. The green mesh in the So and Si sites corresponds to 3.0 σ Fo-Fc electron density calculated with dimethylurea and the displayed water molecule omitted.
Permeability of dvUT to water has not been determined experimentally, however, water permeation has been observed in apUT11 and a mammalian UT22, although there is controversy over the latter10. In the absence of ligand, three positive electron density peaks are visible in selectivity filter, which likely correspond to partially-ordered water molecules and raise the possibility that dvUT is water permeable. Two of the peaks lie directly between the pairs of phenyalanines and are within hydrogen-bonding distance of the oxygen ladders. The third is hydrogen-bonded to the side chain hydroxyl of T294.
Potential urea binding sites
To investigate how selective permeation of urea is facilitated by the physical and chemical properties of the selectivity filter, we co-crystallized dvUT with urea. After initial attempts to obtain high-resolution data in the presence of urea were unsuccessful, we co-crystallized dvUT with DMU. A 2.4 Å dataset was collected on a crystal grown in the presence of 10 mM DMU.
The structure of dvUT-DMU was solved by molecular replacement, and two strong electron densities were observed in a difference map. Each density is flat and triangular in shape, and is located close to an oxygen ladder (Fig. 3c and e, stereo in Fig. S4). The triangle-shaped flat electron density makes the orientation of DMU unambiguous, with its two amide nitrogen atoms facing an oxygen ladder. We define the two sites as So and Si, for their proximity to either the outside or inside of a cell (Fig. 3b).
We hypothesize that a partially dehydrated DMU in the So site is stabilized by four different interactions. First, the two amide nitrogen atoms on DMU are 2.9 and 2.6 Å away from the side chain and backbone oxygen atoms of E187, respectively, poised to form hydrogen bonds. Second, the two side chains of F190 and F243 sandwich DMU so that the two aromatic rings provide stabilization of the partial positive charge on the amide nitrogen by amide-π stacking interactions23. Third, the carbonyl oxygen atom on DMU is 3.0 Å away from a water molecule that in turn hydrogen bonds with a backbone nitrogen atom (Fig. 3c). Fourth, the oxygen ladder is at the very end of Pb (Fig. 3a), and this arrangement conveniently utilizes the partial negative charge created by the α-helix dipole to further stabilize the partial positive charge on the amide nitrogens. The use of helix dipoles to stabilize bound permeants has been noted previously in potassium channels24 and in chloride transporters25.
Binding of DMU in the Si site is similar to that at the So site, but shifted a few tenths of an angstrom towards the center of the membrane so that both of the amide nitrogen atoms are 2.7 and 2.8 Å away from the center oxygen in the ladder (Fig. 3e.). The slight shift is probably because the side chain oxygen atom of Gln24 is not available because it is farther away to coordinate a nearby gold atom.
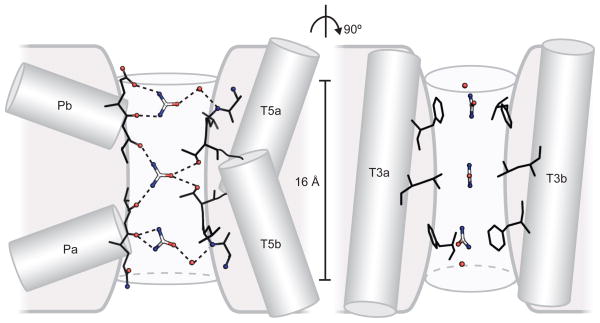
Given the similarities in shape and electrostatics between urea and DMU, urea likely occupies the So and Si sites in a similar manner as DMU. We therefore modelled urea into the two sites using the DMU coordinates (Fig. 4). In order for urea to complete its journey across cell membrane, it must pass through the space between So and Si, which we define as the Sm site because it is in the middle of the membrane (Fig. 3b and 3d). We modelled a urea molecule into this site by positioning the two amide nitrogen atoms within hydrogen bonding distance to the two inner most oxygen atoms from the oxygen ladders (Fig. 4). This arrangement naturally places the carbonyl oxygen atom of urea within hydrogen bonding distance to the hydroxyls from both T130 and T294, two residues that are 100% conserved in all known UT sequences. Unlike urea in the So and Si sites, a urea in Sm is sandwiched by two leucine side chains ~ 6.1 Å apart that also exhibit strong conservation in mammalian UTs. We propose that the Sm site in the middle of the selectivity filter is superbly suited to screen a permeating ligand for the appropriate electrostatic properties as well as a stringent test of planarity. This site however, is likely not accessible to DMU because the methyl groups will clash with the two threonine side chains.
Figure 4. Schematic view of the selectivity filter.
The selectivity filter is shown from two angles. The predicted locations of three urea molecules and their hydrogen bonding partners are on the left. In the perpendicular direction, the filter is compressed by phenylalanine and leucine side chains lining the walls of the pore (right). Helices contributing residues to the selectivity filter are represented as gray cylinders.
Summary
The structure of dvUT showed a continuous solvent accessible pore in the structure, indicating that dvUT, and by extrapolation, UTs in general likely operate by a channel-like mechanism. Selective permeation of urea is achieved by a long and narrow selectivity filter that allows dehydrated urea molecules to permeate in single-file. Compensation for dehydration energy is provided by continuous coordination of urea with hydrogen bonds as it goes through the selectivity filter, and by well-placed α-helix dipoles and amide-π interactions (Fig. 4). Furthermore, as a urea molecule progresses through the filter, its orientation is rigorously maintained by closely spaced hydrophobic residues, so that optimal hydrogen bonding and single-file conduction can be achieved. This mechanism is consistent with the ability of UTs to exclude molecules larger than urea, but allow smaller analogs such as formamide to pass through.
It is not immediately apparent how universal the trimeric complex is among urea transporters. A recent study on apUT argued that that particular homolog was a dimer11. The stoichiometry of mammalian UT remains unknown. The existence of the mammalian UT-A1 isoform, which consists of two tandem copies of UT sequences, suggests that UT-A1 forms oligomers with even numbers of UT domains. However, UT-A1 co-expresses in the same cells with UT-A326, which only has one UT domain, thus raising the possibility that UT-A1 and UT-A3 assemble to form a trimer-like complex, although attempts to isolate a UT-A1/UT-A3 complex by co-immunoprecipitation have not been successful27. Given that UTs appear to conduct urea constitutively, and that each subunit contains its own complete pore, it is entirely possible that differences in native oligomeric state between homologs would not have a large effect on their basic function.
Similar to other channels that conduct neutral molecules, such as ammonia28, 29, water30, 31, or glycerol32, UT is constructed by two oppositely oriented homologous halves. The arrangement of the UT transmembrane helices T1a-T5a and T1b-T5b bears a resemblance to the fold of the Amt/Rh familiy of ammonia channels28, 29 (Fig. S5a–b). Together with the weak sequence identity between the two proteins (22% between dvUT and E. coli AmtB), this would suggest that the two families are evolutionarily related. However, there is little similarity between the two proteins in the structure of the permeation pathway itself. The most significant difference is that the tilted helices Pa and Pb that contribute the residues forming the oxygen ladders are not present in AmtB. Their place is taken in the AmtB structure by mostly hydrophobic side chains from helices T1 and T6 (equivalent to T1a and T1b in dvUT, which are not involved in the pore) (Fig S6a-b). The selectivity filter of AmtB is overall narrower than dvUT (Fig S6c), and is largely hydrophobic save for a pair of central histidine residues that invite comparison to T130 and T294 in dvUT (Fig S6d–e). In addition, dvUT lacks the 11th transmembrane segment possessed by AmtB, and the trimer interface is constructed by different transmembrane helices (Fig S5c–d).
The structure of dvUT has revealed the physical and chemical principles governing selective permeation of urea in UTs. The structure also provides a framework for further studies to understand how the rate of urea permeation is determined and how regulation of UT function, non-covalently by small-molecule compounds9, 33 and covalently by phosphorylation34, 35, is achieved.
Methods Summary
An N-terminal His-tagged SUMO-dvUT fusion protein was expressed in E. coli BL21(DE3) cells and purified on a cobalt affinity column. After cleavage of the affinity tag and SUMO domain with SUMO protease and a second round of chromatography on a Superdex 200 10/300 GL gel filtration column, the protein was concentrated to ~8 mg/mL in buffer containing 300 mM NaCl, 20 mM HEPES pH 7.5, 5 mM β-mercaptoethanol, and 40 mM n-octyl-β-D-maltoside. Crystals were obtained by sitting-drop vapour diffusion in mother liquor containing 22% PEG1500, 100 mM sodium cacodylate, pH 6.5, and 10 mM thiomersal or KAu(CN)2. A 2.5 Å mercury-derivatized and a 2.3 Å gold-derivatized dataset were collected on beamlines X25 and X29 at the National Synchrotron Light Source at Brookhaven National Laboratory. The mercury dataset was used for location of heavy atom sites and SAD phasing. After density modification and automated and manual model building, the resulting partial structure was used as a molecular replacement model for the gold dataset. Iterative rounds of building and refinement produced a final model with 330 out of 337 residues resolved and R and R-free values of 17.9 and 20.4%. High resolution DMU-dvUT crystals were obtained by co-crystallizing the protein with 10 mM DMU as well as KAu(CN)2, and the 2.4 Å ligand-bound structure was solved by molecular replacement. For the SPA binding assays, 100 μL of 2.5 mg/mL Cu2+-coated YSi SPA beads (GE # RPNQ00096), 181 μM 14C-urea (55mCi/mmol), 50–250 ng dvUT, and appropriate concentrations of cold urea or DMU were combined per assay in clear bottomed/while-wall 96-well plates in buffer containing 150 mM Tris/Mes, pH 7.5, with 50 mM NaCl, 20% glycerol, 1 mM tris(2-carboxyethyl)phosphine, and 0.1% n-dodecyl-β-D-maltoside (DDM). Reactions performed in the presence of 400 mM imidazole served as negative controls for all conditions tested.
Full Methods
Homology screen, protein purification and crystallization
A total of 14 UT genes were amplified by PCR from the genomic DNA of the following bacteria: Actinobacillus pleuropneumoniae, Bacteroides fragilis, Colwellia psychrerythraea, Desulfovibrio vulgaris, Nitrosomonas europaea, Ochrobactrum anthropi, Pseudomonas aeruginosa, Pseudomonas fluorescens, Pseudomonas putida, Staphylococcus epidermidis, Staphylococcus saprophyticus, Yersinia frederiksenii, Yersinia mollaretti, and Yersinia pseudotuberculosis. Each gene was first cloned into modified pET plasmids (Invitrogen Inc) that produced either an N- or C-terminal his-tagged protein. Small scale (2 Liter) test purifications were conducted, and Desulfovibrio vulgaris UT (dvUT) and Yersinia frederiksenii UT yielded stable detergent-solublized protein as judged by elution as a single mono-dispersed peak from a size-exclusion column. The identity of the purified protein was verified by mass spectrometry. Only dvUT yielded diffracting crystals, and was the focus of further experiments.
To increase the expression level, dvUT was cloned into a modified pET-SUMO plasmid (Invitrogen) with an N-terminal polyhistidine tag and a SUMO domain. The protein was expressed in E. coli BL21(DE3) cells, solubilized with 30 mM n-dodecyl-β-D-maltoside (DDM), and purified on a cobalt affinity column (Clontech Inc). After cleavage of the His tag and SUMO domain by incubation with SUMO protease, the protein was exchanged into a buffer of 300 mM NaCl, 20 mM HEPES pH 7.5, 5 mM β-mercaptoethanol, and 40 mM low-purity n-octyl-β-D-maltoside (OM, Sol-Grade from Anatrace) on a Superdex 200 10/300 GL gel filtration column (GE Health Sciences). Each liter of cell culture yielded 0.2–0.3 mg of dvUT after the gel filtration step. The final protein concentration was ~8 mg/mL as approximated by UV absorbance. Native crystals were grown by vapour diffusion in unmixed sitting drops formed by combining 2 μL of the protein solution with an equal volume of well solution containing 22% polyethylene glycol PEG1500 and 100 mM sodium cacodylate, pH 6.5. Derivatized crystals were grown by including 10 mM thiomersal or potassium gold cyanide in the drop solution. Before flash-freezing in liquid nitrogen, the crystals were cryoprotected by gradually increasing the concentration of PEG1500 in the well solution to 45% over a period of 30 hours. The native crystals diffracted to resolutions of up to 3.8 Å and indexed to the P3(1) space group, with unit cell dimensions a and c of 102.9 and 141.7 Å. Curiously, crystals grown from protein purified in high-purity OM (AnaGrade from Anatrace) uniformly failed to diffract to better than 4.5–5 Å. Inclusion of heavy atoms changed the space group to P6(3) with unit cell dimensions a and c equal to 110.13 and 84.86, and improved resolution to up to 2.3 Å. The DMU-bound crystals were obtained by incubating the protein with 10 mM ligand at 20ºC for 30 min prior to setting up drops in the same condition used to obtain the gold-derivatized crystals.
Data collection and structure solution
Diffraction data were collected on beamlines X25 and X29 at the NSLS and on beamline 24ID-E at the Advanced Photon Source. Two datasets were used for structure solution: a 2.5 Å mercury dataset collected at a wavelength of 1.01 Å, and a 2.3 Å gold dataset collected at a wavelength of 1.04 Å. The data were indexed, integrated and scaled using the HKL2000 software suite36. The mercury dataset exhibited a stronger anomalous signal and was therefore used to obtain the initial phases. Eight heavy atom sites were located by SAD using the programs SHELX37 and phenix.hyss38. The programs Phaser39 and RESOLVE as run by phenix.autosol were used to calculate experimental phases, carry out density modification, and build a partial model containing 213 of the total 337 residues. An additional 32 residues were added manually in Coot40, and the improved model was used to calculate an improved map using combined experimental and model phases in PHASER. After density modification in DM41, ARP/wARP42 was able to dock 189 residues in sequence. Iterative rounds of map calculation, density modification and manual building were used until the model was roughly 85% complete, and then it was used as a molecular replacement model for the gold dataset with Molrep43. The structure was completed using Refmac544 and Coot. For the later cycles of refinement, 4 TLS groups identified by TLSMD45 were included, and the model geometry was analyzed using Molprobity. The final model was complete except for three C-terminal residues and four residues from a disordered loop. The final structure was then used as a model for molecular replacement for a 2.5 Å dataset collected on the DMU-bound crystals. After rigid-body refinement of the molecular replacement solution, the DMU-bound model was refined by simulated annealing in CNS46 and automatic and manual refinement in Refmac5 and Coot.
Tracer uptake in Xenopus laevis oocytes
dvUT and human UT-B (acc NM_015865) were cloned into a modified pBluescript vector for in vitro transcription and the mRNAs were purified by the Trizol reagent (Invitrogen). For each measurement, 10 oocytes were transferred to a transport vial that contained 360 μl transport buffer that completely covers the oocytes and that is composed of 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, and 5mM HEPES, pH 7.6, and supplemented with 181 μM of 14C-urea at 55 mCi/mmol. A timer was started immediately after oocyte transfer. The reaction was stopped at the desired time point by adding 4 ml of ice-cold buffer without radiotracer and aspirated after a brief mixture by swirling the vial. The oocytes were washed three times with the same buffer. After the last wash each oocyte was transferred into a separate scintillation vial that contained 200 μl of 10% SDS, and vortexed vigorously. After an oocyte was dissolved, 5 ml of scintillation cocktail was added to each vial and vortexed. Radioactivity in each vial was determined by liquid scintillation counting. For standards, 3 × 10 μl of each transport buffer was used plus 200 μl of 10 % SDS and 2 ml scintillation cocktail.
SPA-based binding assay
Cu2+-coated YSi SPA beads (GE # RPNQ00096) were diluted to 2.5 mg/mL in 150 mM Tris/Mes, pH 7.5, with 50 mM NaCl, 20% glycerol, 1 mM tris(2-carboxyethyl)phosphine (TCEP, Sigma Co), 0.1% n-dodecyl-β-D-maltopyranoside (Anatrace Inc.) with purified His-tagged dvUT (50 – 250 ng per assay) and radiolabeled urea. 100 μL of SPA-bead/protein/radiotracer solution was added to individual wells of clear-bottom/white-wall 96-well plates. For isotopic dilution and competition experiments depicted in Fig. 1e the final concentration of 14C-urea was kept constant at 181 μM [55 mCi/mmol] whereas the concentration of non-labelled urea or DMU was increased from 0 – 50 mM. Saturation binding (as shown in Fig. 1d) was performed with increasing concentrations of 14C-urea at a specific activity of 2 mCi/mmol. Binding was performed in the dark for 30 min at 4ºC with vigorous shaking on a vibrating platform. To determine the non-specific background binding activity 400 mM imidazole was added to the wells because imidazole competes with the His-tag of the recombinant protein. Plates were read in the SPA mode of a Wallac 1450 MicroBeta™ plate PMT counter. Total counts per minute (cpm) were corrected for non-specific binding by subtracting the cpm of the samples in the presence of imidazole, yielding the specific cpm. To prevent radioligand depletion in the saturation binding experiments 250 ng of purified dvUT, a protein amount substantially below the capacity of the SPA beads, was used per assay. The efficiency of detection was calculated with a standard curve of known amounts of radioactivity (determined by adding of scintillation cocktail to the samples), and the slope of this linear relation was used to transform cpm into mol/L17. All experiments were performed at least in duplicate with replicas of ≥3 and data are expressed as mean ± standard error. Data fits of kinetic analyses were performed using non-linear regression algorithms in Sigmaplot (SPSS Inc). and errors represent the S.E.M. of the fit.
Supplementary Material
Acknowledgments
We thank Dr. R. MacKinnon for advice and support throughout the project, Drs. C. Miller and E. Gouaux for comments on the manuscript, Dr. J. Love for cloning and initial screen of protein expression levels, Dr. Y. Pan for mRNA preparation and oocyte injection, and Drs J. Weng and Y. Cao for crystal screening and data collection at the synchrotrons. Data for this study were measured at beamlines X4A, X4C, X25, and X29 of the National Synchrotron Light Source and the NE-CAT 24ID-E at the Advanced Photon Source. This work was supported by the US National Institutes of Health (HL086392 to M.Z. and T32HL087745 to EJL), the New York Consortium for Membrane Protein Structure which is supported by the NIH Roadmap and Protein Structure Initiatives PSI-II (GM075026 to W. A. Hendrickson), and the American Heart Association (0630148N to M.Z.). M.Z is a Pew Scholar in Biomedical Sciences.
Footnotes
Atomic coordinates and structure factors have been deposited with the Protein, Data Bank under accession IDs 3K3F and 3K3G.
Supplementary Information accompanies the paper.
Author Contributions E.J.L. and M.Z. conceived and designed the experiments. E.J.L. purified and crystallized the protein; M.Q. performed and analyzed the radiotracer flux and SPA assays; E. J. L. and M. Z. collected and processed the X-ray data, solved the structure, and wrote the paper.
Reference List
- 1.Sebbane F, et al. The Yersinia pseudotuberculosis Yut protein, a new type of urea transporter homologous to eukaryotic channels and functionally interchangeable in vitro with the Helicobacter pylori UreI protein. Mol Microbiol. 2002;45:1165–1174. doi: 10.1046/j.1365-2958.2002.03096.x. [DOI] [PubMed] [Google Scholar]
- 2.Weeks DL, Eskandari S, Scott DR, Sachs G. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 3.Hediger MA, et al. Structure, regulation and physiological roles of urea transporters. Kidney Int. 1996;49:1615–1623. doi: 10.1038/ki.1996.235. [DOI] [PubMed] [Google Scholar]
- 4.Sands JM. Mammalian urea transporters. Annu Rev Physiol. 2003;65:543–566. doi: 10.1146/annurev.physiol.65.092101.142638. [DOI] [PubMed] [Google Scholar]
- 5.Bagnasco SM. Role and regulation of urea transporters. Pflugers Arch. 2005;450:217–226. doi: 10.1007/s00424-005-1403-9. [DOI] [PubMed] [Google Scholar]
- 6.Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J Gen Physiol. 1976;68:127–135. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valladares A, Montesinos ML, Herrero A, Flores E. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol Microbiol. 2002;43:703–715. doi: 10.1046/j.1365-2958.2002.02778.x. [DOI] [PubMed] [Google Scholar]
- 8.Kojima S, Bohner A, von Wiren N. Molecular mechanisms of urea transport in plants. J Membr Biol. 2006;212:83–91. doi: 10.1007/s00232-006-0868-6. [DOI] [PubMed] [Google Scholar]
- 9.You G, et al. Cloning and characterization of the vasopressin-regulated urea transporter. Nature. 1993;365:844–847. doi: 10.1038/365844a0. [DOI] [PubMed] [Google Scholar]
- 10.MacIver B, Smith CP, Hill WG, Zeidel ML. Functional characterization of mouse urea transporters UT-A2 and UT-A3 expressed in purified Xenopus laevis oocyte plasma membranes. Am J Physiol Renal Physiol. 2008;294:F956–F964. doi: 10.1152/ajprenal.00229.2007. [DOI] [PubMed] [Google Scholar]
- 11.Raunser S, et al. Oligomeric structure and functional characterization of the urea transporter from Actinobacillus pleuropneumoniae. J Mol Biol. 2009;387:619–627. doi: 10.1016/j.jmb.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao D, Sonawane ND, Levin MH, Yang B. Comparative transport efficiencies of urea analogues through urea transporter UT-B. Biochim Biophys Acta. 2007;1768:1815–1821. doi: 10.1016/j.bbamem.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Mannuzzu LM, Moronne MM, Macey RI. Estimate of the number of urea transport sites in erythrocyte ghosts using a hydrophobic mercurial. J Membr Biol. 1993;133:85–97. doi: 10.1007/BF00231880. [DOI] [PubMed] [Google Scholar]
- 14.Minocha R, Studley K, Saier MH., Jr The urea transporter (UT) family: bioinformatic analyses leading to structural, functional, and evolutionary predictions. Receptors. Channels. 2003;9:345–352. doi: 10.3109/714041015. [DOI] [PubMed] [Google Scholar]
- 15.Chou CL, Knepper MA. Inhibition of urea transport in inner medullary collecting duct by phloretin and urea analogues. Am J Physiol. 1989;257:F359–F365. doi: 10.1152/ajprenal.1989.257.3.F359. [DOI] [PubMed] [Google Scholar]
- 16.Quick M, Javitch JA. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc Natl Acad Sci U S A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter: sodium symporter - inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–677. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 19.Shayakul C, Steel A, Hediger MA. Molecular cloning and characterization of the vasopressin-regulated urea transporter of rat kidney collecting ducts. J Clin Invest. 1996;98:2580–2587. doi: 10.1172/JCI119077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford AD, et al. 97- and 117-kDa forms of collecting duct urea transporter UT-A1 are due to different states of glycosylation. Am J Physiol Renal Physiol. 2001;281:F133–F143. doi: 10.1152/ajprenal.2001.281.1.F133. [DOI] [PubMed] [Google Scholar]
- 21.Lucien N, et al. Antigenic and functional properties of the human red blood cell urea transporter hUT-B1. J Biol Chem. 2002;277:34101–34108. doi: 10.1074/jbc.M205073200. [DOI] [PubMed] [Google Scholar]
- 22.Yang B, Verkman AS. Analysis of double knockout mice lacking aquaporin-1 and urea transporter UT-B. Evidence for UT-B-facilitated water transport in erythrocytes. J Biol Chem. 2002;277:36782–36786. doi: 10.1074/jbc.M206948200. [DOI] [PubMed] [Google Scholar]
- 23.Imai YN, Inoue Y, Nakanishi I, Kitaura K. Amide-pi interactions between formamide and benzene. J Comput Chem. 2009;30:2267–2276. doi: 10.1002/jcc.21212. [DOI] [PubMed] [Google Scholar]
- 24.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 25.Dutzler R, Campbell EB, Cadene M, Chait BT, MacKinnon R. X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of anion selectivity. Nature. 2002;415:287–294. doi: 10.1038/415287a. [DOI] [PubMed] [Google Scholar]
- 26.Terris JM, Knepper MA, Wade JB. UT-A3: localization and characterization of an additional urea transporter isoform in the IMCD. Am J Physiol Renal Physiol. 2001;280:F325–F332. doi: 10.1152/ajprenal.2001.280.2.F325. [DOI] [PubMed] [Google Scholar]
- 27.Blount MA, Klein JD, Martin CF, Tchapyjnikov D, Sands JM. Forskolin stimulates phosphorylation and membrane accumulation of UT-A3. Am J Physiol Renal Physiol. 2007;293:F1308–F1313. doi: 10.1152/ajprenal.00197.2007. [DOI] [PubMed] [Google Scholar]
- 28.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata K, et al. Structural determinants of water permeation through aquaporin-1. Nature. 2000;407:599–605. doi: 10.1038/35036519. [DOI] [PubMed] [Google Scholar]
- 31.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 32.Fu D, et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 33.Levin MH, de la Fuente R, Verkman AS. Urearetics: a small molecule screen yields nanomolar potency inhibitors of urea transporter UT-B. FASEB J. 2007;21:551–563. doi: 10.1096/fj.06-6979com. [DOI] [PubMed] [Google Scholar]
- 34.Shayakul C, Hediger MA. The SLC14 gene family of urea transporters. Pflugers Arch. 2004;447:603–609. doi: 10.1007/s00424-003-1124-x. [DOI] [PubMed] [Google Scholar]
- 35.Zhang C, Sands JM, Klein JD. Vasopressin rapidly increases phosphorylation of UT-A1 urea transporter in rat IMCDs through PKA. Am J Physiol Renal Physiol. 2002;282:F85–F90. doi: 10.1152/ajprenal.0054.2001. [DOI] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 38.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project number 4 The CCP4 suite: programs for protein cystallography. Acta Crystallog sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Potterton L, et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 43.Lebedev AA, Vagin AA, Murshudov GN. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr D Biol Crystallogr. 2008;64:33–39. doi: 10.1107/S0907444907049839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 45.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 46.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallog sect D. 1998;54 (Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.