Abstract
SUMMARY
Pyruvate dehydrogenase kinase (PDK) isoforms are molecular switches that downregulate the pyruvate dehydrogenase complex (PDC) by reversible phosphorylation in mitochondria. We have determined structures of human PDK1 or PDK3 bound to the inhibitors AZD7545, dichloroacetate (DCA), and radicicol. We show that the trifluoromethylpropanamide end of AZD7545 projects into the lipoyl-binding pocket of PDK1. This interaction results in inhibition of PDK1 and PDK3 activities by aborting kinase binding to the PDC scaffold. Paradoxically, AZD7545 at saturating concentrations robustly increases scaffold-free PDK3 activity, similar to the inner lipoyl domain. Good DCA density is present in the helix bundle in the N-terminal domain of PDK1. Bound DCA promotes local conformational changes that are communicated to both nucleotide-binding and lipoyl-binding pockets of PDK1, leading to the inactivation of kinase activity. Finally, radicicol inhibits kinase activity by binding directly to the ATP-binding pocket of PDK3, similar to Hsp90 and Topo VI from the same ATPase/kinase superfamily.
INTRODUCTION
The pyruvate dehydrogenase complex (PDC) is a member of the highly conserved mitochondrial α-ketoacid dehydrogenase complexes comprising the PDC, the branched-chain α-ketoacid dehydrogenase complex (BCKDC), and the α-ketoglutarate dehydrogenase complex (Patel and Roche, 1990; Reed, 2001). PDC catalyzes the oxidative decarboxylation of pyruvate to give rise to acetyl-CoA, linking glycolysis to the Krebs cycle. The 9 × 106 dalton mammalian PDC is organized around a 60-meric dodecahedral core consisting of acetyltransferase (E2p) and E3-binding protein (E3BP) (Hiromasa et al., 2004). Through N-terminal peripheral domains of the two constituent proteins, the E2p/E3BP core binds multiple copies of pyruvate dehydrogenase (E1p), dihydrolipoamide dehydrogenase (E3), pyruvate dehydrogenase kinase (PDK), and pyruvate dehydrogenase phosphatase (PDP) (Reed, 2001).
The mammalian PDC is tightly regulated by reversible phosphorylation. The phosphorylation of specific serine residues in E1p by PDK results in the inactivation of PDC, whereas the dephosphorylation by PDP restores PDC activity (Harris et al., 2001; Holness and Sugden, 2003). Phosphorylation of E1p occurs at three serine residues (Ser264, site 1; Ser271, site 2; and Ser203, site 3) (Sale and Randle, 1981; Teague et al., 1979; Yeaman et al., 1978), although phosphorylation of each site alone inactivates PDC (Kolobova et al., 2001; Korotchkina and Patel, 2001a). To date, four PDK isoforms (1–4) in human mitochondria have been identified (Popov et al., 1997). Each PDK isoform exhibits different site specificity: all four isoforms phosphorylate sites 1 and 2 at different rates (for site 1, PDK2 > PDK4 ≈ PDK1 > PDK3; for site 2, PDK3 > PDK4 > PDK2 > PDK1), but only PDK1 modifies site 3 (Kolobova et al., 2001; Korotchkina and Patel, 2001b).
PDKs are recruited to the PDC by preferentially binding to the inner lipoyl (L2) domain of the E2p subunit in the E2p/E3BP core (Liu et al., 1995; Patel and Roche, 1990). Binding of PDKs to the L2 domain requires a lipoyl group covalently attached to Lys173 of L2 (Radke et al., 1993). Colocalization of PDK with the E1p substrate bound to the E1-binding domain (immediately downstream of L2) of E2p results in enhanced PDK activity. Individual isoforms exhibit different binding affinities for L2, with PDK3 > PDK1 ≈ PDK2 > PDK4 (Tuganova et al., 2002). PDK3, which binds to L2 most tightly, is robustly activated by the E2p/E3BP core; the majority of this activation is achieved by binding to isolated L2 (Baker et al., 2000; Roche et al., 2003). PDK1 and PDK2 activities are augmented by the E2p/E3BP core by 2.5-fold and 10-fold, respectively, but not by binding to L2 alone (Baker et al., 2000; Roche and Hiromasa, 2007). PDK4 has the lowest affinity for L2, and is only minimally stimulated by the E2p/E3BP core (Roche and Hiromasa, 2007).
Mitochondrial protein kinases comprising PDKs and the related BCKD kinase (BCK) constitute a novel family of protein kinases (Popov et al., 1997), in which motifs that normally occur in eukaryotic Ser/Thr/Tyr kinases (Hanks et al., 1988) are absent. Structural studies of PDK and BCK have revealed that these kinases consist of two distinct domains: the N- and C-terminal domains (Kato et al., 2005; Machius et al., 2001; Steussy et al., 2001). The N-terminal domain of PDK (or B domain in BCK) consists of eight a helices with a four-helix bundle-like structure forming the core. The C-terminal domain (or K domain in BCK) contains the phosphotransfer catalytic site whose architecture is conserved in the GHKL ATPase/kinase superfamily including DNA gyrase B, Hsp90, histidine kinases, and MutL (Dutta and Inouye, 2000). Members of this superfamily share four conserved motifs (N, G1, G2, and G3 boxes), which form a unique ATP-binding fold (Alex and Simon, 1994; Bergerat et al., 1997; Dutta and Inouye, 2000; Smirnova et al., 1998). This fold includes a common structural element known as the “ATP lid,” whose conformational change is coupled to both ATP hydrolysis and protein-protein interactions (Ban et al., 1999; Machius et al., 2001; Wigley et al., 1991). The crystal structure of the PDK3-L2 complex has revealed that L2 binds to the lipoyl-binding pocket in the N-terminal domain of PDK3. This binding causes the open conformation of the active-site cleft in PDK3, in contrast to the closed conformation of rat PDK2 free from L2 (Kato et al., 2005; Steussy et al., 2001). The open active-site cleft destabilizes the ATP lid located inside the ATP-binding pocket of PDK3, which facilitates an ADP/ATP exchange to remove product inhibition exerted by ADP.
PDK isoforms exhibit tissue-specific expression; PDK1 is detected in heart, pancreatic islets, and skeletal muscles; PDK2 is expressed in all tissues; PDK3 is present in testes, kidney, and brain; and PDK4 is abundant in heart, skeletal muscle, kidney, and pancreatic islets (Bowker-Kinley et al., 1998). The expression of PDK2 and PDK4 is induced in starvation and diabetes, which is reversed by insulin treatment (Harris et al., 2001; Wu et al., 1998). Impaired insulin-induced downregulation of PDK4 (due to the lack of insulin or insensitivity to insulin) leads to the overexpression of PDK4 and shuts off glucose oxidation in diabetic animals (Holness and Sugden, 2003; Roche et al., 2003). Therefore, PDK4 is a potential drug target for the treatment of type II diabetes. Furthermore, very recent studies from three different groups showed that PDK1 is specifically upregulated by HIF-1 in hypoxic cancer cells, which in turn reduces PDC activity (Kim et al., 2006; Koukourakis et al., 2005; Papandreou et al., 2006). Thus, PDK1 was suggested to be a potential target for antitumor therapies (Pan and Mak, 2007; Simon, 2006). Selective inactivation of PDK isoforms by the specific inhibitor dichloroacetate (DCA) has been shown to derepress a mitochondrial potassium-ion channel axis, trigger apoptosis in cancer cells, and inhibit tumor growth (Bonnet et al., 2007; Cairns et al., 2007).
Among known synthetic inhibitors for PDK, the pyruvate analog DCA (Figure 1) is the most common classic inhibitor for PDK isoforms (Bersin and Stacpoole, 1997; Whitehouse et al., 1974), and has been shown to have beneficial effects in diabetes, lactic acidosis, and myocardial ischemia (Bersin and Stacpoole, 1997; Henderson et al., 1997; Stacpoole, 1989). Biochemical studies indicated that DCA and ADP synergistically inhibit activities of PDK isoforms (Bao et al., 2004; Pratt and Roche, 1979). Recent mutational analyses and the PDK2 structure in complex with DCA revealed that DCA binds to the N-terminal domain of PDK2 (Klyuyeva et al., 2007; Knoechel et al., 2006). However, the inhibition mechanism of DCA is not known, as no structural change in PDK2 was observed upon DCA binding. The glucose-lowering compound AZD7545, (2R)-N-{4-[4-(dimethylcarbamoyl)phenylsulfonyl]-2-chlorophenyl}-3,3,3-trifluoro-2-hydroxy-2-methylpropanamide (Figure 1), manufactured by AstraZeneca, UK, has been shown to inhibit PDK2, resulting in enhanced glucose breakdown through the PDC flux in tissues from diabetic animals (Mayers et al., 2005; Morrell et al., 2003). The related inhibitors AZ12 and Nov3r were shown to bind to the lipoyl-binding pocket in the N-terminal domain of PDK2 (Knoechel et al., 2006). Therefore, it would appear that this group of compounds indirectly exerts its inhibitory effect by preventing PDK from binding to the E2p/E3BP core. Finally, the antitumor compound radicicol (Figure 1) inhibits the activities of Hsp90 and Topo VI by blocking ATP binding to these enzymes (Corbett and Berger, 2005; Roe et al., 1999). PDK2 and related BCK are also inhibited by radicicol (Besant et al., 2002; Tuganova et al., 2001). As PDK and BCK are members of the GHKL ATPase/kinase superfamily, which includes Hsp90 and Topo VI, it has been proposed that radicicol inhibits PDK isoforms in the same manner as it inhibits Hsp90 and Topo VI (Tuganova et al., 2001).
Figure 1.
Molecular Structures of PDK Inhibitors AZD7545, Dichloroacetate, and Radicicol
To provide the structural basis for the inhibitory mechanism of PDKs by these three classes of synthetic inhibitors, we have determined the crystal structures of the human PDK1-AZD7545, PDK1-DCA, and PDK3-L2-radicicol complexes.
RESULTS AND DISCUSSION
Inhibition of Human PDK Isoforms by Specific Inhibitors
We studied the inhibition of SUMO (small ubiquitin modifier)-tagged PDK1 and SUMO-PDK3 activities by AZD7545, DCA, and radicicol (Figure 2). To estimate the half-maximal inhibitory concentration (IC50) for AZD7545, PDK1 or PDK3 along with the E2p/E3BP core and the pyruvate dehydrogenase (E1p) substrate were incubated with increasing stoichiometric excesses of AZD7545 in 2% dimethyl sulfoxide (DMSO), followed by assays for residual kinase activity. Under our assay conditions, AZD7545 shows an IC50 of 87 nM for PDK1 and 600 nM for PDK3 (Figure 2A). The IC50 for inhibition of PDK1 estimated here is greater than 2-fold higher than that reported previously (Morrell et al., 2003), but a lower kinase concentration (40 nM versus 50 nM in the present study) was used in the earlier work (R.M. Mayers, personal communication), and IC50 values are enzyme concentration dependent. When measured similarly, radicicol exhibits an IC50 of 230 mM for the inhibition of PDK1 activity (at 0.56 μM kinase concentration) and an IC50 of 400 μM for PDK3 (at 0.28 μM concentration) (Figure 2B).
Figure 2. Inhibition of PDK1 and PDK3 Activities by AZD7545, Radicicol, and DCA.
(A) IC50 for AZD7545, SUMO-PDK1, or SUMO-PDK3 (50 nM each) in the presence of E1p bound to the E2p/E3BP core was titrated with increasing concentrations of AZD7545 in 2% DMSO as indicated. Following an incubation of 30 min, residual kinase activity was assayed as described in Experimental Procedures. PDK activity is expressed as % maximal (8.2 nmol/min/mg for PDK1; 63.6 nmol/min/mg for PDK3; both measured in 2% DMSO). Inhibition curves were fitted and IC50 values were obtained using the Prism program (GraphPad Software, San Diego, CA, USA).
(B) IC50 for radicicol. For titrations with radicicol, 0.56 μM SUMO-PDK1 or 0.28 μM SUMO-PDK3 was used. The remaining conditions are also described in Experimental Procedures.
(C) Inhibition of PDK1 and PDK3 by AZD7545, DCA, and radicicol with the E1p substrate on the E2p/E3BP core. Inhibitor concentrations used were 5- to 40-fold higher than the respective IC50 or Ki values. Following incubation for 30 min, kinase activity was assayed as in (A), and residual activity levels are expressed as % control. Control activities for PDK1 and PDK3 in 2% DMSO are as in (A). Control activity without DMSO for DCA inhibition is 9.7 nmol/min/mg for PDK1 and 91.9 nmol/min/mg for PDK3. The bars in (C) and (D) represent averages of two independent experiments.
(D) Inhibition of PDK1 and PDK3 in the absence of the E2p/E3BP core. Basal kinase activities were assayed with free E1p as a substrate, with the remaining conditions as in (A). Control activities in 2% DMSO for AZD7545 and radicicol inhibitions are 6.3 nmol/min/mg for PDK1 and 5.7 nmol/min/mg for PDK3. Control activities without DMSO for DCA inhibition are 6.4 nmol/min/mg for PDK1 and 4.9 nmol/min/mg for PDK3. Note that scales on the ordinates (relative activity, % control) are different between (C) and (D).
We next determined the degree of inhibition of PDK1 and PDK3 activities by saturating concentrations of the inhibitors, with either E1p bound to the E2p/E3BP core (scaffold-dependent kinase activity) or free E1p (scaffold-free kinase activity) as substrate. The concentrations of the inhibitors AZD7545 and radicicol used were at least 10-fold higher than the respective IC50 values for PDK1 and PDK3 (Figures 2A and 2B). In the presence of the E2p/E3BP core, AZD7545 at 10 μM reduces PDK1 and PDK3 activities to 28% and 31% of the control, respectively (Figure 2C). These data are consistent with an earlier proposition that AZD7545 inhibits the E2p-dependent stimulation of PDK2 activity (Morrell et al., 2003; Tuganova et al., 2007). Paradoxically, when the E2p/E3BP core is absent, AZD7545 stimulates scaffold-free basal PDK1 and PDK3 activities to 1.3-fold and 10-fold, respectively (Figure 2D). The 10-fold increase in basal PDK3 activity by AZD7545 is comparable to the fold increase of basal PDK3 activity by the isolated L2 domain (Baker et al., 2000; Kato et al., 2005). The significance of the robust stimulation of basal PDK3 activity by AZD7545 will be discussed.
With the E2p/E3BP core, DCA at 40 mM decreases PDK1 and PDK3 activities to 4% and 28% of the control, respectively (Figures 2C). A saturating DCA concentration was used in the inhibition assay, based on the IC50 for DCA of greater than 1 mM for PDK isoforms (Baker et al., 2000; Bebernitz et al., 2000) and the apparent Ki of 1 mM and 8 mM for PDK1 and PDK3, respectively (Bowker-Kinley et al., 1998). PDK3 with E2p was previously reported not to be significantly inhibited by DCA; however, a nonsaturating DCA concentration (1 mM) was used in those studies (Baker et al., 2000; Roche and Hiromasa, 2007). When the E2p/E3BP scaffold-free activity was measured, DCA similarly reduces PDK1 and PDK3 activities to 4% and 17% of the control, respectively (Figure 2D). In parallel, radicicol at 4 mM inhibits the E2p/E3BP core-dependent PDK1 and PDK3 activities to 4% and 2% of the control, respectively (Figure 2C). With free E1p as a substrate, the same concentration of radicicol reduces the scaffold-free activities of PDK1 and PDK3 to 5% each of the control. The similar degrees of inhibition by DCA and radicicol both in the absence and presence of the E2p/E3BP core suggest that these compounds mainly inhibit the catalytic activity of the kinase. To provide the structural basis for the mechanisms of inactivation by these inhibitors, we have determined the crystal structures of apo-PDK1 as well as the PDK1-AZD7545, PDK1-DCA, and PDK3-L2-radicicol complexes.
Crystal Structure of Human PDK1
The human PDK1 structure was determined maximally at 1.9 Å resolution (Figure 3A; Table 1; see Figure S1 in the Supplemental Data available with this article online). The three structures of PDK1 (apo, AZD7545-bound, and DCA-bound) are virtually the same except that the structure of PDK1-DCA shows a small conformational change in the N-terminal domain (see below). The chain fold of apo-PDK1 is similar to those of rat PDK2 (root-mean-square deviation [rmsd] = 1.2 Å for 320 Cα atoms), human PDK2 (rmsd = 1.3 Å for 329 Cα atoms), and human PDK3 in the PDK3-L2 complex (rmsd = 1.0 Å for 343 Cα atoms) (Kato et al., 2005; Knoechel et al., 2006; Steussy et al., 2001). A noticeable difference between PDK1 and the other PDK isoforms is observed at the ATP-binding pocket, where apo-PDK1 has a completely ordered ATP lid (residues 336–356), whereas its equivalents in PDK2 and PDK3 are disordered to different extents depending on the bound nucleotide state. The ATP lid in PDK1 projects outward, with Glu345 in the ATP lid forming hydrogen bonds (H bonds) with Tyr403′ and Lys92′ in a symmetry-related molecule (Figure 3C). Arg348 in the ATP lid also makes H bonds with Asp241′ and Asn246′ in a second symmetry-related molecule. Therefore, when a nucleotide binds to the ATP-binding pocket, the ordered ATP lid must be reorganized to accommodate the phosphate group of the nucleotide. Significant cracking of PDK1 crystals occurred upon soaking with ATP or ADP, indicating that the above interactions are critical for the crystal packing of apo-PDK1.
Figure 3. Structures of PDK1 and PDK3-L2 in Complex with Inhibitors.
(A) The dimeric structures of apo-PDK1 with superimposed AZD7545 and DCA bound to PDK1. Each PDK1 subunit is represented as a ribbon model (green and cyan). AZD7545 and DCA molecules bound to each PDK1 subunit are shown in space-filling models. The fully ordered ATP lid is colored in yellow. The partially ordered C-terminal tails in the crossed configuration are colored in magenta. (B) The PDK3-L2 dimer with radicicol bound to the ATP-binding pocket. Both PDK3 subunits are shown in green and cyan. The bound L2 domains are colored in yellow, with the lipoyl group shown in a space-filling model. Radicicol molecules bound to each PDK3 subunit are also shown in the same model. The fully ordered C-terminal tails are in magenta. The red arrows in (A) and (B) indicate the active-site clefts of these kinases. Stereo figures of the Cα traces for the PDK1 and PDK3-L2 structures are shown in Figure S1.
(C) The close-up view of the fully ordered ATP lid (yellow) from one PDK1 subunit (cyan). Two symmetry-related PDK1 molecules are shown in gray and pink, respectively. The bound ATP in the structure of PDK3-L2-ATP published earlier (PDB ID code 1Y8P; Kato et al., 2005) is superimposed. Key residues for holding the ordered conformation of the ATP lid are shown in stick models.
(D) The interaction between the C-terminal tail from one PDK1 subunit (shown as a stick model in cyan) and the other subunit (shown as an electro-static surface) in the dimer. The surface of the latter is colored in blue for the positive charge and in red for the negative charge. Dotted lines represent disordered regions of the C-terminal tail.
Table 1.
Data Collection and Refinement Statistics
| Apo-PDK1 | PDK1-AZD7545 | PDK1-DCA | PDK3-L2-Radicicol | |
|---|---|---|---|---|
| Data Collectiona | ||||
| Space group | P41212 | P41212 | P41212 | P6522 |
| Unit cell (Å) | ||||
| a = b | 98.19 | 96.98 | 98.32 | 120.87 |
| c | 110.40 | 111.35 | 110.60 | 239.64 |
| Wavelength (Å) | 0.98 | 0.98 | 0.98 | 0.98 |
| Resolution (Å) | 2.03 | 1.90 | 2.00 | 2.60 |
| Unique reflections | 34,783 | 42,222 | 37,074 | 32,612 |
| Completeness (%) | 97.6 (96.7) | 99.3 (100) | 99.4 (100) | 100 (100) |
| Rmerge (%)b | 4.5 (53.6) | 6.3 (43.4) | 4.0 (56.1) | 5.4 (69.4) |
| <I>/<σ(I)> | 41.9 (2.6) | 29.6 (4.7) | 38.1 (3.2) | 33.4 (2.6) |
| Multiplicity | 9.2 (5.8) | 9.1 (8.7) | 7.4 (7.8) | 7.7 (7.1) |
| Refinementa | ||||
| Number of reflections (work/test) | 33,007/1,737 | 38,495/2,058 | 34,295/1,802 | 30,887/1,647 |
| Number of atoms (mean B value) (Å2) | ||||
| Protein | 2,969 (53.6) | 2,990 (30.7) | 2,972 (43.6) | 3,731 (55.6) |
| Solvents | 212 (46.3) | 315 (39.1) | 242 (44.3) | 81 (45.4) |
| Inhibitor | — | 31 (35.9) | 6 (68.0) | 25 (42.3) |
| Missing residue regions | 30–40, 68–70, 168–169, 204–214, 415–416, 424–436 | 30–40, 168–169, 204–214, 415–416, 424–436 | 30–40, 68–70, 168–169, 204–214, 415–416, 424–436 | 9, 12, 172–175, 307–324, 402–406 in PDK3; 126–127, 221–233 in L2 |
| Rwork (%)c | 19.0 (24.2) | 21.12 (36.3)d | 23.0 (37.2)d | 20.5 (33.3) |
| Rfree (%)c | 21.2 (31.5) | 24.9 (42.1)d | 27.7 (44.1)d | 23.8 (39.4) |
| Rmsd | ||||
| Bond length (Å) | 0.021 | 0.019 | 0.014 | 0.020 |
| Bond angle (°) | 1.527 | 1.696 | 1.378 | 1.820 |
| Ramachandran plot | ||||
| Most favored (%) | 95.0 | 94.1 | 92.7 | 91.6 |
| Allowed (%) | 5.0 | 5.9 | 7.3 | 8.4 |
| Disallowed (%) | 0 | 0 | 0 | 0 |
Values in parentheses refer to data in the highest resolution shell unless otherwise indicated.
Rmerge = ΣhklΣj|Ij — <I>|/ΣhklΣjIj, where <I> is the mean intensity of j observations from a reflection hkl and its symmetry equivalents.
Rwork = Σhkl||Fobs| — k|Fcalc||/Σhkl|Fobs|. Rfree = Rwork for 5% of reflections that were omitted from refinement.
We did not back-soak these crystals to remove excess AZD7545 and DCA. Because the scattering factor of the chloride atom is relatively high, it is likely that excess AZD7545 or DCA, which contain one and two chloride atoms, respectively, results in higher Rwork and Rfree values of the PDK-inhibitor crystals than those of the apo-PDK1 crystal.
Another noticeable conformational change is in the C-terminal tail of PDK1. The segment of residues 418–423 including the conserved residues Asp419 and Trp420 in PDK1 (designated as a “DW” motif) hovers over the other subunit of the dimer (Figures 3A and 3D). Both the side chains and main chains of Asp419 and Trp420 interact with residues Tyr179′, Arg183′, and Arg399′ in the other subunit (Figure 3D). The DW motif appears to be the linchpin that enables the C-terminal tail to anchor to the other subunit in the PDK1 dimer. Several residues preceding this segment and all the downstream residues are disordered. Due to crystallographic two-fold symmetry, the two C-terminal tails, each from one PDK1 subunit, cross over to form a “crosstail” configuration in the dimer (Figure 3A). A similar configuration was first observed in the structure of the PDK3-L2 complex, in which the fully ordered C-terminal tail from one PDK3 subunit interacts with the L2 bound to the lipoyl-binding pocket of the other PDK3 subunit (Figure 3B) (Kato et al., 2005). This fully ordered crosstail configuration locks PDK3 in the open conformation, in which the active-site clefts of the both PDK3 subunits are wider than the closed conformation present in the rat PDK2 structure with completely disordered C-terminal tails (Steussy et al., 2001). As described before, PDK3 in the open conformation is far more active than in the closed conformation, through an increased rate of ATP/ADP exchange and presumably enhanced affinity for the substrate E1p (Kato et al., 2005). Despite a lack of bound L2, the partially ordered crosstails maintain the open conformation in apo-PDK1 (Figure 3A). A similar open conformation with the partial crosstail configuration is also present in the apo and inhibitor-bound structures of human PDK2 without L2 (Knoechel et al., 2006). We suggest that in apo-PDK1 and apo-PDK2, the active sites exist in equilibrium between the closed and open conformations. The presence of bound L2 presumably locks the active sites in these PDK isoforms exclusively in the open conformation. The binding of nucleotide and/or pyruvate to human PDK2 reversibly displaces the DW motif, which in turn hampers L2 binding to PDK2 (Hiromasa et al., 2006). A loss of the crosstail configuration may promote the transformation of the PDK structure from the open to the closed conformation.
Extensive Interactions between AZD7545 and PDK1
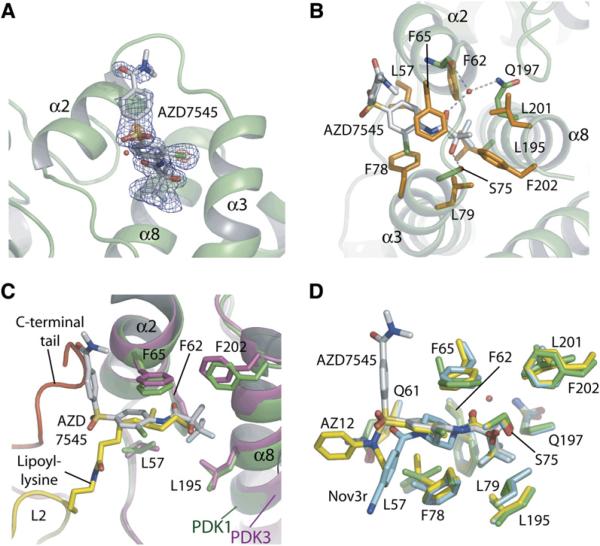
The structure of the PDK1-AZD7545 complex shows that the inhibitor AZD7545 binds to the lipoyl-binding pocket in the N-terminal domain of PDK1 (Figures 4A–4C). The loop between helices α2 and α3 is disordered in the apo-PDK1 structure, and AZD7545 binding imparts the ordering of this loop (see Figure 5D). The interior of the lipoyl-binding pocket in PDK1 is lined with highly conserved hydrophobic residues (Leu57, Phe62, Phe65, Phe78, Leu79, Leu195, Leu201, and Phe202), which form a hydrophobic interface with AZD7545 (Figure 4B). The two conserved phenylalanine residues (Phe65 and Phe78) that sandwich the 2-chlorophenyl group of the bound AZD7545 are apparently important for this interaction. Indeed, Popov's group has recently reported that the corresponding phenylalanine residues in PDK2 (Phe31 and Phe44) are essential for the inhibition of PDK2 activity by AZD7545 (Tuganova et al., 2007). The conserved hydrophobic residues in PDK3 interact exclusively with the lipoyl prosthetic group of L2 (Kato et al., 2005). The superposition of the structures of the PDK1-AZD7545 and PDK3-L2 complexes shows that AZD7545 binds to the lipoyl-binding pocket of PDK1 deeper than the lipoyl group of L2 in the PDK3-L2 structure (Figure 4C). The trifluoromethyl group of AZD7545 appears to be a mimesis of the acetyl group condensed with the lipoyl group of L2 by E1p-catalyzed reductive acetylation. The acetylated lipoyl L2 shows the highest affinity for PDK2 among the three different forms of the L2 domain (oxidized, reduced, and acetylated) (Roche et al., 2003). AZD7545 binding is further fostered by H bonds that were not observed between the lipoyl group of L2 and PDK3 in the PDK3-L2 structure (Kato et al., 2005). The conserved Ser75 in PDK1 forms an H bond with the hydroxyl group of the ligand (Figure 4B), whereas in PDK2, the corresponding serine residue (Ser41) forms H bonds with AZ12, a compound related to AZD7545 and Nov3r (Knoechel et al., 2006). These H bonds are likely a common feature for this class of inhibitors for recognition by the lipoyl-binding pockets from different PDK isoforms. A water molecule is trapped inside the lipoyl-binding pocket of PDK1, which coordinates an H-bonding network involving Phe62, Gln197, and the amide oxygen of AZD7545 (Figure 4B). An equivalent water molecule was also observed in the structure of PDK2 in complex with AZ12 (Knoechel et al., 2006). Therefore, it is tempting to suggest that a modified AZD7545 that directly forms H bonds with Phe62 and Gln197 may show a significantly increased binding affinity for PDK1. The portions of AZ12 and Nov3r that protrude outside of the lipoyl-binding pocket are partially disordered and project in different directions (Figure 4D) (Knoechel et al., 2006). In contrast, the corresponding portion in AZD7545 is fully ordered except for the two methyl groups at the amide end. This portion of AZD7545 is kinked upward almost 90° at the sulfonyl group, and contacts the molecular surface of PDK1. The corresponding surface on PDK3 interacts with the C-terminal tail from the other PDK3 subunit (Figure 4C).
Figure 4. Structure of AZD7545 Bound to the Lipoyl-Binding Pocket of PDK1.
(A) The omit electron density map of the bound AZD7545. The density is superimposed on the refined model of AZD7545 at a 3σ level in blue and a 9σ level in red, with PDK1 shown in a ribbon model. A water molecule trapped in the lipoyl-binding pocket is shown as a red ball. (B) Interactions of AZD7545 with PDK1 residues in the lipoyl-binding pocket. Hydrophobic residues of PDK1 are shown in orange. H bonds between AZD7545 and PDK residues are indicated by dashed lines. The trapped water molecule is shown as a red ball.
(C) Superimposition of bound AZD7545 in PDK1 (green) on the lipoyl lysine residue of L2 (yellow) bound to PDK3 (magenta). The C-terminal tail from the other PDK3 subunit is shown in red. For clarity, helix α3 is removed.
(D) Superimposition of bound AZD7545 in PDK1 (green) with AZ12 (yellow) and Nov3r (cyan) bound to PDK2 (Knoechel et al., 2006). Only residue numbers for PDK1 are indicated. Stereo figures of (B) and (D) are provided in Figure S2.
Figure 5. Structure of the DCA-Bindingn Site in PDK1.
(A) The DCA-binding site in the central core of the helix bundle in the PDK1 N-terminal domain. The omit electron density map is superimposed on the DCA model at the 3σ level.
(B) Structural comparison of the DCA-binding site between PDK1-DCA (green) and apo-PDK1 (pink). Residues in the unwound region are shown in stick models.
(C) Superimposition of DCA-binding sites in the structures of PDK1-DCA (green) and PDK2-DCA (yellow) (PDB ID code 2BU8; Knoechel et al., 2006). Residues surrounding the bound DCA are shown in stick models. Only residue numbers for PDK1 are shown. A stereo version of this figure is provided in Figure S3.
(D) Interactions between helix α6 and the lipoyl-binding pocket in the PDK1-DCA structure (green) superimposed with the apo-PDK1 (pink) and PDK1-AZD7545 (cyan) structures. The bound DCA and AZD7545 are shown in space-filling models. Interacting residues from helix α6 and the lipoyl-binding pocket are shown in stick models. AZD7545 binding orders the loop region between helices α2 and α3.
It is significant that in the absence of the E2p/E3BP core, AZD7545 stimulates basal PDK3 activity by 10-fold, similar to that observed with the L2 domain (Figure 2D) (Baker et al., 2000; Kato et al., 2005). The present results suggest that the binding of the trifluoromethylpropanamide group of AZD7545 to the lipoyl-binding pocket alone is able to markedly enhance scaffold-free basal PDK3 activity. However, it is presently unclear how the binding of AZD7545 robustly stimulates basal PDK3 activity, as no significant conformational changes are observed in the PDK1-AZD7545 structure compared to the apo-PDK1 structure, except for the stabilization of the lipoyl-binding pocket (see Table 2). The absence of significant reconfigurations in the PDK1-AZD7545 structure is consistent with a marginal stimulation (1.3-fold) of basal PDK1 activity by AZD7545 (Figure 2D). On the other hand, under physiological conditions, PDK3 is tightly bound to the E2p/E3BP scaffold of the mammalian PDC (Kato et al., 2005; Tuganova et al., 2002). The intercalation of AZD7545 in the lipoyl domain-binding pocket impedes the anchoring of PDK3 to the E2p/E3BP core, and prevents colocalization of the kinase to the bound E1p substrate on the same scaffold. This results in a significant inhibition of E1p phosphorylation due to substrate inaccessibility, despite the stimulation of basal PDK3 activity by this compound. Thus, AZD7545 appears to be a “dual-role” ligand for PDK3 by serving both as an inhibitor and as a potent activator for this PDK isoform, dependent on the presence and absence of the E2p/E3BP core, respectively. The activator role of AZD7545 likely makes it a useful small molecule for investigating the mechanism underlying the allosteric stimulation of basal PDK3 activity.
Table 2.
Averaged B Factors of the PDK1 Structures
| Apo-PDK1 |
PDK1-DCA |
PDK1-AZD7545 |
||||
|---|---|---|---|---|---|---|
| B Factor (A2) | Differencea (%) | B Factor (A2) | Differencea (%) | B Factor (A2) | Differencea (%) | |
| Overall (residues 41–423) |
52.7 |
—
|
43.9 |
—
|
29.5 |
—
|
| Helix 6a (residues 132–146) |
59.2 |
12.3 |
56.9 |
29.7 |
31.6 |
7.0 |
| Helix 7a (residues 152–166) | 51.1 | –2.9 | 42.6 | –2.8 | 30.0 | 1.6 |
B factors are calculated for the main-chain atoms of the indicated residue range.
Relative differences of the averaged B factors against the overall value are calculated by the following equation: difference (%) = (overall B factor – helix B factor)/overall B factor × 100.
DCA Binding Promotes Conformational Changes at the Active-Site Cleft
PDK1 crystals soaked with DCA produced unambiguous electron density for DCA in the Fo – Fc map of the PDK1-DCA complex (Figure 5A). The DCA-binding site is located inside the helix bundle in the N-terminal domain of PDK1 and is completely shielded from solvent in the apo-PDK1 structure. The shielded DCA-binding site may be responsible for the low affinity of DCA for PDK isoforms (Bowker-Kinley et al., 1998). When DCA enters the buried binding site, a large conformational movement of the helix bundle has to occur transiently to accommodate the ligand. The side chain of His149 resides in the vicinity of the DCA-binding site in apo-PDK1; but in the PDK1-DCA structure, the bound DCA dislocates the His149 side chain more than 6 Å toward the outside of the helix bundle (Figure 5B). The movement of His149 promotes an unwinding of the helical structure in the short segment between Arg148 and Asp151. Consistent with the current model for DCA binding, a very recent report has shown that Arg114 and His115 in PDK2, which correspond to Arg148 and His149 in PDK1, are crucial for the interaction of PDK2 with DCA (Klyuyeva et al., 2007). Because this segment connects helices α6 and aα7, the conformational change in this segment results in slight shifts ( 0.5 Å) of these helices in comparison to the corresponding helices in apo-PDK1 (Figure 5B; Movie S1). The electron density of helix α6 in the PDK1-DCA structure is poorer than that of this helix in apo-PDK1 (data not shown). The averaged B factor for helix α6 (residues 132–146) is 30% higher than the overall B factor, whereas a parallel match-up in the apo structure shows only a 12% increase in B factor for this region (Table 2). As helix α6 is not involved in crystal packing, these results suggest that helix α6 is destabilized by DCA binding. Significantly, the N-terminal region of helix α6 interacts with the lipoyl-binding pocket through an H-bonding network involving Thr139 on helix α6, Glu72 and Ser75 on helix a3, and His198 on helix α8 (Figure 5D). These interactions could conceivably link the stability state of helix α6 to that of the lipoyl-binding pocket. In contrast with DCA, in the PDK1-AZD7545 structure, the increase in the averaged B factor for helix α6 from the overall B factor is smaller (7%) than that in the apo structure (Table 2), which suggests that AZD7545 binding stabilizes the lipoyl-binding pocket and thereby helix α6. Unlike helix α6, the stability of helix α7 (residues 152–166) is not affected by DCA binding, based also on B factor comparisons (Table 2).
We have shown that L2 binding promotes the opening of the active-site cleft, which destabilizes the ATP lid and facilitates ADP dissociation in the PDK3-L2 complex (Kato et al., 2005). Kinetic studies have suggested that the binding of DCA hinders the dissociation of ADP from the active site, resulting in product inhibition (Bao et al., 2004). The slower ADP release may be explained by the slight relocation of helix α7 toward the ATP-binding site in the PDK1-DCA structure, which likely restricts the movement of the ATP lid (Movie S1). ADP binding to PDK2 decreases the affinity of this kinase for L2 by 3-fold, and the presence of both ADP and DCA synergistically reduces the affinity of PDK2 for L2 by over 130-fold, compared to PDK2 alone (Hiromasa et al., 2006; Hiromasa and Roche, 2003). Helices α6 and α7 in apo-PDK1 are spatially juxtaposed and exist as a long helical rod that connects the ATP-binding site and the lipoyl-binding pocket (Movie S1). The signal of ADP binding at the ATP-binding site can be transmitted through this helical rod. As the DCA-binding site is located in the middle of this rod, DCA may play the role of an amplifier for the signal triggered by the bound ADP. The segment between helices α6 and α7 forms part of the active-site cleft of PDK1; therefore, the unwound conformation in this segment, as a result of DCA binding, may also decrease kinase activity by thwarting the binding of the E1p substrate to the kinase. In this context, the structure of an asymmetric PDK3-L2 complex harboring only one bound L2 domain has been recently reported (Devedjiev et al., 2007). In this structure, a corresponding unwound “looping-out” segment is present in the L2-bound subunit of the PDK3 dimer, resulting in narrowing of the active-site cleft by 6 Å in the same subunit. These authors hypothesize that the L2-bound PDK3 subunit is inactivated by this perturbation of the active-site cleft, which impedes the access of the E1p substrate. Reciprocally, the L2-free PDK3 subunit in the PDK3 dimer is activated through the opening of the active-site cleft promoted by the interactions between the fully ordered C-terminal tail from this monomer and the lipoyl-binding pocket in the L2-bound PDK3 subunit.
Radicicol Binds to the Nucleotide-Binding Site of Human PDK3
As shown in Figure 6 and Figure S1, radicicol binds to the ATP-binding site in the C-terminal domain of PDK3, but the bound radicicol does not cause any structural change in the PDK3 and L2 molecules of the PDK3-L2 complex. The ATP lid is largely disordered, similar to that seen in the structure of the PDK3-L2-ADP complex (Kato et al., 2005). These results establish that radicicol inhibits PDK3 activity by competing with ATP for the nucleotide-binding site of the kinase (Figure 2).
Figure 6. Structure of the Radicicol-Binding Site in PDK3-L2.
(A) The omit electron density map of radicicol bound to PDK3. The density is superimposed on the refined model of radicicol at the 3σ level in blue and the 9σ level in red.
(B) Interactions between PDK3 residues and radicicol. H bonds are indicated by dashed lines. Water molecules are depicted as red balls.
(C) Comparison between radicicol and ATP bound to PDK3. The structure of PDK3-L2-ATP (PDB ID code 1Y8P; Kato et al., 2005) (magenta) is superimposed on PDK3-L2-radicicol (cyan).
(D) Superimposition of radicicol molecules bound to PDK1 (cyan), Hsp90 (PDB ID code 1BGQ; Roe et al., 1999) (pink), and Topo VI (PDB ID code 2HKJ; Corbett and Berger, 2005) (yellow). The three structures were superimposed based on the corresponding residues shown in the figure. Stereo figures of (B)–(D) are provided in Figure S4.
(E) Comparison of the shape of the radicicol-binding pockets of PDK3, Hsp90, and Topo VI. The electrostatic surface of each protein is shown with the negative charge in red and the positive charge in blue.
Sequence identities between the C-terminal domain of PDK3 and the GHKL domains of Hsp90 and Topo VI are only 7%–12%. Despite the low sequence identities, the folding of these GHKL domains is similar (Figure 7). Out of 150–160 residues in these GHKL domains, 85 residues from PDK3 are superimposable with the corresponding residues in Hsp90, with an rmsd of 1.6 Å. For Topo VI, 84 residues from PDK3 are superimposable, with an rmsd of 1.5 Å. In the PDK3-L2-radicicol structure, all hydroxyl and carbonyl groups of the bound radicicol participate in forming direct or indirect H bonds with residues in the ATP-binding site (Figure 6B). In particular, an H-bonding network involving three water molecules and Asp287 as well as Thr352 of PDK3 is conserved in the GHKL proteins (Figure 6D). This conserved H-bonding network is important for PDK3 to recognize the adenine base of the bound ATP (Figure 6C). A substitution of Asp288 in PDK2 corresponding to Asp287 of PDK3 abolishes ATP binding by PDK2 (Bowker-Kinley and Popov, 1999). Thus, the present results establish that this H-bonding network is a key determinant for radicicol binding to GHKL proteins.
Figure 7. Radicicol-Binding Sites in PDK3, Hsp90, and Topo VI.
Structures of PDK3 (cyan), Hsp90 (PDB ID code 1BGQ; Roe et al., 1999) (pink), and Topo VI (PDB ID code 2HKJ; Corbett and Berger, 2005) (yellow) are superimposed based on the conserved residues in the nucleotide-binding site (see Figure 6D). The sole radicicol molecule from PDK3-L2-radicicol is shown in a stick model. Residues discussed in the text are shown as stick models.
Radicicol inhibits Hsp90 effectively, with an estimated IC50 value of less than 1 μM (Roe et al., 1999). By comparison, an apparent IC50 of radicicol for Topo VI is much higher, approximated at 100 μM with the maximal inhibition achieved at 500 μM (Corbett and Berger, 2005). The latter IC50 value for radicicol is in the range of those estimated for PDK1 (400 μM) and PDK3 (230 μM) (Figure 2B) and the apparent Ki of 23 μM determined for PDK2 (Tuganova et al., 2001). These results indicate that the affinity of radicicol for Hsp90 is at least 100-fold higher than that for Topo VI and PDK isoforms. From a structural viewpoint, the side chain of Lys44 in Hsp90 is present as an overhang outside the radicicol-binding pocket and forms a direct H bond with the epoxide oxygen of radicicol, which locks the inhibitor in place (Figure 6E) (Roe et al., 1999). In contrast, Topo VI has only an indirect H bond with this oxygen from residue His50 of this protein (Corbett and Berger, 2005), whereas an equivalent H bond is absent in PDK3 (Figure 7). The binding-site entrances in both proteins are wide open (Figure 6E), which is likely to facilitate the dissociation of bound radicicol. Thus, it is plausible that Lys44 in Hsp90 serves as the specific determinant that confers the higher affinity of Hsp90 for radicicol over Topo VI and PDK3. Along the same lines, Arg254 in PDK3 that projects over the bound radicicol may play a similar role to Lys44 in Hsp90 (Figure 6E). This arginine residue is completely conserved in the PDK family, but is not present in other GHKL proteins (Figure 7). Thus, a modified radicicol with the epoxide oxygen translocated within the macrocyclic ring to be close to the invariant Arg254 for H-bond formation may show higher affinity for kinases of the PDK family.
EXPERIMENTAL PROCEDURES
Assays for Inhibition of PDK Activities
To determine the IC50 of AZD7545 for PDK1 and PDK3, a mixture containing 50 nM SUMO-PDK1 or SUMO-PDK3, 1.5 μM E1p, 50 nM E2/E3BP core, and different amounts of AZD7545 was incubated at 25° C for 30 min in a phosphorylation buffer of 20 mM potassium phosphate (pH 7.5), 50 mM KCl, 2 mM MgCl2, and 2 mM dithiothreitol (DTT). A slightly different condition was used during determination of the IC50 of radicicol for PDK1 and PDK3, in which 0.56 μM SUMO-PDK1 or 0.28 μM SUMO-PDK3, 6.4 μM E1p, 2.3 μM E2/E3BP core, and different amounts of radicicol were incubated. The AZD7545 and radicicol stock solutions were prepared in 100% DMSO and diluted into the assay mixture to a 2% final DMSO concentration. The presence of a SUMO tag is necessary to maintain the solubility of PDK1 and PDK3 and has no effect on kinase activity. The phosphorylation reaction was initiated by adding [γ-32P]ATP (specific activity 0.5 μCi/nmol ATP) to a 50 μM (for AZD7545 IC50 determination) or 100 μM (for radicicol IC50 determination) final concentration in a total reaction volume of 60 μl. The reaction was terminated at 1 min intervals by withdrawing 15 μl of the reaction mixture and mixing it immediately with 5 μl of the 4× concentrated SDS-PAGE loading buffer containing 100 mM EDTA. The reaction mixture was heated at 65° C for 10 min and then applied to an SDS-PAGE gel, followed by phosphorimaging analyses. The phosphorylation level was quantified using a Typhoon 9200 variable-mode imager (Pharmacia, Piscataway, NJ, USA) by comparison with standards. PDK activities with the inhibitors were normalized against control activities without inhibitors. Data analyses and curve fittings were carried out using the Prism program (GraphPad Software, San Diego, CA, USA). To determine the maximal inhibitory levels of PDK inhibitors in the absence and presence of the E2p/E3BP core, a saturating concentration of each inhibitor (10 μM AZD7545, 40 mM DCA, and 4 mM radicicol) was incubated with 50 nM SUMO-PDK1 or SUMO-PDK3, and 1.5 μM E1p with or without 50 nM E2/E3BP core at 25° C for 30 min. The reaction was initiated by adding [γ-32P]ATP to a 50 μM final concentration in a total volume of 60 μl. The reaction was terminated at 60 s, followed by SDS-PAGE and phosphorimaging analyses as described above.
Crystallization and Structure Determination
Crystals were obtained using the hanging-drop vapor-diffusion method by mixing 2 μl of PDK1 solution (50 mg/ml untagged PDK1 in 50 mM potassium phosphate [pH 7.5], 250 mM KCl, 800 mM lysine, 5% glycerol, and 20 mM DTT) with 2 μl of well solution (0.42 M Na-K tartrate, 0.1 M Na citrate [pH 5.6]) and kept at 20° C. The high PDK1 concentration required for crystallization was achieved by the inclusion of 800 mM lysine in the protein solution. Dilution seeding was used to induce nucleation. Single crystals grew to 500 × 500 × 500 mm in size within a week. Crystals were transferred to fresh well solution for 18–24 hr and soaked in well solution containing 50 mM DCA or 0.1 mM AZD7545 for 30 min. These crystals were serially transferred to well solution containing 25% glycerol and flash-frozen in liquid propane. Crystals of the PDK3-L2-radicicol complex were prepared by soaking those of the apo-PDK3-L2 complex (Kato et al., 2005) with 1 mM radicicol. Diffraction data were collected at beamlines 19ID and 19BM at the Structural Biology Center at the Advanced Photon Source (Argonne, IL, USA). The data were processed with HKL2000 (Otwinowski and Minor, 1997).
The PDK1 structures were solved by molecular replacement using the program Phaser (Read, 2001) in the CCP4 package (CCP4, 1994). A PDK1 model based on the published PDK3 structure (Protein Data Bank [PDB] ID code 1Y8N; Kato et al., 2005) was constructed using a homology-modeling technique in the SWISS-MODEL program (Schwede et al., 2003). After rigid-body refinements with REFMAC5 (Murshudov et al., 1997), the electron density map was improved using DM (Cowtan and Main, 1996). The Fo – Fc maps unambiguously showed the electron density of the inhibitors in individual crystals soaked with AZD7545 and DCA. After modeling the inhibitors, water molecules were gradually added in subsequent refinement cycles. The structure of PDK3-L2-radicicol was solved by the difference Fourier method using the published structure of PDK3-L2 (PDB ID code 1Y8P; Kato et al., 2005) as a starting model. The refinement was carried out as described for the PDK1 refinement. Data processing and refinement statistics are summarized in Table 1. Molecular graphics for structural representations were drawn by PyMOL (DeLano Scientific LLC, Palo Alto, CA, USA).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Rachel Mayers at AstraZeneca for the generous supply of the compound AZD7545 and critical reading of the manuscript. We also thank Dr. Kirill Popov at the University of Alabama at Birmingham for sharing with us the human E1p and E2p/E3BP expression plasmids for the PDK activity assays. We are indebted to Drs. Mischa Machius, Diana Tomchick, and Chad Brautigam in the Structural Biology Laboratory of the University of Texas Southwestern Medical Center for the collection of synchrotron data. This work was supported by grants DK62306 and DK26758 from the National Institutes of Health and grant I-1286 from the Welch Foundation. The use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research under contract W-31-109-ENG-38. The authors declare that there is no financial conflict of interest related to this work.
Footnotes
Supplemental Data Supplemental Data include four figures, one movie, and Supplemental Experimental Procedures for protein expression, purification, and SUMO-tag removal for crystallization and can be found with this article online at http://www.structure.org/cgi/content/full/15/8/992/DC1/.
Accession Numbers Coordinates and structure factors have been deposited in the Protein Data Bank under ID codes 2Q8F, 2Q8G, 2Q8H, and 2Q8I for the apo, AZD7545-bound, and DCA-bound PDK1 structures and the radicicol-bound PDK3-L2 structure, respectively.
REFERENCES
- Alex LA, Simon MI. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- Baker JC, Yan X, Peng T, Kasten S, Roche TE. Marked differences between two isoforms of human pyruvate dehydrogenase kinase. J. Biol. Chem. 2000;275:15773–15781. doi: 10.1074/jbc.M909488199. [DOI] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Bao H, Kasten SA, Yan X, Roche TE. Pyruvate dehydrogenase kinase isoform 2 activity limited and further inhibited by slowing down the rate of dissociation of ADP. Biochemistry. 2004;43:13432–13441. doi: 10.1021/bi049488x. [DOI] [PubMed] [Google Scholar]
- Bebernitz GR, Aicher TD, Stanton JL, Gao J, Shetty SS, Knorr DC, Strohschein RJ, Tan J, Brand LJ, Liu C, et al. Anilides of (R)-trifluoro-2-hydroxy-2-methylpropionic acid as inhibitors of pyruvate dehydrogenase kinase. J. Med. Chem. 2000;43:2248–2257. doi: 10.1021/jm0000923. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bersin RM, Stacpoole PW. Dichloroacetate as metabolic therapy for myocardial ischemia and failure. Am. Heart J. 1997;134:841–855. doi: 10.1016/s0002-8703(97)80007-5. [DOI] [PubMed] [Google Scholar]
- Besant PG, Lasker MV, Bui CD, Turck CW. Inhibition of branched-chain α-keto acid dehydrogenase kinase and Sln1 yeast histidine kinase by the antifungal antibiotic radicicol. Mol. Pharmacol. 2002;62:289–296. doi: 10.1124/mol.62.2.289. [DOI] [PubMed] [Google Scholar]
- Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Bowker-Kinley M, Popov KM. Evidence that pyruvate dehydrogenase kinase belongs to the ATPase/kinase superfamily. Biochem. J. 1999;344:47–53. [PMC free article] [PubMed] [Google Scholar]
- Bowker-Kinley MM, Davis WI, Wu P, Harris RA, Popov KM. Evidence for existence of tissue-specific regulation of the mammalian pyruvate dehydrogenase complex. Biochem. J. 1998;329:191–196. doi: 10.1042/bj3290191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Papandreou I, Sutphin PD, Denko NC. Metabolic targeting of hypoxia and HIF1 in solid tumors can enhance cytotoxic chemotherapy. Proc. Natl. Acad. Sci. USA. 2007;104:9445–9450. doi: 10.1073/pnas.0611662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (Collaborative Computational Project, Number 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure. 2005;13:873–882. doi: 10.1016/j.str.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Cowtan KD, Main P. Phase combination and cross validation in iterated density-modification calculations. Acta Crystallogr. D Biol. Crystallogr. 1996;52:43–48. doi: 10.1107/S090744499500761X. [DOI] [PubMed] [Google Scholar]
- Devedjiev Y, Steussy CN, Vassylyev DG. Crystal structure of an asymmetric complex of pyruvate dehydrogenase kinase 3 with lipoyl domain 2 and its biological implications. J. Mol. Biol. 2007;370:407–416. doi: 10.1016/j.jmb.2007.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Harris RA, Huang B, Wu P. Control of pyruvate dehydrogenase kinase gene expression. Adv. Enzyme Regul. 2001;41:269–288. doi: 10.1016/s0065-2571(00)00020-0. [DOI] [PubMed] [Google Scholar]
- Henderson GN, Curry SH, Derendorf H, Wright EC, Stacpoole PW. Pharmacokinetics of dichloroacetate in adult patients with lactic acidosis. J. Clin. Pharmacol. 1997;37:416–425. doi: 10.1002/j.1552-4604.1997.tb04319.x. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Roche TE. Facilitated interaction between the pyruvate dehydrogenase kinase isoform 2 and the dihydrolipoyl acetyltransferase. J. Biol. Chem. 2003;278:33681–33693. doi: 10.1074/jbc.M212733200. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 2004;279:6921–6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Hu L, Roche TE. Ligand-induced effects on pyruvate dehydrogenase kinase isoform 2. J. Biol. Chem. 2006;281:12568–12579. doi: 10.1074/jbc.M513514200. [DOI] [PubMed] [Google Scholar]
- Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 2003;31:1143–1151. doi: 10.1042/bst0311143. [DOI] [PubMed] [Google Scholar]
- Kato M, Chuang JL, Tso SC, Wynn RM, Chuang DT. Crystal structure of pyruvate dehydrogenase kinase 3 bound to lipoyl domain 2 of human pyruvate dehydrogenase complex. EMBO J. 2005;24:1763–1774. doi: 10.1038/sj.emboj.7600663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Klyuyeva A, Tuganova A, Popov KM. Amino acid residues responsible for the recognition of dichloroacetate by pyruvate dehydrogenase kinase 2. FEBS Lett. 2007;581:2988–2992. doi: 10.1016/j.febslet.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoechel TR, Tucker AD, Robinson CM, Phillips C, Taylor W, Bungay PJ, Kasten SA, Roche TE, Brown DG. Regulatory roles of the N-terminal domain based on crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands. Biochemistry. 2006;45:402–415. doi: 10.1021/bi051402s. [DOI] [PubMed] [Google Scholar]
- Kolobova E, Tuganova A, Boulatnikov I, Popov KM. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem. J. 2001;358:69–77. doi: 10.1042/0264-6021:3580069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotchkina LG, Patel MS. Probing the mechanism of inactivation of human pyruvate dehydrogenase by phosphorylation of three sites. J. Biol. Chem. 2001a;276:5731–5738. doi: 10.1074/jbc.M007558200. [DOI] [PubMed] [Google Scholar]
- Korotchkina LG, Patel MS. Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J. Biol. Chem. 2001b;276:37223–37229. doi: 10.1074/jbc.M103069200. [DOI] [PubMed] [Google Scholar]
- Koukourakis MI, Giatromanolaki A, Sivridis E, Gatter KC, Harris AL. Pyruvate dehydrogenase and pyruvate dehydrogenase kinase expression in non small cell lung cancer and tumor-associated stroma. Neoplasia. 2005;7:1–6. doi: 10.1593/neo.04373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Baker JC, Roche TE. Binding of the pyruvate dehydrogenase kinase to recombinant constructs containing the inner lipoyl domain of the dihydrolipoyl acetyltransferase component. J. Biol. Chem. 1995;270:793–800. doi: 10.1074/jbc.270.2.793. [DOI] [PubMed] [Google Scholar]
- Machius M, Chuang JL, Wynn RM, Tomchick DR, Chuang DT. Structure of rat BCKD kinase: nucleotide-induced domain communication in a mitochondrial protein kinase. Proc. Natl. Acad. Sci. USA. 2001;98:11218–11223. doi: 10.1073/pnas.201220098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers RM, Leighton B, Kilgour E. PDH kinase inhibitors: a novel therapy for type II diabetes? Biochem. Soc. Trans. 2005;33:367–370. doi: 10.1042/BST0330367. [DOI] [PubMed] [Google Scholar]
- Morrell JA, Orme J, Butlin RJ, Roche TE, Mayers RM, Kilgour E. AZD7545 is a selective inhibitor of pyruvate dehydrogenase kinase 2. Biochem. Soc. Trans. 2003;31:1168–1170. doi: 10.1042/bst0311168. [DOI] [PubMed] [Google Scholar]
- Murshudov G, Vagin A, Dodson E. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr., Sweet RH, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pan JG, Mak TW. Metabolic targeting as an anticancer strategy: dawn of a new era? Sci. STKE. 2007;2007:e14. doi: 10.1126/stke.3812007pe14. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Popov KM, Hawes JW, Harris RA. Mitochondrial α-ketoacid dehydrogenase kinases: a new family of protein kinases. Adv. Second Messenger Phosphoprotein Res. 1997;31:105–111. [PubMed] [Google Scholar]
- Pratt ML, Roche TE. Mechanism of pyruvate inhibition of kidney pyruvate dehydrogenase kinase and synergistic inhibition by pyruvate and ADP. J. Biol. Chem. 1979;254:7191–7196. [PubMed] [Google Scholar]
- Radke GA, Ono K, Ravindran S, Roche TE. Critical role of a lipoyl cofactor of the dihydrolipoyl acetyltransferase in the binding and enhanced function of the pyruvate dehydrogenase kinase. Biochem. Biophys. Res. Commun. 1993;190:982–991. doi: 10.1006/bbrc.1993.1146. [DOI] [PubMed] [Google Scholar]
- Read R. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1373–1382. doi: 10.1107/s0907444901012471. [DOI] [PubMed] [Google Scholar]
- Reed LJ. A trail of research from lipoic acid to α-keto acid dehydrogenase complexes. J. Biol. Chem. 2001;276:28329–28336. doi: 10.1074/jbc.R100026200. [DOI] [PubMed] [Google Scholar]
- Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell. Mol. Life Sci. 2007;64:830–849. doi: 10.1007/s00018-007-6380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche TE, Hiromasa Y, Turkan A, Gong X, Peng T, Yan X, Kasten SA, Bao H, Dong J. Essential roles of lipoyl domains in the activated function and control of pyruvate dehydrogenase kinases and phosphatase isoform 1. Eur. J. Biochem. 2003;270:1050–1056. doi: 10.1046/j.1432-1033.2003.03468.x. [DOI] [PubMed] [Google Scholar]
- Roe SM, Prodromou C, O'Brien R, Ladbury JE, Piper PW, Pearl LH. Structural basis for inhibition of the Hsp90 molecular chaperone by the antitumor antibiotics radicicol and geldanamycin. J. Med. Chem. 1999;42:260–266. doi: 10.1021/jm980403y. [DOI] [PubMed] [Google Scholar]
- Sale GJ, Randle PJ. Analysis of site occupancies in [32P]phosphorylated pyruvate dehydrogenase complexes by aspartyl-prolyl cleavage of tryptic phosphopeptides. Eur. J. Biochem. 1981;120:535–540. doi: 10.1111/j.1432-1033.1981.tb05733.x. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MC. Coming up for air: HIF-1 and mitochondrial oxygen consumption. Cell Metab. 2006;3:150–151. doi: 10.1016/j.cmet.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Smirnova IN, Kasho VN, Faller LD. Inferences about the catalytic domain of P-type ATPases from the tertiary structures of enzymes that catalyze the same elementary reaction. FEBS Lett. 1998;431:309–314. doi: 10.1016/s0014-5793(98)00760-1. [DOI] [PubMed] [Google Scholar]
- Stacpoole PW. The pharmacology of dichloroacetate. Metabolism. 1989;38:1124–1144. doi: 10.1016/0026-0495(89)90051-6. [DOI] [PubMed] [Google Scholar]
- Steussy CN, Popov KM, Bowker-Kinley MM, Sloan RB, Jr., Harris RA, Hamilton JA. Structure of pyruvate dehydrogenase kinase. Novel folding pattern for a serine protein kinase. J. Biol. Chem. 2001;276:37443–37450. doi: 10.1074/jbc.M104285200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teague WM, Pettit FH, Yeaman SJ, Reed LJ. Function of phosphorylation sites on pyruvate dehydrogenase. Biochem. Biophys. Res. Commun. 1979;87:244–252. doi: 10.1016/0006-291x(79)91672-3. [DOI] [PubMed] [Google Scholar]
- Tuganova A, Yoder MD, Popov KM. An essential role of Glu-243 and His-239 in the phosphotransfer reaction catalyzed by pyruvate dehydrogenase kinase. J. Biol. Chem. 2001;276:17994–17999. doi: 10.1074/jbc.M009327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuganova A, Boulatnikov I, Popov KM. Interaction between the individual isoenzymes of pyruvate dehydrogenase kinase and the inner lipoyl-bearing domain of transacetylase component of pyruvate dehydrogenase complex. Biochem. J. 2002;366:129–136. doi: 10.1042/BJ20020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuganova A, Klyuyeva A, Popov KM. Recognition of the inner lipoyl-bearing domain of dihydrolipoyl transacetylase and of the blood glucose-lowering compound AZD7545 by pyruvate dehydrogenase kinase 2. Biochemistry. 2007;46:8592–8602. doi: 10.1021/bi700650k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse S, Cooper RH, Randle PJ. Mechanism of activation of pyruvate dehydrogenase by dichloroacetate and other halogenated carboxylic acids. Biochem. J. 1974;141:761–774. doi: 10.1042/bj1410761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley DB, Davies GJ, Dodson EJ, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]
- Wu P, Sato J, Zhao Y, Jaskiewicz J, Popov KM, Harris RA. Starvation and diabetes increase the amount of pyruvate dehydrogenase kinase isoenzyme 4 in rat heart. Biochem. J. 1998;329:197–201. doi: 10.1042/bj3290197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman SJ, Hutcheson ET, Roche TE, Pettit FH, Brown JR, Reed LJ, Watson DC, Dixon GH. Sites of phosphorylation on pyruvate dehydrogenase from bovine kidney and heart. Biochemistry. 1978;17:2364–2370. doi: 10.1021/bi00605a017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.