Abstract
Human senescence marker protein 30 (SMP30), which functions enzymatically as a lactonase, hydrolyzes various carbohydrate lactones. The penultimate step in vitamin-C biosynthesis is catalyzed by this enzyme in non-primate mammals. It has also been implicated as an organophosphate hydrolase, with the ability to hydrolyze diisopropyl phosphofluoridate and other nerve agents. SMP30 was originally identified as an aging marker protein, whose expression decreased androgen independently in aging cells. SMP30 is also referred to as regucalcin and has been suggested to have functions in calcium homeostasis. The crystal structure of the human enzyme has been solved from X-ray diffraction data collected to a resolution of 1.4 Å. The protein has a 6-bladed β-propeller fold and it contains a single metal ion. Crystal structures have been solved with the metal site bound with either a Ca2+ or a Zn2+ atom. The catalytic role of the metal ion has been confirmed by mutagenesis of the metal coordinating residues. Kinetic studies using the substrate gluconolactone showed a kcat preference of divalent cations in the order Zn2+ > Mn2+ > Ca2+ > Mg2+. Notably, the Ca2+ had a significantly higher value of Kd compared to the other metal ions tested (566, 82, 7 and 0.6 µm for Ca2+, Mg2+, Zn2+ and Mn2+, respectively), suggesting the Ca2+-bound form may be physiologically relevant for stressed cells with an elevated free calcium level.
Keywords: SMP30, regucalcin, DFPase, lactonase, divalent metal, structure, kinetic parameters
Senescence marker protein 30 (SMP30) was originally identified as an aging marker protein, whose expression decreases in an androgen independent manner in rat liver and kidney cells (1). Several reports have attempted to establish SMP30 as an anti-aging factor (2–11). SMP30 is also known as regucalcin and was reported in 1988 (12) as a calcium binding protein that does not have an EF-hand calcium binding motif. Since then studies have shown that regucalcin plays a pivotal role in Ca2+ regulation and homeostasis in mammalian cells (13, 14).
The physiological role of SMP30 has been linked to various enzymatic, as well as non-enzymatic functions, including its zinc dependent gluconolactonase activity. In the penultimate step of vitamin-C biosynthesis in non-primate mammals, L-gulonate is converted to L-gulonolactone by SMP30 which functions as a gulonolactonase (15). Primates are unable to synthesize their own vitamin-C due to the absence of gulonolactone oxidase, which is the enzyme that catalyzes the final step oxidation of gulonolactone to ascorbate. This function in vitamin-C biosynthesis in non primates was confirmed in a study that showed that SMP30 deficiency caused scurvy in knockout mice (16).
Although, our primary interest lies in understanding the enzymatic function of SMP30 and its implication to human physiology, we are also exploring its potential as an organophosphorus (OP) hydrolase. In 1999, Billecke et al (17) purified a soluble enzyme from mouse liver that is capable of hydrolyzing the OP compound diisopropylfluorophosphate (DFP), which turned out to be SMP30. Subsequently, it was reported that SMP30 could hydrolyze DFP in the presence of Mg2+ (18). These aforementioned reports as well as distant sequence similarities (10% identity, 16% similarity) and a prediction of being a 6-bladed β-propeller, led us believe that SMP30 is related to two eukaryotic calcium dependent OP hydrolases: human serum paraoxonase-1 (PON1) and squid diisopropylfluorophosphatase (DFPase). PON1, which catalyzes the hydrolysis of various OP nerve agents, such as sarin and soman, is now thought to function as a lactonase (19). DFPase, which is structurally homologous to PON1, was named for its ability to catalyze the hydrolysis of DFP like substrates (20).
As there are no reported crystal structures of a eukaryotic SMP30, our initial goal was to solve the crystal structure of human SMP30 and to obtain an overall architecture of the protein and its active site. We also wanted to see how the active sites of PON1 and DFPase correspond to the SMP30 structure so that we could have a better idea of relating these enzyme families. Secondly, we wanted to understand and establish the role of metal ions in the structure and function of SMP30. Previous studies, with rat and mouse SMP30, have reported that Zn2+ is the metal of choice in gluconolactonase activity (16), Mg2+ in DFPase activity (18) and Ca2+ in cell regulation and homeostasis (14). In contrast to the calcium-dependent PON1 and DFPase enzymes, the catalytic role of Ca2+ in SMP30 has been brought into doubt. Data from a 45Ca2+-binding assay had suggested that rat SMP30 is not a calcium binding protein (18). Also, it was suggested that mouse SMP30 does not have a calcium dependent gluconolactonase activity (16).
Here we report the first crystal structures of the human enzyme SMP30, which are bound with either Ca2+ or Zn2+. Additionally, kinetic characterization of SMP30 using the substrate gluconolactone demonstrates a preference for Zn2+ with a decreasing catalytic efficiency with the Mn2+, Mg2+ and Ca2+ bound forms. An overall comparison of the homologous enzymes, particularly PON1 and DFPase, delineates a unique role of SMP30’s metal binding site and active site. Also, the comparison of the dissociation constants of the metal ions with their free cellular concentrations allow us to speculate that SMP30 may switch to a calcium dependent lactonase under stressed conditions that yield an increased concentration of calcium.
MATERIALS AND METHODS
Expression of Human SMP30
The full-length human SMP30 cDNA (gi-22025639) was amplified by PCR using the pRc/CMV plasmid (kindly supplied by Scott Billecke) as template. The forward and reverse primers were 5’-GCGCGAATTCTTCCATTAAGATTGAG-3’ and 5’-GCGTCGACTCATCATCCCGCATA-3’ which contain the restriction sites for EcoRI and SalI, respectively (in bold). The PCR-amplified DNA fragment was ligated into pGEX-4T-3 vector (GE Healthcare), which has a GST tag at the N-terminus to the full-length human SMP30 gene with an engineered thrombin-cleavage site. The cloned human SMP30 gene was verified by DNA sequencing.
The SMP30-pGEX-4T-3 construct was transformed into E. coli BL21 (DE3) RIL codon plus cells and expressed, in a modified LB media. For protein expression and purification, all chemicals used were obtained from Sigma-Aldrich unless otherwise noted. One liter of Miller LB media (Fisher Sci.) was supplemented by 10 ml of 40% glycerol (filter sterilized), 1 ml of 0.2 M potassium phosphate buffer (pH 7.1), 1 mM CaCl2, and 1 M NaOH was added to bring the final pH to 7.1. Before inoculation, 100 mg/L ampicillin and 34 mg/l chloramphenicol were added to the sterilized media. Ten ml of overnight culture was added per liter of media and grown at 37 °C until the OD600nm reached 2.0 at ~4 h. The cultures were then subjected to a cold shock and kept at 4 °C for 45 min. They were then induced with 1 mM IPTG and grown at room temperature (25 °C) for 12–14 h. Cells were harvested by centrifugation at 5,000 g and stored at −80 °C prior to purification.
Purification of Human SMP30
All purification procedures were carried out at 4 °C. Frozen cells were thawed and suspended in a lysis buffer (50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 5 mM DTT, 1 mM EDTA, 1 mM PMSF, and 10 units/mg DNAse-I) and disrupted by sonication. Detergent extraction was done immediately in 0.1% (v/v) triton X-100 for 15 min and the lysate was centrifuged at 12,000 g for 35 min. Supernatant was incubated with glutathione sepharose 4B (GE Healthcare) beads for 50 min, flow through was discarded and the resin was washed three times with high salt wash buffer (25 mM Tris-HCl, pH 7.5, 1 mM CaCl2, 1 mM DTT, 170 mM NaCl). The GST-SMP30 fusion protein was eluted with elution buffer (10 mM reduced glutathione, 25 mM Tris-HCl, pH 7.5, 1 mM CaCl2, 1 mM DTT, 170 mM NaCl), concentrated with centrifugal membranes with a 10 kDa cutoff (Millipore) and was subjected to overnight proteolysis by thrombin (Enzyme Research Laboratories). The proteolyzed sample was incubated with the glutathione sepharose beads for 30 min and then the supernatant above the settled resin was dialyzed with a low ionic strength buffer (25 mM Tris-HCl, pH 7.5, 1 mM CaCl2, 1 mM DTT) for 3 hours. The sample was then passed through a 5 ml benzamidine column (GE Healthcare) to remove thrombin and the flow through was concentrated to ~ 5 mg/ml. The enzyme activity was confirmed by a gluconolactonase assay (described below) and the purity was checked by SDS-PAGE.
Alanine scanning mutagenesis of the metal binding residues was performed using the Quikchange protocol (Stratagene) to make the site-directed mutations E18A, N103A, N154A and D204A. The procedures used for the expression and purification of the mutants were similar to that of the wild type enzyme.
SMP30 Enzyme Kinetics
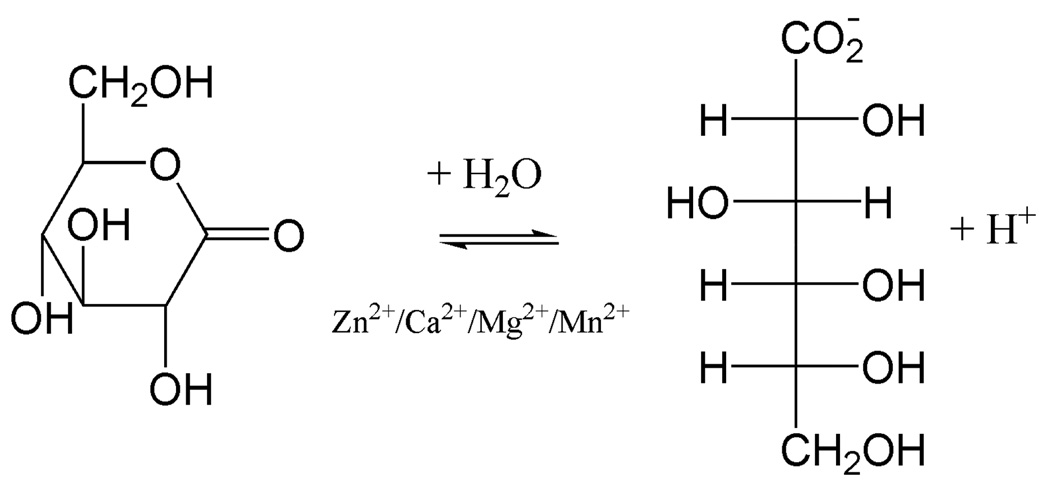
Enzyme activity was measured using the substrate D-glucono-δ-lactone by following the decrease in absorbance of a pH indicator p-nitrophenol (pKa 7.1) caused by the net stoichiometric formation of an H+ (Fig. 1). The pH (6.4) of the reaction mixture was kept below the pKa (7.1) so that the majority of the indicator species would be phenolate ion (yellow) before the start of the reaction and any change in color (Absorbance decrease at 405 nm) would be attributed to the stoichiometric production of H+ due to the enzymatic catalysis. The reaction mixture contained 10 mM PIPES buffer (pH 6.4), 10 mM D-glucono-δ-lactone, 0.25 mM p-nitrophenol, 75 µM ZnCl2 and an aliquot of enzyme in a total volume of 1 ml. The decrease of Absorbance at 405 nm was correlated to a change of [H+] by a standard curve with known amounts of HCl (fresh dilutions from commercial stock). Initial velocities were repeated at a range of substrate concentrations. Kinetic parameters of Vmax and KM were correlated to H+ production by a non-linear fit to v0 = Vmax[S]/(KM + [S]) using the program GraphPad Prism 3.0. The enzyme concentration used in assays was determined using the BCA protein assay (Thermo Scientific) and used in the calculation of kcat values. The experiments were repeated with metal ion concentrations up to 1 mM and the kinetic parameters were calculated. Standard errors for kcat and KM were determined based on the least squares fit of the Michaelis-Menton equation.
Figure 1.
The reaction of gluconolactone to gluconic acid is catalyzed by the enzyme SMP30. This reaction requires a divalent cation in the SMP30 active site such as Zn2+, Ca2+, Mg2+ or Mn2+.
Dissociation Constants of Divalent Cations with SMP30
Metal free enzyme was prepared to study the effects of various divalent cations on gluconolactonase activity. Acid rinsed plastic containers were used for all the purification steps, de-ionized water was passed through a chelex column (Biorad) and the buffers were dialyzed with chelex, before use. To remove residual cationic metal ions, 1 mM EDTA was added to the high salt and elution buffers. To determine the dissociation constants of the metal ions, the initial velocities were repeated using a range of metal ion concentrations keeping the substrate gluconolactone concentration at 10 mM. The metal ion dissociation constant (Kd) was quantified by non-linear fitting of the data to equation [1]
| [1] |
using the program GraphPad Prism 3.0, where R and Rf are the experimentally determined rate and a floating value of rate (automatically calculated by the program during curve fitting), respectively. Since we are fitting the observed enzymatic rates at various metal concentrations and saturating substrate, this treatment allows us to determine the metal Kd of the substrate bound form of the enzyme-metal complex. E and M are the total enzyme and added metal ion concentrations, respectively, and Kd is the metal ion dissociation constant.
Crystallization of Human SMP30
Although a preliminary crystallization report was reported by others for human SMP30 (21), our initial crystallization conditions were screened by commercial crystallization screening kits (crystal screen 1 and 2 and PEG screen, Hampton research). Initial screening hits were then further refined to optimize the protein crystal quality. Crystals were obtained at room temperature (25 °C) at protein concentrations of 3–5 mg/ml using the hanging drop method (22). Diffraction quality crystals were grown from a crystallization solution containing 0.1 M Tris buffer, pH 7.8, 38% PEG 6000, 5 mM CaCl2 and 5 mM DTT. Aliquots of 2 µl of protein and crystallization solutions were mixed to form each hanging drop, and crystals formed in 3–7 days. After screening for cryoprotectant, a suitable stabilizing solution was prepared containing 0.1 M Tris buffer, pH 7.8, 26% PEG 6000, 30% PEG 400, 5 mM CaCl2 and 5 mM DTT. Crystals were soaked into a stabilizing solution for a few minutes and then flash frozen in liquid nitrogen and stored in liquid nitrogen until data collection. Since initial trials to grow crystals with zinc, instead of calcium in the crystallization solution were unsuccessful, we soaked the calcium-grown crystals in the stabilizing solution (without calcium) with a gradual increase in the zinc concentration up to a final concentration of 1 mM. At ZnCl2 concentrations above 1mM the crystals began to crack. One data set was collected from such a zinc soaked crystal that diffracted to 1.9 Å.
X-ray Data Collection and Structure Determination
Crystals were shipped to the Advanced Photon Source (APS) of Argonne National Laboratory, Chicago IL through the express crystallography program. The SGX-CAT beam line was an automated protein crystal data collection program operated by SGX Pharmaceuticals. This beam line is presently operated by Eli Lilly. Datasets were collected at beam line 31-ID-D of the APS facility. The diffraction data were indexed and processed by the programs MOSFLM, SCALA and TRUNCATE of the CCP4 suite of programs (23).
The phases for SMP30 were solved using a molecular replacement search model made using a template (protein data bank (PDB) code: 2ghs), which shows 20% protein sequence identity with the SMP30 sequence. Molecular replacement was performed using the program PHASER (24). The top molecular replacement solution from PHASER was fed into the model-building program ARP/wARP (23), and it successfully built 551 of the 598 residues in the asymmetric unit. After further cycles of refinement and loop building by the programs Refmac5 and COOT (25) a complete model was obtained. Further refinement was done using the program SHELXL (26) to get the final model of SMP30.
RESULTS
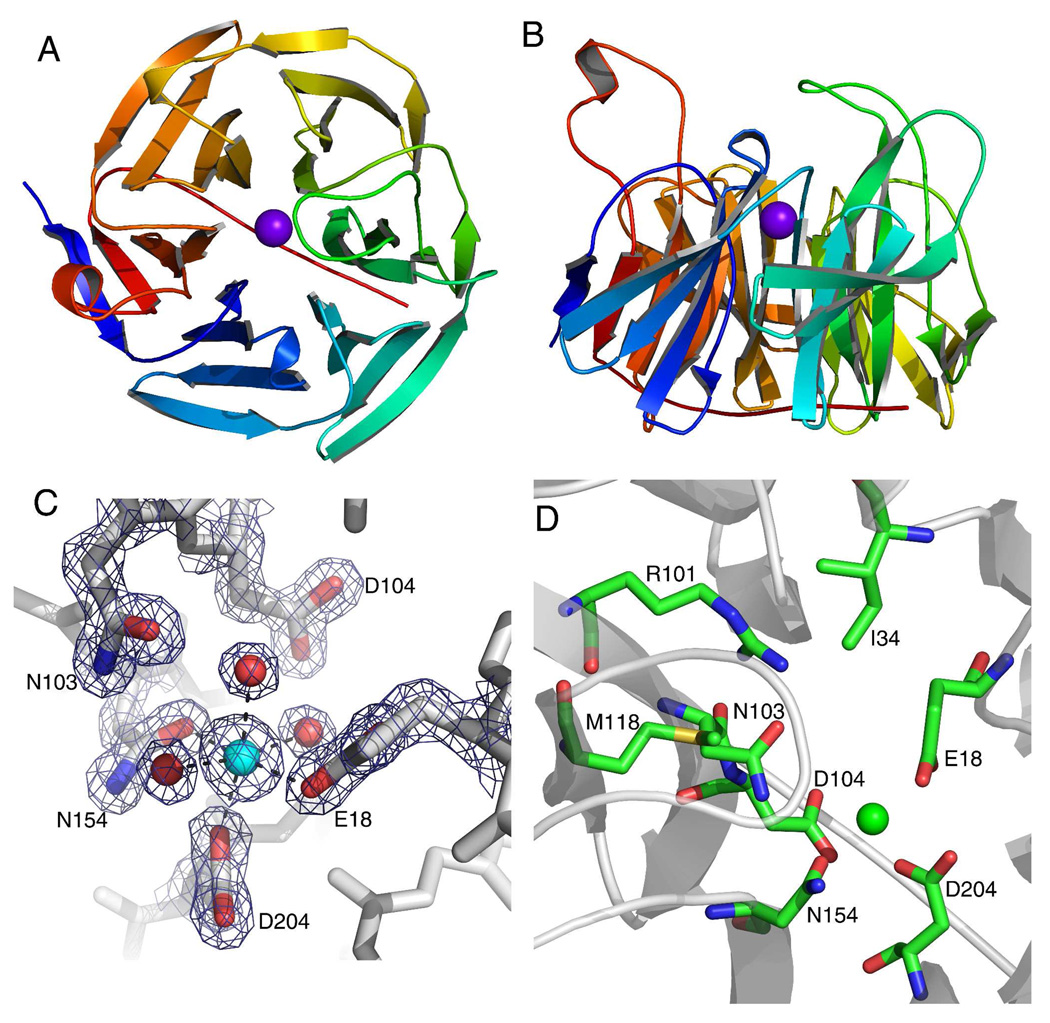
We determined the crystal structure of human SMP30 with Ca2+ or Zn2+ bound at its active site. The six-bladed β-propeller fold and active site are shown for the Ca2+ bound form in Fig. 2. Initially, the crystal structure of Ca2+-SMP30 was solved by molecular replacement using a molecular replacement search model made using a template of a putative lactonase protein from agrobaterium tumefaciens, which has 20% protein sequence identity with the SMP30 sequence. The Ca2+-SMP30 structure was refined against data to a resolution of 1.4 Å, with final R-factor values of 0.12 and 0.19 for Rwork and Rfree, respectively. This structure enabled a structure solution of the Zn2+-bound form of SMP30 using diffraction data to a resolution of 1.9 Å with final R-factor values of 0.22 and 0.25 for Rwork and Rfree, respectively. For each structure there are two protein molecules per asymmetric unit and one metal ion bound to each protein molecule. The structures were validated by the programs Procheck and SF check of the CCP4 program suite (23) and the program WHAT IF (27). A summary of the data collection and refinement of the crystal structures is presented in Table 1.
Figure 2.
The crystal structure of human SMP30 with Ca2+ bound. A, the ribbon structure of SMP30 displays the six-bladed β-propeller fold with each blade displayed in a rainbow color. The active site Ca2+ is shown in the middle of the β-propeller as a purple sphere. B, a 90° x-axis rotation of the view in panel-A displaying the loops which project up from the β-sheets and define the surface around the active site. The active site is located just above the location of the Ca2+ in this view. C, the metal binding site of human SMP30 with an electron density map (coefficients 2Fo-Fc, 2.0σ) around highlighted residues. The residues E18, D204 and N154 are bound to the Ca2+. Three water molecules complete the ligation of the hexa-coordinate Ca2+. The residues N103 and D104 are near the Ca2+, but not within bonding distance. D, The active site residues of SMP30 are shown. In addition to residues surrounding the Ca2+ (also shown in panel-C) three additional residues whose side-chains are exposed to the putative-substrate binding pocket are shown (I34, R101 and M118).
Table 1.
Data collection, phasing and refinement statistics of SMP30
| Crystal parameters | Ca2+ | Zn2+ |
|---|---|---|
| Space group | P21 | P21 |
| Unit cell dimensions | 64.47, 51.00, 86.12 | 64.36, 52.01, 85.83 |
| a, b, c (Å), β (°) | 100.11 | 100.11 |
| Beam line | SGXCAT 31-ID-D | SGXCAT 31-ID-D |
| Data collection | ||
| Wave length(Å) | 0.9793 | 0.9793 |
| Resolution (Å) | 18.4–1.41 (1.49–1.41) a | 26.0–1.90 (2.02–1.90) |
| Completeness (%) | 98.7 (91.9) | 99.0 (94.0) |
| Redundancy | 7.2 (6.3) | 7.0 (6.3) |
| I / σI | 13.4 (3.1) | 10.2 (1.7) |
| Rmerge linear | 0.059 | 0.083(1.18) |
| Refinement statistics | ||
| Refinement program | SHELXL | REFMAC5 |
| Resolution (Å) | 8.0–1.42 | 26.0–1.92 |
| Rwork/Rfree b | 0.122/0.191 | 0.22/0.25 |
| Number of atoms (non-hydrogen) | 4703 | 4745 |
| B-factor main chain | 21.07 | 42.1 |
| B-factor side chain | 29.25 | 45.3 |
| RMSD bond lengths (Å) | 0.01 | 0.03 |
| RMSD bond angles c | 0.03 Å | 2.5° |
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|Io-Ia|/Σ(Ia), where Io is the observed intensity and Ia is the average intensity, the sums being taken over all symmetry related reflections. R-factor = Σ|Fo-Fc|/Σ(Fo), where Fo is the observed amplitude and Fc is the calculated amplitude. Rfree is the equivalent of Rworking, except it is calculated for a randomly chosen set of reflections that were omitted (5%) from the refinement process (41).
The RMSD bond angles are reported as an angle distance using the program SHELXL and as degrees in the program REFMAC5.
Overall Structures of SMP30
The overall structure of SMP30 contains 24 β-strands forming six β-sheets (Fig. 2A,B) that form a closed circular arrangement around a central solvent filled tunnel that has one metal binding site. Each β -sheet can be thought of as a blade of the 6-bladed β-propeller fold. In the majority of previously solved β-propeller proteins the active site is located in this central tunnel. Two regions of higher disorder, encountered while solving the crystal structures, were assigned to a flexible one-turn α-helix (residues 268–275) connected with short loops and a six residue flexible loop (residues 121–127), which is positioned at the top of the central tunnel presenting a lid structure to the putative active site. This unique positioning of the loop may play a role as a gatekeeper to the active site or present interactions that dictate substrate specificity. The C-terminal tail of SMP30 is 12-residues long (residues 287–299), and it traverses across the bottom of the central tunnel. The residues surrounding the front of the metal binding site (R101, M118, I34 and D104) form the putative active site of the enzyme (Fig. 2D).
Single Metal Binding Site
The single metal binding site observed near the active site that is coaxial with the central tunnel of the β-propeller. In the Ca2+-bound crystal structure of SMP30, the Ca2+ atom coordinates with the residues E18, N154 and D204 and three water molecules (Fig. 2C). The average Ca2+ to oxygen distances in subunits A and B are: 2.13 Å for E18, 2.30 Å for N154, 2.18 Å for D204, and 2.25 Å, 2.28 Å and 2.32 Å for the three water molecules. The residue N103 is also present in close proximity to this site, though not in direct coordination. In the Zn2+-bound form of SMP30, the residues involved in coordination with the single Zn2+ atom are identical to those for the Ca2+-bound form. The average distances in subunits A and B between Zn2+ and each coordinated oxygen atom are as follows: 2.12 Å for E18, 2.27 Å for N154 and 2.37 Å for D204. The Zn2+ water distances are 2.46 Å, 2.57 Å and 2.60 Å in chain B, and a single water was modeled into chain A with a distance of 2.24 Å from the Zn2+ atom.
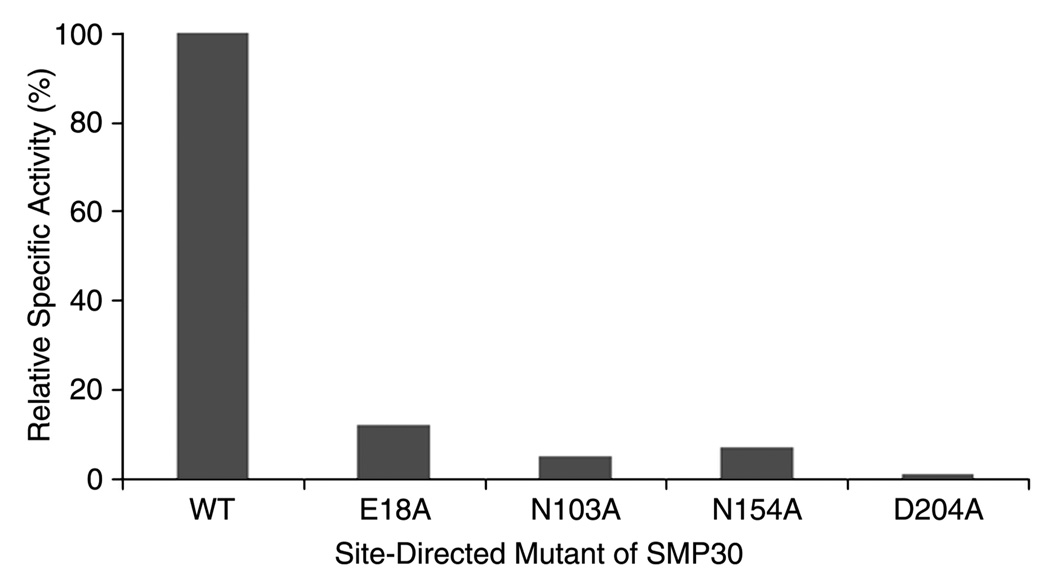
To determine whether the metal ion has a structural or catalytic role we did alanine scanning mutagenesis of the metal residues. The residues E18, N103, N154 and D204 were individually mutated to alanine and the four mutants were expressed and purified to homogeneity. Relative to the wild type specific activity, the metal site mutants each displayed a significant loss of activity (Fig. 3). This assay was performed at a Zn2+ concentration which was 10-fold greater than the Kd measured for the wild type enzyme. The loss of activity seen for these alanine mutations is interpreted to result from a reduced affinity of the mutants for the Zn2+ ion, which is essential for catalysis. The reported values of relative activity are the average of at least two independent experiments performed in triplicate.
Figure 3.
The relative specific activity of wild type versus E18A, N103A, N154A and D204A are shown for the hydrolysis of gluconolactone under standard assay conditions described in the experimental section with 75 µM Zn2+.
Homology to other Six-bladed β-propellers
A structural alignment is shown in Supplemental Figures 1 and 2, Supporting information of SMP30 with known six-bladed β-propeller enzymes. SMP30 displayed structural homology with the following proteins: bacterial drug resistant protein from S. aureus (Drp35), PON1, DFPase, and a protein from Xanthomonas campestris (XC5397) (31) with root mean squared deviation (RMSD) values of 2.14, 2.9, 1.9, and 1.74 Å, respectively. A structural comparison of the metal binding site of SMP30 with that of Drp35, PON1, DFPase and XC5397 reveals similar coordination geometries. The residues E18, N154 and D204 in SMP30 correspond to E53, N224 and D269 in PON1; E48, N185 and E236 in Drp35; E21, N175 and E229 in DFPase; and E48, N191 and D242 in XC5397, respectively. Although the SMP30 residues E18, N154 and D204 are conserved in all four proteins, there are no additional residues coordinating with the catalytic metal ion, unlike the structural homologues PON1, DFPase and XC5397. However, the residue N103 of SMP30 is present at nearly an identical position to N168 of PON1, and the Asn residue of the other homologues.
The side chain of N103 of SMP30 has a unique conformation distinctly different from its structural homologues. The chi1 angle of the side chain is rotated 90° relative to a position that would be able to coordinate with the metal ion. It might be possible that this unique positioning of the N103 residue allows a unique conformation suitable for substrate binding and catalysis. It may also partly explain how SMP30 binds a variety of divalent metal ions at the same site, unlike its structural homologues, that apparently only bind calcium.
Our preliminary enzyme kinetic studies showed that adding Zn2+ to an assay reaction of the enzyme, purified with Ca2+ present, showed a marked increase in the reaction rate. This led to our pursuit of a crystal structure with Zn2+ bound. We wanted to sort out whether the Zn2+ ion displaced the Ca2+ and bound to the same metal binding site. The structure of the Ca2+-SMP30 was superposed with the Zn2+-SMP30 with a RMSD of 0.24 Å and the metal binding site was nearly identical, as described above (Supplemental Figure 2, Supporting information). To confirm the placement of Zn at metal binding site of SMP30 we generated an anomalous difference fourier map using anomalous difference data from the Zn-bound structure of SMP30 collected at 0.9793 Å compared to the Zn-edge of 1.2837 Å. The electron density map was generated using the difference of Bijvoet pairs for amplitude and phases from the final refined Zn-bound structure. As shown in Supplemental Figure 3, Supporting information, a strong signal is displayed for an anomalous difference fourier map generated with a 4.0 sigma cutoff. The Ca-edge is at 3.0704 Å and would not display this signal. Our structures unequivocally show that Zn2+ displaces Ca2+ to occupy the same site and does not occupy a secondary site, confirmed by the lack of a second peak in the anomalous difference fourier map within one subunit of SMP30.
Role of Divalent Cations in Gluconolactonase Activity
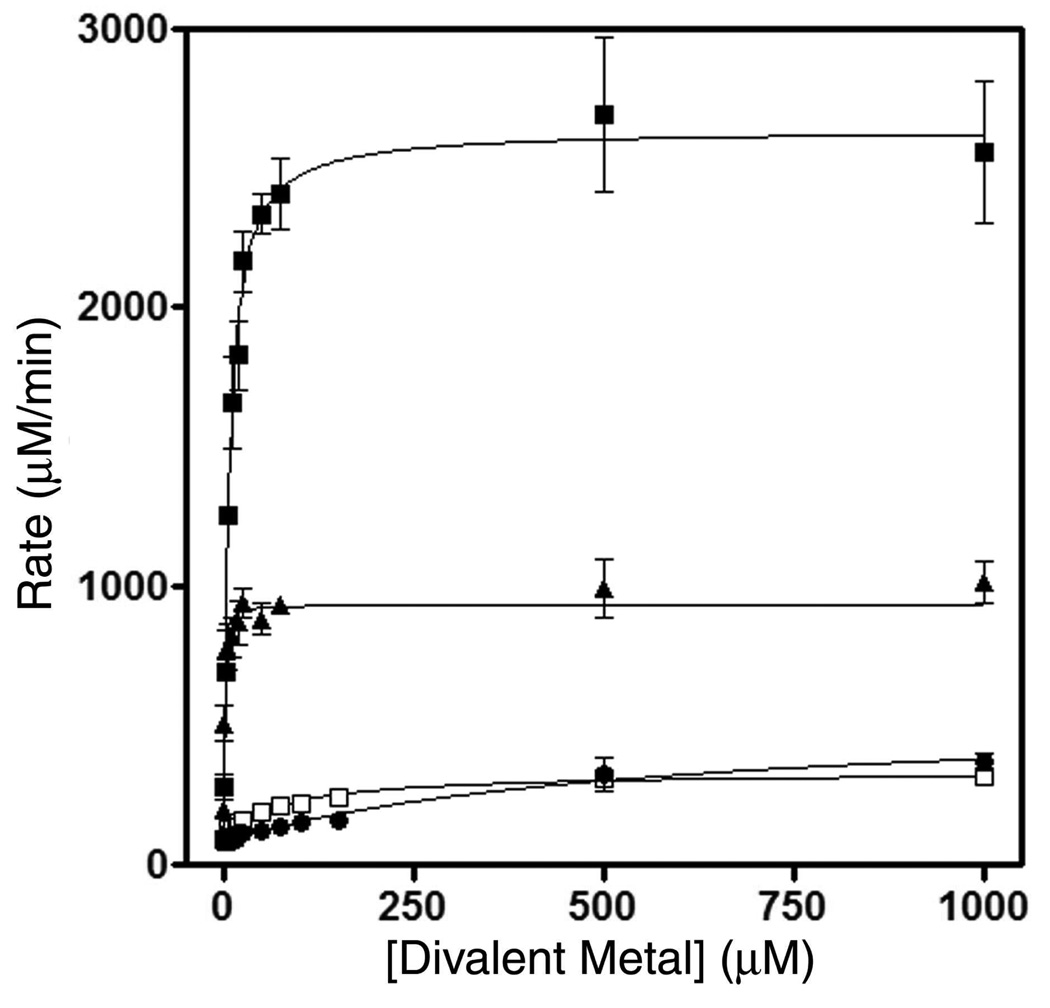
Enzyme kinetic studies of SMP30 with divalent metal ions showed some interesting results. Although maximum gluconolactonase activity was observed in the presence of Zn2+, we observed significant gluconolactonase activity in the presence of Ca2+, Mg2+ and Mn2+ as well. The kinetic parameters kcat and KM of the SMP30 gluconolactonase reaction in the presence of various metal ions is reported in Table 2. To understand the physiological role of various divalent cations in the SMP30 reaction we also calculated the dissociation constants of the divalent cations towards human SMP30, with respect to the activity of the enzyme. Here the Mn2+-bound enzyme showed maximum affinity (lowest Kd value) when compared to the other cations (Fig. 4 and Table 2).
Table 2.
Kinetic characterization of SMP30 with gluconolactone and its metal dependence a
| KM (mM) a | kcat (s−1) a | kcat/KM (mM−1s−1) a | Kd (µM) b | Cellular concentrations c |
|
|---|---|---|---|---|---|
| Zn2+ | 2.7 ± 0.5 | 341 ± 35 | 126 ± 27 | 7.0 ± 0.9 | fM-nM |
| Mn2+ | 0.6 ± 0.1 | 79 ± 8 | 132 ± 26 | 0.6 ± 0.1 | 0.1–10 µM |
| Mg2+ | 1.3 ± 0.3 | 31 ± 4 | 24 ± 7 | 82 ± 21 | 500 µM |
| Ca2+ | 3.7 ± 0.7 | 48 ± 6 | 13 ± 3 | 566 ± 249 | 0.1–1 µM (resting)/ 1000 µM (stress) |
KM and kcat values are reported for human SMP30 using the substrate gluconolactone and a saturating concentration of the divalent metal ion listed using conditions detailed in the materials and methods.
The reported Kd values were obtained using 10 mM gluconolactone and varying the metal concentration to obtain velocities. A fit of Equation 1 to data shown in Figure 4 yielded the reported values of Kd as detailed in material and methods.
Estimates of physiological concentration of divalent cations were taken from the litterature (34–40).
Figure 4.
The metal dependent gluconolactonase activity of SMP30 is compared for Zn2+ (solid squares), Mn2+ (triangles), Mg2+ (open squares), and Ca2+ (circles) bound enzyme. The gluconolactonase assay was performed with saturating gluconolactone (10 mM) with enzyme that had been metal depleted, then incubated with varying concentrations of the indicated divalent cations. Other details of this comparative assay are described in the experimental section.
DISCUSSION
In recent years, there has been considerable interest in the structure and function of SMP30, with several implications of its anti-aging role, predominantly through calcium regulation and homeostasis. It is also interesting to find out that the SMP30 gene is highly conserved among eukaryotes (Supplemental Figure 4, Supporting information). At the same time, studies have shown that SMP30 is an enzyme that catalyzes a lactonase reaction on physiologically occurring substrates like gulonolactone or synthetic OP compounds like DFP. It was clear that a crystal structure would aid in the understanding of the function of this enzyme. On our way towards solving the first crystal structures of the human SMP30, we also characterized enzyme kinetics that varies based on which metal ion is bound at the active site. A comparison of the structural homologues and the metal dependent studies are discussed further.
A 3D BLAST (28) search with SMP30 identified a bacterial homologue from Agrobacterium tumefaciens (PDB code: 2ghs) with the closest matched structure (score of 77 bits). Although the structure of this protein is available in the PDB, further details are not yet published. Earlier we had identified this protein structure as a homologue, and we used it to construct a search model for molecular replacement to solve the structure of SMP30. The RMSD of our SMP30 structure aligned with the structure of this bacterial enzyme was 1.39 Å with an alignment length of 280 residues. However, the sequence identity was only 20% over a stretch of 291 residues. High structural homology together with low sequence homology has been observed for other members of the β-propeller fold (29). Although the overall structure of the bacterial enzyme is very similar to that of SMP30, the absence of a bound metal in the bacterial enzyme is conspicuous. A close comparison of the metal binding residues of SMP30 and this structural homologue reveals that the metal coordinating residues are conserved. Of the three conserved residues, the positions of N154 and E18 of SMP30 correspond well with that of N170 and E40 of the bacterial homologue. However the residue D204 of SMP30 and D221 of the bacterial homologue, though present in roughly the same positions, have different orientations. The residue D221 is shifted away from the metal binding center, thereby making it unsuitable for metal coordination. As both the SMP30 structure (1.4 Å) and the bacterial homologue (1.5 Å) are high-resolution structures, we can infer that a shift of D221 may explain its inability to bind a metal ion. A comparison of the putative active site pocket residues of SMP30 with that of the bacterial homologue showed an overlap of similar residues in the corresponding positions. N103, R101, C16 and M118 of SMP30 correspond well with N125, R123, L38 and M140 of the bacterial homologue. Despite the absence of a metal bound to this bacterial homologue, it is the most structurally similar protein in the PDB to our reported human SMP30 structure.
Other SMP30 homologues include the bacterial enzymes XC5397, a Xanthomonas campestris protein with an ability to hydrolyze D-glucono-δ-lactone (30) and Drp35, a drug resistance protein capable of hydrolyzing lactones (31), from Staphylococcus aureus (Supplemental Figure 5, Supporting information). The RMSD of XC5397 and Drp35 with SMP30 are 1.74 Å and 2.14 Å, respectively. Interestingly both these bacterial enzymes have Ca2+-dependent lactonase activity. Three of the four conserved calcium-coordinating residues E48, N191 and D242 of XC5397, as well as E48, N185 and D236 of Drp35, correspond to E18, N154 and D204 of SMP30. Although the bacterial enzymes Drp35 and XC5397 have both been characterized as lactonases, they have significant differences from SMP30. XC5397 acts as a dimer and has a total of six calcium ions, two in each subunit and two in the dimer interface; in contrast SMP30 has only one metal per molecule and functions as a monomer. Unlike XC5397, which can hydrolyze only gluconolactone, SMP30 can hydrolyze a range of lactones (16). Mouse SMP30 also has been shown to display a DFPase activity in the presence of Mg2+ (18). Furthermore, the gluconolactonase activity of SMP30 displayed maximum activity in the presence of Zn2+, whereas XC5397 required Ca2+ for maximum activity (30). Though the hydrolysis of gluconolactone seems to be an overlapping kinetic function between XC5397 and SMP30 it is farfetched to correlate the functions of the bacterial enzyme XC5397 with those of the mammalian SMP30s. Likewise, a comparison of the active site residues of the bacterial enzyme Drp35 with SMP30 showed a few residues that were placed in similar positions and a few others, which were not. The residues F153, L105, S237 and A90 of Drp35 correspond well with M118, R101 and G205 of SMP30. In contrast, the residues N299, F64, D152 and A90 of the Drp35 active site pocket do not have similar residues in SMP30. This suggests that SMP30 is distinctly different from Drp35 with its Zn2+-dependent gluconolactonase activity. Although the catalytic Ca2+ of Drp35 binds to similar residues as in SMP30, another Ca2+ ion is bound on the surface, which is coordinated to residues T133, S110, D130, Y135 and G112. There are no corresponding residues found in SMP30. Although each of the three bacterial lactonases discussed have overall structures similar to SMP30, they have limited utility to understand the physiological function of mammalian SMP30.
Of the eukaryotic β-propellers, the structure and function of SMP30 most resembles PON1 and DFPase. The RMSD of SMP30 overlays with PON1 and DFPase are 2.9 Å and 1.87 Å, respectively. In addition to the known OP hydrolase activity of PON1 and DFPase, PON1 has been identified as a lactonase (19, 32). Although the aromatic lactones that PON1 prefers to hydrolyze (32) are different from gluconolactone and gulonolactone that are known substrates of SMP30, the comparison of the active site residues of PON1 with our SMP30 putative active-site pocket allows us to predict which residues of SMP30 play a role in determining substrate and metal site specificity.
The conserved PON1 catalytic Ca2+-binding residues E53, N224 and D269 correspond well to E18, N154 and D204 of SMP30 (Supplemental Figure 2, Supporting information). However PON1 has two other residues (N168 and N270) that coordinate with the catalytic calcium. The corresponding residues in SMP30 (N103 and G205) are unable to coordinate with the metal ion. Other critical active site residues of PON1 are H115 and H134, which are in close proximity to the catalytic Ca2+-binding site. From an alignment, the corresponding residue R101 of SMP30 is placed in between these two histidines in such a way that its backbone corresponds to H134 and its side chain nitrogen atoms are very close to H115.
The HDL-associated enzyme PON1 is different from SMP30 with some distinctive features. PON1 is not a water-soluble enzyme like SMP30, and it requires detergents or HDL particles for its function (33). Additionally, there is a second high affinity Ca2+-binding site deep in the tunnel of PON1, which is referred to as the structural calcium. The residues D169, D54 and backbone carbonyl of I117 of PON1 coordinate with this structural calcium. However, with the exception of D104, which corresponds well with D169 of PON1, no other corresponding residues are present in SMP30. We can unequivocally conclude that SMP30 does not contain this second structural metal site homologous to PON1 and DFPase. The three helices of PON1 give it a structure distinct from all other homologues. Helix one is the secreted signal peptide that helps PON1 to bind to HDL, whereas helix two and three form a canopy to give a somewhat closed active site. Interestingly, SMP30 has one α-helix (residues 268–275) located outside its active site and is connected by a flexible loop. Also there is an additional flexible loop (residue 121–127) that resembles a flap over the tunnel (Fig. 2B). Together, the helix and the loop of SMP30 may be functionally analogous to the two helices of PON1 that form a canopy outside its active site.
The squid neuron enzyme DFPase is another member of the OP hydrolase enzymes with a six-bladed β-propeller architecture. Although SMP30 and DFPase have a sequence similarity of 16%, the RMSD of 1.87 Å shows these proteins are structurally quite similar. Notably, it has been reported that SMP30 has a DFPase activity, in the presence of Mg2+ (17). As SMP30 and DFPase can both hydrolyze DFP, it seemed instructive to compare our structure of SMP30 to the active site of DFPase. The conserved Ca2+-coordinating residues of DFPase: E21, D229 and N175 correspond to E18, E204 and N154 of SMP30 (Supplemental Figure 2, Supporting information). The fourth calcium-coordinating residue N120 of DFPase corresponds to N103 of SMP30 and is not coordinated with the metal ion. One of the active site residues of DFPase, H287 aligns with C16 of SMP30, whereas the other DFPase residue E37 does not have a related residue in SMP30. It is interesting to note that the R101 of SMP30 has no corresponding residues in DFPase. Like PON1, DFPase also has a second “structural” calcium that is absent in SMP30. Residues D232, L273 and H274 coordinate with this calcium, and no corresponding residues were observed in SMP30. When comparing the overall structure of SMP30 with DFPase we see the absence of the prominent helix in the later. However, the C-terminal tail of SMP30 and DFPase traverse the tunnel in a similar fashion on the side opposite the enzymes’ active sites. A comprehensive mutagenesis and kinetic characterization of SMP30 is currently underway to elucidate the SMP30 enzyme mechanism and its relationship to these similar, yet distinct, eukaryotic enzymes. From the distant sequence homology, structural homology and related functions, we have reason to believe that SMP30, PON1 and DFPase have evolved from a common protein.
Human SMP30 contains only one metal bound per protein subunit as revealed in our crystal structures. This becomes interesting when compared to the structural homologues described previously, which have at least two metal ions per molecule, one catalytic and the other structural. Our preliminary site-directed mutagenesis studies on the metal-coordinating residues demonstrated that the metal ion in SMP30 is catalytic (Fig. 3). To further probe the catalytic role of the residues we calculated the temperature dependent activity of the metal-free enzyme pre-incubated with, or without the metal ions (Supplemental Figure 6, Supporting information). If the metal ion was structural, incubation of the enzyme with a high concentrations of calcium, prior to exposure to higher temperatures, would be expected to retain wild type activity to a greater extent than the enzyme which was not treated with metal. However, if the metal was catalytic there would be no difference in the temperature-dependent activity-curve, as we found to be the case, thereby supporting a conclusion that the metal is catalytic.
Interestingly, all the reported crystal structures of the homologues have only calcium bound to the protein and are each believed to be calcium dependent enzymes. SMP30, on the other hand, is thought to be a Zn2+-dependent gluconolactonase, a Mg2+-dependent DFPase as well as a Ca2+-binding protein. We have been successful in solving crystal structures bound to Ca2+ and Zn2+. This promiscuity towards various divalent cations becomes more intriguing as the Zn2+ and Ca2+-bound structures have identical metal binding sites and nearly super imposable structures (Supplemental Figures 2, Supporting information). We have calculated the kinetic constants and found that although Zn2+ has the highest kcat for the hydrolysis of gluconolactone; Ca2+, Mn2+ and Mg2+ also catalyze the reaction, albeit at a slower rate (Table 2). Although the gluconolactone KM for the Ca2+-dependent activity is a bit higher (3.7 mM) when compared to the Zn2+-dependent activity (2.7 mM), the kcat for the Ca2+-dependent activity (48 s−1) is only 14% of that found for the Zn2+-bound form (341 s−1).
To understand the physiological relevance of this metal dependent activity we calculated the dissociation constants of the metal ions and found that Mn2+ has the highest affinity for the enzyme. As a result the concentration of Mn2+ needed to catalyze the reaction is lower compared to the other metal ions (Fig. 4, Table 2). Notably, when we compare the reported free concentrations of these divalent cations in the cell (34–38), with our experimentally determined dissociation constants of the metal ions, this promiscuity may have significant functional implications (Table 2). The free cellular concentration of Zn2+ is between nanomolar to femtomolar, and its calculated Kd for gluconolactonase activity is 7 µM. The free cellular concentrations of Zn2+ may not be high enough to be physiologically significant. The experimentally determined Kd of Mn2+ and Mg2+ are 0.6 µM and 82 µM, respectively; whereas their free cellular concentrations are roughly 0.1 µM - 10 µM and 500 µM, respectively. Comparing these values to our experimentally determined Kd values it is evident that both Mn2+ and Mg2+ could comprehensibly bind to SMP30 and catalyze the gluconolactonase reaction in a normal healthy liver or kidney cell.
Apart from the first crystal structures of the human enzyme the most exciting and novel aspect of this research is that we have established that SMP30 has Ca2+ dependent gluconolactonase activity. This directly contradicts previous studies that conclude SMP30 did not show any enzymatic activity in the presence of calcium ions (16, 18). Our measured kcat value of 48 sec−1 (Table 2) shows that the Ca2+-bound enzyme can catalyze the reaction at a reasonable rate, contrary to previous reports (16). Our calculated Kd of Ca2+ (566 µM) provides a possible explanation of why the previous studies did not observe any activity with calcium ions if conditions with sufficient Ca2+ were not tried. Alternatively, the previous samples might not have been metal-free and might have had other high-affinity divalent cations already bound to the metal binding site. In a separate study the association constant of Ca2+ has been reported for the rat liver protein to be 4.19 × 105 M−1 (equivalent to a Kd of 2.4 × 10−6 M) with 6 or 7 calcium binding sites per molecule (13). However our crystal structure clearly shows that there is only one metal binding site in SMP30. The difference between our calculated Kd values for Ca2+ (566 × 10−6 M) and that of the earlier study (2.4 × 10−6 M) might be because of the different experimental approaches. While they measured the association constant by equilibrium dialysis, which does not involve any substrate, we calculated the Kd with respect to the enzymatic activity. We are interested in the role of metal ions with respect to its gluconolactonase activity and hence developed an assay to calculate the Kd in the presence of the substrate, gluconolactone. Hence, this may explain why our Kd values are higher than what have been reported previously (13). However, in our case heterologous expression and purification yielded highly pure SMP30 protein. The presence in previous studies of trace quantities of impurities with very high calcium binding affinity might cause problems when proteins are purified directly from tissues. An impurity with a high calcium affinity could give anomalous results.
To find a possible connection between the gluconolactonase activity and calcium regulation of SMP30 it is instructive to compare the experimentally measured Kd of Ca2+ (566 µM) with that of its free cellular concentrations (0.1–1.0 µM); it is evident that Ca2+ is not likely to be the metal of choice for gluconolactonase activity in a resting cell. Based upon the numerous studies on the mouse and rat proteins, one should consider whether SMP30 functions as an anti-aging molecule by maintaining calcium homeostasis. It is well known that the extracellular free calcium concentration (1–2 mM) is about 10,000 fold higher than the intracellular concentration (0.05–1.0 µM) as has been reported extensively in the context of calcium homeostasis and aging (39, 40). The aging process and reduced cellular function have been linked to, among other factors, alterations in the capacity to maintain a constant low level of Ca2+. The age related loss of critical calcium regulatory channels and pumps have been reviewed extensively (39, 40). It might be possible that a gradual increase in intracellular calcium with gradual decrease of calcium-pumping functions may alter the calcium concentrations during aging. If the concentrations of intracellular calcium increase above the Kd value (566µM), then as per our data, SMP30 might function as a Ca2+ dependent gluconolactonase (Table-2). Also since all the age-related studies have been reportedly done with rodents (which produce vitamin-C) a definitive role of calcium dependence in human SMP30 function could only be established after further complementary and supplementary in vivo studies. Regardless, one can conclude that SMP30 plays an important role in humans due to its high degree of conservation in the SMP30 protein sequence among higher eukaryotes (Supplemental Figure 4, Supporting information).
On the other hand, the promiscuous nature of metal ions and the high Kd values for calcium might actually suggest that SMP30 might function indirectly to regulate calcium levels or have a totally different function other than calcium regulation and homeostasis; especially in the haplorrhini suborder of primates who are unable to produce their own vitamin-C. There might be a yet unknown alternate physiological substrate that SMP30 will hydrolyze with a much higher rate in the presence of calcium. In light of the metal binding studies done previously (13), it will be useful to calculate the calcium Kd in the absence of substrate to see if the substrate influences this value.
In conclusion, along with the first high-resolution Ca2+-bound and Zn2+-bound crystal structures of the human enzyme our studies show several novel and interesting features of SMP30 never reported before. There is only one metal binding site, which is quite different from any of its homologues. Also, both zinc and calcium bind to the same metal-binding site in an identical manner. This is interesting, as normally the coordination of zinc is quite distinct from that of calcium in enzymes. It is possible that the metal coordination is different for the calcium and zinc bound sites only in the presence of the substrate and thus contributes to the difference in enzymatic rate. Our studies show for the first time that the human SMP30 has Ca2+-dependent enzyme activity, and differences to previous studies were discussed. We also compared the physiological concentrations of divalent cations with our calculated metal ion Kd values and conclude that Mn2+ and Mg2+ are the likely functional metal ions for healthy cells. Overall, our kinetic and structural insights of human SMP30 allow further elucidation of the physiological role of this exciting anti-aging molecule.
Supplementary Material
SUPPORTING INFORMATION AVAILABLE
Supplemental Figure 1: The six-bladed β-propeller folds are shown for structural homologs of human SMP30.; Supplemental Figure 2: Superposition of calcium/zinc structures of SMP30 and with homologues PON1 and DFPase; Supplemental Figure 3: Anomalous difference fourier map of Zn-bound SMP30; Supplemental Figure 4: Amino acid sequence alignment of eukaryotic versions of SMP30; Supplemental Figure 5: Structure-based amino acid sequence alignment of human SMP30 structural homologs prepared with the program STAMP; and Supplemental Figure 6: Temperature dependent activity curve of human SMP30. This material is available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGEMENT
Use of the Advanced Photon Source at the Argonne National Laboratory was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract DE-AC02-06CH11357. Use of the SGX Collaborative Access Team (SGX-CAT) beam line at Sector 31 of the Advanced Photon Source was provided by SGX Pharmaceuticals, Inc., who constructed and operated the facility. The authors would like to thank Dr. Randy Read of Cambridge University and Chavas Leo of University of Manchester for their valuable insights. The authors would also like to thank Dr. Danny Ramadan of the Thorpe Lab, for help with the metal binding analysis.
Abbreviations
- APS
Advanced Photon Source
- DFPase
diisopropylfluorophosphatase
- PON1
paraoxonase 1
- RMSD
root mean squared deviation
- SMP30
senescence marker protein 30
- OP
organophosphorus
- PDB
protein data bank
- Kd
dissociation constant
- DFP
diisopropylphosphofluoridate
Footnotes
This work was supported by NIH Grants 2P20RR015588 from the National Center for Research Resources and 1R01HL084366 from the National Heart, Lung, and Blood Institute.
Protein Data Bank entry codes 3g4e and 3g4h are the atomic coordinates and structure factors for the Ca2+ bound and the Zn2+ bound crystal structures of human SMP30, respectively.
REFERENCES
- 1.Fujita T, Uchida K, Maruyama N. Purification of senescence marker protein-30 (SMP30) and its androgen-independent decrease with age in the rat liver. Biochim Biophys Acta. 1992;1116:122–128. doi: 10.1016/0304-4165(92)90108-7. [DOI] [PubMed] [Google Scholar]
- 2.Feng D, Kondo Y, Ishigami A, Kuramoto M, Machida T, Maruyama N. Senescence marker protein-30 as a novel antiaging molecule. Ann N Y Acad Sci. 2004;1019:360–364. doi: 10.1196/annals.1297.062. [DOI] [PubMed] [Google Scholar]
- 3.Fujita T, Inoue H, Kitamura T, Sato N, Shimosawa T, Maruyama N. Senescence marker protein-30 (SMP30) rescues cell death by enhancing plasma membrane Ca(2+)-pumping activity in Hep G2 cells. Biochem Biophys Res Commun. 1998;250:374–380. doi: 10.1006/bbrc.1998.9327. [DOI] [PubMed] [Google Scholar]
- 4.Ishigami A, Fujita T, Handa S, Shirasawa T, Koseki H, Kitamura T, Enomoto N, Sato N, Shimosawa T, Maruyama N. Senescence marker protein-30 knockout mouse liver is highly susceptible to tumor necrosis factor-alpha- and Fas-mediated apoptosis. Am J Pathol. 2002;161:1273–1281. doi: 10.1016/s0002-9440(10)64404-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishigami A, Fujita T, Inoue H, Handa S, Kubo S, Kondo Y, Maruyama N. Senescence marker protein-30 (SMP30) induces formation of microvilli and bile canaliculi in Hep G2 cells. Cell Tissue Res. 2005;320:243–249. doi: 10.1007/s00441-004-1073-5. [DOI] [PubMed] [Google Scholar]
- 6.Ishigami A, Kondo Y, Nanba R, Ohsawa T, Handa S, Kubo S, Akita M, Maruyama N. SMP30 deficiency in mice causes an accumulation of neutral lipids and phospholipids in the liver and shortens the life span. Biochem Biophys Res Commun. 2004;315:575–580. doi: 10.1016/j.bbrc.2004.01.091. [DOI] [PubMed] [Google Scholar]
- 7.Maruyama N, Ishigami A, Kuramoto M, Handa S, Kubo S, Imasawa T, Seyama K, Shimosawa T, Kasahara Y. Senescence marker protein-30 knockout mouse as an aging model. Ann N Y Acad Sci. 2004;1019:383–387. doi: 10.1196/annals.1297.068. [DOI] [PubMed] [Google Scholar]
- 8.Son TG, Kim SJ, Kim K, Kim MS, Chung HY, Lee J. Cytoprotective roles of senescence marker protein 30 against intracellular calcium elevation and oxidative stress. Archives of Pharmacal Research. 2008;31:872–877. doi: 10.1007/s12272-001-1240-3. [DOI] [PubMed] [Google Scholar]
- 9.Son TG, Zou YN, Jung KJ, Yu BP, Ishigami A, Maruyama N, Lee J. SMP30 deficiency causes increased oxidative stress in brain. Mechanisms of Ageing and Development. 2006;127:451–457. doi: 10.1016/j.mad.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 10.Tuder RM. Aging and cigarette smoke: Fueling the fire. American Journal of Respiratory and Critical Care Medicine. 2006;174:490–491. doi: 10.1164/rccm.200607-924ED. [DOI] [PubMed] [Google Scholar]
- 11.Yumura W, Imasawa T, Suganuma S, Ishigami A, Handa S, Kubo S, Joh K, Maruyama N. Accelerated tubular cell senescence in SMP30 knockout mice. Histology and Histopathology. 2006;21:1151–1156. doi: 10.14670/HH-21.1151. [DOI] [PubMed] [Google Scholar]
- 12.Yamaguchi M, Mori S, Kato S. Calcium-binding protein regucalcin is an activator of (Ca2+-Mg2+)-adenosine triphosphatase in the plasma membranes of rat liver. Chem Pharm Bull (Tokyo) 1988;36:3532–3539. doi: 10.1248/cpb.36.3532. [DOI] [PubMed] [Google Scholar]
- 13.Yamaguchi M. Role of regucalcin in calcium signaling. Life Sci. 2000;66:1769–1780. doi: 10.1016/s0024-3205(99)00602-5. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi M. Role of regucalcin in maintaining cell homeostasis and function (Review) International Journal of Molecular Medicine. 2005;15:371–389. [PubMed] [Google Scholar]
- 15.Linster CL, Van Schaftingen E. Vitamin C. Biosynthesis, recycling and degradation in mammals. FEBS J. 2007;274:1–22. doi: 10.1111/j.1742-4658.2006.05607.x. [DOI] [PubMed] [Google Scholar]
- 16.Kondo Y, Inai Y, Sato Y, Handa S, Kubo S, Shimokado K, Goto S, Nishikimi M, Maruyama N, Ishigami A. Senescence marker protein 30 functions as gluconolactonase in L-ascorbic acid biosynthesis, and its knockout mice are prone to scurvy. Proc Natl Acad Sci U S A. 2006;103:5723–5728. doi: 10.1073/pnas.0511225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Billecke SS, Primo-Parmo SL, Dunlop CS, Doorn JA, La Du BN, Broomfield CA. Characterization of a soluble mouse liver enzyme capable of hydrolyzing diisopropyl phosphorofluoridate. Chemico-Biological Interactions. 1999;120:251–256. doi: 10.1016/s0009-2797(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 18.Kondo Y, Ishigami A, Kubo S, Handa S, Gomi K, Hirokawa K, Kajiyama N, Chiba T, Shimokado K, Maruyama N. Senescence marker protein-30 is a unique enzyme that hydrolyzes diisopropyl phosphorofluoridate in the liver. FEBS Lett. 2004;570:57–62. doi: 10.1016/j.febslet.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Gaidukov L, Tawfik DS. The development of human sera tests for HDL-bound serum PON1 and its lipolactonase activity. Journal of Lipid Research. 2007;48:1637–1646. doi: 10.1194/jlr.D600045-JLR200. [DOI] [PubMed] [Google Scholar]
- 20.Scharff EI, Koepke J, Fritzsch G, Lucke C, Ruterjans H. Crystal structure of diisopropylfluorophosphatase from Loligo vulgaris. Structure. 2001;9:493–502. doi: 10.1016/s0969-2126(01)00610-4. [DOI] [PubMed] [Google Scholar]
- 21.Warizaya M, Kinoshita T, Yamaoka M, Shibata T, Saito N, Nakajima H, Fujii T. Expression, purification, crystallization and preliminary X-ray diffraction studies of human liver regucalcin. Acta Crystallogr D Biol Crystallogr. 2004;60:2019–2021. doi: 10.1107/S0907444904020608. [DOI] [PubMed] [Google Scholar]
- 22.Jancarik J, Kim SH. Sparse Matrix Sampling: a Screening Method for Crystallization of Proteins. J. Appl. Cryst. 1991;24:409–411. [Google Scholar]
- 23.CCP4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 24.Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Sheldrick GM, Schneider TR. SHELXL: high-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 27.Vriend G. WHAT IF: A molecular modeling and drug design program. Journal of Molecular Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SFGW, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Jawad Z, Paoli M. Novel sequences propel familiar folds. Structure. 2002;10:447–454. doi: 10.1016/s0969-2126(02)00750-5. [DOI] [PubMed] [Google Scholar]
- 30.Chen CN, Chin KH, Wang AHJ, Chou SH. The First Crystal Structure of Gluconolactonase Important in the Glucose Secondary Metabolic Pathways. Journal of Molecular Biology. 2008;384:604–614. doi: 10.1016/j.jmb.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 31.Tanaka Y, Morikawa K, Ohki Y, Yao M, Tsumoto K, Watanabe N, Ohta T, Tanaka I. Structural and mutational analyses of Drp35 from Staphylococcus aureus - A possible mechanism for its lactonase activity. Journal of Biological Chemistry. 2007;282:5770–5780. doi: 10.1074/jbc.M607340200. [DOI] [PubMed] [Google Scholar]
- 32.Khersonsky O, Tawfik DS. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry. 2005;44:6371–6382. doi: 10.1021/bi047440d. [DOI] [PubMed] [Google Scholar]
- 33.Josse D, Ebel C, Stroebel D, Fontaine A, Borges F, Echalier A, Baud D, Renault F, Le Maire M, Chabrieres E, Masson P. Oligomeric states of the detergent-solubilized human serum paraoxonase (PON1) J Biol Chem. 2002;277:33386–33397. doi: 10.1074/jbc.M200108200. [DOI] [PubMed] [Google Scholar]
- 34.Ash DE, Schramm VL. Determination of Free and Bound Manganese(Ii) in Hepatocytes from Fed and Fasted Rats. Journal of Biological Chemistry. 1982;257:9261–9264. [PubMed] [Google Scholar]
- 35.Costello LC, Guan ZX, Kukoyi B, Feng P, Franklin RB. Terminal oxidation and the effects of zinc in prostate versus liver mitochondria. Mitochondrion. 2004;4:331–338. doi: 10.1016/j.mito.2004.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galeotti T, Palombini G, Vanrossum GDV. Manganese Content and High-Affinity Transport in Liver and Hepatoma. Archives of Biochemistry and Biophysics. 1995;322:453–459. doi: 10.1006/abbi.1995.1488. [DOI] [PubMed] [Google Scholar]
- 37.Sarisa NL, Mervaalaa E, Karppanena H, Khawaja JA, Lewenstam A. Magnesium An update on physiological, clinical and analytical aspects. Clinica Chimica Acta. 2000;294:1–26. doi: 10.1016/s0009-8981(99)00258-2. [DOI] [PubMed] [Google Scholar]
- 38.Williams RJP. Free Manganese(Ii) and Iron(Ii) Cations Can Act as Intracellular Cell Controls. Febs Letters. 1982;140:3–10. doi: 10.1016/0014-5793(82)80508-5. [DOI] [PubMed] [Google Scholar]
- 39.Fujita T. Calcium homeostasis and signaling in aging. In: Mattson MP, editor. Calcium homeostasis and signaling in aging. Elsevier, Amsterdam: 2002. pp. 13–26. [Google Scholar]
- 40.Yamaguchi M. Impact of aging on calcium channels and pumps. In: Mattson MP, editor. Calcium homeostasis and signaling in aging. Elsevier, Amsterdam: 2002. pp. 47–65. [Google Scholar]
- 41.Brunger AT. The free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPORTING INFORMATION AVAILABLE
Supplemental Figure 1: The six-bladed β-propeller folds are shown for structural homologs of human SMP30.; Supplemental Figure 2: Superposition of calcium/zinc structures of SMP30 and with homologues PON1 and DFPase; Supplemental Figure 3: Anomalous difference fourier map of Zn-bound SMP30; Supplemental Figure 4: Amino acid sequence alignment of eukaryotic versions of SMP30; Supplemental Figure 5: Structure-based amino acid sequence alignment of human SMP30 structural homologs prepared with the program STAMP; and Supplemental Figure 6: Temperature dependent activity curve of human SMP30. This material is available free of charge via the Internet at http://pubs.acs.org.