Abstract
The FLT3 receptor tyrosine kinase plays an important role in normal hematopoietic development and leukemogenesis. Point mutations within the activation loop and in-frame tandem duplications of the juxtamembrane domain represent the most frequent molecular abnormalities observed in acute myeloid leukemia. Interestingly these gain-of-function mutations correlate with different clinical outcomes, suggesting that signals from constitutive FLT3 mutants activate different downstream targets. In principle, mass spectrometry offers a powerful means to quantify protein phosphorylation and identify signaling events associated with constitutively active kinases or other oncogenic events. However, regulation of individual phosphorylation sites presents a challenging case for proteomics studies whereby quantification is based on individual peptides rather than an average across different peptides derived from the same protein. Here we describe a robust experimental framework and associated error model for iTRAQ-based quantification on an Orbitrap mass spectrometer that relates variance of peptide ratios to mass spectral peak height and provides for assignment of p value, q value, and confidence interval to every peptide identification, all based on routine measurements, obviating the need for detailed characterization of individual ion peaks. Moreover, we demonstrate that our model is stable over time and can be applied in a manner directly analogous to ubiquitously used external mass calibration routines. Application of our error model to quantitative proteomics data for FLT3 signaling provides evidence that phosphorylation of tyrosine phosphatase SHP1 abrogates the transformative potential, but not overall kinase activity, of FLT3-D835Y in acute myeloid leukemia.
Deregulated tyrosine phosphorylation is a well established molecular hallmark in the development of both solid tumors and hematopoietic malignancies (1, 2). In acute myeloid leukemia (AML),1 mutations of the Fms-like tyrosine kinase 3 (Flt3) gene or its overexpression represent the most frequent molecular abnormalities, observed in ∼35 and ∼90% of patients, respectively (3, 4). Internal tandem duplication (ITD) in the juxtamembrane region occurs in ∼25% of patients, whereas another 7% of cases manifest as point mutations in the activation loop at Asp-835 (typically D835Y). Both mutation classes induce constitutive tyrosine kinase activity and confer IL-3-independent growth of the factor IL-3-dependent BaF3 and 32D cells. Interestingly, the ITD and D835Y mutants respond differently to the current suite of targeted therapeutics (5), and in vitro studies suggest that they can activate different downstream targets, including STAT5a (6–8). Collectively, these observations motivate continued efforts to delineate oncogenic pathways in further detail to expand our understanding of the molecular basis for disease progression and to identify additional therapeutic entry points.
Over the past decade, mass spectrometry-based proteomics has become the technique of choice for large scale identification of peptides derived from various biological contexts (9, 10). Moreover, numerous methodologies now support relative quantification measurements, providing a routine means to monitor protein expression and post-translational modification state as a function of biological perturbation. Multiplexed isotope labels such as the iTRAQ (11) and TMT (12) reagents provide several advantages for quantitative proteomics, including (i) compatibility with the study of proteins derived from in vitro and in vivo systems, (ii) the ability to analyze up to eight samples (e.g. conditions) in a single LC-MS/MS run, and (iii) cumulative MS signal for equivalent peptide precursors derived from each experimental condition and cumulative signal for fragment ions resulting from MS/MS of any given peptide precursor. The additive nature of MS and MS/MS signals is particularly advantageous for analysis of low abundance proteins or rare post-translational modifications.
Upon dissociation during MS/MS analysis, both iTRAQ and TMT tags fragment to yield “reporter” ions that contain different stable isotopes; the respective peak intensities or areas of these signals provide a relative measure of parent peptide abundance in each biological condition. Assuming that individual peptides are correctly assigned to their source protein, then a combination of individual ratios from all detected peptides provides a reasonable estimate for quantification at the protein level (11, 13–17). This approach is applicable for so-called protein profiling studies where multiple peptides are detected and used as supporting evidence for the quantification of a given protein.
The situation is quite different for quantification of rare post-translational modifications such as tyrosine phosphorylation. Here it is common for proteins to carry a single site of tyrosine phosphorylation. Moreover, even when multiple phosphorylation events are detected on the same protein, the relative stoichiometry across sites may vary significantly as a result of their independent regulation via divergent biological processes. As a result, ratios from individual peptides must be analyzed separately outside the context of collective peptide evidence for a specific protein. In addition to these limitations, it is also possible that low abundance peptides are subjected to MS/MS analysis only once in a given LC-MS acquisition. Assignment of statistical significance to peptide ratios measured under these conditions, e.g. single peptides sequenced by only one MS/MS scan, is particularly challenging. To date, estimates of significance for individual scans have relied either on global ratio thresholds, for example ignoring any ratio less than 2-fold above or below a given control (18, 19), or derivation of noise models based on in-depth empirical characterization of individual ion peaks (20).
An additional consideration, less well recognized, is that each quantification strategy is susceptible to different sources of error and hence may require different data analytic strategies to estimate significance boundaries. For example, the co-elution properties of labels based on stable isotopes of carbon, nitrogen, and oxygen eliminate non-systematic measurement errors associated with the instability of electrospray or laser-based ionization. Nonetheless, stable isotope-based methods are subject to other sources of error. For example, measured signal-to-noise values for peptide ratios based on stable isotope labeling by amino acids in cell culture (21) and ICAT (22) may vary across the useable mass-to-charge range due to (i) non-uniform detector response and (ii) time-dependent contribution of the organic effluent to base-line noise levels during the LC gradient. In contrast, quantification via iTRAQ and TMT reagents is based on ratio measurements of reporter ions that are generated during MS/MS and confined to a limited m/z range. In principle, this approach eliminates contributions to measurement error that are due to either m/z-dependent detector response or interference from LC gradient noise and hence facilitates development of an experimentally simple procedure for assignment of statistical significance to peptide ratios.
Here we utilize low and high energy MS/MS scans in tandem on an LTQ-Orbitrap to identify and quantify iTRAQ-labeled phosphotyrosine-containing peptides enriched from cells that express wild type, ITD, or D835Y FLT3 kinase. In total, 371 unique phosphotyrosine peptides on 276 proteins were quantified from ∼2 cells per iTRAQ channel. In addition, we developed an error model that relates the variance of measured ratios to observed reporter ion intensity and provides a p value, q value, and confidence interval for every peptide identified. We also demonstrate that our model is stable over time and can be applied in a manner directly analogous to ubiquitously used external mass calibration routines. Collectively, these figures of merit provide for reproducible quantification even under conditions in which phosphorylation response is modest. Comparison of signaling pathways mediated by FLT3 wild type (WT), ITD, and D835Y suggests that tyrosine phosphorylation of SHP1 induced by FLT3-D835Y, but not by FLT3-ITD, may alter the transformative potential, but not overall kinase activity, of FLT3-D835Y in AML, potentially by suppression of JAK2/STAT5a signaling.
EXPERIMENTAL PROCEDURES
Cell Culture, Cell Lyses, and Proteolytic Digestion
BaF3 cells stably expressing FLT3-ITD and FLT3-D835Y were cultured with 5% CO2 at 37 °C in RPMI 1640 medium with 10% FCS and supplemented with 2 mm glutamine, 100 units/ml penicillin/streptomycin, and 0.8g/liter G418. BaF3 cells expressing wild type FLT3 were similarly cultured with 10% WEHI-3-conditioned medium as a source of IL-3. Cells were placed in 100 ml of fresh medium at 2e5 cells/ml and collected after 12 h at 4–5e5 cells/ml. One flask of FLT3-WT BaF3 cells was treated with 50 ng/ml FL for 5 min before collection. Cell pellets were washed with 10 ml of cold PBS and stored at −80 °C until needed.
Cell pellets were lysed with 2 ml of 8 m urea supplemented with 1 mm activated Na3VO4. Proteins were reduced with 10 mm DTT at 50 °C for 1 h and alkylated with 55 mm iodoacetamide at r.t. for 1 h in the dark. Cell lysates were diluted to 1.6 m urea with 8 ml of 100 mm NH4HCO3. Trypsin was added at a 1:50 (enzyme:substrate) ratio. After incubation at 37 °C overnight, whole cell digests were acidified with 10% TFA to pH 3–4 and centrifuged at 4000 rpm to remove insoluble particles. Peptides were purified on a Waters 100 mg C18 plate and eluted with 25% MeCN, 0.1% acetic acid (HOAc). Eluted peptides were divided into 100-μg aliquots, freeze-dried, and stored at −20 °C until iTRAQ labeling.
iTRAQ Labeling and Phosphotyrosine Peptide Enrichment
One aliquot of each sample was labeled with one isoform of iTRAQ reagent (114 for WT FLT3, 115 for WT FLT3/FL, 116 for FLT3-ITD, and 117 for FLT3-D835Y) according to the manufacturer's protocol. Briefly, iTRAQ reagent was reconstituted with 70 μl of ethanol, and peptides were resuspended with 30 μl of 0.5 m triethylammonium acetate bicarbonate. After mixing, the solution was incubated at r.t. for 1 h. Then all four samples were combined and dried in a SpeedVac.
Phosphotyrosine antibodies (20 μl of pTyr-100 from Cell Signaling Technology (Beverly, MA) and 20 μl of PT-66 from Sigma) were incubated with 16 μl of 50% protein G-agarose beads (Calbiochem) in 100 mm HEPES, 100 mm NaCl, pH 7.4 (IP buffer) at 4 °C for 8 h on a rotator. The protein G-agarose beads were rinsed with 400 μl of IP buffer for 5 min. iTRAQ-labeled peptides were reconstituted with 400 μl of IP buffer. After the pH was adjusted to ∼7.4 with 1 n NaOH, the sample was transferred to 4 μl of rinsed protein G-agarose beads and incubated at 4 °C overnight on a rotator. Protein G-agarose beads were spun down at 4000 rpm and rinsed with 400 μl of IP buffer three times. Peptides were eluted with 60 μl of 100 mm glycine, 7% MeCN, pH 2.5 at r.t. for 15 min. Protein G-agarose beads were spun down at 4000 rpm, and the supernatant was loaded onto a preconditioned TiO2 (Titansphere, GL Sciences) column (200-μm inner diameter, 6 cm long) at 3 μl/min. After loading, the TiO2 column was rinsed with 80% MeCN, 1% TFA at 10 μl/min for 10 min and 0.1% TFA at 10 μl/min for 10 min.
Mass Spectrometry Analysis
Peptides enriched on the TiO2 column were directly eluted onto a C18 trap column (100-μm inner diameter, 4 cm long; POROS 10R2) with 50 μl of 100 mm (NH4)2HPO4, 1 mm EDTA at 2 μl/min. The trap column was rinsed with 1% HOAc at 5 μl/min for 10 min and connected to a C18 analytical column (25-μm inner diameter, 10 cm long, 3-μm Monitor C18 resin) (23). Peptides were eluted at a flow rate of ∼5 nl/min with a 2-h gradient: 1–25% B in 75 min, 25–40% B in 15 min, 40–70% B in 5 min, 70% B for 10 min, and 70–1% B in 5 min (A, 0.2 m HOAc; B, 70% MeCN, 0.2 m HOAc) using a NanoAcquity system with a precolumn split. Eluted peptides were directly electrosprayed into an LTQ-Orbitrap XL for MS/MS analysis with a CAD-HCD dual scan mode. MS spectra were acquired in the Orbitrap with target value at 1e6, maximum fill time at 500 ms, and resolution at 15,000. A precursor was selected for MS/MS if its signal was over 2e4 and its charge state was 2+ to 4+. Up to eight precursors could be selected for each cycle and then were excluded for 20 s after MS/MS spectra were acquired. For each precursor, a CAD MS/MS spectrum was first generated and detected in the linear trap, and an HCD MS/MS spectrum was then generated in the new high energy collision cell with 47% collision energy and 2.4 amu precursor isolation window and detected in the Orbitrap at a resolution of 7500. The low mass cutoff for HCD scans was set at 100 amu. The data files were processed with in-house software to separate CAD and HCD scans into two Mascot-searchable files as described previously (24). The two files were submitted to Mascot (v2.1) and searched against all 55,824 entries in a database that consisted of human FLT3 and the NCBI RefSeq mouse protein database (released on April 3, 2006). The search parameters were set as follows. The mass tolerance values for fragment ions were set at 0.8 amu for the CAD search and 0.05 amu for the HCD search. Other common parameters were as follows: number of missed cleavages permitted, 2; fixed modifications on cysteine (carbamidomethyl), N terminus (iTRAQ4plex), and lysine (iTRAQ4plex); and variable modifications of methionine oxidation and tyrosine, serine, and threonine phosphorylation. The data were also searched against the reverse database with the same search parameters. The search results were filtered to only include peptides that contained tyrosine in the sequence and were phosphorylated. The false discovery rate was estimated to be 1% at a Mascot score of 20 and 4% at a Mascot score of 15. All scans with a Mascot score over 15 were manually inspected to confirm sequences and phosphorylation site assignment. Images of the precursor region (±5 daltons) for each identified peptide were automatically obtained from the raw mass spectrometry data files and then inserted, along with output from the corresponding Mascot search, into Excel spreadsheets using our multiplierz desktop software (25). MS/MS scans were eliminated from further analysis if contaminant peaks were observed within 1.2 amu of the precursor in the preceding MS scan.
iTRAQ Reporter Ion Extraction and Processing
The following steps were performed for all iTRAQ-labeled samples analyzed. The reporter ion intensities were extracted and corrected for injection time and known isotopic impurities in each iTRAQ reagent (13). Finally, all reporter intensities for a given MS/MS scan are bounded from below by the maximum noise level (reported by the instrument data system) for any of the reporter peaks in that scan.
Derivation of Error Model
Tryptic peptides were generated from two equal aliquots of BaF3 cell lysate. Two 100-μg samples from each batch were labeled and mixed as shown in Fig. 2a. A small portion (∼200 ng) was analyzed using the CAD-HCD dual scan mode as described above. For all pairs of reporter ions corresponding to replicate measurements, a common multiplicative constant is applied such that the median ratio was 1:1. Subsequently, the parameters β and α (which specify the relationship between the variance of the peak pairs and their mean as described in Equation 1) were estimated by MACL (26, 27) implemented as an iteratively reweighted least squares algorithm.
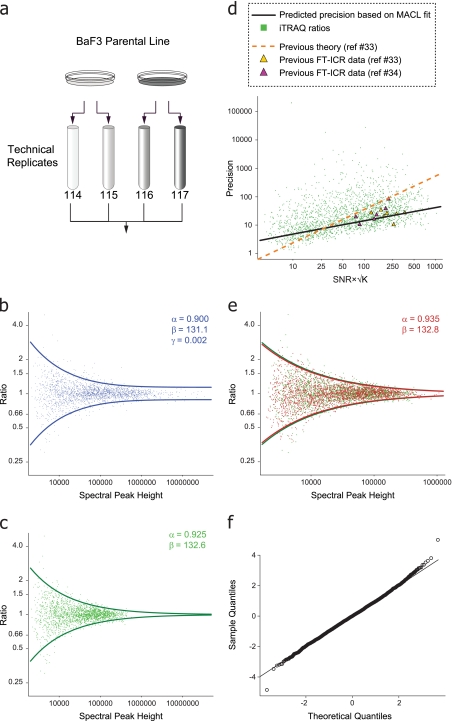
Fig. 2.
a, equal aliquots of tryptic peptides derived from murine BaF3 cells were processed as two sets of technical replicates, iTRAQ labeled as indicated, and subjected to LC-MS/MS using a dual scan CAD-HCD method (Fig. 1a). b, a log-log plot of the reporter ion ratios as a function of their geometric mean spectral peak height for technical replicates yielded a best fit curve with constant variance at high signal intensity. c, 95% acceptance region after MACL-based fit to data normalized for linear trap ion injection time. d, precision of the data in c plotted as a function of measured signal-to-noise ratio (SNR) and number of data points, K, across an iTRAQ reporter ion spectral peak (33, 34). Precision based on MACL-derived variance function (Equation 1, black line) and previous theory (33, 34) (dashed orange line) is shown. Experimental data from previous studies are indicated as yellow (33) and purple (34) triangles, respectively. e, data from a biological replicate generated 3 months later and processed as a second set of technical replicates (red) were generated as described above and overlaid with the corrected data in c (green). f, Q-Q plot of the data in e after transformation to uniform variance confirmed the approximate normality of error in log intensities.
Data Analysis for FLT3 Signaling
Immunoprecipitated, iTRAQ-labeled tyrosine-phosphorylated peptides were analyzed by LC-MS/MS as described above. Reporter ion peak intensities were processed as described above. To correct for minor variations in source protein concentration, a small aliquot of supernatant was analyzed separately; the median iTRAQ reporter ion ratio from these data was used as a normalization factor for analysis of data from the tyrosine-enriched fraction. The resulting intensity pairs were used to calculate p values under the null hypothesis of a 1:1 ratio. These p values can, in turn, be used to estimate q values (28). Finally, a confidence interval was estimated by applying the error model at the observed intensity values of the reporter pairs.
Immunoprecipitation and Immunoblotting Analysis
The mutant FLT3 receptor-expressing BaF3 cells grown at log phase were treated with 100 nm PKC412 (kindly provided by Dr. Johannes Rossel, Novartis Pharma, Basel, Switzerland) or DMSO (as control) for 30 min. The wild type FLT3-expressing cells were treated with DMSO or with either 50 ng/ml FLT3 ligand (Sigma) or 50 ng/ml ligand plus 100 nm PKC412 for 30 min. The cells were then harvested for protein extraction using Nonidet P-40 lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1.0% Nonidet P-40) plus 1× protease inhibitor mixture (Calbiochem) following a standard protocol. The total protein was then subjected either to immunoprecipitation followed by immunoblotting analysis or to immunoblotting analysis using the antibody indicated. Briefly, 500 μg of total protein were incubated with the indicated antibody for 1 h with rotation at 4 °C, and 25 μl of the protein A/G plus-agarose (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) was then added and incubated overnight at 4 °C. The protein complexes were then washed three times in TNN buffer (40 mm Tris-HCl, pH 8.0, 10 mm NaCl, 0.5% Nonidet P-40), the pelleted complexes were resuspended in 1× Laemmli sample buffer, and the supernatant was either stored at −80 °C or subjected to immunoblotting analysis using a standard protocol. Antibodies against phosphotyrosine (pY99), FLT3 (c-20), SHP1 (c-19), and pERK1/2 (E4) were purchased from Santa Cruz Biotechnology, Inc. Antibodies against STAT5 and pSTAT5 were purchased from Transduction Laboratories. Antibodies against pAKT (Ser-473), AKT, and ERK1/2 were purchase from Cell Signaling Technology.
RESULTS
LTQ-Orbitrap Dual Scan Mode Provides High Sensitivity MS/MS and High Resolution iTRAQ Reporter Ion Detection
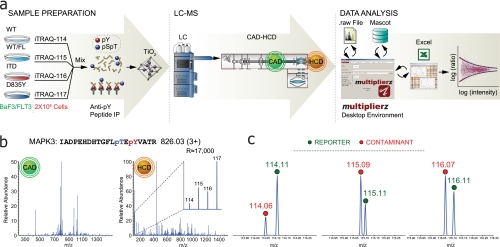
iTRAQ-labeled peptides are typically analyzed with triple quadrupole- or hybrid quadrupole-TOF instruments. Quadrupole ion trap geometries generally produce suboptimal results because the iTRAQ reporter ions often lie below the stability limit as dictated by the precursor peptide mass-to-charge ratio and pseudopotential well parameters used for activation (for example, activation q = 0.23). Recently, Olsen et al. (29) described the performance of a radiofrequency-only multipole collision cell located adjacent to the c-trap on an LTQ-Orbitrap mass spectrometer. This configuration provides for “triple quadrupole-like” fragmentation (HCD) at energies above those typically used in ion trap instruments with efficient trapping of low mass fragment ions. We used a similar strategy for the work described herein (Fig. 1a) but modified the acquisition method to include first a low energy MS/MS scan of every precursor in the linear ion trap (CAD) followed by a higher energy MS/MS scan in the octopole collision cell (HCD). Fig. 1b shows CAD and HCD spectra corresponding to a doubly phosphorylated, iTRAQ-labeled peptide from MAPK3 (see discussion below and “Experimental Procedures”). As reported previously (29), both MS/MS modes yield high confidence sequence assignment based on Mascot searches with the HCD mode providing efficient capture and detection of the iTRAQ reporter ions (Fig. 1b, inset). Further examination of the low mass region revealed the presence of contaminant ions having nominal masses that correspond to those of the iTRAQ reporters (Fig. 1c). At lower resolution, these contaminants can introduce significant error, particularly for the application described herein, where quantification is based on individual peptide ratios. These data illustrate the range of potential errors introduced by isobaric contaminants and hence the value of high mass resolving power in conjunction with iTRAQ-based quantification studies.
Fig. 1.
a, analytical work flow for enrichment and analysis of phosphotyrosine-containing peptides from an AML model system with WT and two oncogenic FLT3 mutants (ITD and D835Y). Tryptic peptides from ∼2e6 cells per biological condition were iTRAQ-labeled and subjected to sequential enrichment of phosphotyrosine-containing peptides, first with anti-phosphotyrosine antibodies and then a TiO2 column. Next, LC-MS/MS was performed on an Orbitrap XL using a dual CAD-HCD scan method. Our multiplierz mzAPI-based desktop environment was used for subsequent data analysis. b, CAD and HCD MS/MS spectra of a doubly phosphorylated peptide from MAPK3. iTRAQ reporter ions are not observed in the CAD scan but are among the most intense peaks in the HCD scan. c, high mass resolving power (m/Δm > 15,000) in the low mass region facilitates discrimination of contaminate ions (red circles) from iTRAQ reporters (green circles). IP, immunoprecipitation; pS, phosphoserine; pT, phosphothreonine; pY, phosphotyrosine.
Observed Relationship between Standard Deviation of iTRAQ Ratios and Intensity of Individual Reporter Ions Suggests a Variance Function
Although high mass resolution within the iTRAQ reporter ion region alleviates potential interference due to isobaric contaminants, other sources of error persist and require the development of an appropriate model to estimate the significance associated with quantification measurements. To better understand the uncertainty associated with measured iTRAQ ratios, we generated tryptic peptides from whole cell lysates of murine BaF3 cells. Peptides from two sets of technical replicates were labeled with iTRAQ reagents (Fig. 2a). LC-MS/MS analysis was performed on an LTQ-Orbitrap with each precursor subjected to CAD and HCD as described above. Peptide iTRAQ ratios were generated using our multiplierz desktop environment (30) and plotted as a function of reporter ion mass spectral peak height (see “Experimental Procedures”). As with other large scale data (31), we observed a clear dependence of variance on spectral peak height (Fig. 2b). Previous reports (13, 14) in proteomics have also noted this relationship, but in most cases simple thresholds were used, for both abundance ratio and intensity, below which data were deemed unreliable. In a recent study, Hundertmark et al. (20) estimated a variance function based on infusion of a small number of iTRAQ-labeled standard peptides. In contrast, we sought to leverage the full benefit of iTRAQ reagents by using equal aliquots of tryptic peptides derived from cell lysate (“calibration mixture”), effectively providing thousands of putatively 1:1 ratios that spanned a wide range of mass spectral peak height. We derived an error model based on the assumption that systematic errors were equivalent for all iTRAQ fragment ions and that detector response was uniform across the reporter ion m/z range. In addition, we assumed that the log of the iTRAQ reporter ion mass spectral peak height follows a simple, two-parameter exponential variance function. In effect, for the analysis of our calibration mixture, we treated the log of each iTRAQ ratio as a difference between replicate measurements from a common underlying log-normal distribution. Consequently, the variance of the log ratios is equal to twice that of the log peak height distribution,
where Ratio refers to the ratio of two independent random variables, Peak1 and Peak2, which represent measured iTRAQ peak heights, both distributed log-normally with a mean log magnitude μ and variance βe−αμ. We next estimated the parameters α and β in Equation 1 by maximum approximate conditional likelihood (26) and finally established a 95% acceptance region. Interestingly, we observed that convergence of parameters in Equation 1 required the addition of a constant term (βe−αμ + γ; Fig. 2b) at odds with previous reports that suggested a continual increase in precision as a function of spectral peak height (32–35). To explore this anomaly further, we next normalized the iTRAQ peak height values reported in the native mass spectrometry data files based on the ion injection times required to reach the preset target value for MS/MS (36). Fig. 2c shows the 95% acceptance region after application of the above procedure to our normalized data. Here the variance asymptotically approaches zero at high spectral peak height values in agreement with Equation 1 (e.g. γ ≈ 0). In addition, the apparent range of quantification is consistent with other reports of single scan dynamic range for the Orbitrap mass spectrometer (37, 38).
Previous work beginning with NMR-based studies (32) and then extended to other resonance techniques such as FT/ICR (33, 34) proposed analytical solutions for spectral peak parameters as a function of observed signal-to-noise ratio and the number of data points (K) across a given peak. Given the similarities in detection between FT/ICR and Orbitrap technologies, we asked whether our results would agree with previous efforts to predict measurement variance. Fig. 2d shows the precision of our iTRAQ ratios (green dots) plotted as a function of signal-to-noise ratio and √K (see Refs. 33 and 34). The black line represents the MACL-based fit to our data, whereas the dashed orange line corresponds to the precision predicted by a theoretical framework described in the context of earlier FT/ICR studies (33, 34). Given the discrepancy between the precision predicted by each approach (black and orange lines, respectively), we next overlaid FT/ICR experimental data taken with permission from Refs. 33 and 34 (yellow and purple triangles). Surprisingly, our model provided a better prediction for the precision of these data as compared with the previously proposed theory (33, 34). This result further validates our approach, particularly given that the FT/ICR data were acquired nearly 20 or more years ago on a different mass spectrometry platform and under different ionization conditions (electrospray versus electron impact).
To test the robustness of our approach, we repeated the sample preparation from cells cultured 3 months after the first acquisition (Fig. 2a). Again, observed spectral peak height values were normalized to ion injection times, and the resulting data were used to refit the parameters α and β in Equation 1. An overlay of both data sets (Fig. 2e), with their respective 95% acceptance regions, shows a very high degree of alignment. Finally, we generated a Quantile-Quantile plot (Fig. 2f) based on variance correction of the data in Fig. 2e. The apparent normality of the transformed data validates our assumption of log normality for spectral peak heights and justifies the assignment of p and q values for all measured peptide ratios contained within a single experiment. In addition, the variance function enables an estimation of confidence intervals for all measured ratios. Collectively, these results suggest that a variance function can be established once and then applied to all subsequent data acquired with identical instrument parameters, similar in concept to the process of external mass calibration routinely used in mass spectrometry.
Ligand Stimulation Induces Small, but Statistically Significant, Up-regulation of WT FLT3
The ability to provide significance values for measured peptide ratios supports experimental designs that more closely recapitulate physiological conditions. For example, serum starvation of cultured cells is commonly used to amplify the signal that results from perturbation of a target pathway (cytokine stimulation, etc.). However, the degree to which this cellular stress negatively impacts the subsequent response is not well understood. The preferred experimental scenario, in which cells are cultured with a full complement of cytokines and growth factors, requires robust measurement of small changes in an otherwise active signaling background. Toward this end, we established murine BaF3 cell lines that stably express WT, point mutation D835Y, or ITD FLT3 kinase, respectively. Cells grown under normal culture conditions were lysed and labeled with iTRAQ reagents (Fig. 1a and “Experimental Procedures”). Next, phosphotyrosine-containing peptides were isolated via antibody-based purification followed by enrichment on titanium dioxide and finally quantified by LC-MS/MS analysis. Non-systematic errors in iTRAQ-based quantitation may result from simultaneous fragmentation of multiple precursors closely spaced within the m/z range selected for MS/MS analysis. This phenomenon has recently been explored in detail (39). Consistent with our focus on tyrosine phosphorylation, we observed potential precursor contamination in only a small number of scans, and the corresponding peptides were eliminated from further consideration (see “Experimental Procedures”). Across two bioreplicates (2e6 cells per iTRAQ channel per bioreplicate), we identified 371 unique tyrosine-phosphorylated peptides, representing 349 unique phosphotyrosine sites on 276 proteins (<5% false discovery rate; see “Experimental Procedures” and supplemental Tables S1 and S2). In total, 148 unique peptides, containing 142 unique phosphotyrosine sites on 117 proteins, were detected across two experiments and formed the basis data set for the remainder of our results. Supplemental Table S3 lists peptide sequences, phosphorylation sites, and quantification data of those commonly identified phosphotyrosine-containing peptides.
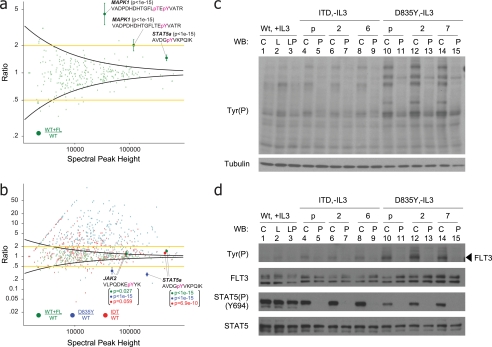
We first asked what pathways were activated when the WT kinase was stimulated with FL. Fig. 3a shows that overall kinase activity changed only modestly upon addition of FL to cells cultured under standard conditions. These data were confirmed by Western blot (Fig. 3c, lanes 1 and 2). Use of a global threshold (e.g. 2-fold change; Fig. 3a, yellow lines) resulted in assignment of 24 phosphotyrosine sites, including singly (Tyr-204) and doubly (Tyr-202/Tyr-204) phosphorylated MAPK1 and singly (Tyr-185) phosphorylated MAPK9, as significantly modulated. However, application of our error model (Fig. 3a, black curves) identified statistically significant regulation of tyrosine phosphorylation on 47 unique peptides, spanning several key signaling molecules. For example, our proteomics data show that phosphorylation of Tyr-694 on STAT5a increased by ∼1.5-fold under ligand stimulation (Fig. 3a); although this modest level does not meet a global, 2-fold threshold, our error model provides a p value <1e−15. Comparison of these data with those obtained by Western blotting (Fig. 3d, STAT5(P)(Y694) panel, lanes 1 and 2) highlights the utility of our approach to monitor subtle variations as compared with standard biochemical techniques. A similar fold-change (1.4×) and significant p value (2.2e−16) were obtained for STAT5a (Tyr(P)-694) in the analysis of a biological replicate (supplemental Table S2). Use of our error model also revealed that 11 of the 24 phosphotyrosine sites assigned based on global threshold logic are likely to be false positives based on the large error associated with measurement of weak signals (Fig. 3a). As mentioned above, the error model allowed us to calculate a confidence interval for every quantified ratio (supplemental Tables S1 and S2). For example, we can estimate that the ratio of STAT5a (Tyr(P)-694) should be between 1.4 and 1.6 as shown in Fig. 3a.
Fig. 3.
a, identification of phosphorylation regulated by WT FLT3 in cells cultured under normal serum conditions. Addition of exogenous FL increases STAT5a phosphorylation (Tyr-694) by only 1.4-fold relative to unstimulated WT FLT3. Yellow lines represent a global 2-fold threshold. Intensity-based variance function, derived from an analysis of technical replicates (black curve and Fig. 2), yields a statistically significant p value (<10e−22) and 95% confidence interval for regulation of Tyr(P)-694 on STAT5a. p values and 95% confidence intervals for phosphorylated peptides from MAPK1 are also shown. b, relative tyrosine phosphorylation levels of WT FLT3 + FL (green), FLT3-ITD (red), and FLT3-D835Y (blue), each normalized against unstimulated WT FLT3. Tyrosine phosphorylation on STAT5a (Tyr-694) and JAK2 (Tyr-1007) is coordinately up-regulated by both FLT3-ITD and stimulated WT FLT3. In contrast, phosphorylation on these sites is diminished in the context of FLT3-D835Y signaling. c, immunoblot of multiple BaF3 clones with anti-Tyr(P) antibody (specific clone or polyclonal) harboring WT FLT3 (lanes 1–3), FLT3-ITD (lanes 4–9), and FLT3-D835Y (lanes 10–15) confirmed data in b that the point mutant exhibited higher kinase activity as compared with WT FLT3 or FLT3-ITD. C, control (no stimulation); L, ligand stimulation at 50 ng/ml for 30 min; P, treatment with PKC412 at 100 nm for 30 min; p, polyclonal (otherwise specific clone designation is listed). d, immunoblot of cells in c with antibodies against phosphotyrosine, total FLT3, phospho-STAT5 (Tyr(P)-694), and total STAT5. Despite the higher constitutive phosphorylation observed for the point mutant, downstream signaling on STAT5 (Tyr(P)-694) is significantly diminished as compared with WT FLT3 and FLT3-ITD. The legend is the same as in c. pT, phosphothreonine; pY, phosphotyrosine; WB, Western blot.
STAT5a Is Differentially Activated by FLT3-ITD and -D835Y
To understand the effect of either ITD or D835Y mutation on the complex signaling network that exists under normal physiological conditions, we next plotted ratios for tyrosine-phosphorylated peptides derived from WT + FL (green), D835Y (blue), and ITD (red), each normalized to the corresponding phosphopeptide derived from cells with unstimulated, WT kinase (Fig. 3b). Both ITD and D835Y show elevated tyrosine phosphorylation levels on many proteins compared with WT, including STAT3, MAPK1, SHC, and PLCγ2, which is consistent with the constitutive kinase activity reported for both mutants (3, 40). This result was confirmed by anti-Tyr(P) Western blotting in multiple BaF3 clones (Fig. 3c, lanes 1, 4, 6, 8, 10, 12, and 14). The majority of up-regulated phosphotyrosine sites were found on proteins involved in nuclear membrane structure, including vimentin, L-plastin (LCP1), and emerin, consistent with enhanced proliferation in the context of each FLT3 mutant (6).
Elevated phosphorylation levels of STAT5 have been correlated with enhanced self-renewal and reduced differentiation of hematopoietic stem cells both in vitro and in vivo (40, 41). Fig. 3b indicates that, in the absence of IL-3, the phosphorylation level of STAT5a (Tyr-694) is sustained in ITD (red) at a level comparable to that in WT stimulated with FL (green) and is markedly lower in D835Y (blue). We confirmed these observations in multiple BaF3 clones by Western blot using an antibody specific for Tyr(P)-694 on STAT5 (Fig. 3d, lanes 1, 4, 6, 8, 10, 12, and 14). These data are consistent with recent reports that described activation of specific targets downstream of FLT3 mutants (6). We next used PKC412, a small molecule inhibitor of FLT3 kinase activity (42), to more specifically link STAT5a phosphorylation to constitutive activation of the receptor by each mutation. Indeed, application of PKC412 had little, if any, effect on STAT5a Tyr(P)-694 in the context of the WT FLT3, whereas phosphorylation on the same site was completely abrogated in the context of both ITD and D835Y mutations (Fig. 3d, lanes 3, 5, 7, 9, 11, 13, and 15).
Direct Comparison of Constitutively Active FLT3 Mutants Suggests That D835Y Down-regulates JAK-STAT5a Signaling via Tyrosine Phosphorylation of SHP1
Although the ITD is correlated with a poor clinical prognosis, our data clearly demonstrate that overall kinase activity is much higher for the D835Y mutant. This result is consistent with recent findings for several BCR-ABL mutations (such as T315I) where phosphorylation levels did not necessarily correlate with oncogenic potential (43). One possible explanation to reconcile the apparent juxtaposition of hyperkinase activity and low transformative potential is that negative regulatory molecules and pathways are among those activated in the context of the D835Y mutant. Interestingly, we observed significant down-regulation of tyrosine phosphorylation on JAK2 (Tyr-1007) in addition to STAT5a (Tyr-694) in the context of D835Y as compared with WT FLT3 (Fig. 3b). Moreover, phosphorylation of JAK2 (Tyr(P)-1007) is known to be essential for downstream activation (44).
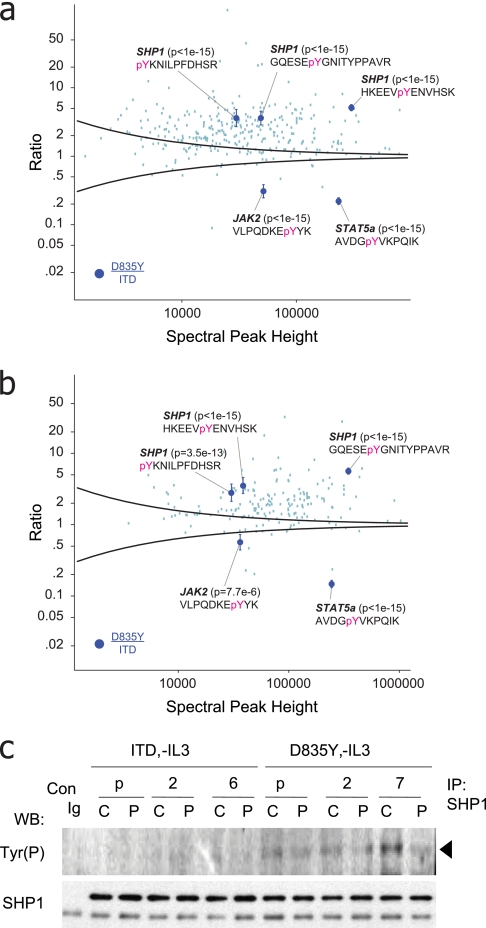
To directly probe for divergent signaling, we next plotted ratios for tyrosine-phosphorylated peptides resulting from D835Y and ITD mutants in two bioreplicates, respectively (Fig. 4, a and b). In the context of D835Y signaling, we observed increased tyrosine phosphorylation on several phosphatases, including PTPN6 (SHP1), PTPN11 (SHP2), PTPN12 (PTPG1), and PTPN18 (BDP1). In particular, SHP1, expressed predominantly in hematopoietic cells (45), negatively regulates the activity of many receptors, including c-KIT (another type III tyrosine receptor), by targeting the JAK-STAT pathway (46–48). We identified three phosphotyrosine sites on SHP1 (Tyr-276, Tyr-536, and Tyr-564), all up-regulated as compared with the ITD and with a significant p value (≤7.5 e-13). Immunoprecipitation of SHP1 followed by Western blot (Fig. 4c) confirmed that (i) protein expression levels were consistent in the context of FLT3-ITD and FLT3-D835Y, and (ii) SHP1 phosphorylation decreased upon specific inhibition of FLT3-D835Y with PKC412 (42). Collectively, our results suggest that, as compared with FLT3-ITD, constitutive activation conferred by D835Y leads to phosphorylation of the negative regulatory phosphatase SHP1 as well as down-regulation of JAK2-STAT5a signaling.
Fig. 4.
a and b, relative tyrosine phosphorylation driven by FLT3-D835Y normalized to FLT3-ITD. Intensity-based variance function, based on an analysis of technical replicates (black curve and Fig. 2), provided for identification of statistically significant points of divergent signaling downstream of oncogenic FLT3 mutants. p values and 95% confidence intervals for phosphorylated peptides from STAT5a (Tyr(P)-694), JAK2 (Tyr(P)-1007), and SHP1 (Tyr(P)-276, Tyr(P)-536, and Tyr(P)-564) were illustrated. c, Western blot analysis of SHP1 immunoprecipitates, from multiple BaF3 clones (as in Fig. 3, c and d), probed with anti-Tyr(P) or anti-SHP1 antibodies. Consistent with the proteomics data in a and b, SHP1 tyrosine phosphorylation was elevated in the context of the point mutant as compared with either WT FLT3 or FLT3-ITD. C, control (no stimulation); P, treatment with PKC412 at 100 nm for 30 min; p, polyclonal (otherwise specific clone designation is listed). pY, phosphotyrosine; WB, Western blot; Con, control; IP, immunoprecipitation.
DISCUSSION
We demonstrated that a combination of linear ion trap CAD with higher energy HCD fragmentation offered an effective means to analyze iTRAQ-labeled peptides on an Orbitrap XL instrument. Significantly, the high mass resolving power (m/Δm >15,000) in the iTRAQ reporter ion region minimizes interference from potential contaminant species that may confound quantification data. We also developed a robust error model that allows estimation of p value, q value, and confidence interval on any measured iTRAQ ratio from a single experiment. Our approach represents a paradigm shift relative to previous methodologies for quantitative proteomics in that we derive a parametric error model from a large number of weak variance estimates (49), all within a single acquisition, rather than extensive, empirical characterization of individual mass spectrometry peaks (20, 33, 34, 50). The ability of our model, developed herein for LC-MS/MS analysis of iTRAQ-labeled peptides on an Orbitrap, to provide improved estimates of precision for data acquired some two decades ago on small molecules ionized by electron impact and analyzed in MS mode on an FT/ICR mass spectrometer strongly supports the validity of our approach.
In practice, our framework is directly analogous to the use of external mass calibration and provides the added benefit that the experimental setup for calibration is identical to that for subsequent quantification analyses. The ability to estimate variance once and then predict the statistical significance of future measurements directly enables proteomics experiments that are designed to monitor specific biological response to perturbation rather than large scale cataloguing of peptides that may be irrelevant to the question at hand. In fact, our observation that overall phosphorylation levels were elevated downstream of the point mutant as compared with the FLT3-ITD (Fig. 4, a and b) nicely illustrates this scenario. Here the vast majority of phosphopeptide ratios deviate significantly from 1:1, and hence these data are not amenable to analysis by methodologies (18, 51–53) that require internal normalization to a large null (e.g. 1:1) distribution.
We envision that application of our error model will be particularly valuable for quantitative analysis of rare or otherwise intractable post-translational modifications, low abundance proteins, primary cells, clinical samples, or in general any scenario in which multiple rounds of technical and biological replicates are not readily available. In this study, we applied quantitative proteomics in conjunction with our error model to identify multiple points of divergent signaling downstream of FLT3 mutants expressed in murine BaF3 cells. Although kinases are attractive drug targets in principle, clinical experience to date, for example in the case of BCR-ABL inhibition by Gleevec for chronic myelogenous leukemia, demonstrates that patients often become refractory to single agent therapeutics (54). These observations verify the need for continued elucidation of oncogenic pathways to identify novel and synergistic molecular targets.
In the context of FLT3 signaling in BaF3 cells, we identified 349 unique phosphotyrosine sites on 276 proteins. Direct comparison of specific phosphorylation events between the two oncogenic mutants and across bioreplicates clearly demonstrated that FLT3-ITD and FLT3-D835Y elicit divergent signaling responses as evidenced for example by differences in phosphorylation on JAK2, STAT5a, and SHP1. The latter plays a critical, negative regulatory role in attenuation of signaling for a variety of receptor tyrosine kinases throughout multiple cell processes (55, 56). In addition, aberrant SHP1 expression has been reported in a number of malignancies (57). Data from Chen et al. (58), in the context of FLT3-ITD-mediated leukemia models, suggested that down-regulation of SHP1, accompanied by a concomitant decrease in phosphatase activity, removed a key negative feedback mechanism for control of kinase activity and hence augmented oncogenic FLT3 cell proliferation and survival. Our work compliments this and other previous studies (6, 7, 59) and suggests additional mechanistic detail that may explain phenotypic differences observed between FLT3-ITD and FLT3-D835Y. A recent report demonstrated that tyrosine phosphorylation at Tyr-536 and Tyr-564 near the C terminus of SHP1 inactivated JAK-STAT signaling by recruitment of SOCS1 via GAB2 (48). These data illustrate that SHP1 can disrupt discrete signaling pathways through a scaffold function independent of catalytic activity or expression level. Our data clearly demonstrate that SHP1 is preferentially phosphorylated at the C terminus in the context of D835Y, although it remains to be determined whether phosphorylation of these sites results from direct interaction with FLT3 or is mediated through a downstream effector; nonetheless, our results are consistent with the intriguing hypothesis that specific activation of SHP1 tyrosine phosphorylation down-regulates JAK2-STAT5a signaling, ultimately abrogating the transformative potential of the point mutant as compared with FLT3-ITD. Collectively, our data provide a broad view of signaling downstream of oncogenic FLT3 mutants and suggest new avenues for further research.
Supplementary Material
Acknowledgments
We thank Eric Smith for valuable discussion and preparation of the figures. In addition, we acknowledge Micha Mandel for critical discussion of the statistical analysis.
* This work was supported, in whole or in part, by National Institutes of Health Grant P50HG004233 from the NHGRI. This work was also supported by the Dana-Farber Cancer Institute.
 This article contains supplemental Tables S1–S3.
This article contains supplemental Tables S1–S3.
1 The abbreviations used are:
- AML
- acute myeloid leukemia
- FLT3
- Fms-like tyrosine kinase 3
- ITD
- internal tandem duplication
- WT
- wild type
- FL
- FLT3 ligand
- IL-3
- interleukin 3
- STAT5a
- signal transducer and activator of transcription 5a
- JAK2
- Janus kinase 2
- SHP1
- protein tyrosine phosphatase, non-receptor type 6
- SOCS1
- suppressor of cytokine signaling 1
- GAB2
- growth factor receptor-bound protein 2-associated binding protein 2
- HCD
- higher energy collisional dissociation
- MACL
- maximum approximate conditional likelihood
- r.t.
- room temperature
- iTRAQ
- isobaric tags for relative and absolute quantitation
- TMT
- tandem mass tag
- LTQ
- linear trap quadrupole
- HOAc
- acetic acid
- CAD
- collisionally activated dissociation
- ERK
- extracellular signal-regulated kinase
- MAPK
- mitogen-activated protein kinase
- pAKT, pERK, and pSTAT5
- phosphorylated AKT, ERK, and STAT5.
REFERENCES
- 1.Blume-Jensen P., Hunter T. (2001) Oncogenic kinase signalling. Nature 411, 355–365 [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. (2000) Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 3.Gilliland D. G., Griffin J. D. (2002) The roles of FLT3 in hematopoiesis and leukemia. Blood 100, 1532–1542 [DOI] [PubMed] [Google Scholar]
- 4.Small D. (2006) FLT3 mutations: biology and treatment. Hematology Am. Soc. Hematol. Educ. Program 1, 178–184 [DOI] [PubMed] [Google Scholar]
- 5.Knapper S. (2007) FLT3 inhibition in acute myeloid leukaemia. Br. J. Haematol 138, 687–699 [DOI] [PubMed] [Google Scholar]
- 6.Choudhary C., Schwäble J., Brandts C., Tickenbrock L., Sargin B., Kindler T., Fischer T., Berdel W. E., Müller-Tidow C., Serve H. (2005) AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood 106, 265–273 [DOI] [PubMed] [Google Scholar]
- 7.Grundler R., Miething C., Thiede C., Peschel C., Duyster J. (2005) FLT3-ITD and tyrosine kinase domain mutants induce 2 distinct phenotypes in a murine bone marrow transplantation model. Blood 105, 4792–4799 [DOI] [PubMed] [Google Scholar]
- 8.Rocnik J. L., Okabe R., Yu J. C., Lee B. H., Giese N., Schenkein D. P., Gilliland D. G. (2006) Roles of tyrosine 589 and 591 in STAT5 activation and transformation mediated by FLT3-ITD. Blood 108, 1339–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domon B., Aebersold R. (2006) Mass spectrometry and protein analysis. Science 312, 212–217 [DOI] [PubMed] [Google Scholar]
- 10.Mann M., Hendrickson R. C., Pandey A. (2001) Analysis of proteins and proteomes by mass spectrometry. Annu. Rev. Biochem 70, 437–473 [DOI] [PubMed] [Google Scholar]
- 11.Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 12.Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 13.Shadforth I. P., Dunkley T. P., Lilley K. S., Bessant C. (2005) i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6, 145–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin W. T., Hung W. N., Yian Y. H., Wu K. P., Han C. L., Chen Y. R., Chen Y. J., Sung T. Y., Hsu W. L. (2006) Multi-Q: a fully automated tool for multiplexed protein quantitation. J. Proteome Res 5, 2328–2338 [DOI] [PubMed] [Google Scholar]
- 15.Boehm A. M., Pütz S., Altenhöfer D., Sickmann A., Falk M. (2007) Precise protein quantification based on peptide quantification using iTRAQ. BMC Bioinformatics 8, 214–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oberg A. L., Mahoney D. W., Eckel-Passow J. E., Malone C. J., Wolfinger R. D., Hill E. G., Cooper L. T., Onuma O. K., Spiro C., Therneau T. M., Bergen H. R., 3rd (2008) Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J. Proteome Res 7, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bantscheff M., Eberhard D., Abraham Y., Bastuck S., Boesche M., Hobson S., Mathieson T., Perrin J., Raida M., Rau C., Reader V., Sweetman G., Bauer A., Bouwmeester T., Hopf C., Kruse U., Neubauer G., Ramsden N., Rick J., Kuster B., Drewes G. (2007) Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol 25, 1035–1044 [DOI] [PubMed] [Google Scholar]
- 18.DeSouza L., Diehl G., Rodrigues M. J., Guo J., Romaschin A. D., Colgan T. J., Siu K. W. (2005) Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res 4, 377–386 [DOI] [PubMed] [Google Scholar]
- 19.Garbis S. D., Tyritzis S. I., Roumeliotis T., Zerefos P., Giannopoulou E. G., Vlahou A., Kossida S., Diaz J., Vourekas S., Tamvakopoulos C., Pavlakis K., Sanoudou D., Constantinides C. A. (2008) Search for potential markers for prostate cancer diagnosis, prognosis and treatment in clinical tissue specimens using amine-specific isobaric tagging (iTRAQ) with two-dimensional liquid chromatography and tandem mass spectrometry. J. Proteome Res 7, 3146–3158 [DOI] [PubMed] [Google Scholar]
- 20.Hundertmark C., Fischer R., Reinl T., May S., Klawonn F., Jänsch L. (2009) MS-specific noise model reveals the potential of iTRAQ in quantitative proteomics. Bioinformatics 25, 1004–1011 [DOI] [PubMed] [Google Scholar]
- 21.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 22.Gygi S. P., Rist B., Gerber S. A., Turecek F., Gelb M. H., Aebersold R. (1999) Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol 17, 994–999 [DOI] [PubMed] [Google Scholar]
- 23.Ficarro S. B., Zhang Y., Lu Y., Moghimi A. R., Askenazi M., Hyatt E., Smith E. D., Boyer L., Schlaeger T. M., Luckey C. J., Marto J. A. (2009) Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal. Chem 81, 3440–3447 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Ficarro S. B., Li S., Marto J. A. (2009) Optimized Orbitrap HCD for quantitative analysis of phosphopeptides. J. Am. Soc. Mass Spectrom 20, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 25.Parikh J. R., Askenazi M., Ficarro S. B., Cashorali T., Webber J. T., Blank N. C., Zhang Y., Marto J. A. (2009) multiplierz: an extensible API based desktop environment for proteomics data analysis. BMC Bioinformatics 10, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadler W. A., Smith M. H. (1986) A reliable method for estimating the variance function in immunoassay. Comput. Stat. Data Anal 3, 227–239 [Google Scholar]
- 27.Davidian M., Carroll R. J. (1987) Variance function estimation. J. Am. Stat. Assoc 82, 1079–1091 [Google Scholar]
- 28.Storey J. D., Tibshirani R. (2003) Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A 100, 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 30.Askenazi M., Parikh J. R., Marto J. A. (2009) mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nat. Methods 6, 240–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y. H., Dudoit S., Luu P., Speed T. P. (2001) Normalization for cDNA microarray data, in Microarrays: Optical Technologies and Informatics (Bittner M. L., Chen Y., Dorsel A. N., Dougherty E. R. eds) pp. 141–152, SPIE, San Jose, CA [Google Scholar]
- 32.Posener D. W. (1974) Precision in measuring resonance spectra. J. Magn. Reson 14, 121–128 [Google Scholar]
- 33.Chen L., Cottrell C. E., Marshall A. G. (1986) Effect of signal-to-noise ratio and number of data points upon precision in measurement of peak amplitude, position and width in Fourier transform spectrometry. Chemom. Intell. Lab. Syst 1, 51–58 [Google Scholar]
- 34.Liang Z. M., Marshall A. G. (1990) Precise relative ion abundances from Fourier transform ion cyclotron resonance magnitude-mode mass spectra. Anal. Chem 62, 70–75 [DOI] [PubMed] [Google Scholar]
- 35.Liang Z., Marshall A. G. (1990) Time-domain (interferogram) and frequency-domain (absorption-mode and magnitude-mode) noise and precision in Fourier transform spectrometry. Appl. Spectrosc 44, 766–775 [Google Scholar]
- 36.Bantscheff M., Boesche M., Eberhard D., Matthieson T., Sweetman G., Kuster B. (2008) Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteomics 7, 1702–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Makarov A., Denisov E., Lange O., Horning S. (2006) Dynamic range of mass accuracy in LTQ Orbitrap hybrid mass spectrometer. J. Am. Soc. Mass Spectrom 17, 977–982 [DOI] [PubMed] [Google Scholar]
- 38.Venable J. D., Wohlschlegel J., McClatchy D. B., Park S. K., Yates J. R., 3rd (2007) Relative quantification of stable isotope labeled peptides using a linear ion trap-Orbitrap hybrid mass spectrometer. Anal. Chem 79, 3056–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ow S. Y., Salim M., Noirel J., Evans C., Rehman I., Wright P. C. (2009) iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”. J. Proteome Res 8, 5347–5355 [DOI] [PubMed] [Google Scholar]
- 40.Choudhary C., Müller-Tidow C., Berdel W. E., Serve H. (2005) Signal transduction of oncogenic Flt3. Int. J. Hematol 82, 93–99 [DOI] [PubMed] [Google Scholar]
- 41.Zheng R., Small D. (2005) Mutant FLT3 signaling contributes to a block in myeloid differentiation. Leuk. Lymphoma 46, 1679–1687 [DOI] [PubMed] [Google Scholar]
- 42.Weisberg E., Boulton C., Kelly L. M., Manley P., Fabbro D., Meyer T., Gilliland D. G., Griffin J. D. (2002) Inhibition of mutant FLT3 receptors in leukemia cells by the small molecule tyrosine kinase inhibitor PKC412. Cancer Cell 1, 433–443 [DOI] [PubMed] [Google Scholar]
- 43.Skaggs B. J., Gorre M. E., Ryvkin A., Burgess M. R., Xie Y., Han Y., Komisopoulou E., Brown L. M., Loo J. A., Landaw E. M., Sawyers C. L., Graeber T. G. (2006) Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc. Natl. Acad. Sci. U.S.A 103, 19466–19471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng J., Witthuhn B. A., Matsuda T., Kohlhuber F., Kerr I. M., Ihle J. N. (1997) Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol. Cell. Biol 17, 2497–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonks N. K., Neel B. G. (1996) From form to function: signaling by protein tyrosine phosphatases. Cell 87, 365–368 [DOI] [PubMed] [Google Scholar]
- 46.Jiao H., Berrada K., Yang W., Tabrizi M., Platanias L. C., Yi T. (1996) Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol. Cell. Biol 16, 6985–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klingmüller U., Lorenz U., Cantley L. C., Neel B. G., Lodish H. F. (1995) Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell 80, 729–738 [DOI] [PubMed] [Google Scholar]
- 48.Minoo P., Zadeh M. M., Rottapel R., Lebrun J. J., Ali S. (2004) A novel SHP-1/Grb2-dependent mechanism of negative regulation of cytokine-receptor signaling: contribution of SHP-1 C-terminal tyrosines in cytokine signaling. Blood 103, 1398–1407 [DOI] [PubMed] [Google Scholar]
- 49.Du P., Stolovitzky G., Horvatovich P., Bischoff R., Lim J., Suits F. (2008) A noise model for mass spectrometry based proteomics. Bioinformatics 24, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 50.Anderle M., Roy S., Lin H., Becker C., Joho K. (2004) Quantifying reproducibility for differential proteomics: noise analysis for protein liquid chromatography-mass spectrometry of human serum. Bioinformatics 20, 3575–3582 [DOI] [PubMed] [Google Scholar]
- 51.Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 52.Wang G., Wu W. W., Zeng W., Chou C. L., Shen R. F. (2006) Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J. Proteome Res 5, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 53.Roy S. M., Becker C. H. (2007) Quantification of proteins and metabolites by mass spectrometry without isotopic labeling. Methods Mol. Biol 359, 87–105 [DOI] [PubMed] [Google Scholar]
- 54.Weisberg E., Manley P. W., Cowan-Jacob S. W., Hochhaus A., Griffin J. D. (2007) Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nat. Rev. Cancer 7, 345–356 [DOI] [PubMed] [Google Scholar]
- 55.Ostman A., Böhmer F. D. (2001) Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol 11, 258–266 [DOI] [PubMed] [Google Scholar]
- 56.Zhang J., Somani A. K., Siminovitch K. A. (2000) Roles of the SHP-1 tyrosine phosphatase in the negative regulation of cell signalling. Semin. Immunol 12, 361–378 [DOI] [PubMed] [Google Scholar]
- 57.Wu C., Sun M., Liu L., Zhou G. W. (2003) The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene 306, 1–12 [DOI] [PubMed] [Google Scholar]
- 58.Chen P., Levis M., Brown P., Kim K. T., Allebach J., Small D. (2005) FLT3/ITD mutation signaling includes suppression of SHP-1. J. Biol. Chem 280, 5361–5369 [DOI] [PubMed] [Google Scholar]
- 59.Choudhary C., Brandts C., Schwable J., Tickenbrock L., Sargin B., Ueker A., Böhmer F. D., Berdel W. E., Müller-Tidow C., Serve H. (2007) Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood 110, 370–374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.