Abstract
SUZ12 is a core component of the polycomb repressive complex 2 (PRC2) and is required for the differentiation of mouse embryonic stem cells (ESCs). PRC2 is associated with transcriptional repression via methylation of H3 Lys-27. We applied quantitative mass spectrometry to investigate the effects of Suz12 deficiency on H3.2 and H3.3 from mouse ESCs. Using high mass accuracy MS combined with CID or electron transfer dissociation (ETD) tandem mass spectrometry, we identified a total of 81 unique modified peptides from H3.2 and H3.3 and assigned 46 modifications at 22 different positions, including distinct coexisting modifications. In certain cases, high mass accuracy LTQ-Orbitrap MS/MS allowed precise localization of near isobaric coexisting PTMs such as trimethylation and acetylation within individual peptides. ETD MS/MS facilitated sequencing and annotation of phosphorylated histone peptides. The combined use of ETD and CID MS/MS increased the total number of identified modified peptides. Comparative quantitative analysis of histones from wild type and Suz12-deficient ESCs using stable isotope labeling with amino acids in cell culture and LC-MS/MS revealed a dramatic reduction of H3K27me2 and H3K27me3 and an increase of H3K27ac, thereby uncovering an antagonistic methyl/acetyl switch at H3K27. The reduction in H3K27 methylation and increase in H3K27 acetylation was accompanied by H3K36 acetylation and methylation. Estimation of the global isoform percentage of unmodified and modified histone peptides (amino acids 27–40) showed the relative distribution of distinct coexisting histone marks. Our study revealed limitations of antibody-based Western blotting methods for detection of coexisting protein modifications and demonstrated the utility of quantitative tandem mass spectrometry for detailed analysis of the dynamics of coexisting post-translational modifications in proteins.
Histone proteins, H1, H2A, H2B, H3, H4, and their variants, constitute the core of nucleosomes that organize DNA into chromatin. Histones can be extensively modified, and various post-translational modifications (PTMs)1 of histones have been reported, including phosphorylation (ph), mono- (me1), di- (me2), trimethylation (me3), acetylation (ac), formylation, citrullination, and ubiquitylation (1–3). Most histone PTMs are located within the N-terminal tails of histones, which contain several basic residues such as lysines and arginines. Histone PTMs influence the transcriptional activity of genes by affecting chromatin structure via modulation of histone-DNA interaction. For instance, reduction of a positive charge on H4K16 upon acetylation leads to decompression of chromatin (4). In addition, histone PTMs create or block the docking sites for non-histone proteins, including transcription factors and epigenetic readers/writers/erasers (1, 5). For instance, H3K27 methylation has been suggested to be recognized by the PRC1 complex, which results in RING1B (a component of the PRC1 complex)-mediated ubiquitylation of H2A, thereby inducing compaction of chromatin and stable repression of genes (6–9).
An increasing amount of data suggests that PTMs of histones are important for regulating transcription and that the enzymes involved in this are required for proper differentiation programs and development. For example, a bivalent regulatory domain of H3 was identified by genome-wide location analysis of histone marks in murine ESCs (10). The transcriptionally active mark H3K4me3 was shown to co-localize with the transcriptionally repressive mark H3K27me3 in promoter regions of many key developmental genes in ESCs. These “bivalent domains” silence developmental genes in ESCs and resolve upon differentiation. Moreover, the combination of different histone PTMs can affect the affinity of different proteins toward histones and chromatin. For instance, the 14-3-3 protein, which binds to H3S10ph, increases its affinity when H3K14 becomes acetylated (11, 12). In contrast, heterochromatin protein 1 (HP1) binds to H3K9me2/me3 and dissociates upon phosphorylation of the adjacent H3S10 residue (13). In addition, several histone-modifying enzymes have specificity for more than one amino acid residue. Finally, cross-talk is observed where multivalent modifications can act in concert to reinforce transcriptional activation or repression (14): for instance, it was found that asymmetric dimethylation on H3R2 in mammals inhibits H3K4 trimethylation by MLL1, thereby repressing the transcription of genes (15, 16).
Mass spectrometry-based approaches are advantageous for mapping the combination of modifications in histones, whereas antibody-based approaches, such as Western blotting, are limited in detection of co-occurring PTMs in a protein (17, 18). High mass accuracy mass analyzers such as the FT-ICR mass analyzer or the orbitrap mass analyzer are very useful in histone research as they allow distinguishing near isobaric modifications such as acetylation (42.011 Da) and trimethylation (42.047 Da) on lysine residues and accurate annotate of coexisting PTMs. The recent advances in technologies for ion-electron reactions including electron capture dissociation and electron transfer dissociation (ETD) methods (19, 20) have improved MS/MS analysis and sequencing of large peptides (>2 kDa). These methods have been used in histone research (21, 22).
Mass spectrometry-based proteomics allows identification of modified peptides/proteins but also quantification of modified peptides via label-free (23) or isotope labeling methods (24, 25). For example, Beck et al. (23) used a label-free LC-MS/MS approach to quantify histone modifications upon treatment of human cells with the histone deacetylase inhibitor PXD101 and demonstrated that acetylations on H2A, H2B, H3, and H4 were affected by the PXD101 treatment. Bonenfant et al. (24) used stable isotope labeling with amino acids in cell culture (SILAC) (26) in conjunction with mass spectrometry for quantification of histone modifications during the cell cycle and measured changes in the global level of histone modifications on all core histones. Moreover, mass spectrometry-based methods have contributed in monitoring the activity of enzymes responsible for adding or removing modifications on histone proteins. LSD1, a nuclear amino oxidase homologue, is the first lysine demethylase discovered, and its demethylase activity was confirmed by analysis of histone peptides by mass spectrometry (27). Histone-modifying enzymes play an important role in epigenetic regulations (1, 28). SUZ12, together with EED and the SET domain-containing protein EZH2, forms the core of the PRC2 complex. EZH2 is a histone methyltransferase that catalyzes the di- and trimethylation of H3K27. The three polycomb group proteins of the PRC2 complex are all required for EZH2 histone methyltransferase activity, and loss of either Suz12 or Eed results in a global loss of H3K27me2 and -me3 (29, 30). Importantly, the activity of the PRC2 complex is required for development, stem cell differentiation, and epigenetic inheritance (8, 31–34). For example, loss of Suz12 causes embryonic death during early postimplantation stages, and the Suz12-deficient embryos contain diminished levels of H3K27me2 and H3K27me3 in vivo (34). However, little is known about how the deletion of Suz12 affects PTMs in addition to H3K27. Here, we demonstrate that SILAC and tandem mass spectrometry using CID and ETD are suitable methods for obtaining qualitative and quantitative information of peptides with a single or a combination of PTMs from H3 variants upon inactivation of Suz12 in mouse ESCs.
EXPERIMENTAL PROCEDURES
Purification of Histones
Establishment of Suz12-deficient (KO) ESCs was described previously (34). Suz12 KO mouse ESCs were cultured for 6 days in SILAC Dulbecco's modified Eagle's medium (Sigma) containing 15% dialyzed fetal bovine serum (Invitrogen), penicillin-streptomycin (Invitrogen), nonessential amino acids (Invitrogen), pyruvate (Invitrogen), 50 mm β-mercaptoethanol, 3.5 g/liter d-glucose, 107 units/ml ESGRO leukemia inhibitory factor (Chemicon), 0.802 mm l-leucine (Sigma), 0.398 mm l-arginine (Sigma), and 0.798 mm l-lysine (Sigma). Lys8 isotope (Cambridge Isotopes, CNLM-291) was used for the Suz12 KO ESCs. Histones were purified with a commercially available histone purification kit (Active Motif, catalogue number 40025) according to the manufacturer's instructions. Briefly, cells were collected in acid extraction buffer with complete protease inhibitor mixture (Roche Applied Science, catalogue number 11873580001) and phosphatase inhibitor mixtures 1 and 2 (Sigma-Aldrich, catalogue numbers P2850 and P5726) and kept at 4 °C for 1–2 h. After centrifugation for 5 min at 14,000 rpm, the supernatants were collected, and pH was adjusted using loading buffer from the histone purification kit. The samples were loaded onto a cation exchange column followed by washing with a low concentration NaCl washing buffer. All histones were eluted in a single step using a high concentration NaCl elution buffer from the histone purification kit. Purified histones were precipitated overnight at 4 °C with 30% trichloroacetic acid. Samples were then centrifuged at 14,000 rpm for 1 h and washed with acetone containing 0.2% HCl followed by a second wash using 100% acetone. After air drying, the pellet was resuspended in ddH2O.
Separation of Intact Histones by Reversed Phase HPLC
The concentration of purified histones was measured using the Q-bit analyzer (Invitrogen). Heavy and light amino acid-labeled histones were mixed in a 1:1 ratio, and a total of 200 μg of histone mixture was separated by reversed phase (RP) HPLC using a C18 column (250 × 2 mm, Jupiter, 300 Å; Phenomenex, Torrance, CA) on an Akta-Basic system (GE Healthcare). The A solvent consisted of 0.06% TFA in ddH2O, and the B solvent was 0.04% TFA + 90% ACN (Sigma). The HPLC gradient increased from 5 to 35% in 10 min, 35 to 60% in 60 min, and 60 to 90% in 2 min.
MALDI Analysis and Enzymatic Digestion
One microliter of each histone fraction was mixed with 0.5 μl of α-cyano-4-hydroxycinnamic acid (10 mg/ml in 70% ACN, 0.1% TFA) on the MALDI target, and the intact masses of proteins were measured using MALDI-TOF MS (Voyager-DE STR, Applied Biosystems) in the positive ion mode (data not shown). All fractions were then freeze-dried and kept at −80 °C until further analysis. Dried samples were then resuspended in 20 μl of 50 mm NH4HCO3, and endoproteinase Arg-C (Calbiochem) was added (about 1:20 enzyme to protein ratio) and incubated at room temperature for 2–6 h.
LC-MS/MS
Peptide mixtures were analyzed by LC-MS/MS using an EasyLC instrument (Proxeon, Odense, Denmark) interfaced to an LTQ-Orbitrap XL instrument (ThermoFisher Scientific, Bremen, Germany). The nanoliter flow LC instrument was operated in a one-column setup with a 25-cm analytical column (100-μm inner diameter, 350-μm outer diameter) packed with C18 resin (ReproSil, Pur C18AQ 3 μm; Dr. Maisch). Solvent A was 0.1% formic acid in ddH2O, and solvent B was 90% acetonitrile (Fisher Scientific) with 0.1% formic acid. Samples were injected in an aqueous 1% TFA solution at a flow rate of 550 nl/min. Peptides were separated with a gradient of 0–40% solvent B over 105–180 min followed by a gradient of 40–60% for 15 min and 60–100% over 5 min at a flow rate of 250 nl/min.
The nanoelectrospray ion source (Proxeon) was used with a spray voltage of 1.5–2.0 kV. No sheath, sweep, and auxiliary gasses were used, and capillary temperature was set to 200 °C. The mass spectrometer was operated in a data-dependent mode to automatically switch between MS and MS/MS acquisition. In the LTQ-Orbitrap, full-scan MS spectra were acquired at a target value of 1 × 106 ions with a resolution of 60,000 (full-width at half-height). Samples were analyzed four times using four different MS/MS methods with a target value of 1 × 104 ions or 4 × 104 ions for linear ion trap (LTQ) MS/MS and 2 × 105 for orbitrap MS/MS. For CID MS/MS methods, singly charged precursor ions were excluded. For ETD MS/MS methods, singly and doubly charged precursor ions were excluded. The following MS and MS/MS scan modes were used. 1) The five most intense ions were isolated for fragmentation in the LTQ using CID with a normalized collision energy setting of 35, and fragment ions were scanned by the LTQ (CID-Hi/Low). 2) The three most intense ions were isolated for fragmentation in the LTQ using CID with a normalized collision energy value of 35, and fragment ions were scanned by the orbitrap (CID-Hi/Hi). 3) The eight most intense ions were isolated for fragmentation in the LTQ using ETD with an activation Q value of 0.25, activation time of 100 ms, and supplementary activation. Resulting fragment ions were scanned by the LTQ (ETD-Hi/Low). 4) The three most intense ions were isolated for fragmentation in the LTQ using ETD with an activation Q value of 0.25, activation time of 100 ms, and supplementary activation. Resulting fragment ions were scanned by the orbitrap (ETD-Hi/Hi). The dynamic exclusion time was set to 40–60 s.
Data Analysis
The raw data from LTQ-Orbitrap were converted to an mgf file using Proteome Discoverer 1.0 software (ThermoFisher Scientific). Database searching was performed against a custom-made database containing mouse histones retrieved from UniProt release 14 using Mascot Daemon version 2.1.0 (Matrix Science). Because of the small size of this database, we did not attempt to calculate false discovery rates, nor did we pursue decoy database searching. MS mass tolerance was set to 6 ppm for all files. Files obtained with MS/MS scans by the LTQ (Hi/Low) were searched with an MS/MS mass tolerance of 0.8 Da. Files obtained with MS/MS scans by the orbitrap (Hi/Hi) were searched with an MS/MS mass tolerance of 0.05 Da. CID data were searched with instrument setting “ESI-trap,” and ETD data were searched with instrument setting “ETD-trap.” Variable modifications included mono- and dimethylation on lysine and arginine residues; trimethylation, acetylation, and formylation on lysines; phosphorylation on serine, threonine, and tyrosine; oxidation on methionine; and acetylation on the N terminus of proteins. In addition, the same mgf file was searched with user-defined modifications that included lysine modifications increased by 8.0142 Da (“heavy PTMs”) to take into account labeling of lysines with stable isotopes. In this way, heavy isotope-labeled peptides with PTMs were identified in the Mascot search. Mascot search results were exported, and low confidence identifications were filtered using Microsoft Office Excel 2007 with the following criteria: peptides with a low score (cutoff score value, 15) and with many putative PTMs (≥5 PTMs/peptide) were removed. Redundant peptides were filtered by selecting the peptide with the highest Mascot score among peptides with the same identification. Filtered data were subjected to manual validation using Qual Browser version 2.0.7 (ThermoFisher Scientific). Peptides (aa 27–40) containing modifications on Lys-27 or Lys-36 were manually quantified using the Qual Browser. Extracted ion chromatograms were constructed for each precursor m/z value with a mass tolerance of 20 ppm and mass precision up to four decimal places. Peak areas for a pair of heavy and light peptides were measured for the same retention time interval. The ratio between the sum of the peak areas from all H3.2 light peptides (aa 27–40) versus the sum of all H3.2 heavy peptides (aa 27–40) was close to 1 (1.0). Also, the SILAC ratios of unmodified peptides (H3.2 and H3.3, aa 53–63) were between 1.1 and 1.2 in the analysis using the four different LC-MS/MS methods. The reproducibility of the determined SILAC ratios over the four different LC-MS/MS methods is shown for two H3.2 peptides (aa 27–40), Kme2SAPATGGVKme2KPHR and Kme3SAPATGGVKme2KPHR, in supplemental Fig. 1.
Immunoprecipitation and Western Blotting
RP-HPLC-purified H3 from mouse ESCs was resuspended in binding buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 5% glycerol, 0.1% Igepal) and incubated for 2 h with protein A-Sepharose beads (Amersham Biosciences) precoupled with H3- and H3K27me2-specific antibodies. Beads were washed six times with binding buffer, and bound proteins were eluted from beads by denaturation at 100 °C for 10 min in Laemmli sample buffer. Eluted proteins were analyzed by standard Western blot analyses using the indicated antibodies.
Antibodies
H3K27me1 (Upstate 07-448), H3K27me2 (Cell Signaling Technology 9728), H3K27me3 (Cell Signaling Technology 9733S), H3K27Ac (Upstate 07–360), H3K36me2 (Cell Signaling Technology 2901), and H3 (Abcam ab1791) antibodies were used.
RESULTS
Mapping Post-translational Modifications of H3. 2 and H3.3 in Mouse ESCs
We isolated histones from wild type and Suz12-deficient ESCs (Fig. 1A) using a commercially available purification kit. This method allowed efficient and fast purification of linker and core histones. The histone preparations were then separated by RP HPLC, and the fractions containing histones H3.2 (Swiss-Prot accession number P84228) and H3.3 (Swiss-Prot accession number P84244) were digested with endoproteinase Arg-C. This protease cleaves at the amide bond C-terminal to arginine residues and generates peptides that may contain multiple lysine residues. In histones, lysine residues frequently carry post-translational modifications, such as acetyl and methyl groups, which are reversible and dynamic marks involved in protein-protein and protein-DNA interactions in chromatin. The peptides were analyzed by nanoliter flow HPLC interfaced to ESI-MS/MS using an LTQ-Orbitrap XL mass spectrometer. A long capillary C18 column (∼25 cm) in combination with a shallow gradient of the organic mobile phase (0.22–0.38% solvent B/min) was used to be able to efficiently separate near identical or isobaric modified peptides prior to MS analysis. Four separate LC-MS/MS methods were used to analyze each sample, all of them utilizing the high mass accuracy orbitrap analyzer for peptide mass determination, either CID or ETD for ion fragmentation, and either the low mass accuracy LTQ or high mass accuracy orbitrap analyzer for MS/MS analysis. This comprehensive LC-MS/MS analysis allowed sequencing of peptides and localization of distinct, coexisting site-specific PTMs within these peptides. Applying this approach to histones H3.2 and H3.3, we identified many previously reported modifications in the N-terminal tails of these proteins (aa 1–50) (Fig. 1, B and C). PTMs that were common to both H3.2 and H3.3 included phosphorylation on Thr-3 and Ser-10; monomethylation on Lys-4, Lys-9, Lys-18, Lys-27, and Lys-36; dimethylation on Lys-9, Arg-26, Lys-27, and Lys-36; trimethylation on Lys-4, Lys-9, and Lys-27; and acetylation on Lys-9, Lys-14, Lys-18, Lys-23, and Lys-27. Modifications found only in H3.2 included acetylation on Lys-4 and dimethylation on Lys-18. Modifications found exclusively in H3.3 included dimethylation on Lys-4, trimethylation on Lys-36, and acetylation on Lys-36.
Fig. 1.
A, quantitative mass spectrometry strategy for histone modifications. Wild type and Suz12-deleted mouse ESCs were grown in the medium containing normal lysine (Lys0) and stable isotope-labeled lysine (Lys8), respectively. Combined intact histones were separated using RP HPLC. Aliquots of intact histones or independently prepared histones were used for Western blotting assays. Histones H3.2 and H3.3 were digested by using endoproteinase Arg-C, and the peptides were analyzed by LC-MS/MS using an ∼25-cm capillary RP HPLC column interfaced to the nano-ESI source of an LTQ-Orbitrap XL instrument. B and C, summary of PTMs that were mapped onto the N-terminal tails of histones H3.2 and H3.3 from mouse ESCs. Most of the previously reported PTMs on H3.2 and H3.3 were identified in the present study. One circle, monomethylation; two circles, dimethylation; three circles, trimethylation; triangle, acetylation; star, phosphorylation.
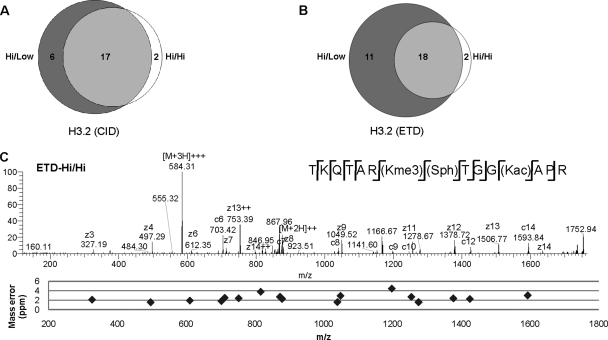
Upon histone sequence database searching and manual interpretation and verification of the MS/MS data, all identified peptides were reduced to a list of unique peptides, each containing no, one, or several modifications. In total, 43 and 38 unique peptides originating from H3.2 and H3.3 samples, respectively, were characterized (supplemental Table 1, supplemental Figs. 3–54, and Figs. 2C, 3C, and 4, C–F). Fig. 2, A and B, show a number of unique H3.2 peptides identified by four different LC-MS/MS methods: CID-Hi/Low, CID-Hi/Hi, ETD-Hi/Low, and ETD-Hi/Hi. Using CID MS/MS, 23 peptides in total were sequenced and assigned using the Hi/Low mode, whereas 19 peptides were assigned by using the Hi/Hi mode. Using ETD MS/MS, 29 peptides were sequenced and assigned by the Hi/Low mode, whereas 20 identifications were obtained by the Hi/Hi mode. The two modes of MS and MS/MS operation generated overlapping data sets, and it is clear that the benefit of using the Hi/Hi mode rather than the Hi/Low mode is limited to peptides with near isobaric coexisting modifications, regardless of the choice of ion dissociation method. Thus, a majority of the peptides were confidently sequenced and annotated by taking advantage of high mass accuracy MS (≤6-ppm mass error) obtained by the orbitrap analyzer followed by sensitive but low mass accuracy MS/MS (<0.8-Da mass error) using the LTQ analyzer. The Hi/Hi mode of operation is very helpful for site-specific assignment of near isobaric modifications such as trimethylation and acetylation because the mass difference between an acetyl and a trimethyl group is only 0.036 Da. For example, the H3 (aa 3–17) peptide contained three lysine residues and two arginine residues, and the measured mass corresponded to the histone peptide containing one acetylation, one phosphorylation, and three methylations (Fig. 2C). To determine which lysine residues were modified, the peptide precursor ion was fragmented by ETD in the LTQ analyzer, and the peptide fragment ions were analyzed in the orbitrap. Interpretation of the resulting ETD MS/MS spectrum allowed annotation of coexisting modifications “H3K9me3/S10ph/K14ac” with an average mass error of 2.46 ppm (S.D. = 0.76). The alternative combination of modified residues “H3K9ac/S10ph/K14me3” exhibited a larger average mass error of 9.81 ppm (S.D. = 32.63) of the peptide fragment ions. We have previously reported an algorithm that takes advantage of the high mass accuracy MS/MS information to assign PTMs (35).
Fig. 2.
MS/MS analysis of histone peptides. The Venn diagrams display the number of observed modified unique H3.2 peptides with one or several PTMs using CID MS/MS (A) or ETD MS/MS (B). The Hi/Low mode generated more peptide identifications than the Hi/Hi mode for both ETD and CID MS/MS. C, annotation of coexisting PTMs by using high mass accuracy orbitrap ETD MS/MS (Hi/Hi). Upper panel, ETD MS/MS spectrum of the H3.3 peptide aa 3–17 TKQTAR(Kme3)(Sph)TGG(Kac)APR (m/z 584.3137, 3+). Lower panel, the distribution of fragment ion mass errors (ppm) for the annotated peptide.
Fig. 3.
ETD and CID MS/MS identified histone peptides with a single or coexisting post-translational modifications. The Venn diagrams display the number of unique identified peptides of H3.2 (A) and H3.3 (B) by CID MS/MS (precursor ion charge state, >1+) and ETD MS/MS (precursor ion charge state, >2+). C, ETD MS/MS spectrum of a phosphopeptide. The H3.2/H3.3 peptide (aa 3–17) TKQTAR(Kme2)(Sph)TGG(Kac)APR (m/z 434.9827, 4+) was identified by ETD MS/MS (upper panel). For comparison, the corresponding CID MS/MS spectrum (m/z 434.9825, 4+) is also shown. The underlined amino acids are potentially modified with phosphorylation (Ser and Thr), dimethylation (Lys), or monomethylation (Lys).
Fig. 4.
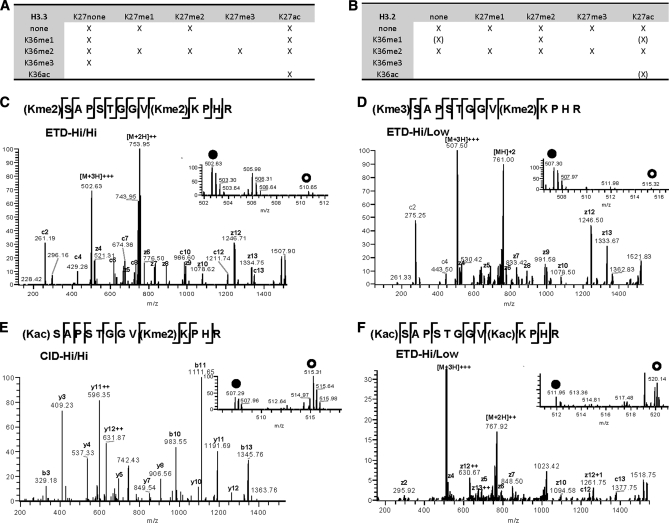
Identification of distinct peptide species (aa 27–40) with one or more coexisting PTMs at Lys-27 and/or Lys-36. The tables display the combinations of modifications that were observed at Lys-27 and Lys-36. All identified unique H3.3 (A) and H3.2 (B) peptides from MS/MS are marked with “X” on the matrixes. The (X) indicates that peptides were not confirmed by MS/MS, but ion signals from the corresponding peptides were observed in the MS spectra. C, identification of the H3.3 peptide containing the K27me2/K36me2 mark by ETD MS/MS (Hi/Hi) of the triply charged precursor ion (m/z 502.6335). D, identification of the H3.3 peptide containing the “K27me3/K36me2” mark by ETD MS/MS (Hi/Low) of the triply charged precursor ion (m/z 507.3048). E, identification of the H3.3 peptide containing the “K27ac/K36me2” mark by CID MS/MS (Hi/Hi) of the doubly charged precursor ion (m/z 760.4384). F, identification of the H3.3 peptide containing the “K27ac/K36ac” mark by ETD MS/MS (Hi/Low) of the triply charged precursor ion (m/z 511.9530). C–F, insets, SILAC peptide ion pairs used for quantification are labeled: the filled circle is wild type (light isotope version), and the empty circle is Suz12−/− (heavy isotope version).
Next, we evaluated the performance of CID and ETD for peptide sequencing and annotation of PTMs. For CID MS/MS, the precursor ion charge state was 2+ or higher, whereas for ETD, the precursor ion charge state was 3+ or higher. Note also that the MS/MS duty cycle is slightly different for these modes of operation (see “Experimental Procedures”). For histone H3.2, in total 25 unique peptide identifications were obtained from CID experiments, whereas 31 unique identifications were obtained from ETD experiments (Fig. 3A). Thirteen peptides were identified by both methods. In the case of histone H3.3, 28 peptide assignments were made by CID experiments, whereas 27 peptide assignments were obtained from ETD experiments (17 peptides were in common) (Fig. 3B). These results suggested that the application of both CID and ETD MS/MS leads to a higher number of annotated peptides as compared with using any one of these dissociation methods. Notably, a nearly 10-fold higher number of phosphopeptides were identified using ETD MS/MS as compared with CID MS/MS. This is partially because of the instability of phosphoserine/threonine peptide ions in CID MS/MS in ion trap analyzers. In contrast, such phosphopeptides often generate useful fragments in ETD MS/MS and allow unambiguous assignment of phosphorylation sites in peptides. For example, the peptide aa 3–17 TKQTARKme2SphTGGKacAPR (m/z 434.9827, 4+) was sequenced and annotated by ETD MS/MS (Fig. 3C, upper panel). The fragment ions covered most of the amino acid sequence, and the phosphate group remained intact so a combination of modifications “K9me2/S10ph/K14ac” could be confidently localized. The corresponding CID MS/MS spectrum (m/z 434.9825, 4+) (Fig. 3C, lower panel) generated a complementary but incomplete y-ion series and signals that exhibited the neutral loss of H3PO4 or H2O from the peptide fragments ion. In summary, our MS/MS results obtained from modified peptides derived from histones H3.2 and H3.3 are consistent with previous reports that ETD is advantageous for site-specific mapping of labile modifications such as phosphorylation on serine, threonine, and histidine residues (36, 37).
Detailed Analysis of Peptides Containing Modifications on Lys-27 and Lys-36
We applied CID and ETD MS/MS to obtain a detailed map of the PTMs in the region that contained the lysine residues Lys-27 and Lys-36 of histones H3.2 and H3.3. We detected distinct combinations of methylation (me1, me2, and me3) and/or acetylation on Lys-27 and/or Lys-36 in the peptide spanning aa 27–40 (Fig. 4, A and B). K27me1 was observed alone or in combination with K36me2. K27me2 was detected either alone or with K36me1 or K36me2 (Fig. 4C). K27me3 was observed alone or together with K36me2 (Fig. 4D). K27ac was observed with unmodified Lys-36 or various Lys-36 modifications (me1, me2, and ac) (Fig. 4, E and F). K36me2 was observed with unmodified Lys-27 or different types of Lys-27 modifications (me1, me2, me3, and ac). Moreover, the diacetylated peptide K27ac/K36ac was detected for the first time and sequenced by MS/MS of the H3.3 peptide from the control ESC samples.
Quantitative Analysis of Histone H3.2 and H3.3 Peptides Containing Lys-27 and Lys-36
SUZ12 is a component of the PRC2 complex that catalyzes methylation of H3K27. We investigated the implications of deleting Suz12 on the PTM status of the lysine residues Lys-27 and Lys-36 of H3.2 and H3.3 in mouse ESCs. Pasini et al. (29) showed that the abundance of both H3K27me2 and H3K27me3 was decreased by deletion of Suz12. We initially confirmed this in the present study by using Western blotting to demonstrate that SUZ12 activity is effectively ablated in our Suz12 KO ESCs as compared with the control ESCs (Fig. 5C). We then applied SILAC and mass spectrometry to obtain quantitative data on unmodified and site-specifically modified histone peptides, including coexisting PTMs on Lys-27 and Lys-36. We focused our analysis on the H3.2 and H3.3 peptides spanning aa 27–40 as they contain a single PTM or a combination of PTMs including the residue Lys-27, which is the substrate site for the PRC2. As mentioned previously, endoproteinase Arg-C generated the appropriate digestion product covering aa 27–40; i.e. no missed cleavage products originating from this part of the histone sequence were detected by LC-MS/MS.
Fig. 5.
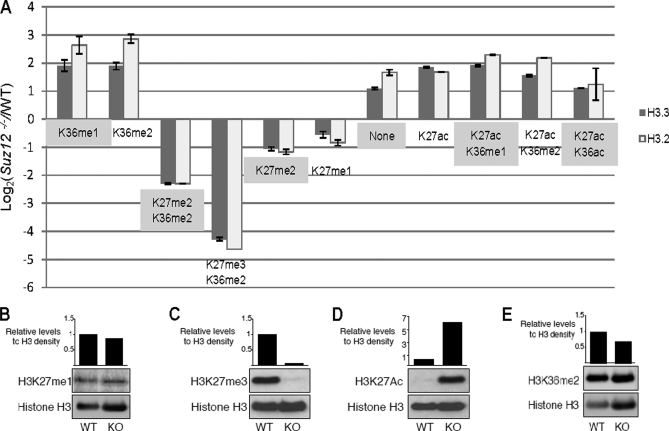
Quantification of distinct H3.2 and H3.3 peptides (aa 27–40). A, SILAC-based relative quantification of H3.2 and H3.3 peptides. Peptides containing the K27me2 and K27me3 marks decreased in abundance, whereas peptides containing the K27ac mark increased in abundance upon Suz12 deletion. Error bars indicate the S.D. calculated from the technical replicates. B–E, Western blotting results using monoclonal antibodies against H3K27me1 (B), H3K27me3 (C), H3K27ac (D), and H3K36me2 (E). H3 (H3 C terminus) was used as a loading control. WT, wild type.
Quantitative mass spectrometry results obtained by SILAC and LC-MS/MS for aa 27–40 peptides from H3.2 and H3.3 are summarized in Fig. 5A. Representative MS and MS/MS spectra of modified histone peptides are shown in Fig. 4, C–F, including H3.3 (aa 27–40) peptides Kme2SAPSTGGVKme2KPHR (Fig. 4C), Kme3SAPSTGGVKme2KPHR (Fig. 4D), KacSAPSTGGVKme2KPHR (Fig. 4E), and KacSAPSTGGVKacKPHR (Fig. 4F). The relative abundance of these modified peptides was obtained from the chromatographic peak area of SILAC-encoded peptide ion pairs and allowed comparison of wild type (light) and Suz12-deficient ESCs (heavy) (Fig. 4, C–F, insets). The K27me3 mark accompanied by K36me2 was decreased in Suz12 KO ESCs (Fig. 4D, inset). K27me2-containing peptides were accompanied by K36me2 or unmodified K36. The abundance of peptides with K27me2/K36me2 (Fig. 4C, inset) marks or K27me2/K36(unmodified) marks was decreased to a lesser extent than that of peptides with K27me3/K36me2. These results are in agreement with the previous report of decreased abundance of H3K27me2 and H3K27me3 upon Suz12 deletion (29). The abundance of K27me2 peptides decreased to a different degree depending on the modification status of Lys-36 upon Suz12 deletion. For example, peptides containing K27me2/K36me2 decreased by 4-fold, whereas K27me2/K36(unmodified) peptides decreased ∼2-fold upon Suz12 deletion. Interestingly, the abundance of H3K27ac peptides was increased upon Suz12 deletion regardless of the nature of the modifications on Lys-36 (Fig. 4, E and F, insets, and Fig. 5, A and D).
We were able to quantify several near identical, isobaric modified peptides, i.e. peptides with identical molecular masses that contain identical modifying moieties that are located at different amino acid residues. For instance, the peptide (aa 27–40) from H3.3 contains three lysine residues, Lys-27, Lys-36, and Lys-37, among which two are known to be methylated. Isobaric peptides often coelute during HPLC, or they elute within a very short retention time window during LC-MS/MS analysis. We utilized a shallow LC gradient and a relatively long (25-cm) chromatographic column to achieve higher chromatographic resolution (“Experimental Procedures”). Thus, isobaric peptides (aa 27–40) from H3.3 that were dimethylated at either Lys-27 or Lys-36 were separated, quantified, and sequenced by LC-MS/MS. The K36me2 peptide eluted first followed by the K27me2 peptide (Fig. 6). Efficient separation in combination with SILAC facilitated the determination of the relative abundance of these highly similar peptide species. The relative abundance of K36me2 increased upon Suz12 deletion, whereas the relative abundance of K27me2 decreased (Fig. 6). Because these modifications are of biological interest, it is extremely useful to be able to quantify and characterize these peptides separately.
Fig. 6.
Separation, sequencing, and quantification of isobaric modified peptides. The extracted ion chromatograms of the doubly charged precursor ion (m/z 739.4333) from the “light” dimethylated peptide H3.3 KSAPSTGGVKKPHR (aa 27–40) (upper) and the corresponding “heavy” peptide m/z 751.4544 (2+) (lower). Separation of these isobaric modified peptides allowed quantification of the two different dimethylated peptide species. The dimethylated peptide “KSAPSTGGVKme2KPHR” eluted prior to the isobaric dimethylated peptide “Kme2SAPSTGGVKPHR.” The filled circle is wild type (light isotope version), and the empty circle is Suz12−/− (heavy isotope version). WT, wild type.
Western blotting results obtained for distinct Lys-27 modifications (H3K27me1, H3K27me3, and H3K27ac) agreed with our quantitative mass spectrometry results (Fig. 5, B–D). We noted that the Western blotting results obtained by using a monoclonal antibody directed toward H3K36me2 showed that H3K36me2 levels were similar between Suz12 KO and control ESCs (Fig. 5E). However, quantitative mass spectrometry produced a more detailed profile of distinct K36me2 peptides: K36me2 in combination with either K27me2 or K27me3 decreased upon Suz12 deletion, whereas H3K36me2 alone or in combination with H3K27ac increased in relative abundance (Figs. 4, C–E, and 5A). These results highlight the advantages and the importance of using unbiased quantitative mass spectrometry for monitoring coexisting transient and dynamic PTMs in proteins. There is an obvious risk of missing detailed quantitative information on coexisting and cooperative PTMs when solely relying on Western blotting for the detection of individual PTMs in histone proteins.
Global Isoform Quantification of Lys-27/Lys-36-modified Peptides
The SILAC-based quantitative mass spectrometry analysis revealed distinct features of individual modified histone peptide species. We also aimed at getting an overall view on the relative abundance profile for unmodified and modified peptides corresponding to aa 27–40, particularly the modification states of the Lys-27/Lys-36 residues and how this profile changed upon deletion of Suz12 in mouse ESCs. A global isoform quantification method (38, 39) was adapted to our data set. For each unique peptide, the global isoform percentage (GP) was calculated according to the following equation.
 |
The GP value for a specific peptide species represents the relative proportion of that peptide isoform/variant among all quantified peptide species sharing the same amino acid sequence. For H3.2 and H3.3 peptides covering the amino acids 27–40, a total of 11 unique peptide species were quantified. GP analysis of these peptides showed that peptides containing “K27me2/K36me2” marks were predominant in H3.2 as well as H3.3 from wild type ESCs (Fig. 7). This result was confirmed by antibody-based assays (immunoprecipitation and Western blotting) (Fig. 7, inset). Higher amounts of H3K36me2 were observed by Western blots of H3K27me2 pulldowns as compared with H3 (C-terminal) pulldowns.
Fig. 7.
GP profile of histone peptides (aa 27–40). Peptides containing the K27me2/K36me2 marks were the most abundant species in histones H3.2 and H3.3 from wild type mouse ESCs. Peptides that exclusively contained K36me2 were most abundant in H3.2 and H3.3 upon Suz12 deletion. Inset, histone detection by immunoprecipitation followed by Western blotting of histones purified from wild type mouse ESCs. A higher abundance of H3K36me2 was observed by Western blotting of H3K27me2 pulldowns as compared with the H3 (C-terminal) pulldowns. Error bars indicate the S.D. calculated from the technical replicates.
Upon Suz12 deletion, peptides containing K36me2 (no modification of Lys-27) were the dominant structure for both H3.2 and H3.3 (Fig. 7), again demonstrating H3K27-directed methyltransferase activity of the PRC2. In general, K27ac peptides were of rather low abundance as compared with K27me or unmodified Lys-27 peptides in both wild type and Suz12 KO ESCs. When all quantified isoforms were categorized according to the Lys-27 modifications (none, me2, and ac), each Lys-27 modification accompanied by K36me2 was most abundant (supplemental Fig. 2). Thus, the GP analysis revealed additional features of the PTM profiles of aa 27–40 peptides from histones H3.2 and H3.3, i.e. features that were missing from the peptide-centric SILAC-based quantification. The combination of peptide-centric relative quantification by SILAC with the “peptide family” relative quantification by GP contributed toward a more detailed interpretation of histone PTMs.
DISCUSSION
The histone modification status influences the transcriptional activity of genes, for example by inducing structural changes in chromatin or by creating docking sites for chromatin-binding effector proteins. Acetylation of lysine residues of histones is mainly linked to transcriptional activation of genes, whereas methylation of specific lysine residues, such as H3K27 and H3K9, is associated with repression of transcription. Coexisting PTMs, PTM cross-talk, and cooperativity of PTMs in protein molecules are of immense importance for the biological activity in cells, tissues, and organisms (14, 40). Thus, by interfering with or by perturbing specific enzymes that modify histones at particular amino acid residues, it is likely that “ripple effects” will be observed, i.e. alterations of modification occupancy at different, adjacent, or remote amino acid residues in the same protein or in closely associated, interacting proteins.
In the present study, we applied quantitative mass spectrometry for mapping and characterization of coexisting PTMs on histones H3.2 and H3.3 from wild type and Suz12 KO ESCs (Fig. 1A). Intact H3 variants were separated by RP HPLC and digested using endoproteinase Arg-C. Peptide mixtures were analyzed by four different LC-MS/MS methods using LTQ-CID/ETD-orbitrap technology. The high mass accuracy and mass resolution achieved by the orbitrap MS combined with sensitive LTQ MS/MS or accurate orbitrap MS/MS allowed the annotation of acetylated, methylated, and phosphorylated histone peptides. A majority of the known PTMs on mammalian H3.2 and H3.3 were observed (Fig. 1, B and C, supplemental Table 1, and supplemental Figs. 3–54), supporting the efficiency, sensitivity, and specificity of our analytical strategy.
Endoproteinase Arg-C was used in this study instead of the more commonly used enzyme trypsin. Digestion of lysine-rich histone proteins with trypsin, endoproteinase Lys-C, or other enzymes that cleave at unmodified lysine residues would have generated many small peptides that are likely to be missed during LC-MS/MS analysis. Endoproteinase Arg-C cleaves only after arginine; hence, it generates longer peptides than trypsin or endoproteinase Lys-C, and it allows quantitative analysis of lysine-containing peptides by mass spectrometry, including acetylated and methylated lysine residues.
LC-MS/MS analysis of H3.2 and H3.3 digested with endoproteinase Arg-C resulted in identification of 43 and 38 unique modified peptides from H3.2 and H3.3, respectively. A majority of modified peptides were found from the peptides aa 3–17, aa 9–17, and aa 27–40. Various isoforms were identified from the region aa 27–40 containing different modifications (none, me1, me2, me3, and ac) at Lys-27 and/or Lys-36 (Fig. 4, A and B). Combinations of PTMs between Lys-27 and Lys-36 were previously observed in HeLa cells (21, 41) except for the H3K36ac species that we report in the present study. A number of putative peptide species (aa 27–40) containing K27me1/K36me1, K27me3/K36me1, K27me1/K36me3, K27me2/K36me3, K27me3/K36me3, and K27ac/K36me3 went undetected, possibly because of limitations of our analytical strategy, the very low abundance of these species, or the fact that histones H3.2 and H3.3 from mouse ESCs may lack these combinations of PTMs.
We report for the first time the co-occurrence of H3K27ac/K36ac in mammalian ESCs. This combination was previously only observed in histones obtained from unicellular organisms, i.e. Tetrahymena thermophila and Saccharomyces cerevisiae (42). H3K36ac has been suggested to be involved in RNA polymerase II-mediated gene transcription (42). We2 and others (43) have recently shown that H3K27ac is associated with transcriptional activation of polycomb group target genes. Thus, the combination of K27ac and K36ac might constitute an “amplifier” that reinforces transcriptional activation in a cooperative manner, much like the coexisting H3K9ac and H3K14ac marks.
We analyzed histone peptides using four different LC-MS/MS methods: CID fragmentation followed by an MS/MS scan using the LTQ (CID-Hi/Low) or the orbitrap (CID-Hi/Hi) or ETD fragmentation followed by an MS/MS scan using the LTQ (ETD-Hi/Low) or the orbitrap (ETD-Hi/Hi). In most cases, the Hi/Low scan mode was sufficient for sequencing peptides and mapping site-specific PTMs (Fig. 2, A and B). The Hi/Hi scan mode suffered from relatively poor ion transmission and reduced duty cycle because ions had to be transferred from the LTQ to the orbitrap for the high mass resolution detection in MS/MS mode. This may be a limitation of the LTQ-Orbitrap XL system used in our study. The recently introduced LTQ-Orbitrap Velos instrument has an improved trapping and transfer geometry that facilitates a higher duty cycle and more efficient ion transmission that will benefit the Hi/Hi mode of operation. Nevertheless, Hi/Hi mode was useful for mapping near isobaric coexisting modifications on multiple candidate PTM substrate sites because of the high mass accuracy determination of fragment ions (Fig. 2C).
We used two different dissociation methods, ETD and CID, to identify as many modified peptides as possible. Indeed, the overall number of identified unique peptides was increased as compared with analyzing samples only by one of the dissociation methods (Fig. 3, A and B) as also reported by others (44). In particular, ETD MS/MS was efficient for detecting a combination of PTMs bearing phosphorylations (Fig. 3C). CID and ETD MS/MS experiments were performed for different charge states of peptides (“Experimental Procedures”). Thus, the results of CID MS/MS and ETD MS/MS experiments are not directly comparable because the fragmentation efficiency of CID and ETD depends on many parameters such as the m/z value, the charge state, and the amino acid composition of peptide ions. In addition, minor variations in the LC separation and the data-directed method for MS/MS of peptides lead to variability. Nevertheless, using both ETD and CID MS/MS extended peptide identifications, achieving comprehensive mapping of PTMs.
The effect of Suz12 deletion was studied by a quantitative proteomics approach using SILAC and LC-MS/MS analysis. Upon Suz12 deletion, we detected a relative decrease of H3K27me2 and H3K27me3 and an increase of H3K27ac (Figs. 4, C–F, and 5A), thereby identifying an antagonistic Lys-27 methyl/acetyl switch. At the same time, we determined the modification status of Lys-36, thereby revealing distinct relationships between Lys-27 and Lys-36 modifications.
Increased acetylation on H3K27 upon Suz12 deletion is in accordance with a very recent study that demonstrated an increase in the abundance of H3K27ac upon knockdown of Su(z)12 or the catalytic subunit of PRC2, E(Z), which are orthologous to mammalian Suz12 and Ezh2 in Drosophila (43). Thus, depletion of methylation at H3K27 by knocking out the Ezh2 or the Suz12 component of the PRC2 leads to antagonistic acetylation of H3K27. A more detailed characterization of the functional role of H3K27ac in regulating polycomb group target gene expression in mouse ESCs will be described separately.2
The mass spectrometry data provided a much more detailed overview of the modification profiles of Lys-27/Lys-36 than was possible with standard Western blotting approaches. For example, K36me2 was identified with K27(unmodified) and K27me1, -me2, -me3, and -ac. K27me2/K36me2-containing peptides decreased to a lesser extent than K27me3/K36me2 peptides upon Suz12 deletion. K27/K36me2 and K27ac/K36me2 peptides were increased upon Suz12 deletion. In contrast, the Western blotting result indicated that H3K36me2 was present at approximately the same level in wild type and Suz12 KO ESCs (Fig. 5E). These observations clearly indicate that Western blotting results that are targeted toward one specific type of PTM have to be carefully interpreted as they may miss important regulatory features that involve multiple, coexisting PTMs. Clearly, mass spectrometry is much better suited to detect and quantify coexisting PTMs in peptides and proteins.
We found that it is important to be able to separate isobaric peptides by LC prior to the MS analysis (Fig. 6) because this allows accurate quantitative analysis of related peptide species by SILAC and GP. Using a long capillary RP HPLC column and a shallow gradient of organic mobile phase, isobaric modified peptides were separated and subsequently quantified by MS and sequenced by MS/MS. Obviously, some low abundance isobaric peptides will be missed because only the most abundant peptide ions are selected for sequencing by data-directed MS/MS approaches.
The GP approach allowed us to estimate the relative abundance profiles of modified peptide species and showed that peptides containing the K27me2/K36me2 marks were the predominant species (55–62%) among all observed H3.2 and H3.3 (aa 27–40) peptides from control ESCs (Fig. 7). This is in excellent agreement with a report demonstrating that the K27me2 species is the most abundant (∼50%) of all Lys-27 species (none, me1, me2, and me3) in mouse ESCs (45).
We found relatively high amounts of H3.3K27me3 (10%) in wild type ESCs, which is possibly due to the wide distribution of H3K27me3 across promoter, protein coding, and intergenic regions (46, 47). In fact, the GP value of the H3.3K27me3/K36me2 marks in our study agrees with a recent report that H3K27me3 covered 11% of chromosome 17 in mouse embryonic fibroblast (47). Furthermore, the GP value of K27me2/K36me2 remained relatively high (∼12%) despite deletion of Suz12. This might suggest that a hitherto undiscovered methyltransferase, in addition to PRC2, can catalyze dimethylation of H3K27 in mammalian ESCs.
In conclusion, quantitative mass spectrometry applied to the identification and quantification of H3.2 and H3.3 revealed most of the known PTMs in wild type and Suz12 KO mouse ESCs. Unique peptides with distinct coexisting PTMs were annotated and quantified, revealing novel features of histone marks at Lys-27 and Lys-36 in the context of PRC2 function and ablation of Suz12. The analytical and computational work flow presented here is suitable for general studies of dynamic coexisting PTMs in proteins to elucidate the effects of environmental, genetic, biochemical, and pharmacological perturbations in cells and organisms.
Supplementary Material
Acknowledgments
We thank Dr. Karen Krzywkowski for providing SILAC materials and for advice. We acknowledge members of the Centre for Epigenetics for technical advice and support. We also thank Dr. Martin Røssel Larsen at the University of Southern Denmark and Drs. Wolfgang Metelmann-Strupat and Claire Dauly at ThermoFisher Scientific for assistance with LTQ-Orbitrap mass spectrometry and associated software-related issues.
* This work was supported in part by a generous grant from the Danish National Research Foundation (to the Centre for Epigenetics).
 This article contains supplemental Table 1 and Figs. 1–54.
This article contains supplemental Table 1 and Figs. 1–54.
2 D. Pasini, M. Malatesta, H. R. Jung, J. Walfridsson, A. Willer, L. Olsson, J. Skotte, A. Wutz, O. N. Jensen, and K. Helin, submitted manuscript.
1 The abbreviations used are:
- PTM
- post-translational modification
- ac
- acetylation
- ESC
- embryonic stem cell
- ETD
- electron transfer dissociation
- GP
- global isoform percentage
- LTQ
- linear quadrupole ion trap
- me1
- monomethylation
- me2
- dimethylation
- me3
- trimethylation
- ph
- phosphorylation
- PRC
- polycomb repressive complex
- RP
- reversed phase
- SILAC
- stable isotope labeling with amino acids in cell culture
- KO
- knock-out
- ddH2O
- double distilled H2O
- aa
- amino acids
- EED
- Embryonic ectoderm development
- Suz12
- Suppressor of zeste 12 protein homolog
- EZH2
- Enhancer of zeste homolog 2
- MLL
- Myeloid/lymphoid or mixed-lineage leukemia protein
- SET
- Su(var)3-9, Enhancer of Zeste, Tri thorax
- NEAA
- Nonessential Amino Acids.
REFERENCES
- 1.Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2.Wisniewski J. R., Zougman A., Mann M. (2008) Nepsilon-formylation of lysine is a widespread post-translational modification of nuclear proteins occurring at residues involved in regulation of chromatin function. Nucleic Acids Res 36, 570–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Wysocka J., Sayegh J., Lee Y. H., Perlin J. R., Leonelli L., Sonbuchner L. S., McDonald C. H., Cook R. G., Dou Y., Roeder R. G., Clarke S., Stallcup M. R., Allis C. D., Coonrod S. A. (2004) Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279–283 [DOI] [PubMed] [Google Scholar]
- 4.Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 5.Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., Brockdorff N. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7, 663–676 [DOI] [PubMed] [Google Scholar]
- 7.Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16, 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasini D., Bracken A. P., Agger K., Christensen J., Hansen K., Cloos P. A., Helin K. (2008) Regulation of stem cell differentiation by histone methyltransferases and demethylases. Cold Spring Harb. Symp. Quant. Biol 73, 253–263 [DOI] [PubMed] [Google Scholar]
- 9.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 [DOI] [PubMed] [Google Scholar]
- 10.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 11.Walter W., Clynes D., Tang Y., Marmorstein R., Mellor J., Berger S. L. (2008) 14-3-3 interaction with histone H3 involves a dual modification pattern of phosphoacetylation. Mol. Cell. Biol 28, 2840–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winter S., Simboeck E., Fischle W., Zupkovitz G., Dohnal I., Mechtler K., Ammerer G., Seiser C. (2008) 14–3-3 proteins recognize a histone code at histone H3 and are required for transcriptional activation. EMBO J 27, 88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mateescu B., England P., Halgand F., Yaniv M., Muchardt C. (2004) Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep 5, 490–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suganuma T., Workman J. L. (2008) Crosstalk among histone modifications. Cell 135, 604–607 [DOI] [PubMed] [Google Scholar]
- 15.Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Lüscher B., Amati B. (2007) Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature 449, 933–937 [DOI] [PubMed] [Google Scholar]
- 16.Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., Bauer U. M. (2007) PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev 21, 3369–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trelle M. B., Jensen O. N. (2007) Functional proteomics in histone research and epigenetics. Expert Rev. Proteomics 4, 491–503 [DOI] [PubMed] [Google Scholar]
- 18.Garcia B. A. (2009) Mass spectrometric analysis of histone variants and post-translational modifications. Front. Biosci 1, 142–153 [DOI] [PubMed] [Google Scholar]
- 19.Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 21.Young N. L., DiMaggio P. A., Plazas-Mayorca M. D., Baliban R. C., Floudas C. A., Garcia B. A. (2009) High throughput characterization of combinatorial histone codes. Mol. Cell. Proteomics 8, 2266–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas C. E., Kelleher N. L., Mizzen C. A. (2006) Mass spectrometric characterization of human histone H3: a bird's eye view. J. Proteome Res 5, 240–247 [DOI] [PubMed] [Google Scholar]
- 23.Beck H. C., Nielsen E. C., Matthiesen R., Jensen L. H., Sehested M., Finn P., Grauslund M., Hansen A. M., Jensen O. N. (2006) Quantitative proteomic analysis of post-translational modifications of human histones. Mol. Cell. Proteomics 5, 1314–1325 [DOI] [PubMed] [Google Scholar]
- 24.Bonenfant D., Towbin H., Coulot M., Schindler P., Mueller D. R., van Oostrum J. (2007) Analysis of dynamic changes in post-translational modifications of human histones during cell cycle by mass spectrometry. Mol. Cell. Proteomics 6, 1917–1932 [DOI] [PubMed] [Google Scholar]
- 25.Hake S. B., Garcia B. A., Duncan E. M., Kauer M., Dellaire G., Shabanowitz J., Bazett-Jones D. P., Allis C. D., Hunt D. F. (2006) Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem 281, 559–568 [DOI] [PubMed] [Google Scholar]
- 26.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 27.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 28.Cloos P. A., Christensen J., Agger K., Helin K. (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22, 1115–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasini D., Bracken A. P., Hansen J. B., Capillo M., Helin K. (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol 27, 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery N. D., Yee D., Chen A., Kalantry S., Chamberlain S. J., Otte A. P., Magnuson T. (2005) The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol 15, 942–947 [DOI] [PubMed] [Google Scholar]
- 31.Ezhkova E., Pasolli H. A., Parker J. S., Stokes N., Su I. H., Hannon G., Tarakhovsky A., Fuchs E. (2009) Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell 136, 1122–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracken A. P., Helin K. (2009) Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 9, 773–784 [DOI] [PubMed] [Google Scholar]
- 33.Hansen K. H., Bracken A. P., Pasini D., Dietrich N., Gehani S. S., Monrad A., Rappsilber J., Lerdrup M., Helin K. (2008) A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol 10, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 34.Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthiesen R., Trelle M. B., Højrup P., Bunkenborg J., Jensen O. N. (2005) VEMS 3.0: algorithms and computational tools for tandem mass spectrometry based identification of post-translational modifications in proteins. J. Proteome Res 4, 2338–2347 [DOI] [PubMed] [Google Scholar]
- 36.Mikesh L. M., Ueberheide B., Chi A., Coon J. J., Syka J. E., Shabanowitz J., Hunt D. F. (2006) The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 1764, 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleinnijenhuis A. J., Kjeldsen F., Kallipolitis B., Haselmann K. F., Jensen O. N. (2007) Analysis of histidine phosphorylation using tandem MS and ion-electron reactions. Anal. Chem 79, 7450–7456 [DOI] [PubMed] [Google Scholar]
- 38.Pesavento J. J., Mizzen C. A., Kelleher N. L. (2006) Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem 78, 4271–4280 [DOI] [PubMed] [Google Scholar]
- 39.Phanstiel D., Brumbaugh J., Berggren W. T., Conard K., Feng X., Levenstein M. E., McAlister G. C., Thomson J. A., Coon J. J. (2008) Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A 105, 4093–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruthenburg A. J., Li H., Patel D. J., Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol 8, 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia B. A., Pesavento J. J., Mizzen C. A., Kelleher N. L. (2007) Pervasive combinatorial modification of histone H3 in human cells. Nat. Methods 4, 487–489 [DOI] [PubMed] [Google Scholar]
- 42.Morris S. A., Rao B., Garcia B. A., Hake S. B., Diaz R. L., Shabanowitz J., Hunt D. F., Allis C. D., Lieb J. D., Strahl B. D. (2007) Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem 282, 7632–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tie F., Banerjee R., Stratton C. A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M. O., Scacheri P. C., Harte P. J. (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters A. H., Kubicek S., Mechtler K., O'Sullivan R. J., Derijck A. A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., Martens J. H., Jenuwein T. (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 [DOI] [PubMed] [Google Scholar]
- 46.Zhao X. D., Han X., Chew J. L., Liu J., Chiu K. P., Choo A., Orlov Y. L., Sung W. K., Shahab A., Kuznetsov V. A., Bourque G., Oh S., Ruan Y., Ng H. H., Wei C. L. (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1, 286–298 [DOI] [PubMed] [Google Scholar]
- 47.Pauler F. M., Sloane M. A., Huang R., Regha K., Koerner M. V., Tamir I., Sommer A., Aszodi A., Jenuwein T., Barlow D. P. (2009) H3K27me3 forms BLOCs over silent genes and intergenic regions and specifies a histone banding pattern on a mouse autosomal chromosome. Genome Res 19, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.