Abstract
The assembly of infectious virus particles is a complex event. For human cytomegalovirus (HCMV) this process requires the coordinated expression and localization of at least 60 viral proteins that comprise the infectious virion. To gain insight into the mechanisms controlling this process, we identified protein binding partners for two viral proteins, pUL99 (also termed pp28) and pUL32 (pp150), which are essential for HCMV virion assembly. We utilized HCMV strains expressing pUL99 or pUL32 carboxyl-terminal green fluorescent protein fusion proteins from their native location in the HCMV genome. Based on the presence of ubiquitin in the pUL99 immunoisolation, we discovered that this viral protein colocalizes with components of the cellular endosomal sorting complex required for transport (ESCRT) pathway during the initial stages of virion assembly. We identified the nucleocapsid and a large number of tegument proteins as pUL32 binding partners, suggesting that events controlling trafficking of this viral protein in the cytoplasm regulate nucleocapsid/tegument maturation. The finding that pUL32, but not pUL99, associates with clathrin led to the discovery that the two viral proteins traffic via distinct pathways during the early stages of virion assembly. Additional investigation revealed that the majority of the major viral glycoprotein gB initially resides in a third compartment. Analysis of the trafficking of these three viral proteins throughout a time course of virion assembly allowed us to visualize their merger into a single large cytoplasmic structure during the late stages of viral assembly. We propose a model of HCMV virion maturation in which multiple components of the virion traffic independently of one another before merging.
Viruses are obligate intracellular parasites that rely on the host cell for many of the processes essential for their replication. Viruses subvert cellular pathways detrimental to virus growth and hijack complex cellular machinery necessary for efficient viral replication. This complex relationship with the host cell is born of necessity; viruses contain relatively little genetic material, which frequently encodes for a limited number of proteins. Understanding the molecular events at the interface of the virus-host relationship is crucial for the identification of targets for future therapeutics.
One of the most complex processes in the viral life cycle is the assembly of infectious viral particles. Multiple events must be coordinated in time and space for the correct incorporation of the viral genetic material into an infectious unit, or virion. For example, the virions of herpesviruses are composed of three essential elements: (i) the viral DNA and its surrounding protein shell, which together form the nucleocapsid, (ii) a layer of proteins surrounding the nucleocapsid termed the tegument, and (iii) a lipid bilayer, or envelope, surrounding the tegumented nucleocapsid in which viral glycoproteins are embedded (1–3). Although the formation of the nucleocapsid occurs in the nucleus, the herpesvirus tegument is acquired as the nucleocapsid moves into the cytoplasm through an undefined mechanism (2, 3). Once in the cytoplasm, the tegumented nucleocapsid acquires its envelope, again through a poorly defined event.
Although it has been known for some time that multiple HCMV1 proteins important for viral assembly localize to a juxtanuclear site termed the assembly zone, only recently have cellular markers for the HCMV assembly zone been described (4–7). This work has led to the view that HCMV virion maturation occurs by progressing through the endoplasmic reticulum to the Golgi apparatus (4, 5) and then via the ESCRT pathway to its site of release into the extracellular space (8). This work represents a major step forward in our understanding of the spatial requirements for virion maturation. However, the molecular events underlying this process remain poorly defined.
The assembly of HCMV particles is a process that has limited homology to cellular events. Therefore, it comes as no surprise that HCMV encodes proteins that are essential for the orchestration of virion assembly. One such protein, pUL99 (also termed pp28), is encoded by the UL99 ORF and resides in the cytoplasm of infected cells (9). pUL99 is required for the envelopment of capsids to generate virions (10, 11). In its absence, tegumented capsids accumulate in the cytoplasm of infected cells (11). This function may require its interaction with the cellular chaperone BiP as pUL99 binds to BiP during infection and disruption of BiP function was shown to inhibit HCMV assembly (12). Despite its critical role in envelopment, pUL99 is dispensable for cell-to-cell spread of HCMV, a process that does not require enveloped nucleocapsids (13).
Another HCMV protein integral to the assembly process is pUL32 (also termed pp150), encoded by the UL32 ORF. Like pUL99, this protein is required for the production of HCMV virions (14–17). pUL32 is capable of directly binding to the nucleocapsid, and this event occurs shortly after nucleocapsid assembly (18). Despite its localization to both the nucleus and the cytoplasm during infection (19, 20), pUL32 acts primarily in the cytoplasm where it may affect the localization and stability of the nucleocapsid (16, 21). In the absence of this viral protein, nucleocapsids are found primarily in the nucleus with few viral particles in the cytoplasm (21).
To generate additional insights into the molecular events controlling HCMV virion assembly, we used a targeted genetic-proteomic approach to identify protein binding partners for pUL99 and pUL32 in the context of viral infection. This work led to the identification of multiple distinct steps in the virion maturation process. The two viral proteins traffic through two separate compartments at the early stages of viral assembly. Although the tegument and nucleocapsid traffic with pUL32 in a clathrin-associated, brefeldin-insensitive vesicle, pUL99 localizes to a distinct brefeldin-sensitive structure that colocalizes with markers of the ESCRT trafficking pathway. The viral envelope glycoprotein B (gB), which is encoded by the UL55 ORF, did not substantially colocalize with pUL99 or pUL32 at early stages of the assembly process. However, we demonstrate that as infection progresses in time these three distinct compartments merge into a single vesicle, which may represent the final site of virion assembly.
EXPERIMENTAL PROCEDURES
Cells, Viruses, and Reagents
Primary human foreskin fibroblasts were used between passages 5 and 15 for all infections. Human foreskin fibroblasts were cultured in medium (Dulbecco's modified Eagle's medium) containing 10% newborn calf serum. The laboratory-adapted wild-type strain AD169, BADwt (23), was used to infect fibroblasts in 150-cm dishes except as indicated. For large scale immunoisolations, cells were infected for 1 h in 5 ml of Dulbecco's modified Eagle's medium + 10% newborn calf serum/dish. After 1 h, the inoculum was removed and replaced with fresh medium.
Recombinant Virus Construction
We used linear recombination in Escherichia coli (14, 22) to generate the BADinUL99GFP and BADinUL32GFP viruses in the BADwt (23) background. Briefly, PCR was used to generate a targeting cassette for recombination. The template for PCR contained a GFP tag followed by a kanamycin cassette flanked by Frt sites. The underlined sequences target the PCR product for homologous recombination into the HCMV genome. The following primers were used: UL99GFP(F), 5′-TCGTGTACACGGCCGGGGAGGGCGACGTGGTACAGATGGTGGTCGTGGTCGCCGGAAGAAGATGGAAAAAG-3′; UL99GFP(R), 5′-ACTACAAAAAAAAAAGCTGAACATGGTCATCTAGCAGCAAAGTTCTCCTTCGTCGTGGAATGCCTTCG-3′; UL32GFP(F), 5′-GCCGTGCAGAACATCCTCCAAAAGATCGAGAAGATTAAGAACACGGAGGAAGCCGGAAGAAGATGGAAAAAG-3′; and UL32GFP(R), 5′-TCACTATCCGATGGTTTCATTAAAAAGTACGTCTGCGTGTGTGTTTCTTAACGTCGTGGAATGCCTTCG-3′. The PCR product was transformed into recombination-competent DY380 E. coli, and chloramphenicol (CamR)- and kanamycin (KanR)-resistant bacteria were selected. Chloramphenicol resistance is provided by the BAC backbone. BAC DNA was purified from CamR, KanR colonies and checked for gross rearrangements by restriction digest. Recombinant BAC DNA was transformed into DH10B to alleviate concerns regarding spurious recombination in DY380. The kanamycin resistance cassette was removed by induction of flp recombinase with arabinose for 1 h at 37 °C. Bacteria were screened for loss of KanR and retention of CamR.
Immunoaffinity Purification of Virus-Host Protein Complexes
Fibroblasts were infected with BADinUL99GFP or BADinUL32GFP at a multiplicity of 3 pfu/cell. Approximately 1 g of infected cells was used per isolation (approximately 30 150-mm dishes). After 72 h, the cells were harvested and frozen in liquid nitrogen as small pellets as described previously (22). Cryogenic lysis was performed using a Retch MM301 Mixer Mill (Retch, Newtown, PA) as described (24, 25), and the resulting frozen cell powders were suspended in an optimized lysis buffer (20 mm K-HEPES, pH 7.4, 0.1 m potassium acetate, 2 mm MgCl2, 0.1% (v/v) Tween 20, 1 μm ZnCl2, 1 μm CaCl2, 1% Triton, 250 mm NaCl, 1:100 (v/v) protease inhibitor mixture (Sigma)) to yield the cell lysate. Anti-GFP rabbit polyclonal antibodies were prepared, affinity-purified, and conjugated as 5 μg of antibodies/mg of M-270 epoxy magnetic beads (Invitrogen Dynal) as described previously (24). 7 mg of conjugated beads were utilized for each immunoaffinity purification. Affinity purifications were performed by 1-h incubation of the cell lysate with magnetic beads at 4 °C, and the isolated complexes were eluted with 700 μl of a freshly made 0.5 n NH4OH, 0.5 mm EDTA solution, frozen in liquid nitrogen, and dried using vacuum centrifugation (SpeedVac Plus SC110A with gel pump GP110, Thermo Scientific) as described (24). The dried sample pellets were suspended in 30 μl of SDS-PAGE buffer with reducing agent (Invitrogen), incubated at 70 °C for 10 min, alkylated with 100 mm iodoacetamide (30 min at room temperature in dark), resolved by one-dimensional gel electrophoresis on a 4–12% NuPAGE Novex Bis-Tris gel (Invitrogen), and stained with SimplyBlueTM SafeStain (Invitrogen).
Mass Spectrometry Analyses of Co-isolated Proteins
To identify the virus and host proteins isolated with pUL99 and pUL32, the entire gel lanes from each isolation were cut into ∼30 slices, diced, and prepared for mass spectrometric analyses as described (26). Briefly, the samples were destained for 10 min in 200 μl of 50% (v/v) acetonitrile in 50 mm ammonium bicarbonate, dehydrated in 100% acetonitrile, and digested with 12.5 ng/μl sequencing grade modified trypsin (Promega, Madison, WI). The resulting peptides were bound to reverse phase resin (POROS 20 R2, Applied Biosystems, Foster City, CA), collected on a ZipTip with C18 resin (Millipore, Billerica, MA), washed twice with 20 μl of 0.1% (v/v) TFA, and eluted with 25 mg/ml 2,5-dihydroxybenzoic acid in 70% (v/v) acetonitrile and 0.1% (v/v) TFA directly onto a magnetic MALDI target (27). Mass spectrometric analyses were performed on a MALDI LTQ Orbitrap XL mass spectrometer (Thermo Electron, Bremen, Germany) (28) as described (26). Instrument calibration was performed by external calibration using standard peptide mixtures for normal and high mass ranges (ProteoMassTM calibration kit, Sigma) as described (26). The instrument was operated in positive mode, and the laser energy set to 15–20 μJ. MALDI LTQ Orbitrap MS analyses were acquired for a mass range of m/z 700–4000 with the following parameters: resolution setting, 60,000 at m/z 400; automated spectrum filter off; 50 scans/step; automated gain control on; allowing storage of 5e5 ions. MS/MS analyses were carried out by MALDI ion trap CID as described (26) using precursor ion isolation in the linear ion trap, a 2-mass unit isolation width, a normalized collision energy of 33%, an activation q of 0.25, and an activation time of 30–300 ms. The resulting MS and MS/MS spectra were inspected and processed using Qual Browser (version 2.0.7, Thermo Fisher Scientific). For the MS data, the list of singly charged monoisotopic masses was generated using Xtract within Qual Browser (XCalibur, version 2.0.7) with the following parameters: MH+, m/z 700–4000 mass range; resolution, 60,000 at m/z 400; and signal-to-noise threshold of peak picking, 2. The lists of putative proteins were obtained by database searching against the National Center for Biotechnology Information nonredundant protein database, version October 16, 2006, using the computer algorithm XProteo. The limited number of co-isolated proteins allowed us to manually inspect the MS/MS spectra and confirm the peptide sequences of the candidate proteins via assessment of the b and y ions and preferential fragmentations at the carboxyl terminus of Asp and Glu or amino terminus of Pro residues (29–32). Search parameters for MS data were: species, viruses (346,953 sequences) and Homo sapiens (152,010 sequences); protein mass, 0–500 kDa; protein pI, 1–14; mixture search, auto; number of candidates displayed, 50; enzyme, trypsin; miscleavages, 1; mass type, monoisotopic; charge state, MH+; mass tolerance, 5 ppm; fixed modification, carbamidomethylation of Cys; and variable modification, oxidation of Met. Additional parameters used for database searching using MS/MS data were: precursor tolerance, 5 ppm; fragment tolerance, 0.5 Da; and instrument: MALDI_I_TRAP. The proteins isolated with UL99 and UL32, their gi numbers, number of peptides detected, protein sequence coverage (%), XProteo scores (d′) following database searching, and the peptide sequences confirmed by MS/MS analyses are given in supplemental Tables S1 and S2. Peptide masses corresponding to trypsin autolysis were excluded. Only the proteins confirmed via the fragmentation of at least two peptides are included in the tables. The XProteo probability score indicates the discriminability (d′) for each candidate protein as a normalized distance between the distribution of the candidate protein score and that of randomly matched proteins (in units of standard deviation) based on an updated version of the ProFound Bayesian algorithm (33). A score of d′ = 4 represents a true positive rate of 0.99 and a false positive rate of 0.05.
Immunofluorescence Microscopy
Fibroblasts were plated on coverslips in 6-well plates. Cells were infected with HCMV at 0.1 pfu/cell. At the indicated times postinfection, cells were washed in PBS, fixed for 15 min in 2% paraformaldehyde, washed again in PBS, and then permeabilized for 15 min in 0.1% Triton X-100. After washing with PBS-T (PBS with 0.2% Tween 20), the cells were incubated for 1 h in PBS-T containing 2% bovine serum albumin. Cells were stained with either a mouse or rabbit primary antibody in PBS-T with 2% bovine serum albumin for 1 h at room temperature. After further washing with PBS-T, slides were incubated for 1 h at room temperature with either goat anti-mouse or goat-anti rabbit secondary antibody conjugated to Alexa Fluor 546 or 633 (Molecular Probes). 4′,6′-Diamidino-2-phenylindole dihydrochloride (DAPI; Molecular Probes) was added with the secondary antibody to visualize the nucleus. The following antibodies were used in this study: anti-pUL99 (11), anti-gB (34), anti-UL48 (35), anti-ubiquitin (Santa Cruz Biotechnology, Inc.), anti-Tsg101 (GeneTex), anti-Hrs1 (Santa Cruz Biotechnology, Inc.), and anti-TGN46 (Abcam). Cells were washed with PBS-T, mounted with Slow Fade (Molecular Probes), and viewed using a Zeiss LSM510 laser scanning microscope.
RESULTS
Targeted Proteomics Approach for Studying Virus-Host Associations
We have previously described a strategy that utilizes a GFP tag to both visualize the dynamic localization of viral proteins and isolate virus-host protein complexes at different times of infection. We successfully incorporated this approach for the study of cellular processes occurring during infections with Sindbis virus (36) and HCMV (22). To gain insights into processes occurring during HCMV virion assembly, we generated two viruses, BADinUL99GFP and BADinUL32GFP, containing the GFP coding region fused to the carboxyl terminus of the pUL99 and pUL32, respectively (Fig. 1A). Using this approach, the GFP-tagged proteins are expressed under the control of their native promoters, thereby ensuring that they are expressed at wild-type levels and with kinetics identical to that of the native protein.
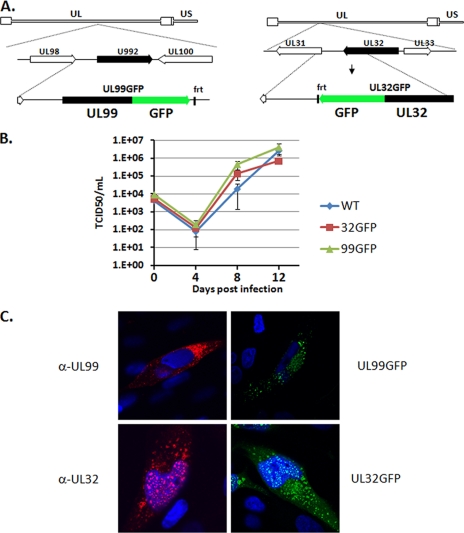
Fig. 1.
Construction and characterization of GFP-tagged HCMV recombinant viruses. A, schematic depiction of the UL99 and UL32 genomic loci of HCMV. GFP was fused to the carboxyl termini of the UL99 and UL32 ORFs to generate the BADinUL99GFP (left panel) and BADinUL32GFP (right panel) strains. B, growth kinetics and viral yield were measured over multiple rounds of viral replication for BADinUL99GFP, BADinUL32GFP, and parental BADwt. Fibroblasts were infected at a multiplicity of 0.1 pfu/cell. Supernatants were harvested at the indicated times, and virus in the supernatants was quantified by the median tissue culture infective dose (TCID50) method. Error bars indicate standard deviations from two experiments titered in duplicate. C, fibroblasts were infected with either the parental strain, BADinUL99GFP, or BADinUL32GFP. At 72 hpi, the localization of pUL99 and pUL32 was determined by immunofluorescence using either antibodies specific for the viral proteins or, in the case of the GFP fusion viruses, the fluorescent signal of the tagged viral protein. Nuclear DNA is stained blue with DAPI. WT, wild type.
Disruption of either the UL99 or UL32 ORF results in a nonviable virus (14–16, 21). Therefore, it was critical to confirm that inclusion of the GFP tag did not affect the growth rate of these strains. Transfection of the BAC DNAs for BADinUL99GFP or BADinUL32GFP into fibroblasts revealed that both viruses were capable of replicating. The minimal impact of the GFP epitope on the growth of these viruses was confirmed by analyzing virus growth over multiple rounds of infection (Fig. 1B). Both viruses replicated with equivalent kinetics and to similar levels as the wild-type strain, confirming that the inclusion of the epitope tag did not disrupt the essential functions of either pUL99 or pUL32 in HCMV assembly.
As an additional confirmation that the GFP epitope did not interfere with the role of the viral proteins during HCMV assembly, we compared the localization of the GFP-tagged and native forms of these proteins during HCMV infection. Using monoclonal antibodies specific for pUL99 and pUL32, we confirmed that the tagged and untagged versions of the viral proteins displayed similar patterns of localization (Fig. 1C). Namely, pUL99 was found exclusively in the cytoplasm, consistent with previous results, whereas pUL32 was found in both the cytoplasm and nucleus during HCMV infection.
Upon confirmation that BADinUL99GFP and BADinUL32GFP behaved like wild-type viruses, we next identified protein binding partners for these two proteins at 72 hpi, a time that marks an early stage of the viral assembly process. We chose this time based upon the observation that pUL99 and pUL32 localize to separate compartments at this time but merge into a single compartment at later times (see below). Fibroblasts were infected with BADinUL99GFP or BADinUL32GFP, cells were harvested 72 h later, and GFP-containing protein complexes were isolated on magnetic beads covalently coupled to anti-GFP antibody. Following extensive washing of the captured complexes, proteins were resolved by gel electrophoresis, the entire gel lanes were sliced, and the proteins in each gel slice were subjected to enzymatic digestion and analyzed by mass spectrometry. Figs. 2 and 3 indicate the proteins isolated with pUL99 and pUL32, respectively. A complete list of the identified proteins, sequence coverage, number of peptides, scores derived from database searching, and peptides confirmed by MS/MS analyses can be found in supplemental Tables S1 and S2.
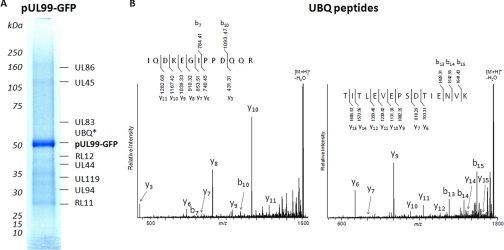
Fig. 2.
pUL99-interacting partners at 72 hpi. A, UL99GFP-interacting proteins were isolated by affinity purification, resolved by gel electrophoresis, and identified by sequential MS and MS/MS analyses. *, peptides corresponding to ubiquitin (UBQ) were detected between 50- and 75-kDa molecular mass markers, and therefore its exact location could not be determined. B, representative MALDI LTQ Orbitrap CID spectra of [M + H]+ ions of peptides corresponding to ubiquitin. Following CID, the generated fragment ions were detected at the linear trap level.
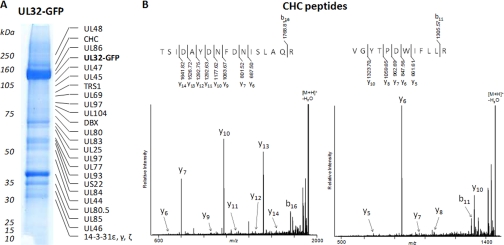
Fig. 3.
pUL32 associates with clathrin and components of viral tegument at 72 hpi. A, UL32-GFP-interacting proteins were isolated via affinity purifications, separated by gel electrophoresis, and identified by mass spectrometry. B, representative MALDI LTQ Orbitrap CID spectra of [M + H]+ ions of peptides corresponding to clathrin (CHC), obtained from the region indicated in A. Following CID, the generated fragment ions were detected at the linear trap level.
pUL99 and pUL32 were not isolated with each other in agreement with their distinct localization at 72 hpi (see Fig. 6). The majority of the proteins isolated with pUL99 and pUL32 are coded by the virus and have previously been shown to be components of the HCMV virion (37). Components of all three elements of the virion (nucleocapsid, tegument, and envelope) were identified in the isolations. This confirmed that the lysis conditions used were suitable for extraction of both lipid-embedded proteins and nucleocapsids. We have not yet confirmed the large number of interactions among virus proteins predicted by this analysis.
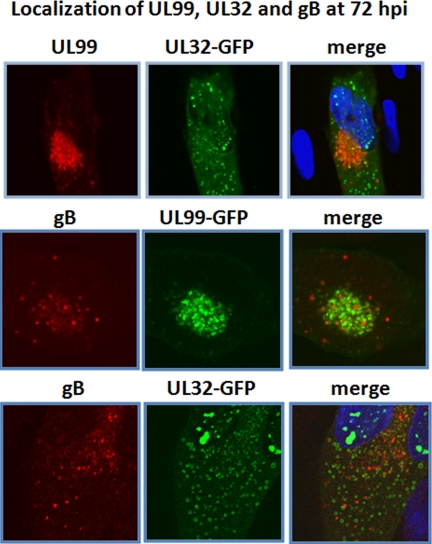
Fig. 6.
HCMV pUL99, pUL32, and gB reside in separate compartments at early stages of viral assembly. Primary human fibroblasts were infected with the indicated virus. 72 h later, cells were fixed and stained with the indicated antibodies. Nuclear DNA is stained blue with DAPI.
pUL99 Colocalizes with Components of ESCRT Pathway
Several cellular proteins were identified in the pUL99 immunoisolation, including peptides corresponding to ubiquitin. Although ubiquitination of proteins is often associated with degradation, ubiquitin modification is also associated with specific trafficking events. Ubiquitinated proteins are sorted and trafficked by the ESCRT pathway (38–40). Ubiquitin-binding proteins in this pathway bind to and initiate a set of vesicular transport events that coordinate traffic from the endoplasmic reticulum/Golgi with components of the endosomal network. Elements of the ESCRT pathway have recently been shown to be important for the assembly of HCMV virions (8). Together these data led us to hypothesize that pUL99 traffics through the ESCRT pathway.
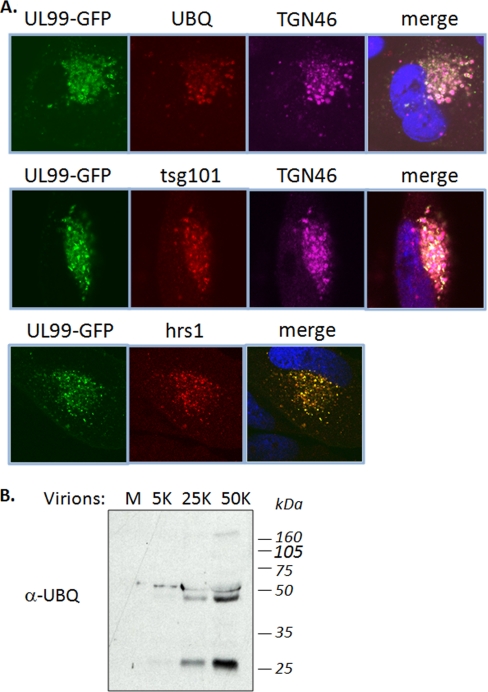
To test whether pUL99 trafficking utilizes the ESCRT pathway, we compared its localization with two ubiquitin-binding proteins of the pathway, Tsg101 and Hrs. Tsg101 is a member of the ESCRT-I complex, and it is the mammalian homologue of the vacuolar protein sorting protein Vps23, which together with Vps28 and Vps27 forms the ESCRT-I protein complex in Saccaromyces cerevisiae. Hrs is the mammalian homologue of Vps27 and is known to be required in the sorting of proteins in the multivesicular body pathway. Both proteins have been implicated in the selection of cargo proteins into the ESCRT pathway by virtue of their ubiquitin binding domains (41, 42). At 72 hpi, pUL99 localized with cytoplasmic ubiquitin in the viral assembly zone during HCMV infection, and we observed substantial overlap in the localization of pUL99 with both Tsg101 and Hrs (Fig. 4A). Furthermore, our analyses of virions by Western blot using an antibody specific to ubiquitin indicated the presence of several putative ubiquitinated proteins (Fig. 4B), including a prominent band consistent with the molecular mass of the posttranslationally modified pUL99 at ∼28 kDa. The absence of Tsg101 and Hrs in our mass spectrometric analyses of pUL99 interactions may be a result of the stringent conditions that we had to utilize to be able to isolate pUL99 from vesicles. Indeed, Tsg101 and Hrs are known to bind ubiquitin via multiple low affinity binding domains. Although determining the identity of the viral proteins that are ubiquitinated requires further study, our findings nevertheless suggest that pUL99 might utilize the ESCRT pathway to facilitate trafficking of viral proteins through the assembly process and provide a potential molecular mechanism behind the observation that the ESCRT pathway is important for HCMV virion assembly (8).
Fig. 4.
HCMV pUL99 colocalizes with ubiquitin-binding proteins of ESCRT pathway. A, at 72 hpi, UL99-GFP colocalized with ubiquitin (UBQ), Tsg101, Hrs1, and TGN46. Nuclear DNA is stained blue with DAPI. TGN46 is a cellular marker of the trans-Golgi complex, previously shown to colocalize with pUL99 during viral assembly. B, Western blot analysis of virion preparations indicating the presence of ubiquitinated proteins at ∼28, 50, and 160 kDa. M, medium alone; 5K, 25K, and 50K, the number (×1000) of infectious virions loaded in each lane.
pUL32 and pUL99 Traffic in Different Pathways
Our analysis pointed to several new interactions between pUL32 and cellular proteins. Of particular interest is the apparent association with the clathrin heavy chain (Fig. 3). Clathrin-coated vesicles originate in multiple locations in the cell, and their formation has long been associated with vesicular trafficking (43). The point of origin of clathrin-coated vesicles can be determined by using the drug brefeldin A (44). Brefeldin A disrupts clathrin-coated vesicle formation from Golgi, whereas it has no effect on clathrin-coated vesicles formed elsewhere in the cell. Clathrin has been found to be required for late events in the HCMV life cycle (45, 46) as treatment with brefeldin A inhibits envelopment. In addition, clathrin is found in highly purified extracellular virions (37). Together these results suggested that HCMV virion assembly involves clathrin-coated vesicle formation from the endoplasmic reticulum or Golgi. We therefore hypothesized that the pUL32-clathrin interaction was important for viral assembly.
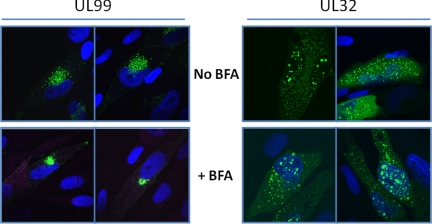
We treated HCMV-infected cells with brefeldin A to assess the impact of inhibition of clathrin-mediated trafficking on pUL99 and pUL32 localization. Treatment with brefeldin A did not result in a significant change in the localization of pUL32, consistent with previous results (6), but it markedly altered the localization of pUL99 (Fig. 5). The protein remained near the nucleus, but it was limited to a more compact domain in the presence of drug. This experiment also confirmed that the two proteins localize quite differently at 72 hpi. These results demonstrate that pUL99 and pUL32 traffic independently of one another during the initial stages of virus assembly: pUL99 utilizes a brefeldin-sensitive pathway, and pUL32 travels a drug-insensitive path. Rather than progressing directly from the nucleus to the assembly zone where pUL99 is located, pUL32 traffics through a clathrin-associated, brefeldin-insensitive pathway.
Fig. 5.
Brefeldin A sensitivity defines pUL99 and pUL32 as residing in separate vesicles at 72 hpi. Fibroblasts were infected with either BADinUL99GFP or BADinUL32GFP. 3 h prior to fixation, cells were treated with brefeldin A (BFA; 3 μg/ml) and fixed, and localization of the pUL99 and pUL32 proteins was determined. Nuclear DNA is stained blue with DAPI.
Nucleocapsids and Tegument Constituents Traffic with pUL32
Examination of the viral proteins predicted to be associated with pUL32 provided additional insight into the process of viral assembly. All of the components of the HCMV nucleocapsid were present among the predicted pUL32 binding partners (supplemental Table S2). This is not surprising given previous reports describing the interaction of pUL32 with the HCMV major capsid protein (18). In addition to major capsid protein, we identified the capsid proteins pUL46, pUL85, and UL80.5, arguing that pUL32 traffics with nucleocapsids. These proteins were not identified in the pUL99 immunoisolation (supplemental Table S1). Numerous tegument proteins (e.g. UL48, TRS1, UL69, UL97, UL25, and US22) were also isolated with pUL32. Coupled with the finding that pUL99 and pUL32 traffic independently through the cytoplasm during the early stages of virion assembly, these results define two independent trafficking events required for the assembly of HCMV virions.
gB Initially Traffics in a Third Compartment, Separate from pUL99 and pUL32
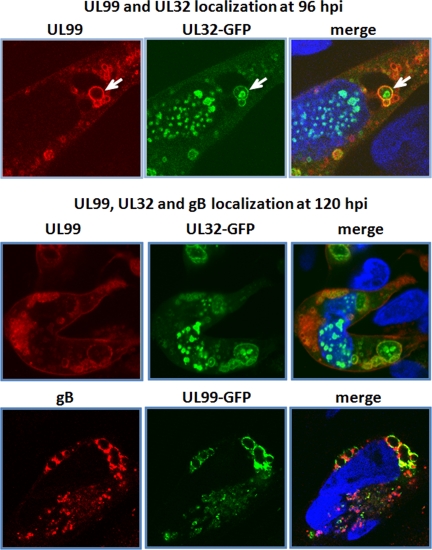
Although we recovered numerous glycoproteins in our isolations, the major viral glycoprotein gB was not detected. This might reflect the fact that isolation of the membrane glycoprotein was impeded by solubility issues. Alternatively, gB may reside in a separate location from the pUL99 and pUL32 compartments described above. To distinguish between these possibilities, we determined the localization of gB in comparison with pUL99 and pUL32 at 72 hpi. In a control experiment, pUL99 and pUL32 were again found to reside in different compartments (Fig. 6, top panels). Although gB exhibited partial colocalization with both pUL99 (Fig. 6, middle panels) and pUL32 (Fig. 6, bottom panels), the vast majority of the gB was localized in distinct small puncta throughout the cytoplasm. These images argue that gB, pUL99, and pUL32 begin the process of viral assembly in three distinct cytoplasmic compartments.
Spatial and Temporal Regulation of Virion Assembly
The finding that three virion proteins reside primarily in separate locations in the cytoplasm at the early stages of the assembly process suggested that these domains likely merge to form a functional assembly compartment. Indeed, at 96 and 120 hpi, we observed numerous instances of vesicles containing either pUL99 plus pUL32 (Fig. 7, top panels) or pUL99 plus gB (Fig. 7, bottom panels). pUL32, pUL99, and gB were found to colocalize along the periphery of these large vesicles, and structures positive for single antigens were often found in the lumen of some large vesicles. In particular, we observed numerous intralumenal structures containing pUL32 (Fig. 7, top panel, arrow). The exact nature of these structures remains unknown.
Fig. 7.
pUL32-, pUL99-, and gB-containing compartments merge as infection progresses. Infected fibroblasts were assayed for the indicated proteins at 96 (top panels) or 120 hpi (bottom panels). Nuclear DNA is stained blue with DAPI. The white arrow points to an example of pUL99 and pUL32 vesicles in the process of merging.
DISCUSSION
pUL99 and pUL32 Associations at 72 hpi
We present here, to our knowledge, the first targeted proteomics study of virus-host protein interactions occurring during HCMV virion maturation. We focused our analyses on pUL99 and pUL32, two viral proteins known to be essential to the virion assembly process. The use of GFP-tagged pUL99 and pUL32 in the context of the viral genome allowed us to integrate information regarding their localizations and interactions during the progression of virion assembly. Using cryogenic cell lysis, affinity purifications on magnetic beads conjugated with anti-GFP antibodies, and mass spectrometry, we identified numerous novel interactions of pUL99 and pUL32 (Figs. 2 and 3). These analyses also confirmed the previously reported interaction of pUL32 with major capsid protein (18). The co-isolations of these virus and host proteins are not definitive proof of direct interactions as these associations may be the result of indirect as well as some potential nonspecific interactions. Noteworthy, the majority of the proteins identified in the isolates of pUL99 and pUL32 are known to be present in cell-free virions (37), representing components of the envelope, tegument, and capsid. In agreement with our observation of the distinct localization of pUL99 and pUL32 at 72 hpi (Figs. 5 and 6), the two proteins were not isolated with each other. Furthermore, the overlap between the viral proteins isolated with pUL99 and pUL32 was limited. In addition to providing temporal interaction maps, these studies also served as a demonstration of the expression of certain viral genes that have not been previously reported, such as RL12.
Temporal Regulation of Intermediate Virion Assemblies
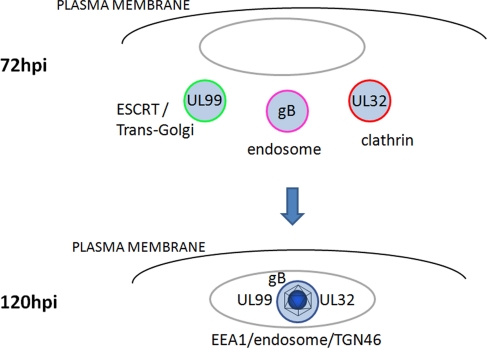
Altogether, the observed associations led to insights into intermediate assemblies of tegument, envelope, and capsid components prior to forming the mature virions (Fig. 8). In particular, the presence of two host proteins, ubiquitin and clathrin in isolates of pUL99 and pUL32, respectively, led us to probe for the presence of parallel distinct processes. We demonstrated that at 72 hpi pUL99 colocalized with ubiquitin as well as Hrs and Tsg101 (Fig. 4A), two ubiquitin-binding proteins of the ESCRT pathway. This observation is in agreement with previous reports demonstrating that the ESCRT pathway is utilized by many RNA (e.g. human immunodeficiency virus, type 1 (47)) and several DNA viruses (including HCMV (8), herpes simplex virus 1 (48), and likely HHV6 (49)) during the envelopment process. Our results indicated the presence of another distinct process occurring in parallel at 72 hpi as pUL32 associated with clathrin and localized in cytoplasmic compartments insensitive to brefeldin A treatment around the periphery of the viral assembly zone. As a third differentially localized process at 72 hpi, the major viral glycoprotein gB localized predominantly to another distinct compartment. These results led us to propose that at least three distinct parallel processes occur at 72 hpi: (i) ubiquitin-mediated ESCRT trafficking of pUL99, (ii) pUL32 trafficking through clathrin-associated vesicles from a non-Golgi source, and (iii) gB trafficking in yet a third compartment (Fig. 8). These distinct compartments proceed to merge as the infection progresses as visualized at 96 and 120 hpi (Figs. 7 and 8), concomitant with an increase in the resulting vesicle size (>2 μm at 120 hpi). These results represent the first observation that the final virion membrane may be derived from the fusion of these three distinct vesicles into a single membrane. The resulting large vesicle may represent the final site of virion assembly.
Fig. 8.
Proposed model of trafficking events involved in HCMV virion assembly.
These results initiate numerous interesting follow-up questions. What drives the fusion of these three vesicles? Regulation of vesicular traffic in the cell is a complex process, involving coat proteins, GTPases, and proteins that regulate vesicular fusion (e.g. soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins) (50). It is unclear which molecular mechanisms are utilized to direct the transport or fusion of vesicles involved in HCMV virion maturation. The differential sensitivity to brefeldin A indicates that distinct types of vesicular transport are involved in virion assembly. However, additional experiments targeted at identifying vesicular components that direct these events are necessary for a complete understanding of these processes. Another question raised by these findings is whether these parallel trafficking events are essential for the production of the mature virion. Although the three observed vesicles may be necessary for the maturation of the virion, these processes may also represent co-evolved alternative mechanisms. A detailed understanding of HCMV virion assembly clearly requires further work on other HCMV virion proteins and associated host factors. This study provides important insights into virion assembly and suggests the existence of parallel processes occurring during virion maturation.
Supplementary Material
Acknowledgments
We thank members of the Cristea and Shenk laboratories for helpful advice, comments, and criticisms. We also thank Joe Goodhouse for assistance with microscopy (Microscopy Core Facility at Princeton University). We also thank the many scientists in both the HCMV and proteomics field upon whose work this study builds.
* This work was supported, in whole or in part, by National Institutes of Health Grant DP1DA026192 from the National Institute on Drug Abuse (to I. M. C.) and Grants CA85786 and CA82396 (to T. S.). This work was also supported by Princeton University start-up funding (to I. M. C.) and an American Cancer Society Postdoctoral Fellowship PF-07073-01-MBC (to N. J. M.).
 This article contains supplemental Tables S1 and S2.
This article contains supplemental Tables S1 and S2.
1 The abbreviations used are:
- HCMV
- human cytomegalovirus
- ESCRT
- endosomal sorting complex required for transport
- GFP
- green fluorescence protein
- BAC
- bacterial artificial chromosome
- pfu
- plaque-forming unit(s)
- hpi
- hours postinfection
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- LTQ
- linear trap quadrupole
- DAPI
- 4′,6′-diamidino-2-phenylindole dihydrochloride
- BiP
- immunoglobulin heavy chain binding protein
- gB
- glycoprotein B.
REFERENCES
- 1.Britt W. J., Boppana S. (2004) Human cytomegalovirus virion proteins. Hum. Immunol 65, 395–402 [DOI] [PubMed] [Google Scholar]
- 2.Gibson W. (2008) Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol 325, 187–204 [DOI] [PubMed] [Google Scholar]
- 3.Kalejta R. F. (2008) Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev 72, 249–265, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das S., Vasanji A., Pellett P. E. (2007) Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol 81, 11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Homman-Loudiyi M., Hultenby K., Britt W., Söderberg-Nauclér C. (2003) Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol 77, 3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez V., Greis K. D., Sztul E., Britt W. J. (2000) Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol 74, 975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo J. Y., Britt W. J. (2007) Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J. Virol 81, 6536–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tandon R., AuCoin D. P., Mocarski E. S. (2009) Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol 83, 10797–10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy E., Yu D., Grimwood J., Schmutz J., Dickson M., Jarvis M. A., Hahn G., Nelson J. A., Myers R. M., Shenk T. E. (2003) Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U.S.A 100, 14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones T. R., Lee S. W. (2004) An acidic cluster of human cytomegalovirus UL99 tegument protein is required for trafficking and function. J. Virol 78, 1488–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva M. C., Yu Q. C., Enquist L., Shenk T. (2003) Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol 77, 10594–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchkovich N. J., Maguire T. G., Paton A. W., Paton J. C., Alwine J. C. (2009) The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the HCMV assembly compartment. J. Virol 83, 11421–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva M. C., Schröer J., Shenk T. (2005) Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc. Natl. Acad. Sci. U.S.A 102, 2081–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu D., Silva M. C., Shenk T. (2003) Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U.S.A 100, 12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn W., Chou C., Li H., Hai R., Patterson D., Stolc V., Zhu H., Liu F. (2003) Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U.S.A 100, 14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tandon R., Mocarski E. S. (2008) Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J. Virol 82, 9433–9444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer H. H., Ripalti A., Landini M. P., Radsak K., Kern H. F., Hensel G. M. (1997) Human cytomegalovirus late-phase maturation is blocked by stably expressed UL32 antisense mRNA in astrocytoma cells. J. Gen. Virol 78, 2621–2631 [DOI] [PubMed] [Google Scholar]
- 18.Baxter M. K., Gibson W. (2001) Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J. Virol 75, 6865–6873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensel G., Meyer H., Gärtner S., Brand G., Kern H. F. (1995) Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol 76, 1591–1601 [DOI] [PubMed] [Google Scholar]
- 20.Sampaio K. L., Cavignac Y., Stierhof Y. D., Sinzger C. (2005) Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J. Virol 79, 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.AuCoin D. P., Smith G. B., Meiering C. D., Mocarski E. S. (2006) Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J. Virol 80, 8199–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moorman N. J., Cristea I. M., Terhune S. S., Rout M. P., Chait B. T., Shenk T. (2008) Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3, 253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu D., Smith G. A., Enquist L. W., Shenk T. (2002) Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol 76, 2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cristea I. M., Williams R., Chait B. T., Rout M. P. (2005) Fluorescent proteins as proteomic probes. Mol. Cell. Proteomics 4, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 25.Cristea I. M., Chait B. T. (2009) Affinity purification of protein complexes, in Proteomics: a Cold Spring Harbor Laboratory Course Manual (Link A. J., LaBaer J. eds.) pp. 48–54, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Y., Li T., Yu F., Kramer T., Cristea I. M. (2010) Resolving the composition of protein complexes using a MALDI LTQ Orbitrap. J. Am. Soc. Mass Spectrom 21, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blethrow J. D., Tang C., Deng C., Krutchinsky A. N. (2007) Modular mass spectrometric tool for analysis of composition and phosphorylation of protein complexes. PLoS ONE 2, e358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strupat K., Kovtoun V., Bui H., Viner R., Stafford G., Horning S. (2009) hybrid mass analyzer. J. Am. Soc. Mass. Spectrom, 1451–1463 [Google Scholar]
- 29.Qin J., Chait B. T. (1995) Preferential fragmentation of protonated gas-phase peptide ions adjacent to acidic amino acid residues. J. Am. Chem. Soc 117, 5411–5412 [Google Scholar]
- 30.Tsaprailis G., Nair H., Somogyi A., Wysocki V. H., Zhong W. Q., Futrell J. H., Summerfield S. G., Gaskell S. J. (1999) Influence of secondary structure on the fragmentation of protonated peptides. J. Am. Chem. Soc 121, 5142–5154 [Google Scholar]
- 31.Schilling B., Wang W., McMurray J. S., Medzihradszky K. F. (1999) Fragmentation and sequencing of cyclic peptides by matrix-assisted laser desorption/ionization post-source decay mass spectrometry. Rapid Commun. Mass Spectrom 13, 2174–2179 [DOI] [PubMed] [Google Scholar]
- 32.Breci L. A., Tabb D. L., Yates J. R., 3rd, Wysocki V. H. (2003) Cleavage N-terminal to proline: analysis of a database of peptide tandem mass spectra. Anal. Chem 75, 1963–1971 [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Chait B. T. (2000) ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem 72, 2482–2489 [DOI] [PubMed] [Google Scholar]
- 34.Britt W. J. (1984) Neutralizing antibodies detect a disulfide-linked glycoprotein complex within the envelope of human cytomegalovirus. Virology 135, 369–378 [DOI] [PubMed] [Google Scholar]
- 35.Bechtel J. T., Shenk T. (2002) Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J. Virol 76, 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristea I. M., Carroll J. W., Rout M. P., Rice C. M., Chait B. T., MacDonald M. R. (2006) Tracking and elucidating alphavirus-host protein interactions. J. Biol. Chem 281, 30269–30278 [DOI] [PubMed] [Google Scholar]
- 37.Varnum S. M., Streblow D. N., Monroe M. E., Smith P., Auberry K. J., Pasa-Tolic L., Wang D., Camp D. G., 2nd, Rodland K., Wiley S., Britt W., Shenk T., Smith R. D., Nelson J. A. (2004) Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol 78, 10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 39.Saksena S., Sun J., Chu T., Emr S. D. (2007) ESCRTing proteins in the endocytic pathway. Trends Biochem. Sci 32, 561–573 [DOI] [PubMed] [Google Scholar]
- 40.Slagsvold T., Pattni K., Malerød L., Stenmark H. (2006) Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol 16, 317–326 [DOI] [PubMed] [Google Scholar]
- 41.Katzmann D. J., Babst M., Emr S. D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 42.Shields S. B., Oestreich A. J., Winistorfer S., Nguyen D., Payne J. A., Katzmann D. J., Piper R. (2009) ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol 185, 213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth M. G. (2006) Clathrin-mediated endocytosis before fluorescent proteins. Nat. Rev. Mol. Cell Biol 7, 63–68 [DOI] [PubMed] [Google Scholar]
- 44.Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eggers M., Bogner E., Agricola B., Kern H. F., Radsak K. (1992) Inhibition of human cytomegalovirus maturation by brefeldin A. J. Gen. Virol 73, 2679–2692 [DOI] [PubMed] [Google Scholar]
- 46.Kregler O., Schilf R., Lander A., Bannert N., Bogner E. (2009) Brefeldin A inhibits expression of DNA packaging proteins and nucleocapsid formation of human cytomegalovirus. FEBS Lett 583, 1207–1214 [DOI] [PubMed] [Google Scholar]
- 47.Bieniasz P. D. (2009) The cell biology of HIV-1 virion genesis. Cell Host Microbe 5, 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pawliczek T., Crump C. M. (2009) Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol 83, 11254–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mori Y., Koike M., Moriishi E., Kawabata A., Tang H., Oyaizu H., Uchiyama Y., Yamanishi K. (2008) Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9, 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spang A. (2008) The life cycle of a transport vesicle. Cell. Mol. Life Sci 65, 2781–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.