Abstract
The inflammatory caspases, human caspases-1, -4, and -5, proteolytically modulate diverse physiological outcomes in response to proinflammatory signals. Surprisingly, only a few substrates are known for these enzymes, including other caspases and the interleukin-1 family of cytokines. To more comprehensively characterize inflammatory caspase substrates, we combined an enzymatic N-terminal enrichment method with mass spectrometry-based proteomics to identify newly cleaved proteins. Analysis of THP-1 monocytic cell lysates treated with recombinant purified caspases identified 82 putative caspase-1 substrates, three putative caspase-4 substrates, and no substrates for caspase-5. By contrast, inflammatory caspases activated in THP-1 cells by mimics of gout (monosodium urate), bacterial infection (lipopolysaccharide and ATP), or viral infection (poly(dA·dT)) were found to cleave only 27, 16, and 22 substrates, respectively. Quantitative stable isotope labeling with amino acids in cell culture (SILAC) comparison of these three inflammatory stimuli showed that they induced largely overlapping substrate profiles but different extents of proteolysis. Interestingly, only half of the cleavages found in response to proinflammatory stimuli were contained within our set of 82 in vitro cleavage sites. These data provide the most comprehensive set of caspase-1-cleaved products reported to date and indicate that caspases-4 and -5 have far fewer substrates. Comparisons between the in vitro and in vivo data highlight the importance of localization in regulating inflammatory caspase activity. Finally, our data suggest that inducers of inflammation may subtly alter caspase-1 substrate profiles.
Proteases are ubiquitous, representing more than 2% of the total human genome, and essential to diverse cellular processes, including catabolism, cell death, and immune function (1–3). Despite their biological importance, the identification of cellular protease substrates remains challenging. Historically, researchers gained insight into protease specificity by assaying enzyme activities against peptide libraries or displays of short peptide sequences on biomolecules (4–7). More recently, we and others have developed technologies to directly identify protease substrates using differential gel mobilities or by enriching for new protein N termini that result from protein cleavage (8–12). These proteomics methods have been applied to the widespread proteolysis that occurs during apoptosis to reveal hundreds of novel caspase substrates. Although this is an important demonstration of the power of these technologies, most proteolysis signaling pathways target a far more limited scope of substrates (13). In this study, we extended an enzymatic N-terminal enrichment technique to study the inflammatory caspases, a family of proteases that enact far more limited proteolysis.
The human caspases (cysteine aspartyl proteases) are a family of 12 homodimeric cysteine proteases that cleave proteins immediately following an aspartic acid residue. Sequence and functional similarities separate the caspases into three evolutionary groups: initiator caspases (caspases-2, -8, -9, and -10), primarily involved in integrating cellular signals and inducing apoptosis; executioner caspases (caspases-3, -6, and -7), the primary effectors of proteolysis during apoptosis; and inflammatory caspases (caspases-1, -4, and -5), discovered as activators of proinflammatory cytokines (14). The details of how inflammatory caspases exert their proinflammatory effects are not fully understood, but recent studies have provided insight into their mechanisms of activation. Proinflammatory stimuli induce multidomain proteins termed nucleotide binding and oligimerization domain (NOD)-like receptors to form multiprotein complexes called inflammasomes (15). Inflammasomes promote oligomerization of inflammatory caspases and self-activation. The diverse potential activators of inflammasomes include combinations of lipopolysaccharide and ATP, which mimic bacterial infections; inorganic crystals such as monosodium urate or calcium phosphate, which mimic gout; and the introduction of cytosolic dsDNA,1 which mimics viral infections (16–20). Although the mechanism of inflammatory caspase activation is an area of active research, the downstream consequences of caspase activation remain poorly understood. Caspase-1 is known to be the key activator of the Interleukin (IL)-1 family cytokines and caspase-7 during inflammation, but several key biological consequences of inflammatory caspase activation, including membrane biogenesis, cell death, and non-classical protein secretion, have not been directly linked to downstream molecular substrates (16, 21–25).
Pyroptosis, a form of caspase-1-dependent proinflammatory cell death, likely requires additional caspase-1-mediated proteolysis (16, 24). Pyroptosis is characterized by activation of caspase-1, which, in turn, activates caspase-7. These activities induce plasma membrane swelling, release of proinflammatory cytokines, DNA damage, and necrotic cell death (26). Inhibition of caspase-1 prevents the morphological characteristics of pyroptosis, including death (27). However, inhibition of plasma membrane swelling and rupture does not prevent release of active IL-1β (27). This suggests that caspase-1 activity is necessary for cell death but that death and caspase substrate release are independent. We sought to discover the role of caspase-1 in inducing these inflammatory responses, including pyroptosis, by globally tracking inflammatory caspase substrates in vitro and in cell culture.
To investigate caspase activities, we applied subtiligase, a bacterial enzyme engineered to directly ligate biotinylated peptides onto protein N termini. This direct affinity enrichment approach coupled with quantitative MS-based identification enabled the discovery of more than 80 cleavage sites following aspartic acids from caspase-1-treated cell lysates. Bioinformatics analysis, quantitative proteomics methods, and biochemical assays confirmed that the majority of these cleavages are catalyzed by caspase-1. This data set represents the largest reported collection of putative caspase-1 substrates including some substrates previously identified in cell biology experiments (13, 28). Interestingly, we observed several caspase-1 cleavage sites that are near known cleavage sites for caspases-3/-7. These nested cleavages suggest that regions of certain proteins are cleaved in both cell death and inflammation. Analysis of caspase-1 activity in cultured monocytes in response to three different inflammatory stimuli confirmed a number of the substrates identified in the lysate studies as well as previous reports of caspase-1-mediated activation of caspase-7 (22, 29, 30). Together, these results represent a step toward gaining a comprehensive understanding of inflammatory caspases in inflammation and pyroptosis.
EXPERIMENTAL PROCEDURES
Materials
Except as noted, all materials were obtained from commercial sources. An optimized subtiligase variant was expressed and purified as described previously though without the use of a helper subtilisin (31, 32). Synthesis of the biotinylated glycolate ester (TEVest2) was conducted using standard Fmoc (fluorenylmethyloxycarbonyl) solid phase peptide synthesis procedures modified for the incorporation of the glycolate ester (9, 33). Tobacco etch virus (TEV) protease and recombinant caspases-1–9 were expressed and purified as described previously (34, 35). Antibodies against caspases-1 (SC1780, Santa Cruz Biotechnology), -3 (9662), and -7 (9492) and cleaved IL-1β (2021s, Cell Signaling Technology) were purchased from the indicated sources.
Cell Culture and Lysate Preparation
THP-1 human monocytic leukemia cells were grown in suspension according to American Type Culture Collection recommendations. In reverse degradomics and background experiments (see “Results”), 0.4–1.0 × 109 cells were pelleted (500 × g, 5 min, rt), washed with 10 ml of PBS (pH 7.4), and pelleted again. Cells (1 × 108 cells/ml) were lysed by resuspending in lysis buffer (100 mm Bicine, 1% Triton X-100, 250 mm KCl, 1 mm EDTA, 1 mm PMSF, 1 mm 4-(2-aminoethylbenzene)sulfonyl fluoride, 0.1 mm E-64, pH 8.0) for 1 h at 4 °C. Background samples were lysed as described with the addition of Z-VAD-fmk (0.1 mm), a general caspase inhibitor. Insoluble membranes and nuclei were pelleted (4200 × g, 5 min, 4 °C), and the supernatant was isolated. For reverse experiments, lysates were treated with 400 nm caspase-1, -4, or -5 for 1 h at rt; treated with 0.1 nm Z-VAD-fmk; and clarified via centrifugation prior to labeling.
For stable isotope labeling with amino acids in cell culture (SILAC) experiments, THP-1 cells were expanded over five passages in media containing [13C6,15N2]lysine and [13C6,15N4]arginine (heavy samples) or [12C6,14N2]lysine and [12C6,14N4]arginine (light samples) to a final count of 5 × 108 cells. Samples were pelleted and lysed as described above. Caspase-1 (400 nm) was added to light samples, both light and heavy samples were incubated at rt for 1 h, and 0.1 μm Z-VAD-fmk was added to the lysates for 10 min. Light and heavy samples were combined and centrifuged prior to labeling.
In forward degradomics samples, THP-1 cells (109) were differentiated by treatment with 100 nm 12-O-tetradecanoylphorbol-13-acetate for 15 h onto three 225-cm2 tissue culture-treated plates. The media were removed, and the plates were washed with 60 ml Opti-MEM (Invitrogen). Caspase-1 was activated in one of three ways. (i) To mimic DNA viral infections, cells in 150 ml of Opti-MEM were transfected with 240 μg of poly(dA·dT) (Sigma) and 600 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturers' recommendations for 6 h. (ii) To mimic inflammation from gout, cells were treated with 300 μg/ml monosodium urate for 3 h. (iii) To mimic bacterial infection, cells were primed with 1 μg/ml LPS (Escherichia coli O55:B5, Sigma) for 3 h followed by activation with 5 mm ATP for 0.5–3 h. Following activation, the supernatant was removed; proteases were inhibited by the addition of Z-VAD-fmk, E-64, 4-(2-aminoethylbenzene)sulfonyl fluoride, PMSF, and EDTA; and the extract was concentrated to ∼5 ml on an Amicon Ultracell (3000 molecular weight cutoff) and labeled as described below. Cell lysates were obtained by adding 10 ml of cold lysis buffer (with Z-VAD-fmk) to the adherent cells and incubated on ice for 30 min. The lysate was clarified by centrifugation prior to labeling.
For forward SILAC experiments, THP-1 cells were expanded over five passages in media containing [13C6,15N4]arginine (heavy samples), [13C6,14N4]arginine (medium samples), and [12C6,14N4]arginine (light samples) (forward experiments) to a final count of 5 × 108 cells. Samples were treated with poly(dA·dT) (light), LPS and ATP (medium), or monosodium urate (MSU) (heavy) as described above. The samples were combined immediately prior to labeling.
N-terminal Labeling and Cleanup
Clarified lysates were treated with 1 mm DTT, 1 mm TEVest2, 1 μm subtiligase, and 10% DMSO for 1 h at rt. Labeled lysates were centrifuged (4200 × g, 5 min) to remove any precipitated protein and filtered through a 0.45-μm cellulose acetate syringe filter. Proteins were separated from detergents, salts, and hydrolyzed ester by gel filtration on a Superdex 30 16/60 column (GE Healthcare), eluting with 100 mm ammonium bicarbonate. Fractions containing proteins were pooled, frozen, and lyophilized.
N-terminal Peptide Isolation
Lyophilized proteins were dissolved in 4 ml of 6 m guanidinium chloride, 100 mm Bicine, 10 mm tris(2-carboxyethyl)phosphine hydrochloride, pH 8.0 and heated to 100 °C for 10 min. Samples were cooled to rt on ice, and cysteine residues were alkylated by the addition of 25 mm iodoacetamide in 500 μl of DMSO for 1 h at rt. To capture biotinylated peptides, samples were diluted with 2 ml of water and gently agitated with 0.5 ml of streptavidin-Sepharose bead slurry (GE Healthcare) for 3 h at rt. The beads were recovered by centrifugation (500 × g, 5 min); washed with 4 m guanidinium chloride (500 μl × 2) and 100 mm NH4CO3H (500 μl × 3) to remove unbound proteins; suspended in 500 μl of 100 mm NH4CO3H, 5 mm CaCl2; and digested overnight with 2 μg of sequencing grade trypsin. The beads were recovered, washed as before to remove residual tryptic peptides, and suspended in 500 μl of 100 mm NH4CO3H. N-terminal peptides were released from the beads via treatment with 30 μg of TEV protease for 2 h at rt. The beads were removed via centrifugation, and the supernatant was concentrated and desalted on C18 ZipTips (Millipore).
Strong Cation Exchange Fractionation
N-terminal peptides were dissolved in 100 μl of 1.0% formic acid and fractionated on a 2.1 × 200-mm polysulfoethyl aspartamide column (PolyLC). Twenty 2-ml fractions were collected over a 40-min elution window (25 mm NH4HCO2, 30% ACN, pH 2.8 to 200 mm NH4HCO2, 30% ACN, pH 2.8 over 32 min, rising to 375 mm NH4HCO2 over the next 7 min). The fractions were dried, desalted on C18 ZipTips (Millipore), dried again, and dissolved in 0.1% formic acid prior to analysis via LC-MS/MS.
MS Analysis and Data Processing
Peptides were separated via reversed phase chromatography on a 75-μm × 15-cm C18 column flowing at 350 nl/min applied directly to a QSTAR Pulsar or QSTAR Elite mass spectrometer (Applied Biosystems). The one or two most intense multiply charged ions in each precursor spectrum were selected for CID fragmentation, and a 3-min dynamic exclusion window was applied to all selected ions. For a portion of the poly(dA·dT)-treated sample and the forward SILAC sample, peptides were also analyzed on an LTQ-FTICR mass spectrometer (ThermoElectron) or an LTQ Orbitrap (ThermoElectron) with the five most intense ions selected for CID fragmentation. MS/MS spectra derived from QSTAR instruments were analyzed using the Analyst software package, and centroid peak lists were generated using the Mascot.dll script. Peak lists were derived from LTQ spectra using Mascot Distiller 2.1.0.0.
Peptide Identification
Peptide identification was performed using Protein Prospector version 5.2.2 (University of California San Francisco (UCSF) Mass Spectrometry Facility), searching against the human December 2008 release of Swiss-Prot (20,333 total proteins). The specified digest protease was trypsin for peptide C termini and nonspecific for peptide N termini, thus allowing for semitryptic peptides. Mass allowances of 50 and 200 ppm were applied to precursor and fragment ions, respectively, for QSTAR instruments and 10 ppm and 0.5 Da, respectively, for the LTQ-FTICR mass spectrometer. Up to two missed tryptic sites were allowed. The peptides were searched with fixed cysteine carbamidomethylation, variable methionine oxidation, and with both fixed and variable addition of an N-terminal SY dipeptide tag to estimate the percentage of unlabeled peptides. Peptides with a score greater than 25.0 and an expectation value below 0.05 were initially considered true hits with a false discovery rate (FDR) of <3.0%. Preliminary analysis of the MSU- and ATP-treated data sets showed unacceptably high FDRs, so an additional protein expectation value of 0.01 was implemented, lowering the FDR to 1.0 and 2.0%, respectively. 95–98% of peptides identified in variable SY tag searches were indeed tagged. Subsequent manual inspection removed peptides with ambiguous tagging (unlabeled peptides with isobaric SY, YS, LH, HL, IH, or HI as the first two residues where a2 and b2 ions may be due to the tag) and peptides of less than seven amino acids excluding the dipeptide tag. Searches against SILAC-labeled peptides fixed the SY tag and allowed variable incorporation of the isotopically labeled amino acids. Protein Prospector identifications were manually verified by examining b and y series ions to confirm the presence of the a2, b2, b3, yn − 1, and yn − 2 ions. Because our data primarily report peptide rather than protein identifications (i.e. one peptide per protein for most proteins), all homologous proteins are reported in the supporting material. To conserve space, peptides in the text are assigned to the protein in the data set with the most unique N termini or, in the case of a tie, alphabetically. Annotated peak lists are provided in the supplemental material. Sequence logo analysis was performed via WebLogo.
MS-based Quantitation and Reproducibility
Three biological replicates of the reverse SILAC samples were analyzed as described. SILAC-based quantitation was performed using the Search Compare function of Protein Prospector. The ratio of light to heavy isotopes was generated by integrating peak areas of light, medium, and heavy precursor ions in survey scan spectra from 10 s prior to MS identification to 30 s after. Data sets were scaled to set the median of peptides not derived from caspase-like cleavages (P1 = Asp) to a SILAC ratio of 1.0 and then combined. We manually inspected spectra for all proposed caspase-derived peptides and any peptide deviating more than 2-fold from the median ratio of non-caspase-derived peptides (20% of all spectra) to investigate the presence of interfering singly charged species or overlapping peptides. Spectra that contained sizeable interfering peaks were discarded. Full details for each peptide including discarded spectra, median ratios, and standard deviations are provided in supplemental Tables 4 and 5.
In Vitro Transcription-Translation
cDNAs encoding synapse-associated protein 1 (SYAP1), zyxin, eukaryotic translation initiation factor 3 subunit J (EIF3J), gasdermin D, splicing factor U2AF 65-kDa subunit (U2AF), and DNA replication licensing factor MCM4 (MCM4) were cloned into pcDNA 3.1(+) (Invitrogen) downstream of the T7 promoter region using EcoRI and XbaI (in the case of MCM4) or HindIII and EcoRI (in all other cases) restriction sites. cDNAs were used as a template for the TnT T7 Coupled Transcription/Translation System (Promega). Proteins were expressed according to the manufacturer's suggestion, incorporating FluoroTect Green (Promega) as a fluorescent label. Post-translation, the reactions were incubated with RNase A (35 ng/μl) at 37 °C for 5 min to degrade residual fluorescent tRNA and incubated with 10 mm N-ethylmaleimide for 10 min at rt to quench any endogenous caspase activity, and residual N-ethylmaleimide was quenched with 20 mm DTT for 10 min at rt. Samples were diluted 4-fold with caspase buffer (25 mm HEPES, 75 mm NaCl, 0.05% CHAPS, 10 mm DTT, pH 7.5), and caspase (50 or 200 nm) was added to the reaction mixture. At the indicated time points, aliquots were taken, added to 4× SDS loading buffer, and frozen on dry ice prior to analysis. Proteins were separated by SDS-PAGE, gels were scanned on a Typhoon 9400 gel scanner (GE Healthcare), and the band intensity was integrated on ImageQuant 5.2 software (GE Healthcare), subtracting for background fluorescence. Kinetic rates were fit via Graphpad Prism 4.03.
Immunoblot Assays of Pyroptotic Cells
The conditioned media and clarified lysates from 107 THP-1 cells were prepared as described for the forward degradomics experiments, transfecting with or without poly(dA·dT) for 0–10 h. Proteins (40 μg per sample) were separated by SDS-PAGE and transferred to a 0.45-μm-pore size Invitralon PVDF membrane (Invitrogen). Membranes were blocked with 2.5% nonfat dry milk, 0.1% Tween, Tris-buffered saline, pH 7.4 (2.5% milk in TBS-T) and incubated overnight in 1:500–1:1000 primary antibody in blocking buffer. The blots were washed with TBS-T (4 × 10 min), incubated with horseradish peroxidase-conjugated anti-mouse, -rabbit, or -goat IgG (1:1000–1:5000), and washed four additional times. Blots were incubated with SuperSignal Femto substrate (Pierce) and imaged via a FluorChem SP (Alpha Innotech).
Lactate Dehydrogenase Release Assay
THP-1 cells (1 × 105) in 96-well plates were differentiated with 100 nm 12-O-tetradecanoylphorbol-13-acetate for 14 h. The cells were washed with Opti-MEM (2 × 50 μl) and transfected with or without 0.2 μg of poly(dA·dT) using Lipofectamine 2000 according to the manufacturers' recommendations. At the indicated time points, the cells were lysed by the addition of 2 μl of 9% SDS and assayed by CytoTox-ONE (Promega) according to the manufacturer's specifications.
RESULTS
Subtiligase-based Biotinylation of N Termini for Positive Enrichment and Proteomics Evaluation
Proteolyis generates new α-amines and α-carboxylates, either of which could be isolated and identified to profile protease activities. However, the ∼80% of protein N termini that are co-translationally acetylated can be excluded from N-terminal analyses, making this approach more attractive (36). Here, we applied a technology developed in our laboratory to characterize the protein N termini of inflammatory caspase-cleaved products (9). Free N termini in THP-1 cell lysates were biotinylated with subtiligase, a variant of the bacterial protease subtilisin BPN′ (Fig. 1a). Subtiligase is engineered to cleave ester but not amide substrates and transfer the N-terminal portion of the ester onto free N termini (31, 37). The enzyme can ligate a broad range of protein and peptide N termini but excludes free amino acids and other amines, including lysine ε-amines. Subtiligase-treated proteins were separated from the spent ester, and labeled species were captured on streptavidin beads. After extensive washing, proteins were trypsinized on the beads, leaving only the N-terminal peptides bound. These were efficiently released by cleavage of a TEV protease site present in the ester. TEV-released N-terminal peptides conveniently retain a residual SY dipeptide tag, allowing virtually unequivocal identification of labeled N termini.
Fig. 1.
Degradomics approach for identifying caspase-cleaved peptides. A, to identify cleavage sites, lysates are N-terminally biotinylated with subtiligase and TEVest2. The labeled proteins are captured on streptavidin beads, trimmed to a single peptide with trypsin, and released from the beads with TEV protease. The resulting labeled peptides are identified by LC/MS/MS. B, two modes are used for identifying caspase-cleaved peptides. i, in reverse degradomics, purified caspase is added exogenously to cellular lysates, generating caspase cleavage events. ii, in forward degradomics, inflammatory (or apoptotic) stimuli induce caspase activation in the cell or conditioned media. These stimulated cells are then lysed and analyzed. C, the overlap of forward and reverse degradomics data sets. D, the overlap between forward degradomics data sets for three different inflammatory stimuli.
We applied this technology to THP-1 monocytes under three different conditions (Fig. 1b). First, untreated cells were analyzed to evaluate the background levels of proteolysis, the distributions of start sites for the identified N termini, and the amino acid residues surrounding cleavage sites. Next, two additional sets of experiments were performed that we term “forward” and “reverse” degradomics. In reverse degradomics experiments, proteolysis is initiated by addition of a known protease to extracts. Conditions can be optimized to maximize substrate identification and minimize secondary proteolysis (through the use of protease inhibitors). However, because cellular structures are homogenized, the observed cleavages may not necessarily reflect those that would occur in an intact cell. In forward degradomics experiments, proteases are activated in their normal cellular context by the addition of specific inflammatory stimuli. Forward degradomics experiments maintain native protein localizations and enzyme concentrations; however, the sensitivity of this technique may be diminished by an inability to regulate the extent of cleavage or abrogate secondary proteolysis. In addition, one does not know a priori the protease responsible for the observed cleavages.
Mass spectral analysis of the N termini tagged in an untreated lysate (the N-terminal “background”) resulted in the identification of 306 N termini from 256 proteins with a false discovery rate of 3.0% (supplemental Table 1). In comparison with a previously compiled background data set from Jurkat T-lymphocytes, fewer of the monocytic N termini observed were the result of full-length protein expression or removal of a short signal sequence (45% of THP-1 versus 72% of Jurkat N termini were found within the first 50 amino acids) (supplemental Fig. 1) (9). This suggests a higher level of back-ground endoproteolytic activity beyond the expected signal peptidase activities. The background data set reveals a large number of internal N termini resulting from cleavages after basic and large aliphatic or aromatic residues, suggesting prevalent cellular tryptic- and chymotryptic-like activities, respectively (supplemental Fig. 2). Of note, only 1.3% (4 of 306) of the identified N termini resulted from proteolysis after an aspartic acid residue, substantially below the natural abundance of the amino acid (5.5%). We have seem similar levels of cleavage after aspartic acids in untreated Jurkat, HeLa S3, and DB cells (9).2 These data suggest that, in the absence of apoptotic or inflammatory stimuli, there is little endogenous caspase activity and few caspase-like substrates. Thus, proteins cleaved after aspartic acid residues in stimulated cells are likely to be caspase substrates.
Identification of Substrates from Caspase-treated Lysates
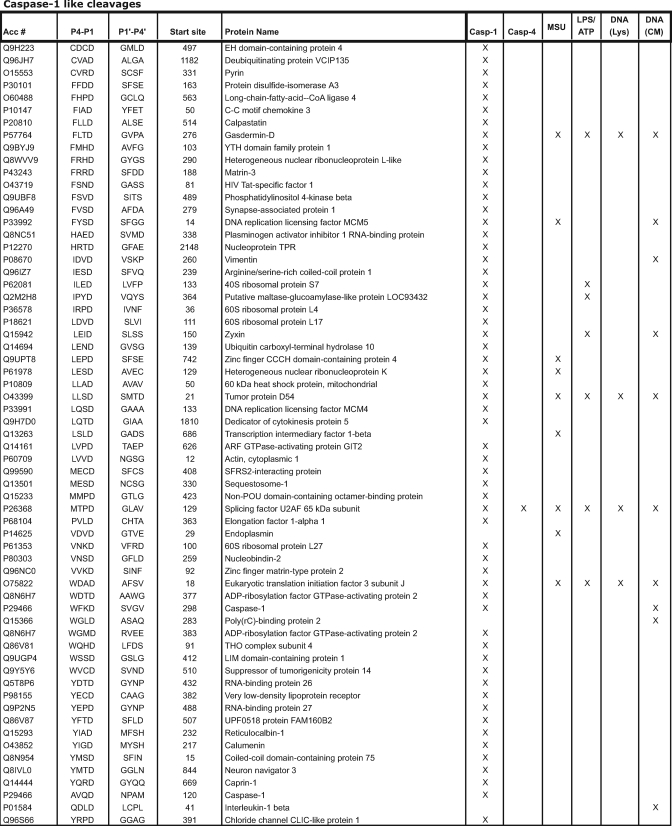
To evaluate the range of potential inflammatory caspase substrates, we conducted reverse degradomics by incubating cell extracts with inflammatory caspases-1, -4, and -5. Lysates from THP-1 cells were incubated with caspase-1, -4, or -5 for 1 h, and N termini were labeled using subtiligase and TEVest2 as described above. Three experimental replicates of caspase-1-treated lysates revealed a total of 55 putative caspase-1 substrates from 834 identified N termini as defined by a P1 aspartic acid (Table I and supplemental Table 2). We analyzed the sequences surrounding the N termini to examine the consequences of caspase addition on amino acid distribution. Caspase-1-treated lysates showed amino acid distributions similar to the untreated lysates except that aspartic acid was 4 times more prevalent at the P1 position (6.6 versus 1.3%) (supplemental Fig. 3). Although the overlap between caspase-treated samples was modest, each replicate showed significant enrichment for cleavage after aspartic acid residues (5.6–9.9% of all peptides) (supplemental Fig. 4). Comparisons between this data set and previously identified caspase substrates revealed an overlap of four caspase-1 substrates (actin, pyrin, calpastatin, and caspase-1) and an additional 13 cleavages that had previously been attributed to other known or unknown caspases (supplemental Table 3) (22, 29, 38, 39). Seven additional caspase-1 cleavage sites were found in proteins with reported caspase cleavages at alternative sites. In three of these, the observed site of cleavage was within four amino acids of a previously identified site for caspase-3 or caspase-7. The remaining 31 cleaved proteins had not previously been reported as substrates for any caspase.
Table I.
In sharp contrast, caspase-4- and -5-treated lysates generated only three and zero caspase-cleaved peptides, respectively, from more than 500 identified N termini (Table I). The general lack of peptide identification for caspases-4 and -5 was not due to these enzymes being inactive because the enzymes had specific activities against synthetic substrates comparable to those reported in the literature (data not shown) albeit 100-fold below caspase-1. Furthermore, subsequent experiments found that caspase-5 has activity against one protein in vitro (see below).
We compared the sequences surrounding the purported caspase-1 cleavage sites with the reported sequence preferences obtained from peptide positional scanning libraries (Fig. 2a) (7). Both sets show similar strong preferences for aromatic and large aliphatic residues at P4 and for small residues at P1′ along with a weak preference for acidic residues at P3. Interestingly, the N-terminal analysis of our data shows a preference for large aliphatic and aromatic residues at P2′, a position not previously investigated via positional scanning libraries. We do not believe these results reflect a bias introduced by subtiligase labeling because subtiligase has little preference at the P2′ position, and apoptotic caspase substrates identified via subtiligase labeling do not show a similar bias (9, 37). Finally, 11% of the cleavage sites contained an acidic residue at P4, consistent with caspase-3/-7 substrate preferences.
Fig. 2.
Caspase-1-cleaved peptides show propensity for hydrophobic residues at P4. A, sequences surrounding the cleavage site of aspartyl-cleaved protein (P1 = Asp) were aligned and analyzed in a sequence logo. Amino acid letter sizes correspond to the frequency; colors correspond to the side chain functionality as follows: acidic (Asp and Glu), red; hydrophobic (Phe, His, Trp, and Tyr), light blue; aliphatic (Leu, Ile, Met, Val, and Pro), dark blue; small (Ala, Gly, Ser, and Thr), green; and other (Cys, Asn, Lys, Gln, and Arg), black. B, SILAC comparisons between caspase-1-treated and untreated lysates were graphed on a double log plot. Aspartyl-cleaved peptides are color-coded according to their P4 residue: putative apoptotic substrates (Asp and Glu), putative inflammatory substrates (Leu, Ile, Met, Val, Pro, Phe, His, Tyr, and Trp), and unknown (Ala, Gly, Ser, and Thr). Note that the putative inflammatory substrates have a high light/heavy ratio, suggesting that they are produced by the addition of caspase-1. Substrates that were selected for further biochemical examination are circled. C, bar graph representation of the caspase-cleaved peptides in B grouped by SILAC ratio. Note that the caspase-derived peptides with the highest light/heavy ratios contained large hydrophobic residues in P4, consistent with caspase-1 specificity.
SILAC-based Quantitation Distinguishes Inflammatory and Apoptotic Substrates
To deconvolute background caspase-like proteolysis in unstimulated cells from that induced by addition of inflammatory caspases, we performed quantitative experiments using SILAC (40). We hypothesized that aspartate-cleaved substrates, present due to low levels of apoptosis in culture or endogenous non-caspase-mediated proteolysis in healthy cells, should be equally abundant in untreated and caspase-1-treated lysates. We evaluated endogenous caspase-like activities by analyzing SILAC-labeled caspase-1-treated and untreated lysates. In three replicate experiments, all caspase-derived substrates were markedly enriched in the caspase-1-treated samples (Fig. 2b and supplemental Table 4). This suggests that the cleavages result from the addition of caspase-1. These data further increased the total number of caspase cleavage sites identified in reverse experiments to 82 (6.0% of N termini). To gain further insight into the contributions of executioner caspase activity and background apoptosis, we separated the caspase-cleaved substrates into three categories based on characteristic substrate preferences at the P4 residue. Acidic residues (Asp and Glu) are typical of apoptotic caspases, whereas hydrophobic and aromatic residues (Leu, Ile, Met, Val, Pro, Phe, His, Tyr, and Trp) are suggestive of inflammatory caspases, and small residues could be indicative of either activity (Fig. 2, b and c) (7). The typical apoptotic-like cleavages that were observed had lower SILAC ratios than the corresponding inflammatory-like cleavages (median ratio of 2.3 versus 14). This suggests that caspase-1 has low activity against the apoptotic substrates or that caspase-1 activates low levels of executioner caspase activities in vitro (e.g. caspase-7).
In Vitro Transcription-Translation Provides Direct Evidence of Caspase Cleavage
To evaluate the ability of caspase-1 to selectively cleave the identified substrates, we expressed a subset of six protein substrates by in vitro transcription-translation (IVT): SYAP1, zyxin, splicing factor U2AF 65-kDa subunit (U2AF), MCM4, EIF3J, and gasdermin D (GSDMD) (Fig. 3 and supplemental Fig. 5). These each had SILAC ratios in the reverse experiments of greater than 4, suggesting that they were efficiently cleaved upon addition of caspase-1. Samples were treated with purified recombinant caspase-1, aliquots were removed and quenched at various times, and the progression of the reaction was tracked via SDS-PAGE to allow determination of the rate of cleavage. All of the tested substrates were cleaved to some degree by caspase-1. Remarkably, the rate of cleavage varied over 500-fold (kcat/Kmapp from <200 to 128,000 m−1 s−1) (Fig. 3, a–c). The most rapidly cut substrate was gasdermin D, which was cleaved ∼50–500-fold faster than the others.
Fig. 3.
In vitro analyses of caspase-1-cleaved substrates show wide variation in rate and selectivity. A, top, individual substrates were expressed by IVT and treated with 50 nm caspase-1 (GSDMD) or 200 nm caspase-1 (all others) (representative data is shown). At the indicated time points, aliquots were diluted with SDS loading buffer and stored flash frozen prior to analysis by SDS-PAGE. Arrows indicate the masses of cleaved products. Bottom, substrates were treated with 50 nm (GSDMD) or 200 nm (all others) caspases-1–9 and incubated for 5 min (GSDMD) or 90 min (all others) prior to analysis by SDS-PAGE. B, fluorescence signals from A were integrated and plotted. Lines were fit to standard first order decay, f = f0 + A0e−kt where f is fluorescence intensity, f0 is background fluorescence, A0 is the fluorescence intensity of the intact protein, k is the rate of the reaction, and t is time. C, apparent kcat/Km was calculated for each substrate by assuming [substrate] ≪ Km and applying k = (kcat/Km)app × [caspase-1]. D, the caspase screen described in A was applied to SYAP1 D281A. E, caspase specificity was analyzed by integrating the fluorescence intensity of the full-length protein. +++, >60% reduction in intensity compared with no caspase control; ++, >40% reduction; +, >20% reduction or visual evidence of a cleaved product; −, >80% reduction of caspase intensity and no visible products. AU, arbitrary units; Casp, caspase; ZYX, zyxin.
To assess whether the IVT substrate cleavages were specific to caspase-1, we screened the proteins cleaved at least 50% at the latest time point (all except U2AF) against a panel of caspases at equal concentrations. The enzymatic activities of the caspases (kcat/Km against optimized substrates) vary over approximately 4 orders of magnitude with the executioner caspases being most active against their optimal substrates (41). Thus, in the absence of relative specificity, we would expect a strong bias toward the executioner enzymes. Surprisingly, only gasdermin D was uniquely cleaved by caspase-1 (Fig. 3, a and e, and supplemental Fig. 5). Of the caspases showing overlapping activity with caspase-1, caspase-8 was the most similar, cleaving three of the tested proteins, two at approximately the same site.
One substrate, SYAP1, was efficiently cleaved by multiple caspases (caspases-1, -3, -5, -7, and -8) with caspases-1, -3, and -7 giving products of approximately the same molecular weight. Despite the high level of similarity and overlap in peptide substrate specificity between the inflammatory caspases, caspase-5 and caspase-1 cleave SYAP1 at distinct sites. Close inspection of the amino acids surrounding the putative caspase-1 cleavage site (Asp278) showed an additional nearby caspase-3/-7 site (DXXD) (FVSD278↓AFD281↓AC). To investigate the role of this second aspartic acid in regulating caspase activity, we repeated the caspase screen with a mutant SYAP1 lacking the second aspartic acid residue (D281A). Consistent with our hypothesis of overlapping but separate cleavages, this mutation abrogated the activity of caspases-3 and -7 but not of caspase-1 (Fig. 3d). Thus, cleavage at Asp278 is unique to caspase-1 in vitro.
Forward Degradomics Experiment Reveals Subset of Reverse Degradomics Substrates
Having identified and verified a number of caspase-1 substrates in vitro, we decided to investigate the proteases in live cells to evaluate the effects of physiologically relevant localizations and concentrations using forward degradomics. To do this, we analyzed cells stimulated with three different inflammatory stimuli that are known to activate caspase-1: MSU, LPS plus ATP, and dsDNA (poly(dA·dT)). Although MSU and LPS/ATP treatments both induced IL-1β release as measured by ELISA (supplemental Fig. 6), we found relatively few caspase-1 substrates using these conditions (Fig. 1d, Table I, and supplemental Table 5). Analysis of both lysates and conditioned media from LPS- and ATP-treated cells yielded 11 total caspase substrates from 1159 total N termini detected (0.9%), including three caspase-3/-7 sequences (P4 = Asp/Glu) and four sequences of indeterminate caspase activity. Proteomics evaluations of MSU-treated cells generated more caspase cleavages (22 of 880 total N termini; 2.5%) but markedly fewer than were identified in reverse degradomics experiments.
To investigate stimuli known to induce more extensive caspase-1 activation, we induced pyroptosis by transfecting dsDNA into the cytoplasm of THP-1 cells. Previous work showed that cytoplasmic dsDNA interacts with absent in melanoma 2 (AIM2) to induce inflammasome formation and pyroptosis in a greater percentage of cells than was observed via LPS/ATP treatment (16, 17, 20, 42, 43). To confirm the ability of dsDNA to induce caspase activity, we induced pyroptosis by transfecting differentiated THP-1 cells with poly(dA·dT) for 0–10 h. The lysates and conditioned media were probed by immunoblot for caspases-1, -3, and -7 along with IL-1β, a known substrate of caspase-1. Consistent with previous reports that caspase-1 activates caspase-7 but not caspase-3, we detected active caspase-1 and its substrates (caspase-7 and IL-1β) in the conditioned media (Fig. 4a) (22). Active caspase-3 was not detected. These substrates appeared synchronously over time and depended on the presence of dsDNA. Interestingly, cleaved IL-1β or active caspases were not detected in the cell lysates, suggesting that these proteins were either unstable or exported outside of the cells.
Fig. 4.
DNA transfection mediates caspase activation and substrate release. A, THP-1 cells were transfected with or without DNA for the indicated times. Cell extracts and conditioned media were prepared and probed by immunoblot for the presence of caspases-1, -3, and -7 and IL-1β. B, release of lactate dehydrogenase (LDH) was assessed for THP-1 cells under the same conditions. Casp, caspase; AU, arbitrary units.
Using this time course to determine a midpoint of caspase-1 activation and cleaved IL-1β release, we analyzed THP-1 cells transfected with poly(dA·dT) for 6 h. Examination of cellular lysates and conditioned media revealed caspase cleavage products primarily in the conditioned media. Two of 299 N termini (0.7%) in the lysate were derived from cleavages after aspartic acid, whereas 17 of 679 N termini (2.5%) were found in the conditioned media (supplemental Tables 6 and 7). These 17 peptides included both the intersubunit linker of caspase-1 and a non-canonical caspase cleavage site for IL-1β (Table I). We evaluated the time course of pyroptosis by tracking lactate dehydrogenase release into the media as a surrogate for cell permeabilization. We found that the kinetics of cell death, as measured by membrane permeabilization, tracked with IL-1β release and caspase cleavage (Fig. 4b). These results demonstrate that caspase activation and pyroptosis are closely linked, consistent with previous experiments showing that inhibition of caspase-1 blocks pyroptosis (24).
SILAC Comparison of Modes of Caspase-1 Activation
To quantitatively assess the roles of inflammatory stimuli in regulating caspase-1 activity, we compared caspase-mediated proteolysis induced by MSU, LPS and ATP, and poly(dA·dT) via SILAC labeling and N-terminal proteomics. THP-1 cells grown in the presence of light (12C6,14N4), medium (13C6,14N4), or heavy (13C6,15N4) arginine were treated with the three activators of caspase-1, and the cell lysates and conditioned media were assessed by our degradomics technology. We identified eight caspase-cleaved sites from a total of 198 N termini (4.0%) in the lysate and five of 188 (2.7%) in the conditioned media (supplemental Table 8). Seven of the nine total caspase-1 substrates were found in all three conditions. Notably, the levels of these substrates were higher in poly(dA·dT)-treated cells, consistent with a greater level of caspase activation under these conditions. By contrast, two caspase-cleaved proteins (elongation factor 1-α and hematopoietic lineage cell-specific protein) were identified solely in the lysate of the poly(dA·dT)-treated cells.
DISCUSSION
Reverse Versus Forward Degradomics Using Subtiligase Tagging
Reverse and forward experimental paradigms each have their advantages and disadvantages but together provide useful insights into targets cleaved by proteases of interest. Reverse experiments control the initiating protease and its dose in a homogenous cell extract but ignore the functions of cellular structures that may sequester substrates. Forward experiments maintain cellular structures, but the proteases involved in processing are not strictly defined. We chose to apply both experimental paradigms to the inflammatory caspases, including models for bacterial infection, viral infection, and cellular stress.
Two recent reverse proteomics investigations targeted caspase-1 but identified only 11 and 41 putative cleavages (22, 44). In our hands, reverse degradomics experiments on caspase-1 identified a comparatively large number of cleavage sites (82), presumably reflecting the sensitivity of the subtiligase labeling technique. However, analogous experiments with caspases-4 and -5 identified zero and three substrates, respectively. The specific activity of caspase-1 is ∼100-fold higher than that of either caspase-4 or -5 (41). This simple difference may explain the dramatically reduced number of identifications for caspase-4 or -5. Not surprisingly, it would appear that caspase-1 is the most active inflammatory caspase effector during inflammation.
Forward degradomics experiments identified fewer substrates than reverse experiments (16–27 substrates, 1.3–3.0% of all peptides in forward mode versus 82 cleavages sites in reverse mode, 5.5% of all peptides). Thus, substrate accessibility appears to severely limit inflammatory caspase cleavages in forward experiments. Although most caspase substrates were identified in all three proinflammatory conditions, two appeared to be specific to the poly(dA·dT) induction of the AIM2 inflammasome. Collectively, 50% of the substrates identified in forward experiments were also identified in reverse experiments (Fig. 1d). This overlap was markedly higher in the canonical caspase-1 substrates (76%) than in non-canonical substrates (33%). These data suggest that additional caspases are active in these cells and that the relative contribution of these activities varies with the inflammatory stimuli.
Caspase Efficiency and Target Specificity
We initially evaluated our reverse degradomics identifications by comparing them with known caspase activities. Our data corroborated positional scanning analyses of caspase-1 specificities and revealed additional determinants of sequence specificity at the P2′ site for large hydrophobic residues (7). To analyze the novelty of our data set, we compared our substrates with a database of annotated caspase cleavage events (CASBAH, the caspase substrate database). Over a third of the proteins we identified are reportedly cleaved by other caspases, often at identical or nested sites. To further investigate this high level of overlapping function, we screened caspases-1 through -9 against IVT expressed substrates. Each of the expressed proteins was cleaved by a unique set of caspases at one or more sites and with different efficiencies (Fig. 3, a and e). The apparent molecular weights of the cut bands were consistent with the cleavage sites predicted by our MS data and previous reports (45). Interestingly, we found that caspase homology and activity against peptide substrates did not necessarily dictate their relative activities against proteins. For example, the inflammatory caspases-1 and -5 cleave SYAP1 at distinct locations (Fig. 3a). By contrast, a protein less genetically and functionally homologous to caspase-5, caspase-8, appears to cleave SYAP1 at the same (or a very similar) site. The sequences of substrates also did not dictate the relative efficiency with which they were cleaved. The primary peptide sequence that best matched the canonical substrate specificity of caspase-1 (EIF3J, WDAD↓AFSV) was cleaved at an intermediate rate. The most efficient caspase-1 substrate, gasdermin D (FLTD↓GVPA), which appeared to be a modest substrate for caspase-1 based on sequence, was also the only substrate cleaved solely by caspase-1. Interestingly, both caspase-1 and gasdermin D are expressed primarily in human immune cells (BioGPS, the gene portal hub). The selectivity, high rate of cleavage by caspase-1, and similar mRNA expression profiles suggest that gasdermin D may be one of the primary targets of caspase-1 in vivo.
The observation that substrates are cleaved by multiple caspases can be rationalized in several ways. First, there is site-specific functional redundancy between caspase-1 and other caspases for some targets, a result that had been observed for other pairs of caspases (e.g. caspases-3 and -7) (45). This in vitro redundancy may be a product of highly accessible cleavage sites (9, 46). Both inflammatory caspases and apoptotic caspases will slowly cleave non-optimal peptide substrates, suggesting that these redundant cleavages may have little or no relevance in vivo. Alternatively, site-specific cleavage of these same proteins may be vital to the multiple cellular caspase functions. The presence of substrates with nested caspase cleavage sites (e.g. SYAP1 cleavage at Asp278 by caspase-1 and Asp281 by caspases-3/-7) supports this hypothesis. Overlapping caspase cleavage sites within proteins imply that cleavage in that segment is conserved and that there is evolutionary pressure to maintain proteolysis by multiple caspases. In fact, in proteins with nested cleavage sites (SYAP1, EIF3J, THOC4, and vimentin), the six amino acids N-terminal of the downstream site are 96% identical between human, mouse, and bovine homologs (supplemental Table 3). Amino acids C-terminal to the downstream site showed greater variation. Interestingly, although the cleavage sites were highly conserved between species, there were considerable variations between proteins with nested sites. These included changes in the amino acid composition, the distance between the cleavage sites, and the order of caspase-1 versus caspase-3/-7 sites. Although we identified and assessed only a few proteins, the high degree of conservation may suggest that these nested sites have important evolutionary roles. Large scale investigations of caspase cleavage events across multiple species could potentially identify more highly conserved sites and help identify key nodes targeted by caspases. In addition to this molecular level functional overlap, gene ablation studies have shown that caspase functions can compensate for one another. Caspase-3-deficient mice are viable in some genetic backgrounds, and caspase-7-deficient mice have only minor phenotypes, but caspase-3/-7-deficient mice die immediately after birth (48). Our detection of substrate overlap is consistent with functional compensation.
Quantitative SILAC analysis of reverse data sets confirmed that putative caspase-1 substrates based on cleavage site specificity were enriched in caspase-1-treated lysates (Fig. 2). Although all N termini with a P1 aspartate were enriched in the caspase-1-treated lysates (seen by SILAC ratios greater than 1.0), the median ratio for caspase-1-like cleavage was substantially higher than for putative caspase-7-like cleavages (16 versus 2.6). Although caspase-7 is activated by caspase-1 during inflammation, our data indicate that the vast majority of proteolysis carried out in this process is in fact mediated by caspase-1. However, without more detailed studies, we cannot determine whether the moderately enriched caspase-7-like substrates are derived from secondary weak activation of caspase-7 or if these substrates are produced directly by low efficiency caspase-1 activity. Virtually all caspase-cleaved substrates with SILAC ratios greater than 6.5 were not found in untreated samples, suggesting a lower boundary ratio for positive identification of caspase-1 cleavage.
Biological Significance of Identified Substrates
There are two key general observations from the reverse and forward degradomics experiments. First and not surprisingly, the total number of caspase-cleaved substrates identified from all the forward experiments was only about half that found in the reverse direction (Fig. 1d). This suggests enzyme-substrate co-localization is critical for cleavage to occur (Table I). However, although half of the substrates found in the forward direction were contained in the reverse data set, half were not. This may result from incomplete sampling or increased caspase-7 activities in forward samples. Second, although most of the caspase substrates are the same irrespective of the inducer of inflammation, two were specific to poly(dA·dT)-treated samples. This may be due to a greater extent of caspase activation in the poly(dA·dT)-treated cells as measured by cleavage of other caspase substrates and release of IL-1β. This is consistent with the onset of pyroptosis inducing breakdown of cellular structures and enabling caspase-1 to access additional cellular compartments. Alternatively, the relevant inflammasomes (AIM2 versus NALP3) may directly regulate caspase-1 substrate accessibility. Previous studies have found that caspase-1 and IL-1β localize differentially in response to diverse stimuli (43, 49, 50); the reduced number of caspase-cleaved substrates in forward experiments imply that active caspase-1 is not delocalized in the cytoplasm or the nucleus. This is also consistent with imaging studies (24, 43). Thus, these experiments further highlight the importance of native cellular context in defining the substrate degradome of a protease.
The mechanism of caspase-1 activation and subsequent secretion of its cleaved substrates are not well understood. Multiple models, including secretory lysosomes, transient membrane permeabilization, and multivesicular bodies, have been proposed, but none account for all experimental observations (49). In a few studies, membrane permeabilization has been observed, consistent with our observation of lactate dehydrogenase release in transfected cells (Fig. 4b). However, these same studies have shown permeabilization to be downstream and independent of IL-1β release (27, 51). We believe the resolution to these data may be that sporadic induction of pyroptosis results in rapid substrate release followed by slower membrane permeabilization. Thus, in our non-synchronized bulk measurements, release of substrates and lactate dehydrogenase can appear to be coupled. Single cell analyses would be necessary to deconvolute these possibilities.
Conclusion
This work extends the scope of our N-terminal labeling technology by identifying in vitro and cellular substrates of the inflammatory caspases. Our ability to detect substrates for caspase-1, but not caspase-4 or -5, sets a benchmark for the levels of proteolytic activity and enzyme selectivity that can be identified via this technique. Moreover, the addition of SILAC to our N-terminal enrichment strategy validates the identified substrates, and the magnitude of the SILAC ratio helps to identify preferred caspase-1 substrates. Sequence analysis of large numbers of caspase-1 substrates provides insight into the primary biochemical specificity of caspase-1. Furthermore, the identification of substrates shared between the inflammatory and executioner caspases may be useful in identifying highly conserved cleavage events. Finally, a series of cell culture experiments gave the first proteomic look at caspase-1 mediated-proteolysis in live cells and provided insight into the variations caused by different inducers of caspase-1 activity.
Although this set of caspase-1 substrates is not comprehensive, it has identified a number of proteins with known roles in immune function and demonstrated the utility of our N-terminal isolation method for profiling moderately active proteases. Interestingly, the differences in number and type of cleaved proteins in the forward and reverse data sets highlight the potential pitfalls of reverse type experiments. Future studies using multiple reaction monitoring-based quantitation or single cell-based analyses should help to dissect the details of caspase-1 activity by different inflammatory signals (52).
Supplementary Material
Acknowledgments
We thank Dr. A. L. Burlingame and Dr. J. C. Trinidad for invaluable assistance in developing MS-based experiments; J. A. Zorn, M. Lopez, and Dr. D. W. Wolan for generous gifts of caspases; and members of the Wells and Burlingame groups for helpful discussions. Mass spectrometry was performed at the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A. L. Burlingame, Director) supported by the Biomedical Research Technology Program of the National Institutes of Health National Center for Research Resources under Grants NIH NCRR P41RR001614 and NIH NCRR RR015804.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grants F32AI077177 (to N. J. A.), R01 GM081051, and R01 AI070292 (both to J. A. W.).
 This article contains supplemental Figs. 1–6 and Tables 1–8.
This article contains supplemental Figs. 1–6 and Tables 1–8.
2 H. Nguyen, N. J. Agard, and J. A. Wells, unpublished data.
1 The abbreviations used are:
- dsDNA
- double-stranded DNA
- IL
- interleukin
- SILAC
- stable isotopic labeling with amino acid in cell culture
- TEV
- tobacco etch virus
- Z-VAD-fmk
- carbobenzoxy-valyl-alanyl-aspartyl-fluoromethyl ketone
- LPS
- lipopolysaccharide
- LTQ
- linear ion trap
- AIM2
- absent in melanoma 2
- SYAP1
- synapse-associated protein 1
- EIF3J
- eukaryotic translation initiation factor 3 subunit J
- GSDMD
- gasdermin D
- U2AF
- splicing factor U2AF 65-kDa subunit
- MCM4
- DNA replication licensing factor MCM4
- FDR
- false discovery rate
- IVT
- in vitro transcription-translation
- MSU
- monosodium urate
- rt
- room temperature
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1.Brasier A. R. (2006) The NF-kappaB regulatory network. Cardiovasc. Toxicol 6, 111–130 [DOI] [PubMed] [Google Scholar]
- 2.Müntz K. (2007) Protein dynamics and proteolysis in plant vacuoles. J. Exp. Bot 58, 2391–2407 [DOI] [PubMed] [Google Scholar]
- 3.Pop C., Salvesen G. S. (2009) Human caspases: activation, specificity, and regulation. J. Biol. Chem 284, 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backes B. J., Harris J. L., Leonetti F., Craik C. S., Ellman J. A. (2000) Synthesis of positional-scanning libraries of fluorogenic peptide substrates to define the extended substrate specificity of plasmin and thrombin. Nat. Biotechnol 18, 187–193 [DOI] [PubMed] [Google Scholar]
- 5.Baggio R., Burgstaller P., Hale S. P., Putney A. R., Lane M., Lipovsek D., Wright M. C., Roberts R. W., Liu R., Szostak J. W., Wagner R. W. (2002) Identification of epitope-like consensus motifs using mRNA display. J. Mol. Recognit 15, 126–134 [DOI] [PubMed] [Google Scholar]
- 6.Matthews D. J., Wells J. A. (1993) Substrate phage: selection of protease substrates by monovalent phage display. Science 260, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 7.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 8.Dix M. M., Simon G. M., Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald L., Robertson D. H., Hurst J. L., Beynon R. J. (2005) Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat. Methods 2, 955–957 [DOI] [PubMed] [Google Scholar]
- 11.Timmer J. C., Enoksson M., Wildfang E., Zhu W., Igarashi Y., Denault J. B., Ma Y., Dummitt B., Chang Y. H., Mast A. E., Eroshkin A., Smith J. W., Tao W. A., Salvesen G. S. (2007) Profiling constitutive proteolytic events in vivo. Biochem. J 407, 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. (2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771–777 [DOI] [PubMed] [Google Scholar]
- 13.auf dem Keller U., Doucet A., Overall C. M. (2007) Protease research in the era of systems biology. Biol. Chem 388, 1159–1162 [DOI] [PubMed] [Google Scholar]
- 14.Fuentes-Prior P., Salvesen G. S. (2004) The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J 384, 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pétrilli V., Dostert C., Muruve D. A., Tschopp J. (2007) The inflammasome: a danger sensing complex triggering innate immunity. Curr. Opin. Immunol 19, 615–622 [DOI] [PubMed] [Google Scholar]
- 16.Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009) AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 18.Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K. L., Superti-Furga G. (2009) An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol 10, 266–272 [DOI] [PubMed] [Google Scholar]
- 19.Cassel S. L., Eisenbarth S. C., Iyer S. S., Sadler J. J., Colegio O. R., Tephly L. A., Carter A. B., Rothman P. B., Flavell R. A., Sutterwala F. S. (2008) The Nalp3 inflammasome is essential for the development of silicosis. Proc. Natl. Acad. Sci. U.S.A 105, 9035–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. (2009) AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J., et al. (1992) A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356, 768–774 [DOI] [PubMed] [Google Scholar]
- 22.Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., Núñez G. (2008) Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics 7, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurcel L., Abrami L., Girardin S., Tschopp J., van der Goot F. G. (2006) Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell 126, 1135–1145 [DOI] [PubMed] [Google Scholar]
- 24.Fernandes-Alnemri T., Wu J., Yu J. W., Datta P., Miller B., Jankowski W., Rosenberg S., Zhang J., Alnemri E. S. (2007) The pyroptosome: a supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ 14, 1590–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nickel W., Rabouille C. (2009) Mechanisms of regulated unconventional protein secretion. Nat. Rev. Mol. Cell Biol 10, 148–155 [DOI] [PubMed] [Google Scholar]
- 26.Fink S. L., Bergsbaken T., Cookson B. T. (2008) Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A 105, 4312–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verhoef P. A., Kertesy S. B., Lundberg K., Kahlenberg J. M., Dubyak G. R. (2005) Inhibitory effects of chloride on the activation of caspase-1, IL-1beta secretion, and cytolysis by the P2X7 receptor. J. Immunol 175, 7623–7634 [DOI] [PubMed] [Google Scholar]
- 28.Doucet A., Butler G. S., Rodríguez D., Prudova A., Overall C. M. (2008) Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol. Cell. Proteomics 7, 1925–1951 [DOI] [PubMed] [Google Scholar]
- 29.Chae J. J., Wood G., Richard K., Jaffe H., Colburn N. T., Masters S. L., Gumucio D. L., Shoham N. G., Kastner D. L. (2008) The familial Mediterranean fever protein, pyrin, is cleaved by caspase-1 and activates NF-kappaB through its N-terminal fragment. Blood 112, 1794–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K. K., Posmantur R., Nadimpalli R., Nath R., Mohan P., Nixon R. A., Talanian R. V., Keegan M., Herzog L., Allen H. (1998) Caspase-mediated fragmentation of calpain inhibitor protein calpastatin during apoptosis. Arch. Biochem. Biophys 356, 187–196 [DOI] [PubMed] [Google Scholar]
- 31.Abrahmsén L., Tom J., Burnier J., Butcher K. A., Kossiakoff A., Wells J. A. (1991) Engineering subtilisin and its substrates for efficient ligation of peptide bonds in aqueous solution. Biochemistry 30, 4151–4159 [DOI] [PubMed] [Google Scholar]
- 32.Atwell S., Wells J. A. (1999) Selection for improved subtiligases by phage display. Proc. Natl. Acad. Sci. U.S.A 96, 9497–9502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braisted A. C., Judice J. K., Wells J. A. (1997) Synthesis of proteins by subtiligase. Methods Enzymol 289, 298–313 [DOI] [PubMed] [Google Scholar]
- 34.Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng 14, 993–1000 [DOI] [PubMed] [Google Scholar]
- 35.Stennicke H. R., Salvesen G. S. (1999) Caspases: preparation and characterization. Methods 17, 313–319 [DOI] [PubMed] [Google Scholar]
- 36.Brown J. L., Roberts W. K. (1976) Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J. Biol. Chem 251, 1009–1014 [PubMed] [Google Scholar]
- 37.Chang T. K., Jackson D. Y., Burnier J. P., Wells J. A. (1994) Subtiligase: a tool for semisynthesis of proteins. Proc. Natl. Acad. Sci. U.S.A 91, 12544–12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnoy S., Kosower N. S. (2003) Caspase-1-induced calpastatin degradation in myoblast differentiation and fusion: cross-talk between the caspase and calpain systems. FEBS Lett 546, 213–217 [DOI] [PubMed] [Google Scholar]
- 39.Prasad S. C., Thraves P. J., Kuettel M. R., Srinivasarao G. Y., Dritschilo A., Soldatenkov V. A. (1998) Apoptosis-associated proteolysis of vimentin in human prostate epithelial tumor cells. Biochem. Biophys. Res. Commun 249, 332–338 [DOI] [PubMed] [Google Scholar]
- 40.Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Calvo M., Peterson E. P., Rasper D. M., Vaillancourt J. P., Zamboni R., Nicholson D. W., Thornberry N. A. (1999) Purification and catalytic properties of human caspase family members. Cell Death Differ 6, 362–369 [DOI] [PubMed] [Google Scholar]
- 42.Roberts T. L., Idris A., Dunn J. A., Kelly G. M., Burnton C. M., Hodgson S., Hardy L. L., Garceau V., Sweet M. J., Ross I. L., Hume D. A., Stacey K. J. (2009) HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science 323, 1057–1060 [DOI] [PubMed] [Google Scholar]
- 43.Gao J., Sidhu S. S., Wells J. A. (2009) Two-state selection of conformation-specific antibodies. Proc. Natl. Acad. Sci. U.S.A 106, 3071–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shao W., Yeretssian G., Doiron K., Hussain S. N., Saleh M. (2007) The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem 282, 36321–36329 [DOI] [PubMed] [Google Scholar]
- 45.Walsh J. G., Cullen S. P., Sheridan C., Lüthi A. U., Gerner C., Martin S. J. (2008) Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. U.S.A 105, 12815–12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McStay G. P., Salvesen G. S., Green D. R. (2008) Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ 15, 322–331 [DOI] [PubMed] [Google Scholar]
- 47.Deleted in proof
- 48.Lakhani S. A., Masud A., Kuida K., Porter G. A., Jr., Booth C. J., Mehal W. Z., Inayat I., Flavell R. A. (2006) Caspases 3 and 7: key mediators of mitochondrial events of apoptosis. Science 311, 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eder C. (2010) Mechanisms of interleukin-1beta release. Immunobiology, in press [DOI] [PubMed] [Google Scholar]
- 50.Nour A. M., Yeung Y. G., Santambrogio L., Boyden E. D., Stanley E. R., Brojatsch J. (2009) Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect. Immun 77, 1262–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgeworth J. D., Spencer J., Phalipon A., Griffin G. E., Sansonetti P. J. (2002) Cytotoxicity and interleukin-1beta processing following Shigella flexneri infection of human monocyte-derived dendritic cells. Eur. J. Immunol 32, 1464–1471 [DOI] [PubMed] [Google Scholar]
- 52.Yocum A. K., Chinnaiyan A. M. (2009) Current affairs in quantitative targeted proteomics: multiple reaction monitoring-mass spectrometry. Brief. Funct. Genomic. Proteomic 8, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.