Abstract
Pseudomonas chlororaphis phage 201φ2-1 produces a large structurally complex virion, including the products of 89 phage genes. Many of these proteins are modified by proteolysis during virion maturation. To delineate the proteolytic maturation process, 46 slices from an SDS-polyacrylamide gel were subjected to tryptic digestion and then HPLC-electrospray ionization-tandem mass spectrometry analysis. The scale of the experiment allowed high sequence coverage and detection of mass spectra assigned to peptides with one end produced by trypsin and the other end derived from a maturation cleavage (semitryptic peptides). Nineteen cleavage sites were detected in this way. From these sites, a cleavage motif was defined and used to predict the remaining cleavages required to explain the gel mobility of the processed polypeptide species. Profiling the gel with spectrum counts for specific polypeptide regions was found to be helpful in deducing the patterns of proteolysis. A total of 29 cleaved polypeptides derived from 19 gene products were thus detected in the mature 201φ2-1 virion. When combined with bioinformatics analyses, these results revealed the presence of head protein-encoding gene modules. Most of the propeptides that were removed from the virion after processing were acidic, whereas the mature domain remaining in the virion was nearly charge-neutral. For four of these processed virion proteins, the portions remaining in the mature virion were mutually homologous. Spectrum counts were found to overestimate the relative quantity of minor polypeptide species in the virion. The resulting sensitivity for minor species made it possible to observe a small amount of general proteolysis that also affected the virions.
There has been a resurgence in interest in bacteriophages because of their potential for use in phage therapy (1, 2), their role at the base of the food chain (3–5), their abundance in metagenomic data (6), and their content of the last great unexplored repository of genetic diversity (7) and as models of complex macromolecular assemblies (8, 9). In bacteriophage genomics, there is a bewildering array of different genomes to characterize. Biochemical or genetic data are available for only a few prototypical phages. Most of the characterization of new phage genomes is derived by sequence comparison with other phages. This is complicated by the fact that phages distribute into a dozen or more groups with intergroup divergence of phage genes being greater than interkingdom divergence of cellular genes (10). Hence, phage-to-phage sequence comparison is often at or beyond the limit of the most advanced profile methods. Yet progress can still be made in modeling the biology of new viruses through observation of structural homology and seeking analogous features between new phage forms and established prototypes. The most abundant bacteriophages are the tailed phages, and the most divergent and challenging of them to characterize at the sequence level have been the giant phages of Pseudomonas (φKZ (11), EL (12), and 201φ2-1 (13)). Sequence comparison within this group is able to align many genes between 201φ2-1 and φKZ, reflecting their assignment to a separate phage genus (14). Although many genes in the third phage, EL, are detectably homologous to genes of the φKZ-like phages, others exhibit a low degree of similarity that makes assigning homology questionable (13). This three-phage group has not been given a taxonomic name, so we refer to them as the “φKZ-related” phages. That there is any prototype among well characterized phages for this group is mainly established by structural homology stemming from the physical characterization of the φKZ virion (9).
Cryoelectron microscopy (cryo-EM)1 studies of the φKZ virion have indicated a distant structural similarity of the capsid shell to the prototypical enterobacteria phage T4 (9), and electron microscopy studies of all three φKZ-related phages reveal a complex contractile tail for which T4 is also the structural paradigm. However, the φKZ-related phages have much more complex virions than T4 or any other characterized phage. Thirty proteins have been cataloged by MS in a tailless mutant of φKZ (15) in contrast to the 13 proteins that form the T4 head (16). Our previous MS study of phage 201φ2-1 identified at least nine proteins that had apparently been cleaved to a smaller size from their initial translation products. This is also analogous to T4, which expresses a prohead protease that cleaves the major capsid protein, a scaffold protein, a vertex protein, and seven other proteins, including itself, during the phage maturation process (16). Four of the processed T4 proteins are injected into the cell during the infection process (16, 17). The virion proteins of 201φ2-1 include subunits of an RNA polymerase (13). Hence, it appears that at least some of the processed proteins of the φKZ-related phages are also likely to be injected into the cell. However, our initial MS survey (13) was insufficient to establish the mature ends of the processed proteins, which is critical information, for example, in fitting the proteins into cryo-EM electron density maps or for defining domain boundaries for protein family construction, and we suspected that there was additional processing that had not yet been detected. Therefore, we mounted a more extensive MS study of the 201φ2-1 virion to completely define the proteins present within it.
Complicating the task of defining the mature ends of the virion proteins is the realization that phages mature in a hostile environment full of other proteases expressed by the host cells. Unlike a recombinant protein preparation, phages are not produced in genetically engineered bacteria that lack a cytoplasmic stress response protease. Moreover, protease inhibitors cannot be added prior to cell lysis to protect the virion proteins from the many proteases of the periplasmic space because intracellular phage maturation is dependent on protease action. Phages have evolved to resist generic protease attack, and most virologists ignore the possibility of general proteolysis of their phage preparations altogether. For example, protease inhibitors are never included in phage preparations because they are toxic to bacteria and may, therefore, interfere with phage infectivity. However, when analyzing a large number of mass spectra as we have done here, evidence of general proteolysis is mixed in with evidence of cleavage by the prohead protease. To separate these different kinds of cleavages, we used sequence profile methods to infer the cleavage site specificity of the prohead protease and plotted spectrum counts (SCs) for each protein fragment in each slice to visualize the distinction between completely cleaved sites and sites cleaved in only a small fraction of proteins. Finally, GBrowse (18) was used to visualize sequence coverage and to plot cleaved segments along a genomic coordinate system in the context of other information about the corresponding genes. By this combined approach, the observed cleavage patterns provided additional insight into the possible functions of the head proteins in these phages.
EXPERIMENTAL PROCEDURES
The phage 201φ2-1 virion preparations were generated as described (13), including use of step and buoyant density centrifugations in CsCl. SDS-PAGE was performed as described (19) with the exception that the phage was first heated to 75 °C for 4 min and then treated with 0.1 μg/ml DNase. Preliminary experiments were conducted to determine the maximum SDS gel protein load that did not overtly smear the banding pattern. Phage proteins (isolated from ∼1 × 1011 particles) were fractionated by one-dimensional SDS-PAGE and then stained with Coomassie Blue. The gel lane was manually dissected to 46 slices with care taken to avoid transecting visible Coomassie-stained bands. After digestion in situ with trypsin (Promega), the digests were analyzed by HPLC-ESI-tandem MS on a Thermo Fisher LTQ fitted with a New Objective PicoView 550 nanospray interface. On-line HPLC separation of the digests was accomplished with an Eksigent NanoLC micro-HPLC system: column, PicoFritTM (New Objective; 75-μm inner diameter) packed to 11 cm with C18 adsorbent (Vydac 218MS; 5 μm, 300 Å); mobile phase A, 0.5% acetic acid (HAc), 0.005% TFA; mobile phase B, 90% acetonitrile, 0.5% HAc, 0.005% TFA; gradient, 2–42% B in 30 min; flow rate, 0.4 μl/min. MS conditions were as follows: ESI voltage, 2.9 kV; isolation window for MS/MS, 3; relative collision energy, 35%; scan strategy, survey scan followed by acquisition of data-dependent CID spectra of the seven most intense ions in the survey scan above a set threshold. Dynamic exclusion was used with a repeat count of 3 and a duration of 30 s. Mascot (Matrix Science, London, UK) was used to search the uninterpreted CID spectra against a locally generated 201φ2-1 protein database (13) that had been concatenated with the Swiss-Prot (version 51.6) database, totaling 216,849 sequences. Methionine was considered as a variable modification, and semitrypsin was selected as the proteolytic specificity. Mass tolerances for the Mascot searches were 1.0 Da for precursor ions and 0.8 Da for fragment ions. Determination of probabilities of protein identifications and cross-correlation of the Mascot results with X! Tandem were accomplished by Scaffold (Proteome Software, Inc., Portland OR). Unless otherwise noted, a 95% confidence limit was used for assignment of peptides.
The tandem MS results obtained from the digest of each gel slice were searched separately using Mascot, and the data files were either evaluated individually or combined into data sets for processing by Scaffold (“MudPIT”). Searches for mass spectra matching unpredicted open reading frames were carried out with DNA ProteoIQ (BioInquire, Athens, GA). Spectrum count profiles were produced by Microsoft Excel from a spectrum count data file exported from Scaffold. For more detailed SC gel profiling, a Perl script was written to process the spectrum report written by Scaffold and produce a similarly formatted spectrum count data file featuring the profile of peptides belonging to specific subregions of the gene product. The program may be obtained from S. C. Hardies. A cleavage site motif logo was produced using WebLogo (20). A hidden Markov model (HMM) corresponding to the logo alignment was prepared with a local implementation of the Sequence Analysis and Modeling System (21, 22) using prior information to set the probability of novel residues occurring in the motif as specified in the associated w0.5 model building script. The resulting HMM was manually edited to set gapping probabilities to zero and to make a glutamate at position −1 mandatory based on the observed invariance of this residue. With gaps disallowed, the HMM scoring algorithm essentially becomes a simple profile matching engine that scores for consistency with the profile as visualized by the logo. GBrowse was implemented as described (13, 18). Perl scripts were written to read Scaffold spectrum reports and Sequence Analysis and Modeling System output files and to graph peptide coverage and cleavage motifs within GBrowse as sequence features.
The SC/molecular mass where indicated for each observed gene product (gp) was calculated and taken as a crude indicator of abundance as described (13). Proteins of known abundance in the virion exhibited SC/molecular mass as follows: gp200 (capsid protein, 1,570 copies/virion), 31.4; gp30 (sheath, 264 copies/virion), 16.5; and tape measure/cell-puncturing device (three copies/virion), 1.2. The efficiency of detection of SC from different proteins was assumed to vary widely; hence, SC/molecular mass was used only to infer that proteins with SC/molecular mass in the range of 1 or below are likely to be present in few copies or even less than one copy per virion, and proteins with SC/molecular mass greater than 10 are probably present in over 100 copies per virion. Results were generally consistent with our previous study (13) with only one protein previously considered to be abundant, gp146, falling markedly below this threshold in the current study.
RESULTS
From analysis of the HPLC-ESI-MS/MS data, 14,643 mass spectra were assigned to 1,567 201φ2-1 tryptic peptides. A breakdown of the data by slice can be found in supplemental Table 1. Peptides were detected from 13 additional virion-associated proteins beyond those reported previously (13). The newly observed virion-associated proteins included the following: gp4, gp71, gp118, gp122, gp127, gp161, gp249, gp270, gp272, gp355, gp383, gp406, and gp429. This brings the total number of genes encoding virion-associated proteins in 201φ2-1 to 89. All of the previously undetected virion proteins were observed in slices consistent with their unprocessed molecular masses, and all were found with relatively low SC/molecular mass, suggesting a low copy number per virion. SC/molecular mass is tabulated for these and other unprocessed virion proteins in supplemental Table 2. Observation of these additional low abundance proteins is consistent with the larger number of gel slices having permitted detection of a wider dynamic range of protein quantity. Because these newly observed proteins were apparently present in low abundance, we cannot exclude that they might be nonspecifically absorbed to the virions.
Semitryptic Spectra and SC Gel Profiles
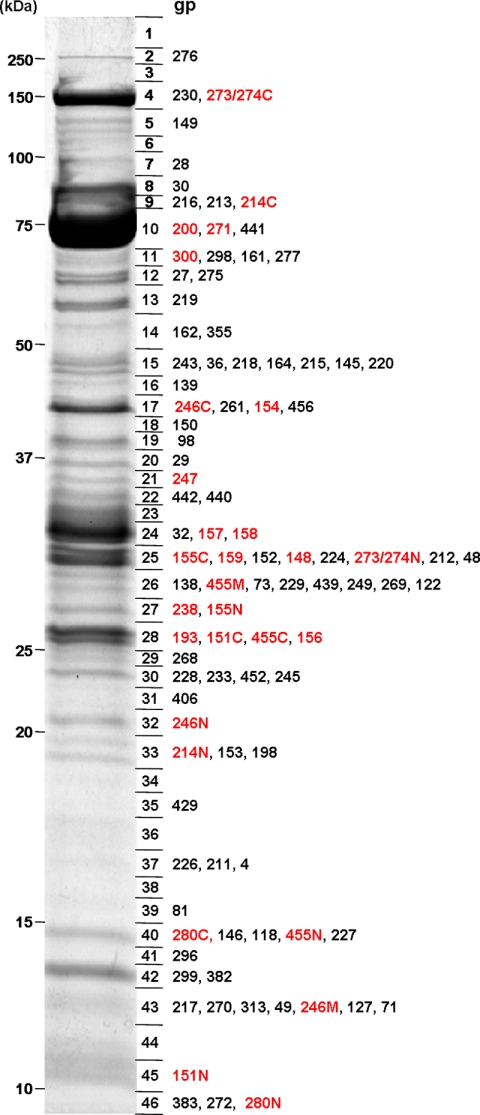
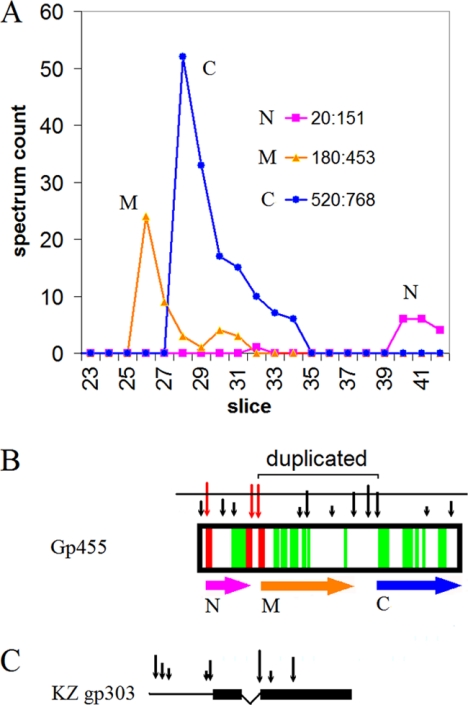
In addition to detecting new virion proteins, substantial additional evidence was recovered to clarify the cleavage patterns of the known virion proteins. Fig. 1 shows a Coomassie-stained image of the gel prior to MS analysis with polypeptide species labeled in the slices where they have maximum SC. Each polypeptide species indicated in red was concluded to have been posttranslationally cleaved based on the analysis discussed below. Fig. 2 illustrates the approach used to determine that one gene product, gp455, had been cleaved to three mature polypeptides. In our previously reported experiment (13), gp455 migrated on the SDS-PAGE gel as if it were only half its predicted size, but there was coverage of peptides over most of the protein sequence. In the experiment described here, three semitryptic peptides were identified originating from gp455 (Fig. 2B, red), revealing that gp455 was not simply cut in half. Included in the processing pattern was a small, N-terminal fragment (corresponding to residues 20–151) that had been cleaved from the initial translation product, and small propeptides on either side had apparently been removed from the virion. However, it was necessary to use the approach described below to elucidate the remainder of the processing pattern of this protein.
Fig. 1.
SDS-PAGE of 201φ2-1 virion proteins. The boundaries of slices that were digested and analyzed by MS are shown immediately to the right of the gel. Each virion protein is listed next to the slice containing its maximum SC. For virion proteins listed in red, the polypeptide has been altered by proteolytic cleavage by the prohead protease. In cases where more than one polypeptide segment remained in the virion, the segments have been annotated with N, M, or C to indicate N-terminal, middle (if present), and C-terminal segments, respectively.
Fig. 2.
Analysis of gp455 proteolytic processing. A, SC gel profile. Detected peptides have been subdivided into N-terminal, middle, and C-terminal regions covering the residue ranges indicated. No peptides were detected in other slices or outside of the indicated ranges. B, peptide coverage. Semitryptic peptides are indicated in red. Above the plot, the bit score of prospective cleavage sites is indicated by the height of a vertical arrow. A horizontal line is drawn, dividing cleavage motifs that were considered high scoring from those considered low scoring as described in the text. The two cleavage sites indicated as “duplicated” arose through a domain duplication. Horizontal arrows indicate the inferred gp455N, -M, and -C polypeptide segments. C, correspondence of the φKZ homolog gp303. Black boxes indicate regions of similarity as detected by Blast. Vertical arrows indicate bit scores of prospective cleavage motifs. The thin line to the left is an extended N-terminal region that is not present in 201φ2-1 gp455. The connector between the two boxes indicates that this region is deleted relative to 201φ2-1 gp455.
Isolation of protein from a larger number of phage particles in the current study compared with our previous report (13) permitted confident assignment of substantially more tandem mass spectra. This made it possible to plot the number of mass spectra assigned to peptides for each protein (the SC) across the gel slices to form a robust gel profile revealing the distribution of the polypeptide (Fig. 2A). In experiments with many fewer spectra or without an attempt to count the number of spectra per slice, the gene products often appear to be smeared over a broad range of slices. In those cases, it is easy to be fooled into believing that the main peak of the protein had migrated in the slice corresponding to its full-length translation product when it actually had migrated like a shorter polypeptide. As a result, internal cleavages can be overlooked as was the case with the N-terminal gp455 fragment described above. However, with the ability to examine an SC profile with significant depth in the SC dimension, a clear peak position emerges for each polypeptide species. With knowledge from the semitryptic peptide data that residues 20–151 of gp455 comprise a separate polypeptide fragment, we plotted SC from that range as shown in Fig. 2 (magenta). The number of assigned peptides for residues 20–151 peak in slice 40 was consistent with the calculated size for this N-terminal segment (gp455N). Another semitryptic fragment from gp455 defined the end of an additional mature polypeptide that begins at residue 180. But a polypeptide from 180 to the C terminus is too large to be migrating in and below slice 25 where the gp455 peptides from this region of the sequence were found. Dividing the region roughly in half and separately plotting the corresponding peptides revealed that the gel has resolved a middle (gp455M) and C-terminal (gp455C) segment (Fig. 2A). However, there were no semitryptic peptides assigned to indicate exactly which cleavage sites define these two segments.
Cleavage Motif
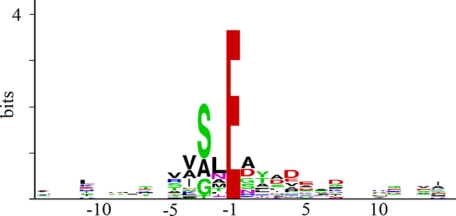
To understand more exactly the ends of the gp455M and gp455C segments and similar instances in others of the processed proteins, it was necessary to devise a method to recognize the cleavage motif. The three semitryptic peptides detected in gp455 exhibited cleavage after a glutamate. The bacteriophage T4 prohead protease cleaves the major T4 capsid protein and others after glutamate (7, 18). The major capsid proteins of the related phages φKZ and EL are also cleaved after glutamate (11, 12). Nineteen semitryptic peptides detected in 201φ2-1 virion proteins (including gp455) exhibited cleavage after glutamate (Table I). However, because of the abundance of glutamate residues in the 201φ2-1 proteome, simply considering every glutamate to be a prospective prohead protease cleavage site was not helpful in further defining the ends of the cleaved species. Therefore, a more restrictive prohead protease motif was sought by examining the alignment of the 19 established 201φ2-1 cleavage sites.
Table I. Cleavage sites of 201φ2-1 proteins determined by detection of semitryptic peptide.
| gp | Residuea | Siteb |
|---|---|---|
| 148 | 113 | ISQE ATLAc |
| 151 | 166 | ASLE DGADc |
| 155 | 13 | VSTE DFADc |
| 155 | 327 | AALE ATAV |
| 158 | 60 | LSLE ATDQ |
| 193 | 139 | GSLE GYAD |
| 238 | 64 | ISTE ALSI |
| 246 | 16 | IANE SVSH |
| 246 | 128 | TSVE EHSD |
| 246 | 203 | ASIE RYIE |
| 246 | 258 | VSLE ATGV |
| 247 | 50 | TALE NIDPc |
| 271 | 61 | VAVE DHFD |
| 274/3 | 275 | VTFE TYQD |
| 280 | 15 | KGLE SADG |
| 280 | 72 | VSAE SATV |
| 455 | 20 | VSNE DAFDc |
| 455 | 151 | PGME HYNP |
| 455 | 179 | ISNE GLIAc |
a The coordinate of the conserved glutamate immediately preceding the cleavage.
b Sequence identified in a semitryptic fragment is underlined.
c Peptide assigned with a minimum peptide identification probability <95% according to Scaffold.
The cleavage motif defined by the 19 semitryptic peptides is represented by the sequence logo in Fig. 3. The logo is scaled to reveal the frequency at which individual residues appear beyond the expectation for a random sequence of the same composition (20). Fig. 3 shows that besides the requirement for glutamate at −1 there are several other preferences in the residues at the cleavages sites. Most prominently, there is a preference for a short side chain amino acid at −3. The logo, besides giving a visual impression of the preferences of the protease, carries quantitative information as well. By definition, a logo is scaled such that the likelihood of matching one of the listed letters in a random sequence is 2 to the power of the height of the stack of letters (23). Summing the heights of the logo from −5 to +7 gives a total bit score of 11.5 counting the glutamate at −1 or 7.7 without counting the glutamate. This corresponds to one false positive per ∼3,500 residues or one false positive per ∼200 glutamate residues. These numbers indicate that there is enough information available to use sequence recognition to aid finding the cleavages sites. Actual search strategies can be expected to produce more false positives than this theoretical limit, but the theoretical limit provides a performance target for the design of the search strategy. We were able to implement a search strategy described below that has some flexibility for including residues in the motif that are not in the training set and still limited false positives to one per 1,200 residues. Because there is a large number of glutamates in the phage proteome, this degree of specificity will still produce many false matches. Motif matching alone probably can never eliminate false positives because access to the prospective cleavage site within the tertiary structure of the protein must be a factor. However, by limiting the search to sites in appropriate positions to explain the SDS gel mobility of the respective polypeptide species and to regions consistent with the tryptic peptide coverage, the specificity embodied in the logo was sufficient to allow the recognition of a likely cleavage site in every case.
Fig. 3.
Sequence logo representing 19 cleavage sites confirmed by detection of semitryptic peptide.
To establish a probabilistic scoring strategy for the prospective cleavage sites, the aligned cleavage sites represented in the logo from positions −5 to +7 were converted to an HMM. The HMM was used to search 201φ2-1 protein sequences for consistency with the logo. The bit scores of matching sites in gp455 are represented by the heights of the vertical arrows in Fig. 2B. A threshold was drawn (Fig. 2B, horizontal line) that separated the high scoring sites in positions to explain the detected polypeptides from lower scoring sites that were distributed more randomly. To estimate the confidence in a high scoring site, the frequency of high scoring sites that were presumed to arise by random chance in non-virion 201φ2-1 proteins was determined as was the frequency of high scoring sites in the proteins known to be processed. High scoring sites appeared 6 times more often in processed proteins (one per 200 residues) than in non-virion proteins (one per 1,200 residues). From these numbers, the confidence from the sequence match alone that a high scoring site is an authentic cleavage site can be set at 5/6 or about 80%. In the case of gp455, the two sites indicated in Fig. 2B were favored as the boundaries of the mature gp455M and gp455C polypeptides. This would indicate that there is an internal propeptide between these two segments that is also removed from the virion. An additional high scoring cleavage motif occurred in the middle of that prospective propeptide. Clustering of high scoring cleavage sites within prospective propeptides was a common occurrence among the processed set of 201φ2-1 genes (not shown). Hence, we hypothesize that these sites are also cleaved. The biological importance could be that redundancy in sites assures that important cleavages take place or that cutting the propeptides to smaller pieces helps them to clear out of the virion. Evidence for heterogeneous cleavage at clustered cleavage motifs is given below for the major capsid protein.
The graphic in Fig. 2B is a snapshot from a dynamic GBrowse display through which we were able to collate other analytical information with the MS results. Fig. 2C shows a comparison for the φKZ homolog KZ gp303 and 201φ2-1 gp455. This comparison reinforces the cleavage site predictions in several ways. φKZ has two segments of BlastP similarity to 201φ2-1 gp455 (Fig. 3C, black boxes) that correspond to the projected mature N and M segments of gp455. High scoring cleavage motifs are predicted for the N segment and the beginning of the M segment of φKZ gp303, although the propeptide separating segments N and M appears to have been deleted. The φKZ gp303 reading frame ends at the site projected for cleavage of the M segment in 201φ2-1. This supports the conclusion that the projected C terminus of gp455M is at the end of a self-contained protein domain. That projected cleavage site scored well by the HMM search algorithm even though it contained a residue (Gly) in position −2 that was absent in the training set. Probabilistic scoring methods can allow the accumulated bit score of well matching positions to overrule mismatching positions in this way. Without this property, it would not be possible to incorporate weakly preferred residues into a motif description. Using Psi-Blast the φKZ gp303 C-terminal segment matched to both gp455M and gp455C segments. This revealed that the M and C segments of gp455 are derived by a domain duplication. The site projected by the HMM search for the beginning of the gp455C segment, therefore, is homologous to the site confirmed by semitryptic peptide data at the beginning of the gp455M segment.
In some cases, the φKZ sequence that aligned with the established 201φ2-1 cleavage did not conform to the cleavage motif, but instead there was a new high scoring motif nearby. Both cleavages defining gp455N and the homologous region in φKZ gp303 are examples of this effect. We suspect that redundant cleavage sites often come and go during evolution, thus allowing drift in the exact cleavage position while conserving the overall processing pattern.
Heterogeneity in Programmed Processing
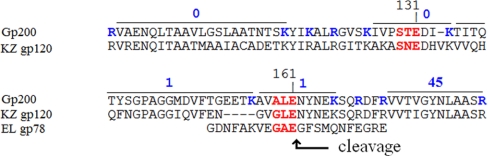
The major capsid protein, gp200, is the 201φ2-1 protein with the most prior knowledge about proteolytic processing. gp200 is the homolog of the major capsid proteins of φKZ and EL for which prohead protease processing sites have been identified by Edman degradation (11, 12). There were no semitryptic spectra observed to indicate where the prohead protease cleavage site is located in 201φ2-1 gp200. In general agreement with φKZ and EL, the gp200 tryptic peptides with high SC were located downstream of the homologous prospective cleavage site. For example, the first peptide positioned downstream of the prospective cleavage at residue 161 that was assigned with high confidence was detected 45 times in the combined MudPIT data set (Fig. 4). However, there were two assigned peptides (detected once each) upstream of residue 161 that extended peptide coverage to residue 135. These results would be consistent with a situation in which the majority of mature gp200 molecules start at position 162, similar to the case in φKZ and EL, with a small fraction of mature capsid polypeptides extending upstream. This hypothesis is supported by recognition of both positions 131 and 161 as high scoring cleavage motifs by the cleavage motif HMM. Similarly, both sites were conserved as high scoring sites in φKZ. Although the EL sequence is not objectively alignable in this region, the sequence immediately upstream of the established mature end was also a high scoring cleavage motif. We propose that positions 131 and 161 are redundant cleavage sites that assure that this critical maturation event goes to completion. In a small minority of molecules, cleavage at 161 is missed by the prohead protease, resulting in a polypeptide that extends to residue 132.
Fig. 4.
Heterogeneous processing of major capsid protein. The spectrum count of tryptic peptides assigned within slice 10 is given above the gp200 sequence. The count of 45 listed for VVTVGYNLAAS includes 20 spectra for peptides having additional residues to the left or right due to missed trypsin cleavages. Residue numbers refer to gp200. High scoring cleavage motifs are in red. Residues after which trypsin should cleave in gp200 are in blue. Cleavage at the indicated position for φKZ gp120 and EL gp78 was established by Edman degradation (11, 12).
A second element of heterogeneity was found at the C terminus of the major capsid protein in the form of a C-terminal extension created by terminator suppression. Although the initial gene predictions had not indicated a reading frame in this region (13), tblastn detected sequence similarity with the φKZ genome past the gp200 and φKZ gp120 C termini and into a genomic region with no other assigned function. To seek any missed translated regions, all acquired mass spectra were searched versus all 201φ2-1 sequences translated in all six frames. This resulted in detection of 10 spectra assigned to three peptides encoded downstream of the capsid stop codon. A plot of those 10 spectra by slice revealed a peak one slice above the peak slice for the major capsid protein (not shown). To be in this gel position indicates that the peptides belong to an extended version of gp200 and are not the product of an independent translational initiation. C-terminal extension through frameshifting is a known strategy for decorating a fraction of a major capsid protein with an extra exterior domain (24). In gp200, the extension consists of 29 residues and is generated by terminator read-through rather than programmed frameshifting.
Partial Cleavages in Exposed Regions of Major Capsid Protein
The SC gel profile of the major capsid protein (Fig. 5A) reveals a great deal more heterogeneity than expected from interaction with the prohead protease. gp200 is expected to be present in 1,560 copies per virion (13), far higher than any other 201φ2-1 protein, and is the predominant component of the intense band in slice 10 (Fig. 1). However, spectra assigned to gp200 are found in all slices of the gel. The down-gel SC profile for gp200 exhibits several major shoulders, and a number of the mass spectra were assigned to semitryptic peptides. Carryover in the HPLC is not an explanation for these down-gel smears because the slice digests were analyzed from the bottom of the gel to the top. There is no precedent for a major capsid protein to be proteolytically processed beyond removal of an N-terminal region, yet the shoulders are superficially like the separate peaks indicating processing in gp455. As discussed below, we have concluded that these shoulders are caused by cleavage of a fraction of the gp200 molecules by proteases other than the prohead protease. Most of the virion proteins exhibited down-gel smears of SC, and many exhibited semitryptic spectra that did not conform to the criteria discussed above for prohead protease processing. We explored all of the patterns for all of the proteins carefully to distinguish these other kinds of proteolytic attack from the actions of the prohead protease. The results for gp200 are discussed further below.
Fig. 5.
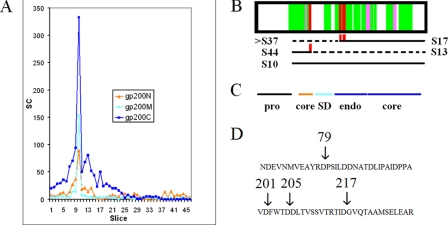
Processing of gp200 (major capsid protein). A, SC gel profile. The protein sequence has been subdivided into the following regions: N (before the stabilization domain), M (stabilization domain including linker regions), and C (after stabilization domain). B, sequence coverage for peptides assigned at ≥95% confidence. Semitryptic peptides are shown in red and pink. The pink peptides migrated in slice 10, which would indicate a derivation from an intact gp200 molecule rather than from a cleaved species. The slice numbers of species produced by the respective cleavages are indicated below the sequence coverage plot. S17 and S44 are slices where the indicated semitryptic peptides are found. For each cleavage, the slice number of the remainder of gp200 that can be inferred to have been produced is indicated next to a dashed line. The inferred partner to the S17 species is expected to be in slice 30 if intact, but instead its components are scattered below slice 37. S10 is the slice number of gp200 cleaved only by the prohead protease. C, position of gp200 domains. The core domains are those in common with T4 and HK97 major capsid proteins, whereas the SD domain is in common with T4 but not HK97 major capsid proteins (26). Endo refers to an endothiapepsin domain peculiar to φKZ-related phages (9). D, location of non-prohead protease cleavages within the gp200 sequence.
The gp200 semitryptic peptides shown in Fig. 5B can be divided into two categories. Semitryptic peptides of the first type (Fig. 5B, pink) are found in slice 10 where the intact protein migrates, which is inconsistent with putative cleavage having occurred prior to SDS-PAGE. These are presumed to be either misassigned spectra or nonspecific trypsin cleavages. These were all assigned with confidence >95% by Scaffold. It is apparent that relying on high confidence assignments of semitryptic spectra to indicate a cleavage without some other form of corroboration can be misleading.
The other gp200 semitryptic peptides (Fig. 5B, red) each appeared in a lower slice, consistent with the expected migration of a polypeptide ending at the inferred non-trypsin cleavage site. These cleavages occurred at residue 79 and in a cluster at positions 201, 205, and 217. These sites are at the boundaries of a domain called the stabilizing domain (SD; Fig. 5C) recognized by sequence similarity to the T4 major capsid protein (9, 26). SD is a small domain that contacts an adjacent capsid protein and is connected to its parent capsid protein by long extended linker chains that drape across the exterior surface of the capsid. The presence of these semitryptic peptides suggests that cleavages have occurred within these exposed linker regions. The cleavage at residue 79 appears to generate the semitryptic peptides found in slice 44 and the major shoulder found in slice 13 (Fig. 5A). The cleavages in the 201–217 region appear to generate the peak in slice 17. The N-terminal companion fragment to the species in slice 17, if intact, should appear in slice 30. Instead, peptides almost exclusively derived from the N-terminal region are scattered below slice 37, and peptides from the SD domain itself are relatively scarce outside of the main peak in slice 10. This leads to the impression that gp200 molecules that were cleaved once in the SD region were often then cleaved again and that some fraction of the gp200 molecules has lost the SD domain altogether. To have been physically lost from the sample, the SD domain would have to have been removed before the CsCl gradient, implying that the virion can sustain some degree of proteolytic damage without releasing its DNA.
The cleavages within gp200 discussed above were all after an aspartate (Fig. 5D), and there was no relationship between the sequences in these cleavage sites and the prohead protease motif. This indicates that another protease, presumably produced by the host and probably acting after lysis, attacked the virions from the exterior. The down-gel “smearing” suggests that there was cleavage at a heterogeneous collection of sites, not just at those sites indicated by the assigned semitryptic peptides. In extending this analysis to others of the abundant virion proteins, the preference for cleavage after aspartate was not universal. However, the tendency to cleave at heterogeneous sites clustered at domain boundaries was observed in other proteins (not shown). A defining property of non-prohead protease cleavages was that they were always partial. In all cases, the major peak of SC from all subregions was in the slice with the intact major polypeptide. Hence, a large proportion of each virion protein was not cleaved at all by a non-prohead protease. We believe that the non-prohead protease cleavages represent proteolytic damage inflicted on the virion by protease it encounters in its microenvironment. As expanded under “Discussion,” we believe that the amount of non-prohead protease cleavage is exaggerated by SC and that most of the protein in the virion survives in an undamaged form.
DISCUSSION
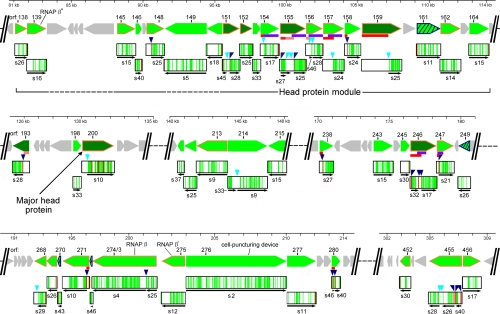
The principles derived from the study of gp455 and gp200 were applied to all 201φ2-1 virion proteins, revealing 29 processed polypeptides derived from 19 different initial translation products (Fig. 6 and Table II). As illustrated under “Results” for gp455, assignment of a processed polypeptide fragment was based on a combination of two criteria: 1) the SC gel profile for tryptic peptides from a subregion of the gene product exhibited a peak in a slice consistent with a segment of lower molecular mass than the full-size gene product and 2) the required cleavage(s) to produce such a segment could be justified based on either the observation of a semitryptic peptide or a match to the prohead protease cleavage motif. We believe that this process for cleavage assignment is substantially more accurate than just correlating peptide coverage with apparent molecular mass from gel migration. The motif matching criterion caused us to reject the processing hypothesis in a number of cases where a gene product lacked peptide coverage on an end. Such regions were often either lacking or very rich in arginine and lysine, explaining why they would have been poorly sampled by MS of trypsin-generated peptides. The higher molecular mass resolving power of the 46-slice gel was less helpful than one might have expected for rejecting the hypothesis that a polypeptide had been cleaved to a smaller species. A plot of molecular mass versus peak slice number of all unprocessed polypeptides showed a variability of two slices relative to the regression line (not shown). Hence, reliance on gel mobility alone to predict removal of a region lacking tryptic peptide coverage could easily generate many incorrect indications of proteolytic processing.
Fig. 6.
Regions of 201φ2-1 genome encoding processed virion proteins. Genomic segments from the full GBrowse display of 201φ2-1 were juxtaposed, and protein features were mapped onto the corresponding genes. Green arrows, proteins detected by mass spectrometry; dark green arrows, virion proteins that were determined to be abundant (13); diagonally striped green arrows, virion-associated proteins newly detected in this study; orange outlined arrows, proteins for which the φKZ homologs were found to be present in a tailless mutant (15); gray arrows, non-virion proteins; purple horizontal bars, a region of sequence homology found in several 201φ2-1 proteins; red horizontal bars, a region with greater than one net negative charge per 10 residues; pink horizontal bars, a region with greater than one net negative charge per 20 residues. Peptide coverage is indicated as follows in the boxed regions below each gene glyph: green, tryptic peptide coverage; red, semitryptic peptide. Triangles above the peptide coverage boxes indicate prohead protease cleavage sites: dark blue, established by a semitryptic peptide; cyan, predicted by motif searching. Thin black arrows below the peptide coverage boxes correspond to the position of a mature polypeptide and corresponding slice number. Two alternative interpretations are shown for gp155N. RNAP, RNA polymerase.
Table II. Prohead protease-processed segments of 201φ2-1 virion proteins.
| gpa | Slice | Residues |
Molecular massb | SCc | SC/molecular massd | |
|---|---|---|---|---|---|---|
| Start | Stop | |||||
| kDa | ||||||
| 148 | 25 | 114e | 414 (C)f | 34.9 | 134 | 3.8 |
| 151N | 45 | 1 | 112 | 11.9 | 91 | 7.6 |
| 151C | 28 | 167e | 400 (C) | 26.4 | 262 | 9.9 |
| 154 | 17 | 80 | 426 (C) | 40.3 | 46 | 1.1 |
| 155N | 27 | 14e | 275 | 27.8 | 67 | 2.4 |
| 155Na | 27 | 14e | 139 | 13.1 | 67 | 5.1 |
| 155C | 25 | 328e | 608 (C) | 30.7 | 378 | 12.3 |
| 156N | 46 | 86 | 157 | 8.0 | 23 | 2.9 |
| 156C | 28 | 158 | 387 (C) | 25.5 | 135 | 5.3 |
| 157 | 24 | 208 | 489 (C) | 31.4 | 225 | 7.2 |
| 158 | 24 | 61e | 378 (C) | 34.9 | 48 | 1.4 |
| 159 | 25 | 631 | 922 (C) | 31.3 | 369 | 11.8 |
| 193 | 28 | 140e | 381 (C) | 27.2 | 472 | 17.4 |
| 200 | 10 | 162 | 746 (C) | 65.1 | 2045 | 31.4 |
| 214N | 33 | 1 | 208 | 22.9 | 23 | 1.0 |
| 214C | 9 | 217 | 917 (C) | 78.6 | 64 | 0.8 |
| 238 | 27 | 65e | 316 (C) | 28.6 | 165 | 5.8 |
| 246N | 32 | 17e | 128e | 11.7 | 95 | 8.1 |
| 246M | 43 | 129 | 203e | 7.8 | 35 | 4.5 |
| 246C | 17 | 258e | 613 (C) | 38.7 | 761 | 19.7 |
| 247 | 21 | 51e | 397 (C) | 38.8 | 191 | 4.9 |
| 268 | 29 | 1 | 225 | 24.7 | 99 | 4.0 |
| 271 | 10 | 61e | 629 (C) | 63.6 | 396 | 6.2 |
| 274/3N | 25 | 1 | 275e | 32.5 | 35 | 1.1 |
| 274/3C | 4 | 276 | 1500 (C) | 137.2 | 327 | 2.4 |
| 280N | 46 | 16e | 72 | 6.1 | >7 | >1.1 |
| 280C | 40 | 73e | 199 (C) | 13.5 | 100 | 7.4 |
| 455N | 40, 41 | 21e | 151e | 15.0 | 17 | 1.1 |
| 455M | 26 | 180e | 453 | 30.7 | 44 | 1.4 |
| 455C | 28 | 521 | 768 (C) | 28.5 | 140 | 5.3 |
a The gene product name is appended with N (for N-terminal), M (for middle), or C (for C-terminal) when more than one segment of a gene product was detected in the virion. gp155N and gp155Na are alternative interpretations of the N-terminal gp155 segment (see text).
b The molecular mass was calculated from the sequence.
c SCs were summed over all slices. The peak of gp280N may be off the bottom of the gel.
d SC/molecular mass values were calculated as a rough indicator of abundance of the polypeptides per virion according to Ref. 13. Based on comparisons of values for the major capsid protein, sheath protein, and tape measure protein, a value of SC/molecular mass over 10 probably represents more than 100 molecules per virion.
e Confirmed by a semitryptic spectrum. Semitryptic spectrum assignments were above 95% confidence except as noted in Table I.
f A coordinate is appended with (C) for an end defined by the stop codon.
Because we used SDS gel mobility to define where to look for prospective cleavage sites, the variability in SDS gel mobility was a factor in determining the confidence in the cleavage site predictions. The range of possible end points for a fragment based on gel migration and peptide coverage could usually be confined to about 30 residues. The properties of the cleavage prediction model with respect to false positive and false negative predictions (see “Results”) should usually produce a correct prediction given that the cleavage site is known to within 30 residues. However, some false predictions are possible, and in particular, propeptides less than 30 residues may have been missed. The strength of the cleavage site predictions was mainly drawn from the large number of sites defined by semitryptic fragments that were used to train the prediction HMM. In phages with fewer processed proteins, it would be necessary to draw on cleavage sites from proteins in homologous phages to derive a well trained model. The most critical property of the cleavage model is the rate of false positive prediction; hence, we believe that the negative control performed by screening all of the nonstructural proteins of the phage to establish this rate is crucial for using this strategy.
As illustrated for gp200 (see “Results”), non-prohead protease cleavages were found to also produce down-gel peaks and shoulders and generate semitryptic peptides. Non-prohead protease cleavages were easily distinguishable from prohead protease cleavages because they were “partial,” that is most of the SC from the entire protein sequence migrated with the intact polypeptide, not the fragmented polypeptide. Moreover, the cleavages suggested by the those semitryptic fragments did not conform to the prohead protease cleavage motif. We considered the possibility that some of the major down-gel peaks and shoulders may have been derived from partial cleavages by the prohead protease at low scoring cleavage motifs. However, we could find no correlation between the apparent sizes of the fragments represented by down-gel peaks and shoulders and the positions of low scoring motifs. Instead, as described in detail for gp200 (see “Results”), the down-gel material appeared to be consistent with partial proteolysis by other proteases presumably not encoded by the virus. Some degree of general proteolysis is hardly surprising because the virus is released on lysis into a soup of periplasmic proteases. The periplasm is the digestive compartment of the bacteria where proteins are degraded to amino acids for transport into the cell.
In this study, the patterns of generalized proteolysis form a background from which the prohead protease cleavages had to be distinguished. For the major capsid protein, the pattern of these cleavages correlates with the domain structure and exposed surfaces of the protein. One could design an experiment to use such information to clarify the structure of virion proteins whose conformation is not known; it could also be used to study the inactivation of virions in the environment. In such studies, protease inhibitors should be used starting in the buoyant density step of the virus preparation. That step separates intact viruses from empty heads, which would logically carry a different cleavage pattern. Protease inhibitors would prevent subsequent development of empty heads that might be acted upon by proteases still contaminating from the cell lysate. We explored whether the gp200 SC migrating down-gel of its main peak (Fig. 5A) might correspond to the fraction of degraded virions in our virus preparation. The ratio of Coomassie staining in low abundances slices versus the intensely staining slice 10 (Fig. 1) was much less than the ratio of total SC among the same slices (not shown). We, therefore, believe that the virus preparation was not substantially degraded after purification but rather that spectrum counting is disproportionately sensitive to sampling low abundance polypeptide species. The use of dynamic exclusion was probably a major contributor to this effect.
Assignment of the processed polypeptides through correlation of SC gel profiles and termini indicated by semitryptic peptides or matches to the prohead protease cleavage motif appeared unambiguous, except for peptide fragments mapping in the N-terminal regions of gp155 and gp246. Both ends of gp246N were defined by semitryptic peptides, but the peptides were found in a slice associated with a much larger molecular mass than that polypeptide would have. The reason for this discrepancy did not become clear until after GBrowse was used to correlate other properties of the sequences with the peptide coverage (Fig. 6). The relevant feature was the content of high negative charge (i.e. high concentration of acidic residues) indicated by the red or pink bars in Fig. 6. Searching for this pattern was motivated by the hunt for a likely scaffold protein (see below). Polypeptides of high negative charge are known to exhibit abnormally slow migration in SDS-PAGE (27). gp264N has greater than one net negative charge per 10 residues, thus explaining its slow migration. With the realization that a number of other regions within the 201φ2-1 virion proteins were highly acidic, we examined those regions for assignments that might have been misled by a similar slow migration phenomenon. Another possible case occurs for gp155N. The peptide coverage is confined to a small negatively charged region. There are prohead protease motifs that could produce a gp155N polypeptide consistent with normal SDS gel migration. But if the fragment migrates anomalously slowly like gp246N, then there is a different C-terminal protease motif that could produce a smaller fragment and still include all the observed peptides. Both possibilities are indicated in Fig. 6.
Understanding Function of 201φ2-1 Protein Processing
If all of the 201φ2-1 proteins that are processed by the prohead protease are involved in head maturation, there should be a correspondence between these proteins and the φKZ homologs that were detected in a tailless mutant (Fig. 6, genes with orange outlines, and Ref. 15). There is a reasonable, although not complete, agreement between these two sets of gene products. For one of the processed 201φ2-1 proteins, gp193, there is no similar sequence in φKZ at all. However, there were processed 201φ2-1 proteins that do have φKZ homologs that did not appear in the tailless mutant data. Although it is possible that the tailless mutant is missing some head proteins in addition to its tail proteins, we think that this is not the case. The processed 201φ2-1 proteins whose homologs are missing from the φKZ tailless mutant data (gp154, gp158, gp214, gp238, and gp280) are of low abundance in the 201φ2-1 virion, and the abundance of their φKZ homologs may have been below the threshold of detection by MS in the analysis of the tailless mutant. We feel it is likely that the head proteins are similarly extensively processed in the other φKZ-related phages. In examining the sequences of the homologous φKZ proteins, we found that about half of the cleavage sites have a high scoring cleavage motif in the homologous position, and the others have a high scoring cleavage motif nearby (not shown). We, therefore, believe that the φKZ-related phages as a group have an unprecedented number of processed head proteins and that this follows from having an unprecedented total number of head proteins. Electron microscopy (28), atomic force microscopy (29), and cryo-EM (9) studies have suggested that the φKZ-like phages have an inner head body, which should account for at least some of the head proteins. It is unclear at this time whether the inner body plays a structural role in organizing the capsid or represents a bolus of proteins prepared for injection into the cell. Whereas it was once thought that only the phage DNA entered the cell on infection, it is now appreciated that many phages also introduce proteins (30).
In a study of T4 head proteins (31), it was found that N-terminal propeptides often contained a capsid targeting sequence that was responsible for assembling the protein into the prohead. The subsequent removal of these propeptides prepares the proteins for injection into the cell. Although we have not yet identified a capsid localization sequence, the abundance of head proteins with propeptides suggests that a similar assembly principle is at work in φKZ-related phages. Our initial characterization of phage 201φ2-1 indicated that the virion contained several subunits of an RNA polymerase (13). One of these subunits (Fig. 6, gp274/3) has now been found to be processed, consistent with the use of a propeptide to attain an interior head location. However, other virion-associated 201φ2-1 RNA polymerase subunits (gp139 and gp275) do not appear to have propeptides. So, there may be more than one way to assemble a protein into the head of 201φ2-1 and its related phages.
Connection between Head and Tail Assembly
There is one major difference in proteolytic processing between 201φ2-1 and φKZ. The results of Edman degradation indicated that the φKZ gp181 protein, which includes the tail lysozyme domain, is processed (11); however, we found no evidence for processing of the 201φ2-1 homolog (gp276). The tail lysozyme is on the end of the tail that is distal from the head. The prohead protease is thought to be confined to the head before it becomes activated and then to proteolytically inactivate itself and mostly leave the head during maturation (32). Prohead protease activity presumably has to be tightly confined to the head because leaking into the cytoplasm and cleaving other head proteins prior to assembly would stop the assembly process. Yet we found that the cleavage site for tail protein φKZ gp181 is a high scoring match to the prohead protease processing motif (not shown), implying that it is cut by the prohead protease. This would seem to require that φKZ gp181 associates with the head to be cleaved. We believe that is exactly what happens. The lysozyme domain of the tail protein in both phages accounts for only a small portion of the C terminus of a very large protein. We assign the rest of KZ gp181 and its homologs to be the tape measure protein, which extends the length of the tail. This assignment is supported not only by the size of KZ gp181 but also by a significant content of predicted α helical and coiled coil structure, both characteristic of tape measure proteins (33, 34). A fusion of the tail lysozyme with the tape measure has been reported for several other phages (35, 36). The cleavage site in φKZ gp181 is near the N terminus of the tape measure domain, which must come into close proximity to the head in the final stage of assembly. Hence, we propose that the cleavage of the φKZ tail lysozyme/tape measure protein occurs during tail attachment to the prohead.
Clustering of Head Structure Genes
One strategy for characterizing a new phage genome that has many novel genes is to use any clues available to organize the novel genes into functional clusters. Genes of an underlying common functionality often cluster in phage genomes. The usual explanation for clustering is that genes that are coadapted to function together stay together and under a regimen of horizontal transfer become clustered together to transfer together. The φKZ-related phages lack the single global head structure gene organization common to many tailed phages (25). However, Fig. 6 reveals that the head structure genes of 201φ2-1, including those inferred by the detection of prohead protease processing or by the presence of the φKZ homolog in the tailless mutant, are found in several clusters. Of these, a homolog of gp193 is not found in φKZ (13), and the clusters containing gp243–249 and gp452–456 are not found in the related phage EL. So the genes within those regions may be considered to encode auxiliary functions rather than proteins that participate in the conserved head assembly program of the φKZ-related phages. The largest remaining cluster is enriched in genes encoding proteins with extensively negatively charged N-terminal propeptides (gp154, gp155, gp156, gp157, gp158, and gp159). This property is typical of scaffold proteins. These propeptides also have patches of predicted coiled coil-like structure, which is also a characteristic of scaffold proteins. Hence, the scaffold function may be performed by one or more of these proteins. Of these, the portion of gp155 and gp159 that remains in the capsid is relatively abundant (Table II). The amount of propeptide that would have been present in the prohead would have also been abundant as appropriate for carrying out a scaffold function.
Summary
The current study takes an important step toward inferring the functions of the 201φ2-1 head proteins. Much work remains to be done. Defining the mature polypeptides aids making better protein profiles and hidden Markov models. Homologs of several known proteins are expected to be within the φKZ-related head and should eventually become ascertainable by these methods. These include identification of the portal protein, the rest of the RNA polymerase subunits, and the protease itself. The masses of several domains making up the φKZ capsid vertex have been estimated by cryo-EM (9). These proteins are known to be present in 55 copies per virion. So a combination of defining the mature polypeptides and better copy number estimates should allow assignment of the vertex proteins. An interesting set of genes is gp154, gp155, gp156, gp157, gp246, and gp247. The mature segments of these genes are all homologous to each other, although profile methods are required to produce an alignment (Fig. 6, purple bars). We have aligned these domains and their homologs from the other φKZ-related phages and submitted the family to the Pfam protein family database (PF12699). At this time, that profile does not match any other sequence. However, as additional genomes and metagenomes are sequenced and scanned against Pfam, we expect that additional homologs will be found that help to clarify the nature of this novel gene family. Finally, the now well established processing pattern in phage 201φ2-1 is available to predict the mature polypeptides formed in the other φKZ-related phages.
Supplementary Material
Acknowledgments
MS analyses were conducted in The University of Texas Health Science Center at San Antonio (UTHSCSA) Institutional Mass Spectrometry Laboratory. We acknowledge Efrem Holley for assistance in generating a Pfam entry for the gp155 family. We thank the UTHSCSA Bioinformatics Center for assistance with computational aspects of the project.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant GM24365. This work was also supported by grants from The Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation, the Welch Foundation (Grant AQ-764), and a UTHSCSA Research Council Pilot Grant.
 This article contains supplemental Tables 1 and 2.
This article contains supplemental Tables 1 and 2.
1 The abbreviations used are:
- cryo-EM
- cryoelectron microscopy
- HMM
- hidden Markov model
- SC
- spectrum count
- MudPIT
- multidimensional protein identification technology
- gp
- gene product
- SD
- stabilizing domain.
REFERENCES
- 1.Skurnik M., Strauch E. (2006) Phage therapy: facts and fiction. Int. J. Med. Microbiol 296, 5–14 [DOI] [PubMed] [Google Scholar]
- 2.Hanlon G. W. (2007) Bacteriophages: an appraisal of their role in the treatment of bacterial infections. Int. J. Antimicrob. Agents 30, 118–128 [DOI] [PubMed] [Google Scholar]
- 3.Wilhelm S. W., Suttle C. A. (1999) Viruses and nutrient cycles in the sea—viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49, 781–788 [Google Scholar]
- 4.Chibani-Chennoufi S., Bruttin A., Dillmann M. L., Brüssow H. (2004) Phage-host interaction: an ecological perspective. J. Bacteriol 186, 3677–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura M., Jia Z. J., Nakayama N., Asakawa S. (2008) Ecology of viruses in soils: past, present and future perspectives. Soil Sci. Plant Nutr 54, 1–32 [Google Scholar]
- 6.Williamson S. J., Rusch D. B., Yooseph S., Halpern A. L., Heidelberg K. B., Glass J. I., Andrews-Pfannkoch C., Fadrosh D., Miller C. S., Sutton G., Frazier M., Venter J. C. (2008) The Sorcerer II global Ocean sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3, e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoenfeld T., Liles M., Wommack K. E., Polson S. W., Godiska R., Mead D. (2010) Functional viral metagenomics and the next generation of molecular tools. Trends Microbiol 18, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostyuchenko V. A., Leiman P. G., Chipman P. R., Kanamaru S., van Raaij M. J., Arisaka F., Mesyanzhinov V. V., Rossmann M. G. (2003) Three-dimensional structure of bacteriophage T4 baseplate. Nat. Struct. Biol 10, 688–693 [DOI] [PubMed] [Google Scholar]
- 9.Fokine A., Kostyuchenko V. A., Efimov A. V., Kurochkina L. P., Sykilinda N. N., Robben J., Volckaert G., Hoenger A., Chipman P. R., Battisti A. J., Rossmann M. G., Mesyanzhinov V. V. (2005) A three-dimensional cryo-electron microscopy structure of the bacteriophage φKZ head. J. Mol. Biol 352, 117–124 [DOI] [PubMed] [Google Scholar]
- 10.Serwer P., Hayes S. J., Zaman S., Lieman K., Rolando M., Hardies S. C. (2004) Improved isolation of undersampled bacteriophages: finding of distant terminase genes. Virology 329, 412–424 [DOI] [PubMed] [Google Scholar]
- 11.Mesyanzhinov V. V., Robben J., Grymonprez B., Kostyuchenko V. A., Bourkaltseva M. V., Sykilinda N. N., Krylov V. N., Volckaert G. (2002) The genome of bacteriophage φKZ of Pseudomonas aeruginosa. J. Mol. Biol 317, 1–19 [DOI] [PubMed] [Google Scholar]
- 12.Hertveldt K., Lavigne R., Pleteneva E., Sernova N., Kurochkina L., Korchevskii R., Robben J., Mesyanzhinov V., Krylov V. N., Volckaert G. (2005) Genome comparison of Pseudomonas aeruginosa large phages. J. Mol. Biol 354, 536–545 [DOI] [PubMed] [Google Scholar]
- 13.Thomas J. A., Rolando M. R., Carroll C. A., Shen P. S., Belnap D. M., Weintraub S. T., Serwer P., Hardies S. C. (2008) Characterization of Pseudomonas chlororaphis myovirus 201φ2-1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 376, 330–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavigne R., Darius P., Summer E. J., Seto D., Mahadevan P., Nilsson A. S., Ackermann H. W., Kropinski A. M. (2009) Classification of Myoviridae bacteriophages using proteins sequence similarity. BMC Microbiol 9, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecoutere E., Ceyssens P. J., Miroshnikov K. A., Mesyanzhinov V. V., Krylov V. N., Noben J. P., Robben J., Hertveldt K., Volckaert G., Lavigne R. (2009) Identification and comparative analysis of the structural proteomes of φKZ and EL, two giant Pseudomonas aeruginosa bacteriophages. Proteomics 9, 3215–3219 [DOI] [PubMed] [Google Scholar]
- 16.Black L. W., Showe M. K. (1983) Morphogenesis of the T4 head, in Bacteriophage T4 (Mathews C. K., Kutter E. M., Mosig G., Berget P. B. eds) pp. 219–245, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 17.Black L. W., Showe M. K., Steven A. C. (1993) Morphogenesis of the T4 head, in Molecular Biology of Bacteriophage T4 (Karam J. D. ed) pp. 218–258, American Society for Microbiology Press, Washington, D. C. [Google Scholar]
- 18.Stein L. D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J. E., Harris T. W., Arva A., Lewis S. (2002) The generic genome browser: a building block for a model organism system database. Genome Res 12, 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas J. A., Hardies S. C., Rolando M., Hayes S. J., Lieman K., Carroll C. A., Weintraub S. T., Serwer P. (2007) Complete genomic sequence and mass spectrometric analysis of highly diverse, atypical Bacillus thuringiensis phage 0305φ8–36. Virology 368, 405–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughey R., Krogh A. (1996) Hidden Markov models for sequence analysis: extension and analysis of the basic method. Comput. Appl. Biosci 12, 95–107 [DOI] [PubMed] [Google Scholar]
- 22.Karplus K., Barrett C., Hughey R. (1998) Hidden Markov models for detecting remote protein homologies. Bioinformatics 14, 846–856 [DOI] [PubMed] [Google Scholar]
- 23.Schneider T. D., Stephens R. M. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18, 6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Condron B. G., Atkins J. F., Gesteland R. F. (1991) Frameshifting in gene 10 of bacteriophage T7. J. Bacteriol 173, 6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casjens S. (2003) Prophages and bacterial genomics: what have we learned so far?. Mol. Microbiol 49, 277–300 [DOI] [PubMed] [Google Scholar]
- 26.Fokine A., Leiman P. G., Shneider M. M., Ahvazi B., Boeshans K. M., Steven A. C., Black L. W., Mesyanzhinov V. V., Rossmann M. G. (2005) Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U.S.A 102, 7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matagne A., Joris B., Frère J. M. (1991) Anomalous behavior of a protein during SDS/PAGE corrected by chemical modification of carboxylic groups. Biochem. J 280, 553–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krylov V. N., Smirnova T. A., Minenkova I. B., Plotnikova T. G., Zhazikov I. Z., Khrenova E. A. (1984) Pseudomonas bacteriophage φKZ contains an inner body in its capsid. Can. J. Microbiol 30, 758–762 [DOI] [PubMed] [Google Scholar]
- 29.Matsko N., Klinov D., Manykin A., Demin V., Klimenko S. (2001) Atomic force microscopy analysis of bacteriophages φKZ and T4. J. Electron Microsc 50, 417–422 [DOI] [PubMed] [Google Scholar]
- 30.Molineux I. J. (2006) Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it? Virology 344, 221–229 [DOI] [PubMed] [Google Scholar]
- 31.Mullaney J. M., Black L. W. (1996) Capsid targeting sequence targets foreign proteins into bacteriophage T4 and permits proteolytic processing. J. Mol. Biol 261, 372–385 [DOI] [PubMed] [Google Scholar]
- 32.Showe M. K., Isobe E., Onorato L. (1976) Bacteriophage T4 prehead proteinase. II Its cleavage from the product of gene 21 and regulation in page-infected cells. J. Mol. Biol 107, 55–69 [DOI] [PubMed] [Google Scholar]
- 33.Katsura I. (1987) Determination of bacteriophage lambda tail length by a protein ruler. Nature 327, 73–75 [DOI] [PubMed] [Google Scholar]
- 34.Casjens S., Hendrix R. (1988) Control mechanisms in dsDNA bacteriophage assembly, in The Bacteriophages (Calender R. ed) Vol. 1, pp. 15–91, Plenum Press, New York [Google Scholar]
- 35.Crutz-Le Coq A. M., Cesselin B., Commissaire J., Anba J. (2002) Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148, 985–1001 [DOI] [PubMed] [Google Scholar]
- 36.Piuri M., Hatfull G. F. (2006) A peptidoglycan hydrolase motif within the mycobacteriophage TM4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol 62, 1569–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.