Abstract
Investigations of the molecular mechanisms underlying responses to nerve injury have highlighted the importance of axonal transport systems. To obtain a comprehensive view of the protein ensembles associated with axonal transport in injured axons, we analyzed the protein compositions of axoplasm concentrated at ligatures following crush injury of rat sciatic nerve. LC-MS/MS analyses of iTRAQ-labeled peptides from axoplasm distal and proximal to the ligation sites revealed protein ensembles transported in both anterograde and retrograde directions. Variability of replicates did not allow straightforward assignment of proteins to functional transport categories; hence, we performed principal component analysis and factor analysis with subsequent clustering to determine the most prominent injury-related transported proteins. This strategy circumvented experimental variability and allowed the extraction of biologically meaningful information from the quantitative neuroproteomics experiments. 299 proteins were highlighted by principal component analysis and factor analysis, 145 of which correlate with retrograde and 154 of which correlate with anterograde transport after injury. The analyses reveal extensive changes in both anterograde and retrograde transport proteomes in injured peripheral axons and emphasize the importance of RNA binding and translational machineries in the axonal response to injury.
Peripheral nerve injuries elicit a cascade of axonal responses that are required for a successful regenerative response. The lesioned axons must signal retrogradely to their cell bodies to activate intrinsic neurite outgrowth mechanisms (1–3) and then overcome physical barriers and inhibitory cues in the extracellular environment to achieve functional regeneration (4, 5). The first indications of a breach in axonal integrity upon injury are most likely abnormal generation of action potentials and/or waves of calcium propagating from the lesion site toward the intact portions of the cell (6, 7). At a later stage, signals carried by motor-driven transport systems start to affect the cell body. This phase includes both an interruption of the normal supply of retrogradely transported molecules such as trophic factor signals (8) and arrival of new signals elicited at the injury site (3, 9). The latter interact with a variety of dynein-associated carriers including importins (10, 11) and kinase family scaffolds (12, 13).
In mammalian neurons, the distances between axonal lesion sites and the nucleus can reach many centimeters (up to 1 m in humans); hence, retrograde signaling events within the first few hours after injury must be independent of new transcription in the cell body. Proteolysis, local protein synthesis, and post-translational signaling modifications such as phosphorylation have all been implicated in generation of the retrograde signaling ensemble (1). This prominent role for post-transcriptional processes suggests that comprehensive characterization of retrograde signaling will require proteomics approaches. In a previous study we used two-dimensional PAGE and mass spectrometry to analyze retrogradely concentrated axoplasm from injured mollusc nerve, identifying a vesicular ensemble blocked by the lesion and an up-regulated ensemble highly enriched in calpain cleavage products of an intermediate filament (14, 15). Follow-up studies in rodent sciatic nerve showed that the mammalian intermediate filament vimentin is produced by local translation of axonal mRNA upon axonal injury and then undergoes calpain-mediated proteolysis, generating a cleavage product that interacts with importins bound to dynein and enables protected retrograde transport of phosphorylated forms of the mitogen-activated protein kinases Erk1 and Erk2 (16, 17). Here, we extend our efforts to determine the components of the retrograde injury signaling ensemble in lesioned nerve by using LC-MS/MS coupled with iTRAQTM11 labeling to directly analyze mammalian axoplasm samples after nerve injury. The analyses reveal extensive changes in both anterograde and retrograde transport proteomes in injured peripheral axons and highlight the importance of RNA binding and translational machinery in the axonal response to injury.
EXPERIMENTAL PROCEDURES
Sample Preparation
8–12-week-old Wistar rats were subjected to crush lesion in the distal part of the sciatic nerve, and ligatures were applied 1 cm distally to the femoral joint. 24 h later animals were sacrificed, and nerve segments distal and proximal to the ligature site were processed separately for axoplasm extraction (18). Briefly, nerve segments were manually dissected to remove connective tissue, incubated for 2 h in 0.2× PBS to lyse glia, washed, and then extracted in 1× PBS (300 μl/five sciatic nerves) for 40 min at room temperature with subsequent centrifugation at 21,800 × g for 10 min at 4 °C. All solutions were supplemented with protease and phosphatase inhibitors (protease inhibitors (Roche Applied Science, catalog number 14696200; two tablets/50 ml of 1× and 0.2× PBS solutions), 1 mm sodium fluoride, 1 mm sodium vanadate, 1 mm sodium molybdate, 1 mm sodium tartrate, 100 μm fenvalerate, 250 nm okadaic acid, and 1 nm calyculin A) and lyophilized. Dried samples were resuspended in 6 m guanidine hydrochloride in 25 mm ammonium hydrocarbonate with 2.1 mm tris(2-carboxyethyl)phosphine hydrochloride and incubated at 57 °C for 1 h after which iodoacetamide was added to 4.2 mm for an additional 45-min incubation at room temperature in darkness. The reduced and alkylated samples were diluted to 1 m guanidine hydrochloride, and side chain-protected trypsin (Promega, Madison, WI) was added at 1:50 (trypsin:protein sample) for digestion to a final concentration of about 2–2.5 × 10−7 m. The pH was adjusted to 8 with 1 m ammonium bicarbonate, and digestion was allowed to proceed at 37 °C overnight. Digests were then acidified by addition of 0.1% trifluoroacetic acid and desalted with C18 Sep-Pak cartridges (Waters, Milford, MA). Quadruplex iTRAQ (Applied Biosystems, catalog number 4352135) derivatization was performed for quantitative analysis according to the vendor's protocol. Mass tags were assigned to samples as follows: m/z 114 for non-injured distal (NID), m/z 115 for injured distal (IND), m/z 116 for non-injured proximal (NIP), and m/z 117 for injured proximal (INP). Derivatized samples were concentrated to about 10 μl of solution/tube and then fractionated by strong cation exchange chromatography (Polysulfoethyl A, 200 × 2.1 mm, 5-μm bead, 200-Å pores (PolyLC) on an ÄKTA HPLC system (GE Healthcare). Peptides were eluted in a gradient of 0–350 mm KCl in 5 mm KH2PO4, 30% acetonitrile; concentrated by SpeedVac (Thermo Electron, San Jose, CA); desalted over a C18 macrotrap peptide reverse phase column (Michrom Bioresources, Inc., Auburn, CA); dried; and redissolved in 10 μl of 0.1% HCOOH/fraction.
LC-MS/MS
Reverse phase separation of each strong cation exchange fraction was carried out on a nanobore 75-μm × 15-cm C18 column at a flow rate 350 nl/min, developing a gradient of 5–50% acetonitrile in 0.1% HCOOH over 60 min. The HPLC system (Agilent 1100, Agilent Technologies, Palo Alto, CA) was connected on line to a quadrupole-orthogonal acceleration-TOF (QSTAR Pulsar or Elite, Applied Biosystems, Foster City, CA) or a linear ion trap-Orbitrap (LTQ-Orbitrap, Thermo Fisher Scientific Inc., Waltham, MA) mass spectrometer. Data were acquired in an information-dependent fashion. For the QSTAR, MS acquisitions (1 s) were followed by CID experiments. The two most abundant multiply charged ions were selected for subsequent CID. First, a regular CID acquisition was performed where the collision energy was automatically adjusted according to the peptide charge and m/z value. Then, with a high resolution precursor ion selection and preset collision energy, only the reporter ion region was monitored. Precursor ions already analyzed were excluded from the selection process for 1 min.
LC-MS analysis on the LTQ-Orbitrap consisted of MS survey scans that were followed by higher energy collision dissociation scans on the three most abundant multiply charged ions in the survey scan. Both precursor ions and higher energy collision dissociation fragments were measured in the Orbitrap. Dynamic exclusion was enabled. Peak lists were generated with Analyst software using Mascot.dll version 1.6b20 and Mascot Distiller version 2.1.0.0 with the QSTAR and the Orbitrap data, respectively.
Initial automated analysis of the obtained spectra was carried out with ProteinProspector 5.0, searching against the UniProt.2007. 12.04.random.concat database (152,718 of 10,725,064 entries searched). Only tryptic peptides were considered, and up to two miscleavages were allowed. Fixed modifications were carbamidomethylation of Cys residues and iTRAQ 4-plex modification of Lys side chains and N termini. Variable modifications considered in the analysis included N-terminal acetylation of proteins, cyclization of N-terminal Gln residues, methionine oxidation, and serine/threonine/tyrosine phosphorylation. Three variable modifications per peptide were permitted. Mass tolerance was set to 300 and 100 ppm for the fragment ions and precursor ions, respectively, in the QSTAR experiments and to 0.8 Da and 25 ppm, respectively, for the Orbitrap experiments. Acceptance criteria were as follows: E-value was set to 0.01, minimal peptide score was 15, and protein score was 25.
The threshold set was supported by two methods used for false positive detection rate (FPR) and false discovery rate (FDR) estimation. FPR was calculated based on the hits detected in the decoy database versus the total number of peptide hits, whereas FDR was calculated via estimation of the ratio of the number of true null hypotheses (H0) from the total number of tests (19). For these parameters, average FPR was 1.64% (range, 0.51–4.70%), whereas average FDR was 0.72% (range, 0.34–2.25%).
These criteria led to the identification of 10,632 peptides in all biological replicates. Application of a stricter setting of 25 for the peptide score threshold decreased the accepted number of peptides to 9267. After removal of duplicate and redundant entries, the number was further reduced to 4954 unique peptides.
Data Analysis and Quantification
Quantitative data for each peptide were obtained from iTRAQ labels at 114.1-, 115.1-, 116.1-, and 117.1-amu peaks by assessment of area under the peak curve for QSTAR-based data or by assessment of peak intensity for Orbitrap-generated spectra. Proteins were reconstituted from multiple peptides data. Accession numbers of identified proteins were mapped onto the corresponding UniGene entries using the Uniref90 clustering database. Peptides that corresponded to proteins from more than one UniProt entry or with multiple UniProt entries were not used for quantification. Redundant entries were condensed as single proteins for quantification and identification purposes if they matched to the same UniGene/UniProt record. Proteins were used for further analyses only if they were identified in at least two of three biological replicates. In cases of protein identifications based on two biological replicates, the requirement was that the protein should appear in both analytical replicates of the sample. The UniProt filters focused attention to 4754 quantifiable peptides from three biological replicates.
To handle analytical and biological variability, we produced box-whisker plots for each multipeptide reconstituted protein in each analytical replica. Box upper and lower values corresponded to the S.E.; a range within the S.D. was considered as non-outlying. The outlier range was defined as
where OVU is the outlier value from the upper limit and OVL is the outlier value from the lower limit. UBV and LBV (upper and lower box values, respectively) were calculated as mean ± S.E. The outlier coefficient (OC) was defined as 1.5. Extreme values were calculated using the same equation except the outlier coefficient was set to 3 (StatSoft). Additional outlier analyses were carried out using Mahalanobis distance assessment. Data obtained by the two methods were compared, and proteins with extreme values according to both analysis methods were excluded.
Clustering
Hierarchic and non-hierarchic cluster analyses were performed on the data. Data standardization for hierarchic clustering utilized Euclidean metrics for distance assessment and Ward minimum variance for linkage (20, 21). Amalgamation curves were used to estimate cluster number and to supervise reclustering. Data were validated using the root mean square deviation of the cluster at each step in hierarchic clustering and pseudo-F ratio, pseudo-T-square estimation, and Dunn's cluster separation maximum group assessment methods. Non-hierarchic clustering was carried out using k-mean and expectation maximization approaches (21). Euclidean distance was used for metrics, and k-values were seeded randomly. Silhouette plot estimation was used for cluster number determination. Multiple replication of clustering was used to avoid local minima. The minimum increase of log likelihood was set to 0.001 for the expectation maximization algorithm. For cluster number validation purpose, the v-fold cross-validation method was used (21).
Principal Component and Factor Analyses
Correlation matrixes were used to find correlations between six variables generated from the iTRAQ data. The number of components was defined based on eigenvalue estimation on scree plots (22). Two extraction methods were used for factor analysis: principal component and then maximum likelihood factor method for validation. Correlations between factor loading and variables higher than 0.7 were considered as significant. Communality analysis was used to estimate the contribution of factors to variance of variables. Quartimax proved to be the best fitting orthogonal rotation method. In addition, oblique rotation was used to exclude secondary factors (23). Factor score data were used to compare per protein covariances of the protein level changes in accordance to factors. We examined the factor score distribution for each protein for data extraction, using multiple comparisons to exclude proteins with higher absolute value scores for factors not affecting the analyzed variable. For example, if factor 2 correlates with the variable IND/NID, proteins assigned to the factor 2 grouping should have factor 2 scores with higher absolute values than their scores for factors 1 and 3. Because the partitions thus obtained were not normally distributed, we applied Kruskal-Wallis one-way analysis of variance on ranks with Dunn's post hoc analysis, and the p value cutoff was set to 0.001. Proteins that met the statistical criteria were assigned to corresponding factors for further clustering. For further details on factor analysis as applied here, please see the supplemental material.
Bioinformatics and Statistics
Factor-assigned protein lists were subjected to gene ontology (GO) and tissue distribution analyses using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (24) with the Expression Analysis Systematic Explorer (EASE) score set to 0.05 and threshold of protein per category set to 3. Data were analyzed against Rattus rattus, Rattus norvegicus, and Mus musculus databases. Settings for functional annotation clustering were as follows: classification stringency, high; similarity gene overlap, 3; similarity threshold for κ statistics, 0.85; initial and final group membership, 3; multiple linkage threshold, 0.5; and FDR threshold, 0.5. Software packages used included Matlab 2008a, Systat 12, Systat SigmaPlot, R, and Excel.
RESULTS
Data Collection
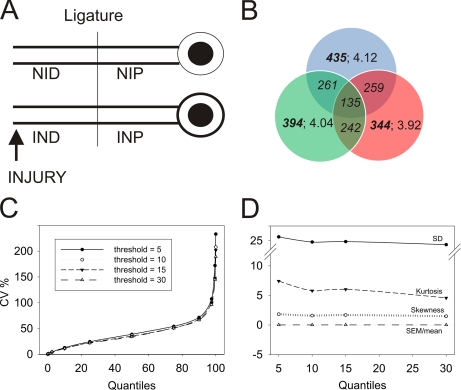
Nerve ligatures provide a physical obstacle allowing for accumulation of retrogradely and anterogradely transported components in axons distal and proximal to the ligature, respectively. We ligated rat sciatic nerves concomitantly with or without crush lesions 2–2.5 cm distal to the ligature and extracted axoplasm from proximal and distal sides of the ligature 24 h later (Fig. 1A), thus generating four experimental sample types: IND, INP, NID, and NIP. 5-mg axoplasm protein samples (corresponding to 100 animals each) were subjected to reduction, alkylation, and tryptic digestion followed by iTRAQ labeling, fractionation by strong cation exchange, and LC-MS/MS analysis. Three independent biological experiments (3 × 100 animals) were interrogated on the QSTAR, and two of these samples were also analyzed using the LTQ-Orbitrap instrument, generating a data set of more than 150,000 spectra overall. After removal of false positives by filtering against the UniProt decoy database, these spectra corresponded to 4754 unique peptides from 1173 unique proteins in all experimental groups from the three experiments, averaging 4.05 peptides per identified protein. The average overlap between two biological replicates was about 65%, whereas overlap of all three biological replicates was much lower (Fig. 1B). Peptide per protein distribution was fairly uniform for all replicates (Fig. 1B).
Fig. 1.
Preprocessing of iTRAQ-based peptide quantification data. A, experimental model used for this study. B, Venn diagram presenting overlap of unique proteins in three biological replicates. Bold fonts represent the total number of proteins in the group. Italics denote the number of shared proteins between the indicated replicates. Regular fonts detail the peptide per protein distribution for a specific replicate. C, distribution of CV values (%) for peptides per protein based on the iTRAQ label area (QSTAR) or intensity (Orbitrap) threshold. The graph depicts the fraction of CV values over the entire peptide per protein distribution. Proteins with at least three available peptide identifications were used for S.D. calculations. D, descriptive statistics parameters for peptide distribution per protein at the designated thresholds. S.D., S.E., skewness, and kurtosis are shown. Neither skewness nor S.E. were affected by intensity threshold change.
To reduce noise contribution to the quantitative data, we assessed the coefficient of variation (CV) of peptide distribution per corresponding proteins at different threshold levels of reporter ion areas corresponding to 5, 10, 15, and 30 integrated counts (henceforth referred to as counts). This analysis did not show any significant differences between the thresholds tested, although data kurtosis was lower for thresholds above 10 counts (Fig. 1, C and D, and supplemental Fig. S1A). We therefore set the threshold to 10, discarding signal values below this for peak area (QSTAR) or peak intensity (Orbitrap), thus further narrowing down the data set to 3940 unique peptides corresponding to 972 unique proteins (supplemental Fig. S1B). All comparisons revealed positive correlations between the data sets, albeit with significant variability in distributions with correlation coefficients ranging from 0.3 to 0.95 (supplemental Fig. S2). The number of proteins identified in different replicas for QSTAR-based experiments were within a 20% difference. The difference was even lower for LTQ-Orbitrap experiments, although the number of proteins identified from LTQ-Orbitrap data was lower than from QSTAR-based experiments. The average standardized Cronbach α value (25) for subsets of two or more biological replicas was ∼0.68, indicating high data reliability (data not shown).
Data Set Complexity
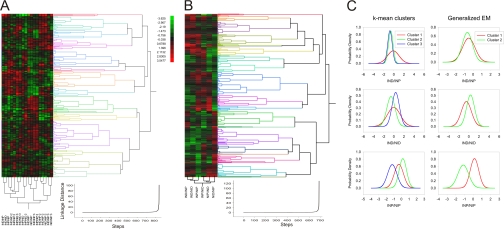
We tried to use hierarchic and non-hierarchic clustering analyses to look for covariance in the data. Hierarchic clustering was based on the assumption that changes in protein level will correlate with the directional transport processes occurring in the nerve; thus for example, proteins clustering closest to dynein retrograde motor components should be those involved in retrograde signaling. However, the resulting clusters did not fit such simple assumptions (Fig. 2, A and B), and increasing the number of variables only complicated the picture. We then attempted to use non-hierarchic cluster models, based on distributions of protein level change, to reveal protein subsets showing similar patterns of change in all three experiments. Both k-mean and generalized expectation maximization clustering did not allow for useful resolution of these data, and furthermore v-fold-based cross-validation analyses did not converge to the same number of clusters in different experiments (Fig. 2C). This suggested that retrograde axonal transport and anterograde axonal transport are not the sole and dominant processes affecting the data set. A number of additional influences might have to be taken into account, including local effects on the ligature itself, leakage through the ligation to the other side, and tissue inflammation. Principle component analysis (PCA) provides an approach for determining the number of independent parameters (called principal components) that can account for the vast majority of variance in a complex data set.
Fig. 2.
Cluster analyses of differentially represented proteins over complete data set. A, hierarchic clustering of proteins differentially represented after injury in all three experiments. Hierarchic clusters were generated using Euclidean distance estimation together with Ward's linkage method. Even spacing was used for dendrogram scaling. Clustering metrics are depicted below. Clusters were validated using root mean square deviation estimation, pseudo-F ratio, pseudo-T ratio, and Dunn's tests. B, a hierarchic cluster example for a single experiment (all procedures are as in A). C, probability density functions of non-hierarchic cluster analyses for variables IND/NIP, IND/NID, and INP/NIP in all three experiments using k-mean and expectation maximization (EM) clustering methods. Initial numbers of clusters were calculated using silhouette plot analysis. We used random seeding for k-calculation. To avoid local minima, we ran 50 replicas and chose the solution with the lowest total sum of distances over all replicas. Matlab implementation of v-fold cross-validation was used to validate correct cluster numbers with v-value set to 10.
PCA and Factor Analyses Enable Extraction of Relevant Proteins
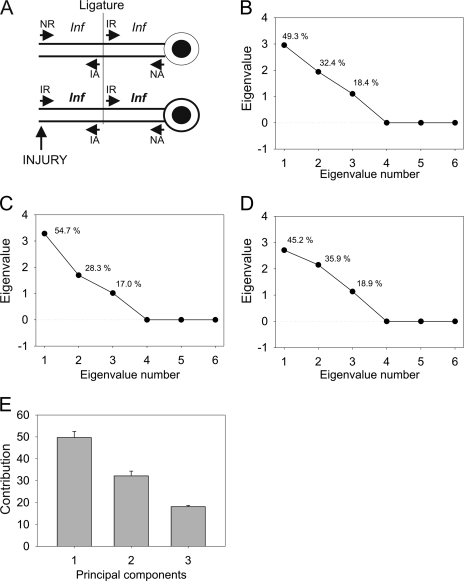
Fig. 3A shows a model of the putative contributions to transport ensembles before and after injury. To extract the contributions of the different experimental groups to the processes shown in the model, we carried out PCA on all four iTRAQ label ratios in six different combinations: IND/NID, INP/NIP, IND/NIP, IND/INP, NID/NIP, and INP/NID. This analysis revealed three eigenvalues with values higher than 1, indicating three principal components that appeared to suffice to account for the complete data set in all three experiments. Eigenvalue scree plots showed similar contributions of each individual principal component in all three independent experiments with small deviations (Fig. 3, B–D). Averaging these contributions indicated an ∼50% contribution of the first component and about 30 and 20% for the second and the third components, respectively (Fig. 3E). We then sought to refine the approach by applying factor analysis.
Fig. 3.
PCA analysis for iTRAQ data. A, schematic map of components contributing to protein ensembles in different compartments of the injury-ligation model. NR, non-injured retrograde; IR, injured retrograde; NA, non-injured anterograde; IA, injured anterograde; Inf, inflammation. B–D, eigenvalues and corresponding component contribution for ITRAQ ratio data for each experiment. All six variables were used for PCA. Numbers above each eigenvalue coordinate depict data variance accounted for by that eigenvalue. E, averaged eigenvalues over three biological experiments. Contribution denotes average variance accounted for by each component. Error bars denote standard error of mean.
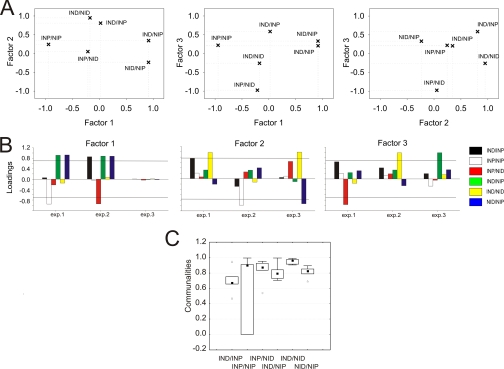
The principal component-based extraction method revealed three statistically significant factors with eigenvalues similar to PCA (Fig. 4). Quartimax rotation optimally stabilized factor loading onto variables with only IND/NIP (Fig. 4, green) and IND/NID (Fig. 4, yellow) being unequivocally loaded by a distinct and dominant factor in all experiments (Fig. 4, A and B). For example, Fig. 4A shows two-dimensional plots of relative loading of the different variables on two factors in each plot. Variables with higher absolute values for a specific factor have stronger correlation with that factor. For IND/NID, the left panel in Fig. 4A shows a value of close to 1 on the factor 2 axis compared with a value close to 0 on the factor 1 axis, indicating strong correlation with factor 2 and very weak correlation with factor 1. The middle panel shows weak correlation of IND/NID with both factors 1 and 3, and the right panel again shows strong correlation of IND/NID with factor 2 and weak correlation with factor 3. Hence, IND/NID is very strongly correlated with factor 2 but nonetheless is also weakly influenced by other factors. The model of Fig. 3A shows that IND/NID should define the injury-regulated retrograde ensemble; thus, proteins strongly correlating with factor 2 should fit this category. Communalities for IND/NID reached 0.96 with a very narrow range of distribution, whereas the median of communalities for IND/NIP reached 0.8 with a wider distribution. Communalities of both IND/NID and IND/NIP lacked outliers or extreme values over all the experiments (Fig. 4C).
Fig. 4.
Factor analysis of injury-related variables. A, representative quartimax rotated variable loading for factors retrieved from principal component data (example for a single experiment). Each panel shows relative factor loading for two factors per variable as indicated. B, factor loadings for variables in three experiments averaged over analytical replicas for each experiment. Loadings were calculated separately for each replica and further averaged for each biological experiment. The threshold was set to 0.7. C, communality assessment for each variable over three experiments (including analytical replicas). The boxes show the 25–75% range, and the inner square in each box is the median. Error bars denote non outlier region. Triangles represent outliers. Communalities for variables were calculated as the sum of squared loadings for that variable: .
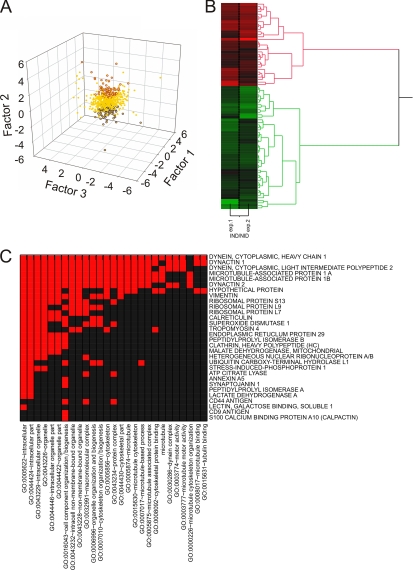
Fig. 3A shows that the IND/NID variable primarily represents the ratio of injured to uninjured retrograde transport-related components; thus, proteins extracted by factor scores for this variable should be involved in injury-regulated retrograde transport. Comparison of all three experiments by factor scores (Fig. 5A and supplemental Fig. S3) revealed that 30 proteins belonging to this category were common to all biological replicates of the injury experiment, whereas a less stringent cutoff taking proteins found in two of the three replicates resulted in a total of 144 proteins correlated positively (71 proteins) or negatively (73 proteins) to the retrograde transport ensemble (Fig. 5B; supplemental Tables S1, S3, and S5; and supplemental supporting spectra). The main positively correlating proteins included dynein heavy chain 1 and dynactin subunit 2, both core components of the retrograde transport machinery (Fig. 5C). The ensemble could be hierarchically subdivided into two distinct and coherent clusters with significant distance between clusters of positively and negatively correlating proteins, thus validating the outcome of the factor analysis (Fig. 5B). The hierarchic clustering pattern was also supported by k-mean clustering analysis (supplemental Fig. S4).
Fig. 5.
Injury-regulated retrograde transport components deduced from factor analysis. A, scatter plot of individual protein scores for all three factors. Red and blue dots represent positive and negative factor scores, respectively, correlating with the IND/NID variable. Yellow dots depict cases not correlating with IND/NID. Factor scores were obtained as a vector of data for the equation Y = μ + Lfi + ε where Y is a value of the case, μ is a population mean, L is a matrix of loadings, ε is a specific error, and f is an estimated vector of factor scores. Statistical significance was estimated with one-way Kruskal-Wallis test with post hoc Dunn's analysis; the p value cutoff was set to 0.001. B, hierarchic cluster analysis for proteins assigned to the injury-regulated retrograde transport ensemble. C, GO annotation functional category clustering for proteins up-regulated in the retrograde transport ensemble after injury. Red indicates a positive relationship; black indicates no relationship.
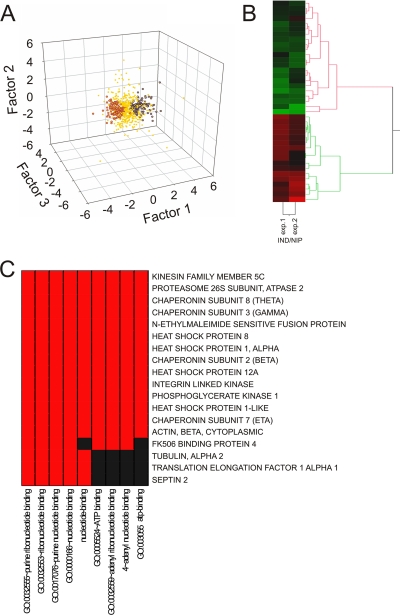
The IND/NIP variable corresponds primarily to the ratio of injured versus uninjured anterograde transport components. We extracted the corresponding protein list by comparing factor scores for each individual protein (Fig. 6A; supplemental Fig. S5; supplemental Tables S2, S4, and S5; and supplemental supporting spectra). Clustering of the data clearly distinguished two groups (Fig. 6B and supplemental Fig. S6). Kinesin heavy chain isoform 5C, one of the major neuronal anterograde transport motor components, was detected in only two of the three biological replicates in negative correlation with factors related to the anterograde transport; thus, all 101 proteins showing similar factor scores were assigned to the anterograde transport ensemble (supplemental Tables S2 and S4). The non-kinesin-containing group was highly enriched for inflammation-related proteins (supplemental Tables S2 and S4), although the inflammation category was assumed to cancel itself out when subtracting transport components in the model of Fig. 3A. This result may indicate a difference in inflammation levels in distal versus proximal sides of the ligation, suggesting non-linear behavior in this aspect of the system and likely increasing the chance of error in linear decomposition-based factor analysis results. As shown in supplemental Fig. S7, the increased error due to such non-linear behavior might call into question ∼7% of the proteins assigned to the anterograde transport ensemble, hence ∼10 proteins of the 150 listed.
Fig. 6.
Injury-related anterograde transport components deduced from factor analysis. A, scatter plot of individual protein scores for all three factors. Red and blue dots represent positive and negative factor scores, respectively, correlating with the IND/NIP variable. B, hierarchic cluster analysis for proteins assigned to the injury-regulated anterograde transport ensemble. C, GO annotation functional category clustering for the main anterograde transport proteins responding to injury. Red indicates a positive relationship; black indicates no relationship.
Gene Ontology Analyses of Injury-regulated Retrograde and Anterograde Transport Ensembles
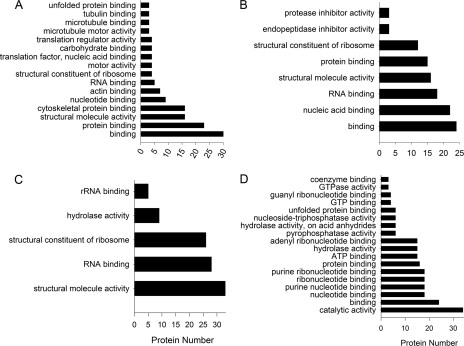
We checked our data against the GO databases for rat and mouse, combining results from both databases to obtain functional insights on the proteins we found. Analyses of all three GO parameter categories, biological processes, cellular compartments, and metabolic functions, revealed that most of the discovered proteins are related to cytoskeleton components, macromolecular complex formation, motor proteins, and biosynthesis (Figs. 5C, 6C, and 7). The GO metabolic function classification was the most useful for obtaining functional insights on these transport ensembles. Proteins up-regulated in the retrograde transport ensemble by injury were highly enriched for protein binding functions (GO:0005515), mostly in the cytoskeletal protein binding (GO:0008092) and microtubule binding (GO:0003777) categories. Other significant groupings for this ensemble included structural molecular activity (GO:0005198), comprising general and microtubule motors (Fig. 7A). Proteins that were less associated with retrograde transport after injury included a significant representation for protein translation, RNA binding, and ribosome machineries (GO:0003723, GO:0003735, and GO:0019843) (Fig. 7B). Metabolic function analysis revealed fewer significant categories among proteins that were more associated with anterograde transport after injury, primarily structural molecular activity, ribosome constituent, and RNA binding categories (GO:0005198, GO:0003735, and GO:0003723) (Fig. 7C). The protein ensemble in reduced association with anterograde transport after injury revealed significant metabolic activity categories, including ATP and GTP binding activity (GO:0005524 and GO:0005525) and multiple catalytic activities (GO:0003824) (Fig. 7D).
Fig. 7.
Gene ontology analyses on injury-regulated axonal transport ensembles. Retrogradely (A and B) and anterogradely (C and D) correlating ensembles are shown. A and C, positively correlating proteins. B and D, negatively correlating proteins.
DISCUSSION
The heterogeneity of cell types and complex morphology and anatomy of neural tissues complicate proteomics studies in the nervous system (26). Contaminations from other tissues and cell types are a prevalent problem; for example, it has been estimated that up to 80% of the proteins in human cerebrospinal fluid are actually contributed by serum contaminations (27). Nerve injury studies must also contend with variations in inflammatory and other responses in injured versus non-injured tissues. Previous studies of axonally transported complexes using proteomics used either affinity-targeted investigations of purified organelles (28–30) or gel-based differential screens (15, 31). Here, we sought to obtain a more comprehensive view of axon transport ensemble changes induced by nerve injury by interrogating ligature axoplasm preparations with iTRAQ quantification and LC-MS/MS. The resulting data set comprised over 1000 proteins, but initial attempts at clustering the candidates showed a high degree of experimental variation, necessitating the use of mathematical and statistical filters.
The iTRAQ analysis provided quantification of a total of 1173 proteins changing upon injury over three trials in all experimental groupings. Attempts to cluster the entire data set were not successful in providing coherent groupings, and in fact, even core components of the retrograde transport machinery such as dynein heavy chain and intermediate chains and dynactin were not clustered together in a reproducible manner. Both biological and technical aspects can influence data variability in iTRAQ experiments (32, 33), and a variety of clustering approaches, including non-hierarchic, k-mean, and expectation maximization clustering, did not solve this problem (Fig. 3). Biological variation was shown to be the principal source of variability in iTRAQ experiments on unicellular organisms (32), and an in vivo multicellular tissue such as the sciatic nerve presents vastly higher complexity and potentially increased variation (34–36). Indeed, there is increasing appreciation of the need for specific approaches to deal with assorted sources of variation in proteomics experiments (37).
In this case, despite pooling of extracts from 100 individual animals per sample and analysis of three independent samples per treatment using two instrument systems, biological or experimental variability did not allow straightforward identification of co-regulated protein ensembles. We therefore resorted to principal component and factor analyses to extract biologically meaningful information because they enable evaluation of the number of factors affecting variance in a data set in the absence of any knowledge or assumptions on the nature of such factors (22). PCA and factor analyses enabled sorting of the data and extraction of 299 proteins in consistent groupings associated with retrograde or anterograde transport. PCA and factor analyses fit well to a simple model describing lesion effects on axonal transport as a combination of anterograde and retrograde transport ensembles and inflammation processes. The ligation paradigm separates between anterograde and retrograde transport components accumulated at either side of the ligature (Fig. 3A). PCA distinguished three components influencing the data set, and their relative contribution was similar in all three experiments (Fig. 3B). Factor analysis in different experiments showed batch variability of factor contributions, apparently arising from biological heterogeneity in the system. Based on factor loadings, we could conclude that the IND/NID and IND/NIP variables were affected by mutually exclusive factors throughout all experiments (Fig. 4C). Comparison of the factor analysis output with the experimental model of Fig. 3A suggested that the IND/NID variable is the most suited to evaluate injury-related changes in retrograde signaling. If inflammation is at equivalent levels throughout and ligature leakages are negligible, retrograde signaling should be the major driver of changes in IND/NID between non-injured and injured states of the sciatic nerve. The high degrees of retrograde transport factor loading for IND/NID and communality support this notion. IND/NIP was the second variable to be unambiguously affected by another factor, and similar considerations suggest that it reflects mainly anterograde transport components. As in the case of IND/NID, this is supported by a high degree of factor loading and communality for IND/NIP in all experiments (Figs. 4C and 6A). Thus, use of factor analysis allowed us to reduce variable numbers from six to two for assessment of injury-related transport, and moreover a single variable sufficed for each type of transport. Importantly, factor analysis based on per case factor scores enables comparison of the contribution of each factor to changes in individual proteins, allowing classification of proteins according to the IND/NID variable for correlation with retrograde transport and according to IND/NIP for correlation with anterograde transport. This decomposition approach significantly reduced data complexity by excluding proteins not clearly correlated with a specific transport direction. Further cluster analysis on the data extracted by factor analysis unambiguously validated the two subsets by forming two clearly distinct clusters (Figs. 5 and 6 and supplemental Figs. S1–S7).
Much of the research effort to date in nervous system proteomics has focused on central nervous system preparations (26, 38), most prominently those related to synaptic transmission (39, 40). Previous characterizations of peripheral nerve proteomes have not distinguished between axonal and glial components of the tissue (36, 41–43) apart from invertebrate preparation analyses from our groups (14, 15) and a very recent study from Cavalli and co-workers (30) on purified Syd1-containing vesicles in sciatic nerve. The transported protein ensembles delineated in this study provide a window on the soluble axoplasm proteome in peripheral neurons, revealing a significant degree of heterogeneity and complexity in this compartment. In addition to providing a basis for future functional analyses, the data already provide intriguing insights on the axonal response to injury. Major categories of proteins associated with axonal transport ensembles in this study include protein synthesis machinery, cytoskeletal and motor proteins, and metabolic proteins (Figs. 5–7 and supplemental Tables S1 and S2). Anterograde transport ensembles are enriched with assorted enzymatic activities indicative of changes in axonal metabolism following injury. Although motor-, cytoskeleton-, and metabolism-associated proteins are predictable components of axonal transport systems, the high preponderance of protein synthetic machinery is striking. A previous study from Twiss and co-workers (31) used in vitro compartmentalized cultures of sensory neurons to analyze axon proteomes. That study, together with other non-proteomics analyses (44–46), highlighted the importance of localized protein translation mechanisms in supporting nerve injury responses and neurite outgrowth (47). However, the occurrence of protein translation machinery in peripheral axons in vivo has been a topic of vigorous debate (48). Most of the reports of ribosomes in mammalian axons have been based on microscopy on static preparations (49), and the results have been open to different interpretations (for reviews, see Refs. 48 and 50). Very recent work from Court et al. (51) has used a combination of electron and fluorescence microscopy techniques to visualize ribosomes in sciatic nerve axons, and these authors suggested that some of the axonal ribosomes translocate from Schwann cells after injury. Our current data provide new support for the occurrence of protein synthetic machinery in axons in vivo because one of the most prominent categories of axonal transport proteins regulated by injury is structural components of ribosomes (Figs. 5 and 7 and supplemental Tables S1 and S2). Both 40S and 60S ribosomal protein components are down-regulated in the retrograde transport ensemble while undergoing up-regulation in the anterograde ensemble after injury along with associated RNA-binding and translation-regulating proteins. This is consistent with dynamic trafficking of ribosomes and associated complexes in axons and suggests that one of the early responses to axonal injury is mobilization of protein synthesis machinery toward the site of the lesion. These data suggest that in contrast to the “opposing camps” of axonal transport versus local translation adherents among human researchers peripheral nerve axons coordinate these two mechanisms to ensure an effective injury response.
Supplementary Material
Footnotes
* This work was supported, in part, by National Institutes of Health Grants NCRR P41RR001614 and NCRR RR012961 from the National Center for Research Resources. This work was also supported by the Israel Science Foundation, the International Foundation for Research in Paraplegia and the Dr. Miriam and Sheldon Adelson Medical Research Foundation.
 This article contains supplemental Figs. S1–S7, Tables S1–S5, and supporting spectra.
This article contains supplemental Figs. S1–S7, Tables S1–S5, and supporting spectra.
1 The abbreviations used are:
- iTRAQ
- isobaric tag for relative and absolute quantitation
- CV
- coefficient of variation
- GO
- gene ontology
- IND
- injured distal
- INP
- injured proximal
- NID
- non-injured distal
- NIP
- non-injured proximal
- PCA
- principal component analysis
- FPR
- false positive detection rate
- FDR
- false discovery rate
- LTQ
- linear trap quadrupole.
REFERENCES
- 1.Hanz S., Fainzilber M. (2006) Retrograde signaling in injured nerve—the axon reaction revisited. J. Neurochem 99, 13–19 [DOI] [PubMed] [Google Scholar]
- 2.Rossi F., Gianola S., Corvetti L. (2007) Regulation of intrinsic neuronal properties for axon growth and regeneration. Prog. Neurobiol 81, 1–28 [DOI] [PubMed] [Google Scholar]
- 3.Abe N., Cavalli V. (2008) Nerve injury signaling. Curr. Opin. Neurobiol 18, 276–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Domeniconi M., Filbin M. T. (2005) Overcoming inhibitors in myelin to promote axonal regeneration. J. Neurol. Sci 233, 43–47 [DOI] [PubMed] [Google Scholar]
- 5.Fawcett J. W. (2006) Overcoming inhibition in the damaged spinal cord. J. Neurotrauma 23, 371–383 [DOI] [PubMed] [Google Scholar]
- 6.Mandolesi G., Madeddu F., Bozzi Y., Maffei L., Ratto G. M. (2004) Acute physiological response of mammalian central neurons to axotomy: ionic regulation and electrical activity. FASEB J 18, 1934–1936 [DOI] [PubMed] [Google Scholar]
- 7.Costigan M., Scholz J., Woolf C. J. (2009) Neuropathic pain: a maladaptive response of the nervous system to damage. Annu. Rev. Neurosci 32, 1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibáñez C. F. (2007) Message in a bottle: long-range retrograde signaling in the nervous system. Trends Cell Biol 17, 519–528 [DOI] [PubMed] [Google Scholar]
- 9.Rishal I., Fainzilber M. (2009) Retrograde signaling in axonal regeneration. Exp. Neurol 10.1016/j.expneurol.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 10.Hanz S., Perlson E., Willis D., Zheng J. Q., Massarwa R., Huerta J. J., Koltzenburg M., Kohler M., van-Minnen J., Twiss J. L., Fainzilber M. (2003) Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 11.Yudin D., Hanz S., Yoo S., Iavnilovitch E., Willis D., Gradus T., Vuppalanchi D., Segal-Ruder Y., Ben-Yaakov K., Hieda M., Yoneda Y., Twiss J. L., Fainzilber M. (2008) Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron 59, 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavalli V., Kujala P., Klumperman J., Goldstein L. S. (2005) Sunday Driver links axonal transport to damage signaling. J. Cell Biol 168, 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindwall C., Kanje M. (2005) Retrograde axonal transport of JNK signaling molecules influence injury induced nuclear changes in p-c-Jun and ATF3 in adult rat sensory neurons. Mol. Cell. Neurosci 29, 269–282 [DOI] [PubMed] [Google Scholar]
- 14.Perlson E., Hanz S., Medzihradszky K. F., Burlingame A. L., Fainzilber M. (2004) From snails to sciatic nerve: retrograde injury signaling from axon to soma in lesioned neurons. J. Neurobiol 58, 287–294 [DOI] [PubMed] [Google Scholar]
- 15.Perlson E., Medzihradszky K. F., Darula Z., Munno D. W., Syed N. I., Burlingame A. L., Fainzilber M. (2004) Differential proteomics reveals multiple components in retrogradely transported axoplasm after nerve injury. Mol. Cell. Proteomics 3, 510–520 [DOI] [PubMed] [Google Scholar]
- 16.Perlson E., Hanz S., Ben-Yaakov K., Segal-Ruder Y., Seger R., Fainzilber M. (2005) Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron 45, 715–726 [DOI] [PubMed] [Google Scholar]
- 17.Perlson E., Michaelevski I., Kowalsman N., Ben-Yaakov K., Shaked M., Seger R., Eisenstein M., Fainzilber M. (2006) Vimentin binding to phosphorylated Erk sterically hinders enzymatic dephosphorylation of the kinase. J. Mol. Biol 364, 938–944 [DOI] [PubMed] [Google Scholar]
- 18.Rishal I., Michaelevski I., Shinder V., Medzihradszky K. F., Burlingame A. L., Fainzilber M. (2010) Axoplasm isolation from peripheral nerve. Dev. Neurobiol Volume 70, pages 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey J. D. (2002) A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol 64, 479–498 [Google Scholar]
- 20.Salvador S., Chan P. (2004) Determining the number of clusters/segments in hierarchical clustering/segmentation algorithms, in ICTAI 2004: Proceedings of the 16th IEEE International Conference on Tools with Artificial Intelligence, Boca Raton, November 15–17, 2004, pp. 576–584, IEEE, Los Alamitos, CA [Google Scholar]
- 21.MacKay D. J. C. (2003) Information Theory, Inference and Learning Algorithms, Cambridge University Press, Cambridge, UK [Google Scholar]
- 22.Lochmuller C. H., Reese C. E. (1998) Introduction to factor analysis. Crit. Rev. Anal. Chem 28, 21–49 [Google Scholar]
- 23.Bernaards C. A., Jennrich R. I. (2005) Gradient projection algorithms and software for arbitrary rotation criteria in factor analysis. Educ. Psychol. Meas 65, 770–790 [Google Scholar]
- 24.Huang D. W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 25.Cronbach L. J., Schonemann P., Mckie D. (1965) Alpha-coefficients for stratified-parallel tests. Educ. Psychol. Meas 25, 291–312 [Google Scholar]
- 26.Tannu N. S., Hemby S. E. (2006) Methods for proteomics in neuroscience. Prog. Brain Res 158, 41–82 [DOI] [PubMed] [Google Scholar]
- 27.Ramström M., Zuberovic A., Grönwall C., Hanrieder J., Bergquist J., Hober S. (2009) Development of affinity columns for the removal of high-abundance proteins in cerebrospinal fluid. Biotechnol. Appl. Biochem 52, 159–166 [DOI] [PubMed] [Google Scholar]
- 28.Kanai Y., Dohmae N., Hirokawa N. (2004) Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 [DOI] [PubMed] [Google Scholar]
- 29.Elvira G., Wasiak S., Blandford V., Tong X. K., Serrano A., Fan X., del Rayo Sánchez-Carbente M., Servant F., Bell A. W., Boismenu D., Lacaille J. C., McPherson P. S., DesGroseillers L., Sossin W. S. (2006) Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics 5, 635–651 [DOI] [PubMed] [Google Scholar]
- 30.Abe N., Almenar-Queralt A., Lillo C., Shen Z., Lozach J., Briggs S. P., Williams D. S., Goldstein L. S., Cavalli V. (2009) Sunday driver interacts with two distinct classes of axonal organelles. J. Biol. Chem 284, 34628–34639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Willis D., Li K. W., Zheng J. Q., Chang J. H., Smit A., Kelly T., Merianda T. T., Sylvester J., van Minnen J., Twiss J. L. (2005) Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci 25, 778–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gan C. S., Chong P. K., Pham T. K., Wright P. C. (2007) Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res 6, 821–827 [DOI] [PubMed] [Google Scholar]
- 33.Hill E. G., Schwacke J. H., Comte-Walters S., Slate E. H., Oberg A. L., Eckel-Passow J. E., Therneau T. M., Schey K. L. (2008) A statistical model for iTRAQ data analysis. J. Proteome Res 7, 3091–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson C. N., Grant S. G. (2006) High throughput protein expression screening in the nervous system—needs and limitations. J. Physiol 575, 367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morón J. A., Devi L. A. (2007) Use of proteomics for the identification of novel drug targets in brain diseases. J. Neurochem 102, 306–315 [DOI] [PubMed] [Google Scholar]
- 36.Jiménez C. R., Stam F. J., Li K. W., Gouwenberg Y., Hornshaw M. P., De Winter F., Verhaagen J., Smit A. B. (2005) Proteomics of the injured rat sciatic nerve reveals protein expression dynamics during regeneration. Mol. Cell. Proteomics 4, 120–132 [DOI] [PubMed] [Google Scholar]
- 37.Oberg A. L., Vitek O. (2009) Statistical design of quantitative mass spectrometry-based proteomic experiments. J. Proteome Res 8, 2144–2156 [DOI] [PubMed] [Google Scholar]
- 38.Liao L., McClatchy D. B., Yates J. R. (2009) Shotgun proteomics in neuroscience. Neuron 63, 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. (2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 40.Emes R. D., Pocklington A. J., Anderson C. N., Bayes A., Collins M. O., Vickers C. A., Croning M. D., Malik B. R., Choudhary J. S., Armstrong J. D., Grant S. G. (2008) Evolutionary expansion and anatomical specialization of synapse proteome complexity. Nat. Neurosci 11, 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu A., Wiœniewski J. R., Mann M. (2009) Comparative proteomic profiling of membrane proteins in rat cerebellum, spinal cord, and sciatic nerve. J. Proteome Res 8, 2418–2425 [DOI] [PubMed] [Google Scholar]
- 42.Huang H. L., Cendan C. M., Roza C., Okuse K., Cramer R., Timms J. F., Wood J. N. (2008) Proteomic profiling of neuromas reveals alterations in protein composition and local protein synthesis in hyper-excitable nerves. Mol. Pain 4, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katano T., Mabuchi T., Okuda-Ashitaka E., Inagaki N., Kinumi T., Ito S. (2006) Proteomic identification of a novel isoform of collapsin response mediator protein-2 in spinal nerves peripheral to dorsal root ganglia. Proteomics 6, 6085–6094 [DOI] [PubMed] [Google Scholar]
- 44.Zheng J. Q., Kelly T. K., Chang B., Ryazantsev S., Rajasekaran A. K., Martin K. C., Twiss J. L. (2001) A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J. Neurosci 21, 9291–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verma P., Chierzi S., Codd A. M., Campbell D. S., Meyer R. L., Holt C. E., Fawcett J. W. (2005) Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J. Neurosci 25, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willis D. E., van Niekerk E. A., Sasaki Y., Mesngon M., Merianda T. T., Williams G. G., Kendall M., Smith D. S., Bassell G. J., Twiss J. L. (2007) Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J. Cell Biol 178, 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W., van Niekerk E., Willis D. E., Twiss J. L. (2007) RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev. Neurobiol 67, 1166–1182 [DOI] [PubMed] [Google Scholar]
- 48.Twiss J. L., Fainzilber M. (2009) Ribosomes in axons—scrounging from the neighbors? Trends Cell Biol 19, 236–243 [DOI] [PubMed] [Google Scholar]
- 49.Koenig E., Martin R., Titmus M., Sotelo-Silveira J. R. (2000) Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J. Neurosci 20, 8390–8400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giuditta A., Kaplan B. B., van Minnen J., Alvarez J., Koenig E. (2002) Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci 25, 400–404 [DOI] [PubMed] [Google Scholar]
- 51.Court F. A., Hendriks W. T., Macgillavry H. D., Alvarez J., van Minnen J. (2008) Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci 28, 11024–11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.