Abstract
Vascular smooth muscle cells maintained in normal (5.6 mm) glucose respond to insulin-like growth factor-I (IGF-I) with increased protein synthesis but do not proliferate. In contrast, hyperglycemia alters responsiveness to IGF-I, resulting in increased SHPS-1 phosphorylation and assembly of a signaling complex that enhances MAPK and phosphatidylinositol 3-kinase pathways. Hyperglycemia also reduces the basal IRS-1 concentration and IGF-I-stimulated IRS-1-linked signaling. To determine if failure to down-regulate IRS-1 alters vascular smooth muscle cell (VSMC) responses to IGF-I, we overexpressed IRS-1 in VSMCs maintained in high glucose. These cultures showed reduced SHPS-1 phosphorylation, transfer of SHP-2 to SHPS-1, and impaired Shc and MAPK phosphorylation and cell proliferation in response to IGF-I. In vitro studies demonstrated that SHPS-1 was a substrate for type I IGF receptor (IGF-IR) and that IRS-1 competitively inhibited SHPS-1 phosphorylation. Exposure of VSMC cultures to a peptide that inhibited IRS-1/IGF-IR interaction showed that IRS-1 binding to IGF-IR impairs SHPS-1 phosphorylation in vivo. IRS-1 also sequestered SHP-2. Expression of an IRS-1 mutant (Y1179F/Y1229F) reduced IRS-1/SHP-2 association, and exposure of cells expressing the mutant to the inhibitory peptide enhanced SHPS-1 phosphorylation and SHP-2 transfer. This result was confirmed by expressing an IRS-1 mutant that had both impaired binding to IGF-IR and to SHP-2 IGF-I increased SHPS-1 phosphorylation, SHP-2 association with SHPS-1, Shc MAPK phosphorylation, and proliferation in cells expressing the mutant. We conclude that IRS-1 is an important factor for maintaining VSMCs in the non-proliferative state and that its down-regulation is a component of the VSMC response to hyperglycemic stress that results in an enhanced response to IGF-I.
Keywords: Insulin-like Growth Factor (IGF), Receptor Tyrosine Kinase, Signal Transduction, Site-directed Mutagenesis, Smooth Muscle
Introduction
Insulin-like growth factor-I (IGF-I)2 binds selectively to the type I IGF receptor, which results in stimulation of several biological functions. IGF-I is a known stimulant of vascular smooth muscle cell (VSMC) proliferation and migration, and these effects are mediated through the phosphatidylinositol 3-kinase and MAPK pathways (1–3). Upon ligand occupancy, the receptor undergoes a conformational change that activates its tyrosine kinase, which phosphorylates downstream signaling molecules. IRS-1 (insulin receptor substrate-1) is one of the important substrates for both the insulin and IGF-I receptor tyrosine kinases (4, 5). IRS-1 is a docking protein that contains multiple domains that mediate protein-protein interactions, including a pleckstrin homology domain, a phosphotyrosine-binding (PTB) domain, and the carboxyl terminus (5, 6). The PTB domain binds directly to the phosphorylated NPXY motif located within the activated insulin, IGF-I, or interleukin-4 receptors (7–10). In addition, the pleckstrin homology domain also couples IRS-1 to activated receptors (11). The carboxyl terminus of IRS-1 contains several phosphotyrosines that interact with Src homology 2 domains of signaling molecules. Following their phosphorylation, several signaling intermediates, such as Grb-2, p85, and SHP-2 (Src homology 2 domain-containing protein-tyrosine phosphatase 2), bind to IRS-1, leading to activation of downstream signaling events (12, 13). IRS-1 has been linked to many of the insulin- or IGF-I-mediated biological responses, including changes in cell size, mitogenesis, and cell migration (14–16). IGF-I-stimulated IGF-IR has been shown to bind to IRS-1 and phosphorylate it in several cell types, including vascular smooth muscle cells (17, 18).
Previous studies have shown that the trans-membrane scaffolding protein SHPS-1 (SHP substrate-1) is a substrate for activated tyrosine kinases (19). Our studies have shown that SHPS-1 phosphorylation and the subsequent assembly of a signaling complex that includes SHP-2, Src, Shc (Src homology 2 domain-containing protein), and Grb2 (growth factor receptor-bound protein 2) is essential for these cells to respond to hyperglycemic stress and that it enhances the ability of IGF-I to activate both MAPK and phosphatidylinositol 3-kinase pathways, leading to increased proliferation and migration (2, 3, 20). Following their tyrosine phosphorylation, IRS-1 and Shc bind independently to Grb2, which increases activation of MAPK/extracellular signal-regulated kinase (ERK) (2, 21, 22). However, the relative contribution of Shc and IRS-1 in regulating this cascade has not been clearly delineated for many different cell types (12, 23). Additionally, IRS-1 concentrations are regulated by several factors, including glucose, insulin, IGF-I, and cell/tissue type (24–30). Several investigators have demonstrated that IRS-1 concentrations are down-regulated in response to hyperglycemia in insulin-sensitive tissues as skeletal muscle and fat (13, 31, 32). Previously, we reported that VSMCs maintained in high glucose concentrations (>12 mm) had a significant decrease in IRS-1. We also showed that the IRS-1 tyrosine phosphorylation response and subsequent Grb2 binding are significantly reduced in response to IGF-I (29). These studies were undertaken to determine the significance of the reduction in IRS-1 and whether failure to reduce the IRS-1 concentration alters assembly of the SHPS-1 complex, thus leading to altered cell proliferation and migration in response to IGF-I.
MATERIALS AND METHODS
Human IGF-I was a gift from Genentech (South San Francisco, CA). Immobilon-P membranes were purchased from Millipore Corp. (Bedford, MA). DMEM containing 4500 mg/liter glucose (DMEM-HG), DMEM containing 900 mg/liter glucose (DMEM-NG), and streptomycin and penicillin were purchased from Invitrogen. Antibodies against phospho-MAPK or total MAPK and β-actin were purchased from Cell Signaling (Beverly, MA). Polyclonal antibodies for IRS-1, SHPS-1, and Shc were obtained from Upstate Biotechnology (Lake Placid, NY). Anti-phosphotyrosine (Tyr(P)99), the hemagglutinin epitope (HA), anti-IRS-1, anti-IGF-IR, anti-SHP-2, and anti-Grb2 antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). SHPS-1 and IGF-IR polyclonal antiserum were prepared in our laboratory. IGF-IR tyrosine kinase inhibitor, PQ401, was purchased from Tocris Bioscience, (Ellisville, MO). Recombinant rat IRS-1 expressed in Sf9 cells with His tag was purchased from Millipore Corp. Synthetic peptides containing the cell permeability sequence of protein transduction domain 4 and the sequence from IGF-IR YARAAARQARA947NPEYFSAADV956 (underlined and hereafter referred to as peptide 299), the tyrosine-phosphorylated form of p299 peptide (corresponding to Tyr950 of IGF-IR), and the sequence from the IRS-1 PTB domain YARAAARQARA206LQLMNIRRCGHS217 (underlined and hereafter referred to as peptide 301, where C is italicized to denote reduction) were synthesized by the Protein Chemistry Core Facility at the University of North Carolina at Chapel Hill. Purity and sequence confirmation were determined by mass spectrometry. We used a heterologous system in this study. It is important to note that the sequences in porcine IGF-IR that bind to IRS-1 are conserved in human IGF-IR. Likewise, the IGF-I receptor tyrosine kinase sequence is completely conserved when human and porcine IGF-IR are compared. Similarly, the PTB domain of porcine IRS-1 has 100% homology to human IRS-1. Finally, porcine IGF-I and human IGF-I have 100% sequence identity. Therefore, we believe that this degree of sequence conservation enables us to study these interactions interchangeably.
Construction, Expression, and Purification of the Cytoplasmic Domain of SHPS-1 (SHPS-1/CD) and IGF-IR
The SHPS-1 cytoplasmic domain containing amino acids 394–503 was PCR-amplified with the SP6 promotor sequence, following which reverse transcription was performed to obtain mRNA (33). Using this template, SHPS-1/CD was generated, employing in vitro translation using wheat germ extract as per the manufacturer's recommendation (Promega).
The mouse fibroblasts (NWTb3) that overexpress human IGF-IR were a kind gift from Dr. Derek LeRoith (Mount Sinai School of Medicine, New York) (34). IGF-IR was purified using NWTb3 cell lysate (25 ml of cell lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl containing protease inhibitors aprotinin (10 μg/ml), leupeptin (1 μg/ml), 4-(2-aminoethyl)-benzenesulfonyl fluoride (1 mm), pepstatin (1 μg/ml), EDTA (2 mm), and Triton X-100 (0.2%)) diluted 1:5 with buffer containing (50 mm HEPES, pH 7.5, 1 m ammonium sulfate, 1 mm 4-(2-aminoethyl)-benzenesulfonyl fluoride, 2 mm EDTA, 1 μg/ml pepstatin, and 1 μg/ml leupeptin). The diluted lysate was applied to a butyl-Sepharose column previously equilibrated with 50 mm HEPES, pH 7.5, containing 1 m ammonium sulfate and 0.02% Triton X-100. After sample loading, the column was washed with equilibration buffer and eluted with 25 mm HEPES, pH 7.5, containing 0.02% Triton X-100. Fractions containing IGF-IR were detected following SDS-PAGE with immunoblotting. The fractions containing the receptor were further purified using concanavalin A chromatography. CaCl2 and MnCl2 were added to final concentrations of 4 and 2 mm, respectively, and the solution was applied to a concanavalin A-Sepharose column, equilibrated with 50 mm Tris, pH 7.2, 0.4 mm CaCl2, 2 mm MnCl2, and 0.02% Triton X-100. The IGF-IR was eluted with the above buffer containing 0.25 m methyl α-d-glucopyranoside and 0.25 m methyl α-d-mannopyranoside.
IGF-IR and SHPS-1 Phosphorylation Measurement
The purified IGF-IR was incubated with 150 ng/ml IGF-I and 10 mm ATP in a buffer containing 50 mm HEPES-NaOH (pH 7.6), 3 mm MnCl2, 10 mm MgCl2, 0.1 mm EGTA, and 0.1 mm Na3O4 and incubated for 15 min for 30 min. IGF-IR activation was determined by immunoblotting using anti-Tyr(P)99 antibody. The activated IGF-IR was used to phosphorylate the cytoplasmic domain of SHPS-1 in a buffer containing 50 mm HEPES-NaOH (pH 7.6), 3 mm MnCl2, 10 mm MgCl2, 0.1 mm EGTA, 1 mm dithiothreitol, 0.1 mm Na3O4, and 0.2 mm ATP and incubation at 30 °C for 30 min. To determine phosphorylation of SHPS-1/CD, the final product was immunoblotted using an anti-Tyr(P)99 antibody.
Cell Culture
Porcine SMCs were isolated from the porcine aortic explants according to the protocol described by Ross (74) and were maintained as described previously (2, 29). Cells were maintained in DMEM-high glucose growth medium (HG) or DMEM-normal glucose growth medium (NG) with 10% fetal bovine serum (Hyclone, Logan, UT), streptomycin (100 ng/ml), and penicillin (100 units/ml). All of the mutant cells were maintained in HG. The SMCs that were used in these experiments were used between passages 4 and 12.
Generation of pLenti Expression Vectors pLenti-HA-IGF-IR Wild Type, pLenti-HA-IRS-1 wild type (IRS-1-WT), pLenti-HA- IRS-1-FF mutant (IRS-FF), and pLenti-HA- IRS-1-R3Q-FF Double Mutant (IRS-R3Q-FF)
Wild type IGF-IR in pBluescript II KS was a kind gift from Dr. Derek LeRoith. Using this as a template, PCR amplification was performed with forward primer 5′-caccatgaagtctggctccggaggagggt-3′, including a Kozak sequence (in boldface type) placed 5′ to the ATG (underlined) start site. The reverse primer 5′-ttaagcgtaatctggaacatcgtatgggtagcaggtcgaagactggggcagcgg-3′ contained the sequence encoding HA epitope (underlined), followed by the stop codon (boldface type). The amplified product was cloned into the pENTR/D-TOPO Gateway entry vector according to the manufacturer's instructions (Invitrogen). The resultant product was confirmed by sequencing (University of North Carolina Genome Analysis Facility, Chapel Hill, NC) and transferred from the entry vector into the pLentiCMV DEST vector using the LR clonase reaction following the manufacturer's instructions (Invitrogen). Full-length human IRS-1 cDNA was purchased from Open Biosystems in pCMV-sport6 vector and after PCR amplification was cloned into the pENTR/D-TOPO Gateway entry vector according to the manufacturer's instructions (Invitrogen). The forward and reverse primers used to generate the PCR product were as follows: forward primer, 5′-caccatggcgagccctccggagagcgat-3′; reverse primer, 5′-ttaagcgtaatctggaacatcgtatgggtactgacggtcctctggctgcttctgg-3′. The forward primer includes a Kozak sequence (boldface type) placed 5′ to the ATG (underlined) start site. The reverse primer contained the sequence encoding HA epitope (underlined) followed by the stop codon (boldface type). The resultant product was sequenced (UNC Genome Analysis Facility, Chapel Hill, NC) to confirm the correct full-length human IRS-1 using M13 forward and reverse primers and five other primers: IRS-1Seq1A, 5′-gggtcagacaaagaacctgatt-3′; IRS-1Seq2A, 5′-cgcccagcctcggtgga-3′; IRS-1Seq3A, 5′-ctaccattacccaccagaag-3′; IRS-1Seq4A, 5′-tgactacatgaacatgtcacca-3′; IRS-1Seq5C, 5′-tgaccatgcagatgagttgtcc-3′.
Using the pENTR IRS-1-WT as template, an IRS mutant was generated where the tyrosines at positions 1179 and 1229 were mutated to phenylalanine (IRS-FF). PCR amplifications were carried out using complementary primer 5′-ggagaatggtcttaactTcatagacctggatttgg-3′ and reverse primer 5′-ccaaatccaggtctatgAagttaagaccattctcc-3′ to generate Y1179F and another complementary primer 5′-gaggatttaagcgcctTtgccagcatcag-3′ and reverse primer 5′-ctgatgctggcaAaggcgcttaaatcctc-3′ to generate Y1229F (where capitalized bases indicate the substitutions). After selection of correct clone based on DNA sequencing, using pENTR IRS-FF as template, a double mutant was constructed where three arginines at positions 212, 213, and 227 were mutated to glutamine (IRS-R3Q-FF) using double-stranded mutagenesis. Initially, PCR amplification was carried out using forward primer 5′-gctgatgaacatcCAgcAAtgtggccactcgg-3′ and reverse primer 5′-ccgagtggccacaTTgcTGgatgttcatcagc-3′ (where capitalized bases indicate the substitutions), by which two of the residues, Arg212 and Arg213, were mutated to glutamine. The resultant DNA was used as a template, and the arginine at position 227 was mutated to glutamine using complementary primers 5′-catcgaggtgggccAAtctgccgtgacgggg-3′ and reverse primer 5′-ccccgtcacggcagaTTggcccacctcgatg-3′ (where capitalized bases indicate the substitution). The products were sequenced (University of North Carolina Genome Analysis Facility) to confirm the correct incorporation of the changes, following which the cDNAs encoding the wild type, single mutant, and double mutant forms of the protein were transferred from the entry vector into pLentiCMV DEST vector using the LR clonase reaction following the manufacturer's instructions (Invitrogen).
Construction of a Plasmid Containing Short Hairpin RNA Template for IRS-1 Silencing
Based on Invitrogen web site design tools, a sequence containing 21 oligonucleotides (GCGAGGATTTAAGCGCCTATG) located within the porcine IRS-1 (accession number EU681268.1; nucleotides 3515–3536) was used to construct the short hairpin RNA template plasmid. The selected sequences were not found in any other mammalian protein searched using the NCBI Blast search program. The oligonucleotides were synthesized by the Nucleic Acids Core Facility at the University of North Carolina, annealed, and ligated into BLOCK-iT U6 RNAi entry vector (catalogue no. K4945-00, Invitrogen), following the manufacturer's instructions. The complete sequence was verified by DNA sequencing. The expression vector was generated using the Gateway LR recombination reaction between the entry vector and BLOCK-iT Lentiviral RNAi Gateway Vector (catalogue no. K4943-00, Invitrogen). The empty vector was used as a control (EVC). After confirmation of the sequence, plasmid DNA was prepared by a Plasmid minikit (Promega, Madison, WI).
Generation of Virus Stocks
293FT cells (Invitrogen) were prepared for generation of virus stocks of each individual pLenti construct. All of the transfections and viral stock generations were carried out as described previously (2, 3).
Establishment of Porcine VSMCs Expressing pLenti Constructs
VSMCs (passages 4 and 5) were seeded at 3 × 105/well in each of two wells of a 6-well plate (catalog no. 353046, Falcon) the day before transduction. The viral stocks were thawed, and the viral complexes were precipitated as described previously (2, 3). The expression of the HA-tagged IRS-1-WT and IRS-1 mutants were detected by immunoblotting with a 1:1000 dilution of anti-HA antibody using 20 μg of protein.
Immunoprecipitation and Immunoblotting
Cells were seeded at 5 × 105 cells/10-cm plate (BD Biosciences) and grown for 5–7 days to reach confluence. The cultures were incubated in serum-free DMEM-HG for 14–18 h before the addition of IGF-I (as indicated). In some experiments, non-transduced SMCs were preincubated with or without the synthetic peptides for 1–2 h before IGF-I was added. Following cell lysis, all immunoprecipitations and immunoblotting were carried out as described previously (2, 3). The proteins were detected using enhanced chemiluminescence (Pierce). The images obtained were also scanned using DuoScan scanning densitometer (Agfa, Morstel, Belgium). Densitometric analyses of the images were determined using National Institutes of Health ImageJ, version 1.37v. To determine differences in MAPK phosphorylation, arbitrary scanning units obtained for phospho-MAPK band intensities were divided by arbitrary scanning units obtained for total β-actin protein band intensities for each time point analyzed. The -fold values or average ratio values obtained from at least three independent experiments for each lane were pooled together.
Cell Proliferation Assay and Wounding and Migration Assay
Assessment of SMC proliferation was performed as described previously (35). IGF-I is known to stimulate smooth muscle cell chemokinesis, and cellular migration was analyzed as previously described (36). Briefly, VSMCs were plated in 6-well dishes and grown to confluence. The monolayer of cells was wounded with a razor blade. After wounding, cells were incubated with SFM plus 0.2% FBS in the presence or absence of IGF-I (50 ng/ml) at 37 °C for 48 h. The wounded monolayers were then fixed and stained (Diff Quick; Dade Behring, Inc., Newark, DE), and the number of cells migrating into the wound area was counted. At least five of the previously selected 1-mm areas at the edge of the wound were counted for each data point.
Statistical Analysis
Student's t test was used to compare the differences between the treatments. The results that are shown in all experiments are representative of at least three separate experiments, and p ≤ 0.05 was considered statistically significant.
RESULTS
Effect of Glucose and IGF-I on Total IRS-1 Protein Levels and IRS-1 Signaling
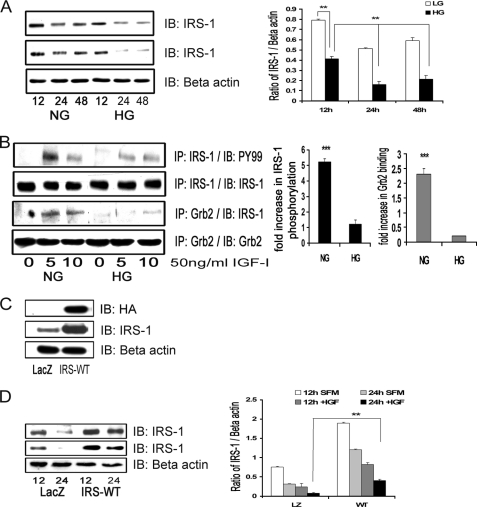
VSMCs that were maintained in 25 mm glucose (DMEM-HG) and then changed to serum-free medium containing the same glucose concentration for 12 h had a significant decrease in total IRS-1 levels (1.8 ± 0.1-fold, n = 3, p < 0.01) compared with cells that were exposed to 5 mm glucose (DMEM-NG). The degree of decrease in IRS-1 was maintained after 24 h (1.9 ± 0.1-fold, n = 3, p < 0.01) and 48 h (2.4 ± 0.2-fold, n = 3, p < 0.01) (Fig. 1A, top). Similar cultures stimulated with IGF-I for 24 and 48 h showed a significant additional decrease (1.9 ± 0.1-fold at 24 h and 1.7 ± 0.2-fold at 48 h, n = 3, p < 0.01) in total IRS-1 in DMEM-HG compared with VSMCs not exposed to IGF-I (Fig. 1A, middle). Consistent with our previous observations, tyrosine phosphorylation of IRS-1 (corrected for changes in total IRS-1) in response to IGF-I was significantly reduced (4.2 ± 0.3-fold, n = 5, p < 0.001) in DMEM-HG cells compared with DMEM-NG cells (Fig. 1B). Grb2 association with IRS-1 was significantly decreased (8.1 ± 0.1-fold, n = 5, p < 0.001) in DMEM-HG cells compared with DMEM-NG cultured cells in response to IGF-I (Fig. 1B).
FIGURE 1.
Hyperglycemia decreases total IRS-1 protein levels and IRS-1-mediated signaling. A, confluent VSMC cultures were maintained in medium containing 5 mm (NG) or 25 mm (HG) glucose prior to these analyses. They were changed to serum-free medium containing the same glucose concentration for 12, 24, or 48 h without (top) or with (second panel) IGF-I (50 ng/ml). The anti-β-actin antibody (loading control) is shown in the lower panel. After cell lysis, aliquots containing equal amounts of protein were separated directly by SDS-PAGE and immunoblotted (IB) with anti-IRS-1 antibody. The bar graph is representative of three independent experiments without IGF-I. Error bars, mean ± S.E. **, p < 0.01. B, the extent of IRS-1 tyrosine phosphorylation was determined by immunoprecipitating (IP) cell lysates with an anti-IRS-1 antibody and then immunoblotting with an anti-Tyr(P)99 (PY99) antibody (first panel), and Grb-2 association with IRS-1 was determined by immunoprecipitating with an anti-Grb2 antibody and then immunoblotting with anti-IRS-1 antibody (third panel). The membranes were stripped and reprobed with either anti-IRS-1 or anti-Grb2 antibody to detect the total levels of IRS-1 (second panel) or Grb2 (fourth panel), respectively. The amount of immunoprecipitate analyzed was adjusted to correct for differences in total IRS-1 between the NG and HG cultures. The bar graph represents three independent experiments. Error bars, mean ± S.E. ***, p < 0.001 when the amount of tyrosine phosphorylation of IRS-1 or the amount of Grb2 associated with IRS-1 at 5 min in response to IGF-I is compared between NG and HG cultures. C, VSMCs expressing either LacZ or IRS-WT were serum-starved in DMEM-HG and analyzed for recombinant protein expression. Cell lysates were immunoblotted using an anti-HA antibody (top), anti-IRS-1 antibody (middle), and an anti-β-actin antibody loading control (bottom). D, confluent VSMCs expressing LacZ or IRS-WT cultured in HG were serum-deprived and stimulated without or with IGF-I (50 ng/ml) for 12 or 24 h. Equal amounts of protein were separated by SDS-PAGE and immunoblotted with anti-IRS-1 antibody (without IGF-I (top) or with IGF-I (middle)) or with anti-β-actin antibody (bottom). The bar graph is representative of three independent experiments. Error bars, mean ± S.E. **, p < 0.01.
In order to determine the effect of maintaining a higher level of IRS-1 than would normally be present in hyperglycemic conditions on IGF-I actions, VSMCs overexpressing HA-tagged IRS-1 (IRS-WT) were constructed. The basal total IRS-1 protein level was higher (5.2 ± 0.1-fold, n = 5, p < 0.001) in IRS-WT cells maintained in DMEM-HG compared with LacZ control cells (Fig. 1C). Following exposure to IGF-I, IRS-1 was reduced to a significantly greater extent in the LacZ cells (2.6 ± 0.2-fold decrease) compared with a 1.9 ± 0.1-fold decrease in IRS-1 WT cells (n = 3, p < 0.01; Fig. 1D).
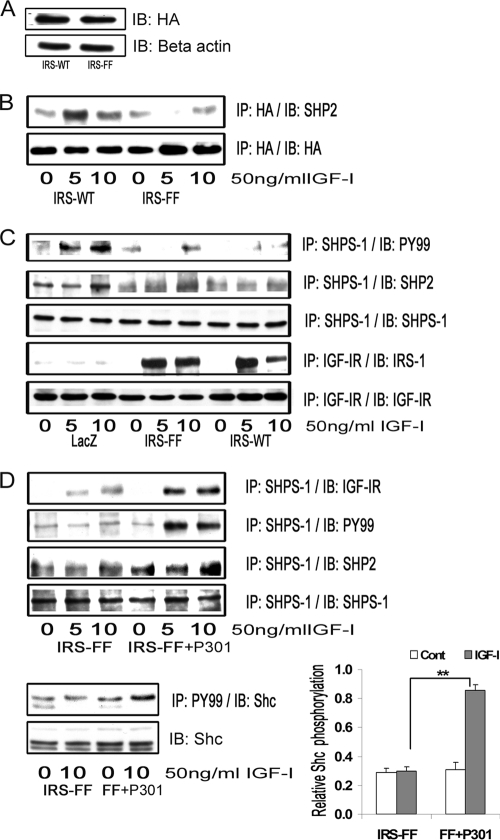
Effect of IRS-1 Overexpression on IGF-I-mediated SHPS-1 Phosphorylation, SHPS-1 Complex Assembly, Downstream Signaling, and Cellular Proliferation
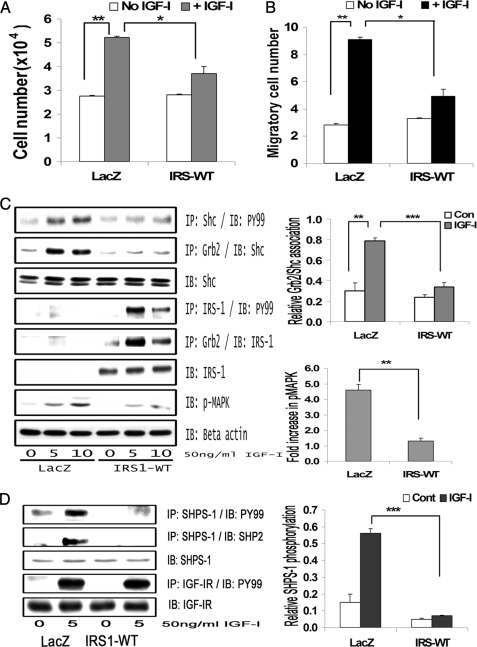
Cellular proliferation in IRS-WT-overexpressing cells maintained in DMEM-HG was significantly impaired (1.1 ± 0.3-fold increase) compared with LacZ control cells (1.98 ± 0.1-fold increase) in response to IGF-I (n = 5, p < 0.05; Fig. 2A). Similarly, VSMC migration in response to IGF-I was significantly reduced (1.97 ± 0.1-fold reduction, n = 4, p < 0.05) in IRS-WT cells maintained in DMEM-HG compared with LacZ control cells (Fig. 2B). Because proliferation of VSMCs in high glucose has been shown to be dependent on Shc phosphorylation and subsequent MAPK activation, we determined the Shc and MAPK phosphorylation responses in SMCs overexpressing IRS-1 that were maintained in DMEM-HG. The Shc phosphorylation response to IGF-I was reduced 2.1 ± 0.2-fold and 2.6 ± 0.1-fold at 5 and 10 min, respectively (n = 4, p < 0.01) in IRS-WT cells compared with LacZ cells, and Shc-Grb2 binding was also reduced 2.7 ± 0.1-fold both at 5 and 10 min after IGF-I stimulation (n = 4, p < 0.001) (Fig. 2C, top panels). Additionally, phosphorylation of MAPK was found to be significantly attenuated (4.1 ± 0.2-fold, n = 5, p < 0.01) in cells overexpressing IRS-WT compared with LacZ cells in response to IGF-I after 10 min (Fig. 2C, bottom panels).
FIGURE 2.
IRS-1 overexpression impairs IGF-I-mediated SHPS-1 phosphorylation and subsequent SHPS-1 complex assembly, leading to reduction in phosphorylation of MAPK and cellular proliferation. A, LacZ and IRS-WT cells were plated (3 × 104 cells) in DMEM-HG with 2% FBS prior to exposure to IGF-I in DMEM-HG with 0.2% platelet-poor plasma. Forty-eight hours after the addition of IGF-I, the cell number was determined. **, p < 0.01 when the change in cell number in response to IGF-I in LacZ control cells is determined; *, p < 0.05 when the number of cells proliferating after exposure to IGF-I is compared between LacZ and IRS-WT cells. Error bars, mean ± S.E. B, LacZ and IRS-WT cells were grown in 6-well dishes in DMEM-HG containing 10% FBS. After wounding, they were allowed to migrate with or without IGF-I in medium containing 0.2% FBS for 48 h. The total number of cells migrating past the wound line in the predetermined areas was counted. The results shown are the mean ± S.E. of at least three independent experiments. **, p < 0.01 when the change in migrating cells in response to IGF-I in LacZ cells is compared with LacZ cells without IGF-I; *, p < 0.05 when the number of cells migrating in response to IGF-I is compared between LacZ and IRS-WT cells. C, confluent LacZ and IRS-WT cells were serum-starved for 16 h in DMEM-HG and then exposed to IGF-I for the times indicated. The extent of Shc and IRS-1 phosphorylation was determined by immunoprecipitating (IP) p52shc (first panel) or IRS-1 (fourth panel) and then immunoblotting (IB) with an anti-phosphotyrosine (Tyr(P)99 (PY99)) antibody. Similarly, the lysates were immunoprecipitated with anti-Grb2 antibody, and the bottom portion of the blot was used for immunoblotting for p52shc (second panel), whereas the top portion of the same blot was immunoblotted for IRS-1 (fifth panel). An equal amount of protein from cell lysates from the same experiment was used to immunoblot for total Shc (third panel) and total IRS-1 (sixth panel). Similarly, an equal amount of protein from the same cell lysates was immunoblotted using anti-phospho-MAPK (seventh panel). The blots were stripped and reprobed using anti-β-actin antibody (bottom panel). The bar graphs are representative of at least three independent experiments and two independent transductions. Error bars, mean ± S.E. ***, p < 0.001 when the amount of Grb2 associated with Shc at 10 min in response to IGF-I is compared between LacZ and IRS-WT cells; **, p < 0.01 when the amount of phosphorylation of MAPK in response to IGF-I at 10 min is compared between LacZ and IRS-WT cells. D, confluent LacZ- and IRS-WT-expressing cells were serum-starved for 16 h in DMEM-HG and then exposed to IGF-I for 5 min, and the extent of SHPS-1 phosphorylation and SHP-2 association was determined by immunoprecipitating with an anti-SHPS-1 antibody and immunoblotting with Tyr(P)99 (first panel) or with anti-SHP-2 antibody (second panel). The extent of IGF-IR tyrosine phosphorylation was assessed by immunoprecipitating IGF-IR and immuoblotting for phosphotyrosine (fourth panel). Lysates from the same experiments were immunoblotted for total SHPS-1 (third panel) or for total IGF-IR (fifth panel). The bar graph shows the relative increase in SHPS-1 phosphorylation for at least three independent experiments. Error bars, mean ± S.E. ***, p < 0.001 when increase in SHPS-1 phosphorylation in response to IGF-I is compared between LacZ- and IRS-WT-expressing cells.
Phosphorylation of SHPS-1 and subsequent assembly of a signaling complex is required for IGF-I stimulation of p52shc phosphorylation and MAPK activation (2). SHP-2 is a key component of that complex that binds directly to phosphorylated SHPS-1 (33); hence, SHPS-1 phosphorylation and SHP-2 recruitment were evaluated. Phosphorylation of SHPS-1 was found to be significantly impaired (5.1 ± 0.2-fold reduction, n = 3, p < 0.001) in IRS-WT cells compared with LacZ cells in response to IGF-I (Fig. 2D). Similarly, the association of SHP-2 with SHPS-1 was found to be reduced (4.2 ± 0.1-fold, n = 3, p < 0.001) in IRS-WT cells compared with LacZ cells in response to IGF-I (Fig. 2D). In contrast, the IGF-IR phosphorylation levels remained the same in both IRS-WT cells and LacZ cells in response to IGF-I (Fig. 2D). Taken together, the above data demonstrate that maintenance of an IRS-1 level in SMCs maintained in high glucose that was similar to the IRS-1 level in VSMCs maintained in normal glucose was associated with a reduction in several of the cellular responses to IGF-I, including SHPS-1 phosphorylation, SHP-2 recruitment to SHPS-1, p52shc phosphorylation, MAPK activation, and cell proliferation.
Effect of IGF-IR and IRS-1 on SHPS-1 Phosphorylation
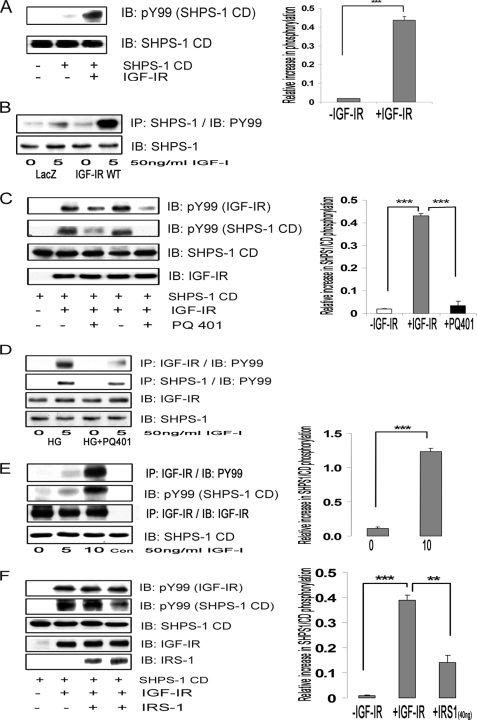
SHPS-1, a transmembrane protein, has been shown to be a substrate of several tyrosine receptor kinases, including the insulin receptor (37). Hence, we investigated whether IGF-IR kinase could phosphorylate SHPS-1. In vitro phosphorylation assays showed that IGF-I-activated IGF-IR directly phosphorylated the SHPS-1/CD (18.7 ± 2.2-fold, n = 3, p < 0.001) compared with SHPS-1/CD without IGF-IR (Fig. 3A) or SHPS-1/CD with inactivated IGF-IR (data not shown). Further, cells overexpressing IGF-IR maintained in HG conditions showed a 2.6 ± 0.1-fold increase in SHPS-1 phosphorylation compared with LacZ cells in response to IGF-I (n = 3, p < 0.001; Fig. 3B). When the IGF-IR inhibitor PQ401 was added in vitro, it inhibited IGF-I receptor autophosphorylation (Fig. 3C, top). More importantly, IGF-IR-mediated phosphorylation of SHPS-1/CD was also inhibited (12.6 ± 1.7-fold decrease, n = 3, p < 0.001; Fig. 3C, second panel). Similarly, when VSMCs cultured in HG conditions were treated with PQ401, IGF-IR phosphorylation and SHPS-1 phosphorylation were decreased (1.7 ± 0.2-fold decrease, n = 3, p < 0.05) in response to IGF-I (Fig. 3D). Furthermore, when IGF-I was used to stimulate IGF-IR phosphorylation in VSMCs overexpressing IGF-IR (Fig. 3E, top) and the IGF-IR immunoprecipitate was analyzed in vitro, SHPS-1/CD phosphorylation was increased to a greater extent compared with the non-IGF-I-stimulated IGF-IR immunoprecipitate (9.1 ± 0.2-fold increase, n = 3, p < 0.001; Fig. 3E, middle). These cell-free in vitro and in vivo data demonstrate that IGF-IR can directly phosphorylate SHPS-1/CD.
FIGURE 3.
IGF-IR kinase phosphorylates SHPS-1 and IRS-1 binding to IGF-IR impairs its ability to phosphorylate SHPS-1 in cells and in a cell-free system. A, SHPS-1/CD generated by in vitro translation using wheat germ extract was incubated in vitro (denoted by the plus or minus sign) with or without activated IGF-IR kinase as described under “Materials and Methods.” The supernatant was analyzed by immunoblotting (IB) for phosphotyrosine (top) or SHPS-1 (bottom). The relative -fold increase in the phosphorylation of SHPS-1/CD is shown in the bar graph. Error bars, mean ± S.E. ***, p < 0.001. B, confluent LacZ and IGF-IR-WT cells were serum-starved for 16 h in DMEM-HG and exposed to IGF-I for 5 min. The extent of SHPS-1 phosphorylation was determined by immunoprecipitating (IP) SHPS-1 and immunoblotting with Tyr(P)99 (first panel). Lysates from the same experiments were immunoblotted for total SHPS-1 (second panel). C, in similar experiments, additional samples that contained IGF-IR and SHPS-1/CD were exposed to the IGF-IR tyrosine kinase inhibitor, PQ401 15 μm (lane 3) or 50 μm (lane 5), and vehicle control (DMSO) was added to lanes 2 and 4. Half of the supernatant was analyzed by immunoblotting using Tyr(P)99 (PY99) antibody representing IGF-IR phosphorylation (top) and SHPS-1/CD phosphorylation (bottom). Both bands were identified by electrophoretic mobility. The other half of the supernatant was analyzed by immunoblotting for total IGF-IR and total SHPS-1 (lower panels). The relative -fold increase in the SHPS-1/CD phosphorylation and suppression by the inhibitor is shown in the bar graph. Error bars, mean ± S.E. ***, p < 0.001. D, VSMCs cultured and maintained in DMEM-HG were serum-starved and then incubated with or without PQ401 (50 μm) for 1 h. IGF-I was added for 5 min. Cell lysates were immunoprecipitated with anti-IGF-IR antibody (first panel) or with anti-SHPS-1 antibody (second panel) and immunoblotted for Tyr(P)99. Cell lysates were immunoblotted with anti-IGF-IR (third panel) or anti-SHPS-1 antibody (fourth panel). E, confluent IGF-IR-overexpressing cells were serum-starved for 16 h in DMEM-HG and then exposed to IGF-I for the times indicated. The cell lysates were immunoprecipitated with anti-IGF-IR antiserum. Some aliquots of reconstituted immunoprecipitate were incubated with SHPS-1/CD in in vitro phosphorylation assays (lanes 1–3) or with buffer alone (Con). After centrifugation, the resultant supernatants were immunoblotted using a phosphotyrosine antibody IGF-IR (top) or phospho-SHPS-1/CD (middle). Similar lysates were immunoprecipitated and immunoblotted with anti-IGF-IR antibody as a loading control (bottom). To ascertain the specificity of the bands, an antibody only control was also used. The bar graph represents the relative increase in SHPS-1/CD phosphorylation in response to IGF-I at 10 min compared with non-stimulated immunoprecipitates; ***, p < 0.001 based on at least three independent experiments. Error bars, mean ± S.E. F, in an in vitro assay, the SHPS-1/CD and activated IGF-IR were incubated with increasing concentrations of recombinant full-length IRS-1, 20 ng (lane 3) and 40 ng (lane 4), for 30 min. An aliquot of each supernatant was analyzed by immunoblotting with an anti-phosphotyrosine antibody that recognizes IGF-IR tyrosine phosphorylation (top) or SHPS-1/CD (middle). The third panel indicates the total SHPS-1/CD. The -fold increase in SHPS-1/CD phosphorylation in response to IGF-IR is shown in the bar graph. Error bars, mean ± S.E. ***, p < 0.001. The -fold decrease in SHPS-1 phosphorylation in the presence of 40 ng of IRS-1 (lane 4) is shown (bar graph) in comparison with no IRS-1 addition (lane 2). Error bars, mean ± S.E.; **, p < 0.01.
Because IRS-1 is a known substrate for both insulin receptor and IGF-IR tyrosine kinases, we wished to determine whether IRS-1 could compete with SHPS-1 as an IGF-IR substrate. Recombinant IRS-1 was added to the in vitro phosphorylation assays. Increasing concentrations of IRS-1 decreased the phosphorylation of SHPS-1/CD (3.9 ± 0.3-fold decrease, n = 3, p < 0.01) (Fig. 3F), but there was no effect on IGF-IR autophosphorylation. These observations, taken together with our results showing that IRS-1-overexpressing cells exhibit impaired SHPS-1 phosphorylation (Fig. 2D), suggest that IRS-1 is competing with SHPS-1 for binding to IGF-IR, thereby altering SHPS-1 phosphorylation.
Effect of Inhibition of IGF-IR/IRS-1 Association on SHPS-1 Phosphorylation
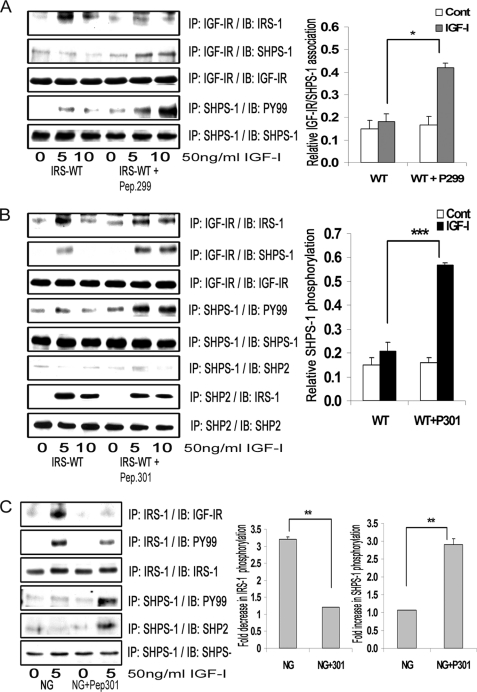
Previous studies have demonstrated that the interaction between the NPXY motif (residues 947–950) of IR/IGF-IR and the PTB domain of IRS-1 mediates its association (7, 9). To determine whether disrupting IGF-IR/IRS-1 association would affect IGF-IR/SHPS-1 association and subsequent IGF-IR-mediated SHPS-1 phosphorylation, we used cell-permeable peptides containing either the NPEY motif of IGF-IR (peptide 299) or a peptide containing the amino acid sequence between residues 206 and 217 of IRS-1 (peptide 301). This sequence contains a region of the PTB domain that has been proposed to mediate its binding to NPXY sequences. IRS-WT cells exposed to either peptide 299 or peptide 301 showed decreased IGF-IR/IRS-1 association compared with IRS-WT cells in response to IGF-I (2.3 ± 0.1-fold, n = 3, p < 0.001 for peptide 299; 1.7 ± 0.2-fold, n = 3, p < 0.05 for peptide 301) (Fig. 4, A and B, top). IRS-1 tyrosine phosphorylation was also significantly impaired (2.5 ± 0.1-fold, n = 3, p < 0.001) in response to IGF-I in IRS-WT cells exposed to peptide 301 compared with non-exposed cells (supplemental Fig. 1B). The reduction of IGF-IR/IRS-1 association was associated with an increase in IGF-IR association with SHPS-1 (2.0 ± 0.3-fold, n = 3, p < 0.05 for peptide 299; 2.6 ± 0.2-fold, n = 3, p < 0.01 for peptide 301). This resulted in a subsequent increase in SHPS-1 phosphorylation in IRS-WT cells exposed to either peptide (3.1 ± 0.2-fold, n = 3, p < 0.001 for peptide 299; 2.6 ± 0.3-fold, n = 3, p < 0.001 for peptide 301) compared with IRS-WT cells not exposed to peptides in response to IGF-I (Fig. 4, A and B). To control for potential differences in tyrosine phosphorylation of peptide 299, the experiment was repeated using a tyrosine-phosphorylated form. Following exposure to this peptide, the amount of IRS-1 associated with IGF-IR and the tyrosine phosphorylation of IRS-1 were significantly impaired (2.3 ± 0.2- and 2.1 ± 0.1-fold, respectively) in response to IGF-I after 5 min in cells exposed to peptide compared with non-exposed cells (n = 3, p < 0.01). Further, the level of association of SHPS-1 with IGF-IR and SHPS-1 phosphorylation was significantly increased in the cells exposed to the peptide in response to IGF-I at 5 min compared with non-exposed cells (3.1 ± 0.1- and 3.3 ± 0.1-fold, respectively) (n = 3, p < 0.001; supplemental Fig. 1A).
FIGURE 4.
Inhibition of IRS-1 binding to IGF-IR increases association of SHPS-1 with IGF-IR and increases SHPS-1 phosphorylation. A and B, confluent cultures expressing WT-IRS-1 were serum-starved overnight and then incubated with or without cell-permeable peptides, IGF-IR (peptide 299) (A) or IRS-1 (peptide 301) (B) at 20 μg/ml for 2 h. IGF-I was added for either 5 or 10 min. Cell lysates were immunoprecipitated (IP) with anti-IGF-IR antibody and then immunoblotted for IRS-1 (top panel) or for SHPS-1 (second panel). The blots were stripped and reprobed with anti-IGF-IR antibody (third panel). Cell lysates from the same experiments were immunoprecipitated with anti-SHPS-1 antibody and immunoblotted with Tyr(P)99 antibody (fourth panel). The blots were stripped and reprobed with anti-SHPS-1 antibody (fifth panel). B, in addition, cell lysates were immunoprecipitated with anti-SHPS-1 antibody and immunoblotted for SHP-2 (sixth panel). Similar lysates were immunoprecipitated with anti-SHP-2 antibody and immunoblotted for IRS-1 (seventh panel) or SHP-2 (eighth panel). The top bar graph shows the relative increase in IGF-IR/SHPS-1 association for at least three independent experiments. Error bars, mean ± S.E. *, p < 0.05 when increase in IGF-IR/SHPS-1 association in response to IGF-I at 10 min is compared between IRS-WT cells with and without peptide 299. The middle bar graph shows the relative increase in SHPS-1 phosphorylation for at least three independent experiments. Error bars, mean ± S.E. ***, p < 0.001 when increase in SHPS-1 phosphorylation in response to IGF-I after 10 min is compared in IRS-WT-expressing cells with or without peptide 301. C, VSMCs cultured and maintained in DMEM-NG were serum-starved and then incubated with or without cell-permeable IGF-IR/IRS-1 peptide 301 at a concentration of 5 μg/ml for 2 h. IGF-I was added for 5 min with and without the peptide. Cell lysates from the above experiments were immunoprecipitated with anti-IRS-1 antibody and immunoblotted for either IGF-IR (upper panel) or Tyr(P)99 (second panel). The blots were stripped and reprobed with anti-IRS-1 antibody (third panel). Cell lysates from the same experiments were immunoprecipitated with anti-SHPS-1 antibody and immunoblotted with Tyr(P)99 antibody (fourth panel) or with anti-SHP-2 antibody (fifth panel). The blots were stripped and reprobed with anti-SHPS-1 antibody (sixth panel). The bar graph shows the relative increase in SHPS-1 phosphorylation for at least three independent experiments. Error bars, mean ± S.E. **, p < 0.01 when the increase in SHPS-1 phosphorylation in response to IGF-I is compared with or without peptide 301.
Formation of the SHPS-1 signaling complex that leads to MAPK pathway activation is dependent upon SHP-2 recruitment to phosphorylated SHPS-1 (2). When we determined the effect of peptide 301 on SHP-2 transfer, it failed to restore the SHP-2 transfer to SHPS-1 despite its ability to increase SHPS-1 phosphorylation. Furthermore, the amount of IRS-1 associated with SHP-2 in the presence of peptide 301 in response to IGF-I was not reduced significantly (1.0 ± 0.1-fold, n = 3, p = not significant; Fig. 4B).
In order to confirm the findings obtained in cells overexpressing IRS-1, we used non-transfected VSMCs cultured in DMEM-NG (Fig. 1). When those cultures were exposed to peptide 301, they showed a significant reduction in IGF-IR/IRS-1 association compared with cells not exposed to the peptide in response to IGF-I (3.6 ± 0.1-fold, n = 3, p < 0.001; Fig. 4C). This reduction in IGF-IR/IRS-1 association led to impairment of tyrosine phosphorylation of IRS-1 in response to IGF-I (2.0 ± 0.2-fold) compared with control cells. Peptide exposure increased the SHPS-1 phosphorylation (2.2 ± 0.3-fold) and SHP-2 transfer to SHPS-1 (1.9 ± 0.1-fold) in response to IGF-I (n = 3, p < 0.05; Fig. 4C). Furthermore, a significant increase was observed in IGF-IR/SHPS-1 association (2.2 ± 0.2-fold, n = 3, p < 0.01), Shc tyrosine phosphorylation (1.9 ± 0.1-fold, n = 3, p < 0.05), and phosphorylation of MAPK (1.9 ± 0.1-fold, n = 3, p < 0.05; supplemental Fig. 1C) in cells exposed to peptide 301 in response to IGF-I compared with non-exposed cells. Concomitant incubation of VSMCs maintained in DMEM-NG with peptide 301 increased cellular proliferation in response to IGF-I (1.8 ± 0.1-fold, n = 3, p < 0.01; supplemental Fig. 1D). Taken together, these results suggest that IRS-1 and SHPS-1 compete for binding to IGF-IR kinase and that the level of IRS-1 that is present in VSMCs maintained in normal glucose is sufficient to suppress SHPS-1 phosphorylation and the subsequent assembly of the SHPS-1-associated signaling complex.
Effect of SHP-2 Sequestration by IRS-1 on SHPS-1 Complex Assembly
IRS-1 contains several YXX(L/I/V) motifs that are known to bind to SHP-2 following stimulation of IRS-1 tyrosine phosphorylation. Therefore, we proposed that IRS-1 overexpression resulted in sequestration of SHP-2 and prevented its recruitment to phosphorylated SHPS-1 (Fig. 4B). To test this hypothesis, we prepared a HA-tagged IRS-1 mutant (IRS-FF), wherein tyrosine residues 1179 and 1229 were mutated to phenylalanine (Fig. 5A). The association of IRS-1 and SHP-2 was reduced 2.7 ± 0.1-fold in the IRS-FF mutant (n = 5, p < 0.01) compared with IRS-WT cells in response to IGF-I after 5 min (Fig. 5B). Recent studies have shown that IRS-1 binding to SHP-2 could potentially be mediated by five other sites, Tyr759, Tyr760, Tyr891, Tyr935, and Tyr1006 (38). This could explain the residual IRS-1 binding to SHP-2 that is detected using the IRS-FF mutant. Despite the reduction in SHP-2 binding to IRS-1, the amount of SHP-2 transferred to SHPS-1 was still impaired in IRS-FF mutant cells in response to IGF-I compared with LacZ cells. This is probably due to the fact that SHPS-1 phosphorylation was still significantly impaired in the IRS-FF cells compared with LacZ cells (Fig. 5C). Furthermore, the levels of SHPS-1 phosphorylation and SHP-2 transfer failed to increase significantly in the IRS-FF cells compared with IRS-WT cells in response to IGF-I.
FIGURE 5.
IRS-1 sequesters SHP-2 and impairs its transfer to SHPS-1. A, VSMCs overexpressing IRS-WT and the IRS-FF mutant were serum-starved overnight and analyzed for recombinant protein expression. Cell lysates were immunoblotted (IB) for HA (top) and for β-actin (bottom). B, confluent cultures of IRS-WT and IRS-FF cells were serum-starved for 16 h and then exposed to IGF-I for the times indicated. Cell lysates were immunoprecipitated (IP) with anti-HA and immunoblotted for SHP-2 (top). The membranes were stripped and probed with anti-HA antibody (bottom). C, confluent cultures of control LacZ cells, IRS-WT cells, and IRS-FF cells were serum-starved overnight and stimulated with IGF-I for 5 or 10 min. The cell lysates were immunoprecipitated with anti-SHPS-1 and immunoblotted for Tyr(P)99 (top panel) or SHP-2 (second panel). They were stripped and reprobed with anti-SHPS-1 antibody (third panel). Lysates from similar experiments were immunoprecipitated with anti-IGF-IR and immunoblotted for IRS-1 (fourth panel). The blots were stripped and reprobed for IGF-IR (bottom panel). D, confluent IRS-FF cell cultures were serum-starved for 14–16 h and incubated with or without cell-permeable IGF-IR/IRS-1 peptide 301 (20 μg/ml) for 2 h followed by IGF-I for either 5 or 10 min. Cell lysates were immunoprecipitated with anti-SHPS-1 and immunoblotted for IGF-IR (first panel), Tyr(P)99 (second panel), SHP-2 (third panel), or SHPS-1 (fourth panel). Lysates were immunoprecipitated with Tyr(P)99 (PY99) and immunoblotted with anti-p52shc (fifth panel) or immunoblotted directly for total Shc (bottom panel). The bar graph shows the relative increase in Shc phosphorylation for at least three independent experiments. Error bars, mean ± S.E. **, p < 0.01 when increase in Shc phosphorylation in response to IGF-I is compared between IRS-FF mutant cells with or without peptide 301.
To determine why inhibiting SHP-2 association with IRS-1 did not alter SHP-2 binding to SHPS-1, we assessed IRS-1/IGF-IR association using the cells that were overexpressing the IRS-FF mutant. When those cells were compared with the IRS-WT cells, there was no difference in IRS-1/IGF-IR association in response to IGF-I after 5 min (1.1 ± 0.1-fold, n = 3, p = not significant; Fig. 5C). The difference in IGF-IR/IRS-1 association after 10 min of IGF-I stimulation could be explained by the ability of SHP-2 to dephosphorylate IRS-1 in WT cells and not in FF mutant cells (data not shown). To determine if inhibiting IGF-IR/IRS-1 interaction by incubating the blocking peptide with the IRS-FF mutant cells could rescue the SHPS-1 phosphorylation and SHP-2 recruitment to SHPS-1, cells expressing the IRS-1-FF mutant were exposed to peptide 301. The addition of peptide 301 resulted in increased IGF-IR association with SHPS-1 in response to IGF-I compared with cells not exposed to the peptide (2.2 ± 0.1-fold, n = 5, p < 0.05; Fig. 5D). Similarly, there was an increase in SHPS-1 phosphorylation (2.6 ± 0.1-fold, n = 3, p < 0.001) and the subsequent SHPS-1/SHP-2 association (2.3 ± 0.3-fold, n = 3, p < 0.05; Fig. 5D). More importantly, tyrosine phosphorylation of p52shc was significantly increased in the IRS-FF cells exposed to peptide 301 (2.9 ± 0.2-fold, n = 3, p < 0.001) compared with cells not exposed to the peptide in response to IGF-I (Fig. 5D). Taken together, these results suggest that disruption of association IGF-IR/IRS-1 association and inhibition of SHP-2 binding to IRS-1 appears to be required for optimal SHP-2 transfer to SHPS-1 and Shc activation.
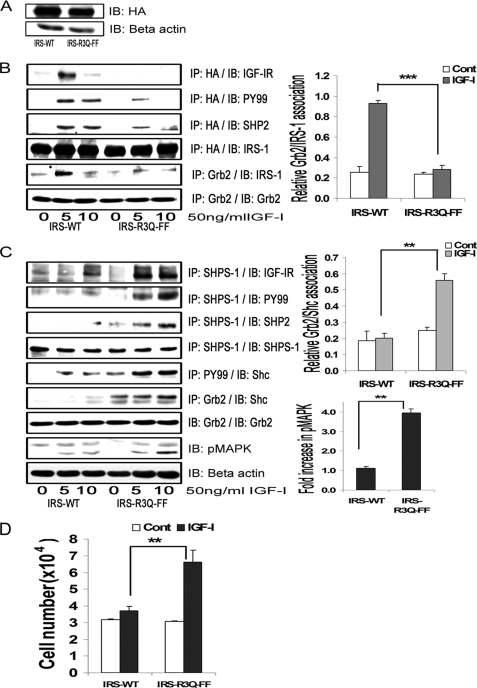
Expression of an IRS-1 Double Mutant That Impairs IGF-IR/IRS-1 Association and IRS-1/SHP-2 Association Restores SHPS-1 Phosphorylation and SHP-2 Association with SHPS-1
To definitively prove that IRS-1 binding to both IGF-IR and SHP-2 accounted for inhibition of SHPS-1 phosphorylation and downstream signaling, we used the IRS-FF mutant as a template and mutated the arginine residues 212, 213, and 217 in the IRS-1 PTB domain to glutamine because these residues have been shown to mediate IRS-1 association with IR/IGF-IR (7, 10). The combined mutant is termed IRS-R3Q-FF. The expressions of HA-tagged IRS-WT and IRS-R3Q-FF were found to be similar when the cell lysates were compared (Fig. 6A). As shown in Fig. 6B, this IRS-1 mutant had significantly impaired binding to IGF-IR in response to IGF-I compared with cells expressing IRS-WT. Further, the tyrosine phosphorylation of IRS-1 and the subsequent IRS-1 binding to Grb2 was found to be significantly reduced in IRS-R3Q-FF compared with IRS-WT cells in response to IGF-I (3.3 ± 0.1-fold, n = 3, p < 0.001). The amount of SHP-2 bound to IRS-1 was found to be reduced (1.9 ± 0.2-fold, n = 3, p < 0.05) but not completely abolished in the double mutant compared with IRS-WT cells (Fig. 6B). Furthermore, following IGF-I stimulation, there was a significant increase in SHPS-1 binding to IGF-IR (2.9 ± 0.4-fold, n = 3, p < 0.01) and an increase in SHPS-1 phosphorylation (3.3 ± 0.1-fold, n = 3, p < 0.001) in the cells expressing the IRS-R3Q-FF mutant cells compared with IRS-WT cells (Fig. 6C). The subsequent transfer of SHP-2 to SHPS-1 was also increased in IRS-R3Q-FF mutant cells compared with IRS-WT cells in response to IGF-I (2.1 ± 0.2-fold, n = 3, p < 0.01; Fig. 6C). When the IRS-R3Q-FF cells were analyzed, p52shc phosphorylation was found to be increased in response to IGF-I compared with IRS-WT cells (3.0 ± 0.2-fold, n = 3, p < 0.001). Cells expressing IRS-R3Q-FF cells showed increased phosphorylation of MAPK (3.3 ± 0.2-fold, n = 3, p < 0.01) compared with cells overexpressing IRS-WT in response to IGF-I (Fig. 6C). Using a lower degree of cellular confluence and increased IGF-I concentration, we observed a decrease in basal Shc phosphorylation response in the mutant cells. Further, in response to IGF-I, the phosphorylation of MAPK increased significantly (3.9 ± 0.1, n = 3, p < 0.001) in the IRS-R3Q-FF cells (supplemental Fig. 2). The proliferation response of cells expressing IRS-R3Q-FF to IGF-I was found to be significantly greater (2.1 ± 0.3-fold, n = 4, p < 0.01) compared with cells expressing IRS-WT (Fig. 6D). These results demonstrate that inhibiting IRS-1 binding to IGF-IR and to SHP-2 attenuated IRS-1 tyrosine phosphorylation and Grb2 binding. Furthermore, these changes resulted in increased SHPS-1 association with IGF-IR kinase, SHPS-1 phosphorylation, SHP-2 transfer, phosphorylation of Shc and MAPK, and cellular proliferation in response to IGF-I.
FIGURE 6.
Inhibition of IRS-1 binding to both IGF-IR and SHP-2 restores SHPS-1 phosphorylation and SHPS-1 complex assembly and enhances the cellular proliferation response to IGF-I. A, VSMCs expressing IRS-WT and the IRS-R3Q-FF mutant were serum-starved overnight and analyzed for recombinant protein expression. Cell lysates were immunoblotted (IB) for HA (top) or β-actin (bottom). B, confluent cultures of IRS-WT and IRS-R3Q-FF cells were serum-starved and then exposed to IGF-I for the indicated time periods. Cell lysates were immunoprecipitated (IP) with anti-HA and immunoblotted for IGF-IR (top panel), Tyr(P)99 (PY99; second panel), SHP-2 (third panel), or total HA (fourth panel). Cell lysates from similar experiments were immunoprecipitated with Grb2 and immunoblotted for IRS-1 (fifth panel) or total Grb2 (sixth panel). The bar graphs are representative of at least three independent experiments. Error bars, mean ± S.E. ***, p < 0.001 when the amount of Grb2 associated with IRS-1 at 5 min in response to IGF-I is compared between IRS-WT and IRS-R3Q-FF cells. C, the cell lysates from the above experiments and similar experiments were immunoprecipitated with anti-SHPS-1 antibody and immunoblotted for IGF-IR (top panel), phosphotyrosine (second panel), SHP-2 (third panel), or total SHPS-1 (fourth panel). Similarly, lysates were immunoprecipitated with Tyr(P)99 antibody and immunoblotted for p52shc (fifth panel). Similar lysates were immunoprecipitated with anti-Grb2 and immunoblotted for Shc (sixth panel) or Grb2 (seventh panel). Ten micrograms of protein from the same cell lysates was immunoblotted directly for phospho-MAPK (eighth panel) or β-actin (bottom panel). The bar graphs are representative of at least three independent experiments. Error bars, mean ± S.E. **, p < 0.01 when the amount of Grb2 associated with Shc (middle graph) or amount of phosphorylation of MAPK (bottom graph) at 10 min in response to IGF-I is compared between IRS-WT and IRS-R3Q-FF cells. D, IRS-WT and IRS-R3Q-FF cells were plated (3 × 104 cells) in DMEM-HG with 2% FBS prior to exposure to IGF-I in DMEM-HG with 0.2% platelet-poor plasma. Forty-eight hours after the addition of IGF-I, cell number was determined. **, p < 0.01 when the change in number of cells proliferating in response to IGF-I is compared between IRS-WT and IRS-R3Q-FF cells. Error bars, mean ± S.E.
Effect of IRS-1 Silencing on SHPS-1 Phosphorylation
To further evaluate the role of IRS-1 in SHPS-1 phosphorylation, we silenced IRS-1 using short hairpin RNA. VSMCs expressing IRS-1 short hairpin RNA showed a 89 ± 1% decrease in IRS-1 (supplemental Fig. 3A) compared with empty vector control. The basal SHPS-1 phosphorylation was significantly increased (4.1 ± 0.2-fold, n = 3, p < 0.01) in IRS-1 Si cells compared with EVC cells, and IGF-I induced an additional SHPS-1 phosphorylation (1.9 ± 0.2-fold, n = 3, p < 0.05) after 5 min (supplemental Fig. 3B). This basal SHPS-1 phosphorylation was due to a basal increase in IGF-IR/SHPS-1 association (2.3 ± 0.2-fold) and an additional (2.4 ± 0.1-fold) increase in response to IGF-I in IRS-1 Si cells compared with EVC. Furthermore, a 1.9 ± 0.1-fold increase in basal MAPK phosphorylation and an additional 2.1 ± 0.2-fold increase in response to IGF-I was observed in IRS-1 Si cells compared with EVC. In addition, cellular proliferation was basally increased in IRS-1 Si cells compared with EVC (1.4 ± 0.2-fold), and it increased an additional 1.7 ± 0.3-fold in response to IGF-I (n = 3, p < 0.01; supplemental Fig. 3C). In addition to increased IGF-IR/SHPS-1 association, the increase in basal SHPS-1 phosphorylation was probably due to an increase in basal IGF-IR phosphorylation (3.9 ± 0.2-fold, n = 2) in the IRS-1 Si cells (supplemental Fig. 3B), which would allow more activated IGF-IR kinase to bind and phosphorylate SHPS-1.
DISCUSSION
IRS-1 is a major substrate for IGF-IR (4, 9) and has been shown to mediate several signaling events, including activation of both the phosphatidylinositol 3-kinase and MAPK signaling pathways (4, 9, 39, 40). Previously, we showed that IGF-I-stimulated IRS-1 phosphorylation and subsequent Grb2 binding were markedly down-regulated in VSMCs that had been maintained in high glucose (e.g. 25 mm) (29). The results from the current study confirm the previous observation that VSMCs in high glucose had a significant reduction in total IRS-1 protein levels at 12, 24, and 48 h compared with cells exposed to normal glucose. Exposure to high glucose has been shown to decrease IRS-1 levels in a variety of cell types (32, 41). IRS-1 degradation has been extensively studied in insulin-sensitive cell types, such as adipocytes and skeletal muscle cells and also in these tissues in animal models (13, 31, 42–44). Hyperglycemia-induced IRS-1 down-regulation has been shown in vascular smooth muscle following PKC activation (45), β-adrenergic stimulation (46), and angiotensin II exposure (47, 48). Exposure to growth factors, including IGF-I, also enhances IRS-1 degradation in a variety of cells (25, 26, 49, 50). In the present study, exposure to high glucose and IGF-I for 24 or 48 h reduced the IRS-1 levels significantly compared with cells maintained in high glucose alone. This observation is supported by the previous studies that showed that the combination of high glucose and insulin reduced IRS-1 levels in several model systems and in humans with type 2 diabetes and obesity (30, 31, 51, 52). Furthermore, in the current study, IRS-1 tyrosine phosphorylation and Grb2 binding were impaired in response to IGF-I, suggesting that exposure to high glucose with subsequent IRS-1 down-regulation attenuates IRS-1-mediated signaling.
In an attempt to determine the consequences of the failure to down-regulate IRS-1, we transduced VSMCs with an IRS-1 cDNA and maintained them in 25 mm glucose. Maintenance of this increased IRS-1 level led to impaired IGF-I stimulated SHPS-1 phosphorylation, but it had no effect on IGF-I-stimulated IGF-I receptor phosphorylation. That this effect is direct is supported by our observation that IRS-1 inhibited IGF-IR-mediated SHPS-1 phosphorylation in vitro in a concentration-dependent manner. In VSMCs exposed to high glucose, IGF-I induces SHPS-1 phosphorylation (53). SHPS-1 has been demonstrated to be a substrate of various growth factor receptor tyrosine kinases, including the insulin and IGF-I receptor (19, 37, 54–58). The role of IRS-1 in inhibiting SHPS-1 phosphorylation in intact SMCs was confirmed by using three independent methods. First, in SMCs maintained in normal glucose, SHPS-1 phosphorylation was suppressed, and the addition of a synthetic peptide that inhibited IGF-IR/IRS-1 association enhanced SHPS-1 phosphorylation. Second, the addition of two synthetic peptides that disrupted IRS-1/IGF-IR association to cells maintained in hyperglycemia that were overexpressing IRS-1 led to enhanced SHPS-1 phosphorylation. Third, expression of a mutant form of IRS-1 that could not bind to IGF-IR at levels that were comparable with WT IRS-1 expression also led to enhanced IGF-I-stimulated SHPS-1 phosphorylation. These findings support the conclusion that IRS-1 functions by binding directly to the IGF-I receptor and suppressing IGF-I receptor-mediated SHPS-1 phosphorylation.
Crystal structure and mutational analysis of the IRS-1 PTB domain showed that arginines 212, 213, and 217 are responsible for binding to the NPXY motif in the receptor, and substitution with glutamine resulted in a complete loss of binding (7, 10). Our findings confirm the importance of these residues because the mutation of the three arginine residues in the PTB domain of the IRS-1 significantly impaired IRS-1/IGF-IR association and IRS-1 tyrosine phosphorylation following IGF-I stimulation. That IRS-1/IGF-IR association was not completely inhibited can be explained by the fact that IRS-1 also binds to the major autophosphorylation sites (Tyr1131, Tyr1135, and Tyr1136) of the IGF-IR (9). In addition, inhibition of IGF-IR/IRS-1 by both phosphorylated and non-phosphorylated forms of peptide 299 suggests that both peptides compete with intact IGF-IR for binding to IRS-1 and supports the conclusion that IRS-1/IGF-IR association is occurring through this interaction.
Following phosphorylation of YXX(L/I/V) motifs within the SHPS-1 cytoplasmic domain, SHP-2 is recruited to SHPS-1, and this is essential for optimal phosphorylation of Shc and MAPK activation in response to IGF-I (2). Our study shows that in addition to inhibiting IGF-IR-mediated SHPS-1 phosphorylation, IRS-1 also sequesters SHP-2, limiting its translocation to SHPS-1 following SHPS-1 phosphorylation. IRS-1 also contains seven YXX(L/I/V) motifs that could bind to the SHP-2 Src homology 2 domains, and substitution of IRS-1 Tyr1172 and Tyr1222 to phenylalanine has been shown to reduce SHP-2/IRS-1 association (59, 60). In this study, we used several approaches to show that IRS-1 functions in part by sequestering SHP-2 and preventing its recruitment to SHPS-1. First, the cells expressing the IRS-1 Y1179F/Y1229F substitution mutant had decreased IRS-1/SHP-2 binding in response to IGF-I. Additionally, exposure of IRS1-FF mutant cells to a peptide that inhibited IRS-1 association with IGF-IR increased SHPS-1 phosphorylation and restored the subsequent SHP-2 transfer. Second, expression of a combined mutant (IRS-R3Q-FF) in which both IRS-1/IGF-IR and IRS-1/SHP-2 binding were decreased led to increases in SHPS-1 phosphorylation and subsequent SHP-2 transfer to SHPS-1. Previous studies have shown that IRS-1/SHP-2 association is differentially regulated in different tissues (61), in response to changes in glucose (62) and state of SMC differentiation (63). These findings suggest that this could be an important mechanism for regulating cellular responsiveness to IGF-I.
In VSMCs, Shc phosphorylation in response to IGF-I depends on SHPS-1 phosphorylation and subsequent SHPS-1 complex assembly (2, 29). Although some studies have shown that IRS-1 and Shc are competitive substrates for receptor tyrosine kinase (8, 9, 64), our results show that competition occurs between SHPS-1 and IRS-1, and this leads to impaired SHPS-1 phosphorylation. Further, IGF-IR overexpression increased SHPS-1 phosphorylation in response to IGF-I. Therefore, attenuation of Shc phosphorylation is not due to direct Shc/IRS-1 competition for access to IGF-IR. Shc has been shown to co-immunoprecipitate with several activated tyrosine kinase receptors, such as epidermal growth factor receptors, erbB-2/neu, and Trk (21, 65–67); however, it does not co-immunoprecipitate with activated IR (68, 69). Therefore, the activation of Shc in VSMCs in response to IGF-I is regulated by SHP-2·Src transfer to SHPS-1 and Src activation (2, 20).
These observations raise the question of the role of IRS-1 in VSMCs maintained in normoglycemic conditions. Our prior studies have shown that when IRS-1 levels are substantially increased (e.g. VSMCs maintained in 5 mm glucose), SHPS-1 phosphorylation, SHP-2 transfer, and Shc/MAPK activation are suppressed (29, 70). Consequently, although the protein synthesis response to IGF-I is maintained, the cell migration and proliferation responses are significantly decreased. Our finding that inhibition of IGF-IR/IRS-1 association in VSMCs maintained in 5 mm glucose resulted in significant enhancement of SHPS-1 phosphorylation suggests that this mechanism is operative in VSMCs exposed to normal glucose. This observation could have major physiologic significance for maintenance of normal vascular function. In normal mice, VSMCs are maintained in the quiescent state (71, 72) and have a minimal proliferative response to IGF-I administration. In contrast, the presence of diabetes cell proliferation is increased substantially (70). Because differentiated VSMCs in blood vessels are maintained in a non-proliferative state, expression of IRS-1 may be one of the important determinants that allow normal protein synthesis and hypertrophic responses to IGF-I and insulin while functioning simultaneously to inhibit cell replication. In contrast, transition to the dedifferentiated phenotype could possibly require a major decrease in IRS-1 in order to permit MAPK pathway activation (17, 73). This conclusion is supported by the results of this study, wherein IRS-1 silencing increased basal IGF-IR phosphorylation, which in turn, in the absence of IRS-1 binding, had increased access to SHPS-1, thereby augmenting basal SHPS-1 phosphorylation. Future studies should investigate the role of IRS-1 in animal models and determine if IRS-1 is regulating the mitogenic response to IGF-I in vascular tissue in vivo.
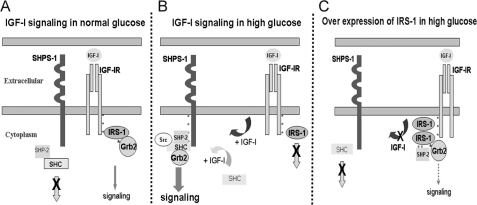
In summary, exposure of VSMCs to high glucose and IGF-I results in down-regulation of IRS-1, thus allowing IGF-I-stimulated SHPS-1 phosphorylation and SHPS-1 complex assembly. The mechanism by which IRS-1 functions is due to its ability to bind to IGF-IR and inhibit IGF-IR mediated SHPS-1 tyrosine phosphorylation as well as sequestration of SHP-2, which inhibits the assembly of the signaling complex on SHPS-1. These changes result in inhibition of IGF-I-stimulated increases in MAPK activation and cell proliferation (Fig. 7). Taken together with the findings from our previous studies, these results have significant implications for understanding the role of IGF-I in stimulating the proliferation of neointimal VSMCs that have been exposed to hyperglycemic stress.
FIGURE 7.
Proposed role of IRS-1 in regulating SHPS-1 phosphorylation under hyperglycemic conditions. A, in VSMCs under normal glucose conditions, in response to IGF-I, competitive binding of IRS-1 to IGF-IR impairs the access of SHPS-1 to IGF-I receptor kinase, thereby decreasing SHPS-1 phosphorylation. Further, IRS-1 sequesters SHP-2, leading to impaired assembly of the signaling complex on SHPS-1. B, in high glucose conditions, reduction in IRS-1 levels allows SHPS-1 improved access to the IGF-I receptor kinase, leading to enhanced phosphorylation of SHPS-1 and subsequent SHP-2 transfer. This facilitates formation of the SHP-2·Src·Shc·Grb-2 signaling complex, which augments phosphatidylinositol 3-kinase and MAPK pathway activation. C, overexpression of IRS-1 decreases SHPS-1 access to the IGF-IR kinase, SHPS-1 phosphorylation, and subsequent complex assembly. The IRS-1 double mutant with impaired binding to IGF-IR and SHP-2 allows increased SHPS-1 access to IGF-IR kinase, thus increasing SHPS-1 phosphorylation and SHP-2 transfer, thereby leading to increased phosphorylation of MAPK and cellular proliferation in response to IGF-I.
Supplementary Material
Acknowledgments
We thank Dr. Lee Graves for suggestions and Laura Lindsey for assistance in preparing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL56580 and AG02331.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- IGF-I
- insulin-like growth factor-I
- IGF-IR
- insulin-like growth factor-I receptor
- VSMC
- vascular smooth muscle cell
- SMC
- smooth muscle cell
- MAPK
- mitogen-activated protein kinase
- SHPS-1/CD
- SHPS-1 cytoplasmic domain
- HA
- hemagglutinin
- PTB
- phosphotyrosine binding
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- WT
- wild type
- HG
- high glucose
- NG
- normal glucose
- EVC
- empty vector control
- Si
- silencing
- RNAi
- RNA interference
- FF
- Y1179F/Y1229F
- R3Q
- R212Q/R213Q/R227Q.
REFERENCES
- 1.Imai Y., Clemmons D. R. (1999) Endocrinology 140, 4228–4235 [DOI] [PubMed] [Google Scholar]
- 2.Ling Y., Maile L. A., Lieskovska J., Badley-Clarke J., Clemmons D. R. (2005) Mol. Biol. Cell 16, 3353–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan Y., Maile L. A., Ling Y., Graves L. M., Clemmons D. R. (2008) J. Biol. Chem. 283, 16320–16331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers M. G., Jr., Sun X. J., Cheatham B., Jachna B. R., Glasheen E. M., Backer J. M., White M. F. (1993) Endocrinology 132, 1421–1430 [DOI] [PubMed] [Google Scholar]
- 5.Sun X. J., Rothenberg P., Kahn C. R., Backer J. M., Araki E., Wilden P. A., Cahill D. A., Goldstein B. J., White M. F. (1991) Nature 352, 73–77 [DOI] [PubMed] [Google Scholar]
- 6.White M. F., Kahn C. R. (1994) J. Biol. Chem. 269, 1–4 [PubMed] [Google Scholar]
- 7.Eck M. J., Dhe-Paganon S., Trüb T., Nolte R. T., Shoelson S. E. (1996) Cell 85, 695–705 [DOI] [PubMed] [Google Scholar]
- 8.Wolf G., Trüb T., Ottinger E., Groninga L., Lynch A., White M. F., Miyazaki M., Lee J., Shoelson S. E. (1995) J. Biol. Chem. 270, 27407–27410 [DOI] [PubMed] [Google Scholar]
- 9.Tartare-Deckert S., Sawka-Verhelle D., Murdaca J., Van Obberghen E. (1995) J. Biol. Chem. 270, 23456–23460 [DOI] [PubMed] [Google Scholar]
- 10.Zhou M. M., Huang B., Olejniczak E. T., Meadows R. P., Shuker S. B., Miyazaki M., Trüb T., Shoelson S. E., Fesik S. W. (1996) Nat. Struct. Biol. 3, 388–393 [DOI] [PubMed] [Google Scholar]
- 11.Vainshtein I., Kovacina K. S., Roth R. A. (2001) J. Biol. Chem. 276, 8073–8078 [DOI] [PubMed] [Google Scholar]
- 12.Ogawa W., Matozaki T., Kasuga M. (1998) Mol. Cell Biochem. 182, 13–22 [PubMed] [Google Scholar]
- 13.Gual P., Le Marchand-Brustel Y., Tanti J. F. (2005) Biochimie 87, 99–109 [DOI] [PubMed] [Google Scholar]
- 14.Baserga R. (2007) Cell Cycle 6, 814–816 [DOI] [PubMed] [Google Scholar]
- 15.Wang L. M., Myers M. G., Jr., Sun X. J., Aaronson S. A., White M., Pierce J. H. (1993) Science 261, 1591–1594 [DOI] [PubMed] [Google Scholar]
- 16.Sadagurski M., Nofech-Mozes S., Weingarten G., White M. F., Kadowaki T., Wertheimer E. (2007) J. Cell Physiol. 213, 519–527 [DOI] [PubMed] [Google Scholar]
- 17.Martin K. A., Merenick B. L., Ding M., Fetalvero K. M., Rzucidlo E. M., Kozul C. D., Brown D. J., Chiu H. Y., Shyu M., Drapeau B. L., Wagner R. J., Powell R. J. (2007) J. Biol. Chem. 282, 36112–36120 [DOI] [PubMed] [Google Scholar]
- 18.Maile L. A., Clemmons D. R. (2002) J. Biol. Chem. 277, 8955–8960 [DOI] [PubMed] [Google Scholar]
- 19.Kharitonenkov A., Chen Z., Sures I., Wang H., Schilling J., Ullrich A. (1997) Nature 386, 181–186 [DOI] [PubMed] [Google Scholar]
- 20.Lieskovska J., Ling Y., Badley-Clarke J., Clemmons D. R. (2006) J. Biol. Chem. 281, 25041–25053 [DOI] [PubMed] [Google Scholar]
- 21.Rozakis-Adcock M., McGlade J., Mbamalu G., Pelicci G., Daly R., Li W., Batzer A., Thomas S., Brugge J., Pelicci P. G. (1992) Nature 360, 689–692 [DOI] [PubMed] [Google Scholar]
- 22.Valverde A. M., Mur C., Pons S., Alvarez A. M., White M. F., Kahn C. R., Benito M. (2001) Mol. Cell Biol. 21, 2269–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guvakova M. A., Surmacz E. (1997) Cancer Res. 57, 2606–2610 [PubMed] [Google Scholar]
- 24.Saad M. J., Araki E., Miralpeix M., Rothenberg P. L., White M. F., Kahn C. R. (1992) J. Clin. Invest. 90, 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rice K. M., Garner C. W. (1999) Biochem. Biophys. Res. Commun. 255, 614–617 [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Hoff H., Sell C. (2000) J. Biol. Chem. 275, 22558–22562 [DOI] [PubMed] [Google Scholar]
- 27.Nakajima K., Yamauchi K., Shigematsu S., Ikeo S., Komatsu M., Aizawa T., Hashizume K. (2000) J. Biol. Chem. 275, 20880–20886 [DOI] [PubMed] [Google Scholar]
- 28.Burén J., Liu H. X., Lauritz J., Eriksson J. W. (2003) Eur. J. Endocrinol. 148, 157–167 [DOI] [PubMed] [Google Scholar]
- 29.Maile L. A., Capps B. E., Ling Y., Xi G., Clemmons D. R. (2007) Endocrinology 148, 2435–2443 [DOI] [PubMed] [Google Scholar]
- 30.Renström F., Burén J., Svensson M., Eriksson J. W. (2007) Metabolism 56, 190–198 [DOI] [PubMed] [Google Scholar]
- 31.Carvalho E., Rondinone C., Smith U. (2000) Mol. Cell Biochem. 206, 7–16 [DOI] [PubMed] [Google Scholar]
- 32.Renström F., Burén J., Eriksson J. W. (2005) Endocrinology 146, 3044–3051 [DOI] [PubMed] [Google Scholar]
- 33.Shen X., Xi G., Radhakrishnan Y., Clemmons D. R. (2009) Mol. Cell Proteomics 8, 1539–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blakesley V. A., Kato H., Roberts C. T., Jr., LeRoith D. (1995) J. Biol. Chem. 270, 2764–2769 [DOI] [PubMed] [Google Scholar]
- 35.Nam T. J., Busby W. H., Jr., Rees C., Clemmons D. R. (2000) Endocrinology 141, 1100–1106 [DOI] [PubMed] [Google Scholar]
- 36.Jones J. I., Prevette T., Gockerman A., Clemmons D. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2482–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujioka Y., Matozaki T., Noguchi T., Iwamatsu A., Yamao T., Takahashi N., Tsuda M., Takada T., Kasuga M. (1996) Mol. Cell Biol. 16, 6887–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanke S., Mann M. (2009) Mol. Cell Proteomics 8, 519–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giorgetti S., Ballotti R., Kowalski-Chauvel A., Tartare S., Van Obberghen E. (1993) J. Biol. Chem. 268, 7358–7364 [PubMed] [Google Scholar]
- 40.Peruzzi F., Prisco M., Dews M., Salomoni P., Grassilli E., Romano G., Calabretta B., Baserga R. (1999) Mol. Cell Biol. 19, 7203–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim B., Feldman E. L. (2009) Apoptosis 14, 665–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein J., Fasshauer M., Ito M., Lowell B. B., Benito M., Kahn C. R. (1999) J. Biol. Chem. 274, 34795–34802 [DOI] [PubMed] [Google Scholar]
- 43.Catalano P. M., Nizielski S. E., Shao J., Preston L., Qiao L., Friedman J. E. (2002) Am. J. Physiol. Endocrinol. Metab. 282, E522–E533 [DOI] [PubMed] [Google Scholar]
- 44.Brozinick J. T., Jr., Roberts B. R., Dohm G. L. (2003) Diabetes 52, 935–941 [DOI] [PubMed] [Google Scholar]
- 45.Motley E. D., Kabir S. M., Eguchi K., Hicks A. L., Gardner C. D., Reynolds C. M., Frank G. D., Eguchi S. (2001) Cell Mol. Biol. 47, 1059–1062 [PubMed] [Google Scholar]
- 46.Kawabe J., Aizawa Y., Takehara N., Hasebe N., Kikuchi K. (2000) J. Hypertens. 18, 1457–1464 [DOI] [PubMed] [Google Scholar]
- 47.Folli F., Kahn C. R., Hansen H., Bouchie J. L., Feener E. P. (1997) J. Clin. Invest. 100, 2158–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taniyama Y., Hitomi H., Shah A., Alexander R. W., Griendling K. K. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 1142–1147 [DOI] [PubMed] [Google Scholar]
- 49.Potashnik R., Bloch-Damti A., Bashan N., Rudich A. (2003) Diabetologia 46, 639–648 [DOI] [PubMed] [Google Scholar]
- 50.Zhande R., Mitchell J. J., Wu J., Sun X. J. (2002) Mol. Cell Biol. 22, 1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carvalho E., Jansson P. A., Axelsen M., Eriksson J. W., Huang X., Groop L., Rondinone C., Sjöström L., Smith U. (1999) FASEB J. 13, 2173–2178 [DOI] [PubMed] [Google Scholar]
- 52.Goodyear L. J., Giorgino F., Sherman L. A., Carey J., Smith R. J., Dohm G. L. (1995) J. Clin. Invest. 95, 2195–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maile L. A., Capps B. E., Miller E. C., Allen L. B., Veluvolu U., Aday A. W., Clemmons D. R. (2008) Mol. Endocrinol. 22, 1226–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stofega M. R., Wang H., Ullrich A., Carter-Su C. (1998) J. Biol. Chem. 273, 7112–7117 [DOI] [PubMed] [Google Scholar]
- 55.Tsuda M., Matozaki T., Fukunaga K., Fujioka Y., Imamoto A., Noguchi T., Takada T., Yamao T., Takeda H., Ochi F., Yamamoto T., Kasuga M. (1998) J. Biol. Chem. 273, 13223–13229 [DOI] [PubMed] [Google Scholar]
- 56.Ochi F., Matozaki T., Noguchi T., Fujioka Y., Yamao T., Takada T., Tsuda M., Takeda H., Fukunaga K., Okabayashi Y., Kasuga M. (1997) Biochem. Biophys. Res. Commun. 239, 483–487 [DOI] [PubMed] [Google Scholar]
- 57.Maile L. A., Badley-Clarke J., Clemmons D. R. (2003) Mol. Biol. Cell 14, 3519–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitsuhashi H., Futai E., Sasagawa N., Hayashi Y., Nishino I., Ishiura S. (2008) J. Neurochem. 105, 101–112 [DOI] [PubMed] [Google Scholar]
- 59.Noguchi T., Matozaki T., Horita K., Fujioka Y., Kasuga M. (1994) Mol. Cell Biol. 14, 6674–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myers M. G., Jr., Mendez R., Shi P., Pierce J. H., Rhoads R., White M. F. (1998) J. Biol. Chem. 273, 26908–26914 [DOI] [PubMed] [Google Scholar]
- 61.Lima M. H., Ueno M., Thirone A. C., Rocha E. M., Carvalho C. R., Saad M. J. (2002) Endocrine 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 62.Bifulco G., Di Carlo C., Caruso M., Oriente F., Di Spiezio Sardo A., Formisano P., Beguinot F., Nappi C. (2002) J. Biol. Chem. 277, 24306–24314 [DOI] [PubMed] [Google Scholar]
- 63.Hayashi K., Shibata K., Morita T., Iwasaki K., Watanabe M., Sobue K. (2004) J. Biol. Chem. 279, 40807–40818 [DOI] [PubMed] [Google Scholar]
- 64.Gustafson T. A., He W., Craparo A., Schaub C. D., O'Neill T. J. (1995) Mol. Cell Biol. 15, 2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pelicci G., Lanfrancone L., Grignani F., McGlade J., Cavallo F., Forni G., Nicoletti I., Grignani F., Pawson T., Pelicci P. G. (1992) Cell 70, 93–104 [DOI] [PubMed] [Google Scholar]
- 66.Obermeier A., Bradshaw R. A., Seedorf K., Choidas A., Schlessinger J., Ullrich A. (1994) EMBO J. 13, 1585–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segatto O., Pelicci G., Giuli S., Digiesi G., Di Fiore P. P., McGlade J., Pawson T., Pelicci P. G. (1993) Oncogene 8, 2105–2112 [PubMed] [Google Scholar]
- 68.Pronk G. J., McGlade J., Pelicci G., Pawson T., Bos J. L. (1993) J. Biol. Chem. 268, 5748–5753 [PubMed] [Google Scholar]
- 69.Yonezawa K., Ando A., Kaburagi Y., Yamamoto-Honda R., Kitamura T., Hara K., Nakafuku M., Okabayashi Y., Kadowaki T., Kaziro Y. (1994) J. Biol. Chem. 269, 4634–4640 [PubMed] [Google Scholar]
- 70.Maile L. A., Capps B. E., Miller E. C., Aday A. W., Clemmons D. R. (2008) Diabetes 57, 2637–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chu A., Ordonez E. T., Hellerstein M. K. (2006) Am. J. Physiol. Cell Physiol. 291, C101–C1021 [DOI] [PubMed] [Google Scholar]
- 72.Neese R. A., Misell L. M., Turner S., Chu A., Kim J., Cesar D., Hoh R., Antelo F., Strawford A., McCune J. M., Christiansen M., Hellerstein M. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15345–15350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rzucidlo E. M., Martin K. A., Powell R. J. (2007) J. Vasc. Surg. 45, Suppl. A, A25–A32 [DOI] [PubMed] [Google Scholar]
- 74.Ross R. (1971) J. Cell Biol. 50, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.