Abstract
This work addresses the biogenesis of heme-copper terminal oxidases in Bradyrhizobium japonicum, the nitrogen-fixing root nodule symbiont of soybean. B. japonicum has four quinol oxidases and four cytochrome oxidases. The latter include the aa3- and cbb3-type oxidases. Although both have a CuB center in subunit I, the subunit II proteins differ in having either a CuA center (in aa3) or a covalently bound heme c (in cbb3). Two biogenesis factors were genetically studied here, the periplasmically exposed CoxG and ScoI proteins, which are the respective homologs of the mitochondrial copper-trafficking chaperones Cox11 and Sco1 for the formation of the CuB center in subunit I and the CuA center in subunit II of cytochrome aa3. We could demonstrate copper binding to ScoI in vitro, a process for which the thiols of cysteine residues 74 and 78 in the ScoI polypeptide were shown to be essential. Knock-out mutations in the B. japonicum coxG and scoI genes led to loss of cytochrome aa3 assembly and activity in the cytoplasmic membrane, whereas the cbb3-type cytochrome oxidase apparently remained unaffected. This suggests that subunit I of the cbb3-type oxidase obtains its copper cofactor via a different pathway than cytochrome aa3. In contrast to the coxG mutation, the scoI mutation caused a decreased symbiotic nitrogen fixation activity. We hypothesize that a periplasmic B. japonicum protein other than any of the identified CuA proteins depends on ScoI and is required for an effective symbiosis.
Keywords: Bioenergetics, Chaperone chaperonin, Copper, Cytochrome Oxidase, Membrane Proteins, Nitrogen Fixation, Periplasm, Symbiosis
Introduction
The common denominator in respiratory heme-copper oxidases is a membrane-integral subunit I that carries as cofactors a low spin heme and a unique high spin heme-copper binuclear center (CuB site) where reduction of O2 to H2O takes place (1–4).
There is diversity regarding the number and cofactor content of the other subunits, which relates to the substrates used as electron donor (5, 6). Reduced c-type cytochromes donate electrons to cytochrome oxidases, whereas reduced quinones deliver electrons to quinol oxidases. The latter possess a cofactor-free subunit II, whereas cytochrome oxidases have cofactors bound to subunit II. In most cases, this is a binuclear Cu-Cu center (CuA site) that is liganded by six highly conserved amino acids (3, 4). The subclass of cbb3-type oxidases is exceptional because its members have a subunit II (CcoO or FixO) that is a monoheme c-type cytochrome instead of the CuA-containing protein (3, 7). Subunit III in cbb3-type oxidases, which is a diheme cytochrome c, is thought to relay the electrons from the cytochrome bc1 complex via CcoO to the redox centers of subunit I (8, 9). Subunit III of all other heme-copper oxidases is cofactor-free, just like the non-conserved small subunit IV (1–4). With few exceptions, the four-subunit composition is typical for prokaryotic heme-copper oxidases, whereas the eukaryotic (i.e. mitochondrial) counterparts are much more complex (10).
Knowledge about subunit and cofactor composition is pivotal for an understanding of the biogenesis of heme-copper cytochrome oxidases. How do subunits assemble in the membrane, and how are the cofactors (hemes, CuA, CuB) inserted? Furthermore, the topology of subunits and redox centers has to be considered in the context of cofactor delivery, which is routed either from the cytoplasm or from the outer side of the membrane. Whereas the low spin heme and high spin heme-CuB are embedded in the membrane-integral segment of subunit I, the CuA center on subunit II lies peripheral to the membrane (4, 10), which is equivalent to the intermembrane compartment in mitochondria and the periplasmic space in Gram-negative bacteria.
Given the complexity of eukaryotic cytochrome oxidase, with possibly more than 30 factors involved in its formation (11, 12), the use of the comparatively simpler bacterial aa3-type oxidases facilitates biogenesis studies. Members of the α- proteobacteria (e.g. Paracoccus denitrificans, Bradyrhizobium japonicum, and Rhodobacter sp.) are attractive model organisms for this purpose because they appear to be the closest extant relatives of a mitochondrial ancestor (2). In fact, a fairly small number of chaperoning proteins have so far been identified as being instrumental in the maturation of bacterial aa3-type cytochrome oxidases: Surf1 for heme A insertion into subunit I (13, 14); CtaG (or CoxG), a homolog of mitochondrial Cox11 (15), for copper trafficking to the CuB site in subunit I (16–18); Sco1 (also called SenC or PrrC), for copper delivery to the CuA site in subunit II (19–23); and TlpA, a protein dithiol:disulfide oxidoreductase with an unknown role in cytochrome aa3 formation in B. japonicum (24, 25). A specialized case appears to be that of PCuAC, which is involved in generating the CuA site of the Thermus thermophilus ba3-type oxidase (26). All of these proteins are membrane-bound, and the active domains of Sco1 and PCuAC face the periplasm, which is consistent with their role in CuA assembly on subunit II. Concerning assembly, the cbb3-type oxidase is again in a class of its own. Because subunits II and III are c-type cytochromes, their synthesis requires the complete set of maturation proteins for the covalent attachment of heme (27, 28). Additional assembly factors are needed whose biochemical functions have not been elucidated (29, 30). Although the cbb3- and aa3-type cytochrome oxidases have in common a conserved subunit I, it was not clear whether they depend on similar assembly factors for that subunit. Part of the work reported here addresses this issue.
B. japonicum, a facultatively symbiotic, nitrogen-fixing bacterium investigated in our laboratory, has eight terminal oxidases, of which two are bd-type oxidases (31) and six are heme-copper oxidases, the latter being further divided into two quinol oxidases and four cytochrome oxidases (Table 1). The coxBACF-encoded cytochrome aa3 is the predominant heme-copper oxidase for aerobic growth. Of particular interest is the fixNOQP-encoded cbb3-type oxidase because it enables endosymbiotic B. japonicum cells (bacteroids) to conserve energy despite the very low free O2 concentration in soybean root nodules (32, 33). Accordingly, fixNOQP mutants do not fix N2 in symbiosis (Fix− phenotype), whereas mutants of all other B. japonicum oxidase genes so far examined are Fix+ (Table 1). This unique trait allows us to test by mutation analysis whether or not candidate biogenesis genes are essential for the maturation of active cbb3-type oxidase.
TABLE 1.
Terminal respiratory oxidases in B. japonicum
| Gene numbera | Gene name | Subunit (SU) | Cytochrome (cyt) type | Copper center | Phenotypeb | -Fold change Bact/Airc | Source/Reference |
|---|---|---|---|---|---|---|---|
| -fold | |||||||
| Heme-copper cytochrome oxidases | |||||||
| blr1170 | coxB | SU II | CuA | Fix+ | − | Ref. 58 | |
| blr1171 | coxA | SU I | aa3 | CuB | −2.1 | ||
| blr1173 | coxF | SU IV | −2.4 | ||||
| blr1175 | coxC | SU III | − | ||||
| bll3785 | coxM | SU II | CuA | Fix+ | 5.1 | Ref. 61 | |
| bll3784 | coxN | SU I | aa3-like | CuB | 3.1 | ||
| bll3783 | coxO | SU IIIA | − | ||||
| bll3782 | coxP | SU IIIB | − | ||||
| bll3781 | coxQ | SU IV | − | ||||
| blr2763 | fixN | SU I | bb3 | CuB | Fix– | 30.6 | Refs. 32 and 60 |
| blr2764 | fixO | SU II | Monoheme cyt c | 26.8 | |||
| blr2765 | fixQ | SU IV | 38.3 | ||||
| blr2766 | fixP | SU III | Diheme cyt c | 23.9 | |||
| bll4481 | SU II | CuA | Fix+ | −2.3 | This workd | ||
| bll4480 | SU I | Diheme protein | CuB | −2.1 | |||
| bll4479e | SU IIIe | Diheme cyt ce | −2.8 | ||||
| Heme-copper quinol oxidases | |||||||
| blr2714 | coxW | SU II | Fix+ | − | Refs. 82 and 83 | ||
| blr2715 | coxX | SU I | bb3 | CuB | − | ||
| blr2716 | coxY | SU III | − | ||||
| blr2717 | coxZ | SU IV | − | ||||
| blr0149 | SU II | NT | −2.8 | Ref. 84 | |||
| blr0150 | SU I | Diheme protein | CuB | −4.1 | |||
| blr0151 | SU III | −4.1 | |||||
| blr0152 | SU IV | −2.5 | |||||
| bd-type quinol oxidases | |||||||
| bll0283 | cydA | SU I | b, bd | Fix+ | − | Ref. 85 | |
| bll0282 | cydB | SU II | − | ||||
| blr3728 | SU I | bd-like | NT | − | Ref. 84 | ||
| blr3729 | SU II | − | |||||
a Gene number according to RhizoBase.
b Symbiotic Fix phenotype of mutants deleted for at least the subunit I and/or subunit II genes. Fix+, 70–100% of wild-type activity; Fix−, 0–5% of wild-type activity; NT, not tested.
c -Fold change of gene expression in soybean bacteroids (Bact; 21 days postinfection) compared with cells grown aerobically (Air). Data were taken from Pessi et al. (86). Positive values, increased expression; negative values, decreased expression; −, within threshold range of ±2.
d The genes had been reported previously (84), but the mutant phenotype was determined in this work.
e Sequence analysis predicts that this cytochrome c transfers electrons obtained from reduced PQQ. Therefore, the oxidase proper (bll4481/4480) is classified here as a cytochrome oxidase.
The purpose of this work was to look for genes in the B. japonicum genome (34) that code for CtaG- and Sco1-like proteins, construct knock-out mutations, and test them for defects in the formation of the aa3-type cytochrome oxidase. If such mutants are also defective in symbiotic N2 fixation, the subsequent test for cbb3-type oxidase presence and activity was thought to reveal a possible role of either protein in the biogenesis of this oxidase. We show here that although the aa3- and cbb3-type oxidases have similar CuB-containing active sites, disparate pathways are used for their biogenesis.
EXPERIMENTAL PROCEDURES
Media, Growth Conditions, and Strains
Escherichia coli was grown in Luria-Bertani (LB) medium (35) containing these concentrations of antibiotics for plasmid selection: ampicillin, 200 μg/ml; kanamycin, 30 μg/ml; tetracycline, 10 μg/ml. B. japonicum was cultivated in peptone salts-yeast extract (PSY)2 medium (36, 37) supplemented with 0.1% l-arabinose. Aerobic cultures (21% O2) were grown in Erlenmeyer flasks containing one-fifth of their total volume of PSY medium and shaken vigorously (160 rpm) at 30 °C. Microaerobic cultures (0.5% O2 in the gas phase) and anaerobic cultures were grown as described previously (38, 39) except that the volume was larger (up to 50 ml for microaerobic conditions, up to 400 ml for anaerobic conditions). Where appropriate, antibiotics were used at these concentrations: spectinomycin, 100 μg/ml; streptomycin, 50 μg/ml; kanamycin, 100 μg/ml; tetracycline, 50 μg/ml (solid media) or 25 μg/ml (liquid media). B. japonicum strains used in this work are listed in Table 2.
TABLE 2.
B. japonicum strains used in this work
| Strains | Relevant genotype or phenotype | Source/Reference |
|---|---|---|
| 110spc4 | Spr wild type | Ref. 36 |
| COX132 | Spr KmrcoxA::Tn5 | Ref. 58 |
| 3613 | SprKmrfixN::Tn5 | Ref. 60 |
| 2575 | Spr Smr ΔscoI::aphII (same orientation) | This work |
| 2576 | Spr Smr ΔscoI::aphII (opposite orientation) | This work |
| 3563 | Spr Kmr ΔcoxB::aphII (same orientation) | This work |
| 3583 | Spr Kmr ΔcoxG::aphII (same orientation)a | This work |
| 3586 | Spr Kmr ΔcoxG::aphII (opposite orientation)b | This work |
| 6532 | Spr Kmr bll4480–81::aphII (same orientation) | This work |
| 6533 | Spr Kmr bll4480–81::aphII (opposite orientation) | This work |
| GRZ3035 | Spr SmrnosZ::Ω | Ref. 64 |
Plant Growth
Soybean seeds (Glycine max (L.) Merr. cv. Williams) were surface-sterilized as described previously except that treatment with 30% H2O2 for 15 min was used (40). The symbiotic phenotype of B. japonicum mutants was determined in infection tests using soybean as host, and whole nodule nitrogenase activity was measured with the acetylene reduction assay (41, 42).
General DNA Biochemistry
Standard techniques were used for plasmid isolation, cloning, transformation, Southern blotting, hybridization, and sequencing (43). E. coli strain DH5α (Invitrogen) was the host for routine clonings, and strain BL21 (DE3) (44) was the host for heterologous protein expression. DNA probes for hybridization were labeled with the digoxigenin DNA labeling kit from Roche Applied Science.
Mutant Construction
Plasmids with the pRJ prefix are from our laboratory collection. Details on their genealogy and DNA content are available from the authors upon request.
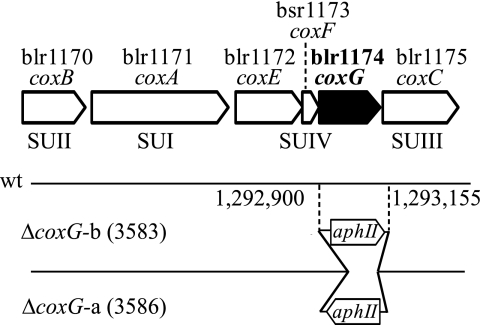
For construction of the coxG deletion mutants, part of the coxG gene on plasmid pRJ3550 was excised, using restriction enzymes BbsI and BamHI, and replaced by the SmaI fragment from pUC4-KIXX (Amersham Pharmacia Biotech AB, Uppsala, Sweden) carrying the aphII gene. The resulting plasmids contained the aphII gene either with the same (pRJ3581) or the opposite (pRJ3582) transcriptional direction as the coxG gene. The two DNA constructs were then cloned into the suicide vector pSUP202pol3 (45) using EcoRI sites, which yielded plasmids pRJ3583 and pRJ3586. Mobilization of these plasmids via E. coli S17-1 (46) into B. japonicum 110spc4 was followed by screening for double recombination events. The resulting two chromosomal coxG deletion mutants carrying the aphII gene in different orientations were named 3583 and 3586 (Table 2 and Fig. 1 with genome coordinates).
FIGURE 1.
Map of the cox gene cluster (blr1170–1178). The arrows show the arrangement of the genes in the cluster. Gene names (if available) are given below the gene numbers. The bottom part of the scheme shows the orientation and precise chromosomal nucleotide position of the kanamycin resistance gene (aphII) inserted in the coxG deletion mutants. wt, wild type; SUI–SUIV, subunits I–IV, respectively.
The first step toward deleting scoI was the PCR-mediated amplification of scoI-flanking DNA using appropriate primers. The 881-bp upstream and 825-bp downstream regions were cloned in pGEM-T Easy (Promega, Madison, WI) and verified by sequencing. Both amplicons were then cloned tail-to-head into pBluescript II KS(+) (Stratagene, La Jolla, CA), resulting in pRJ2572. In two separate constructions, a 1,206-bp PstI kanamycin resistance cassette (aphII) of pBSL86 (47) was inserted in either orientation into the unique PstI site between the scoI-upstream and -downstream regions on pRJ2572. A 2,872-bp EcoRI-XbaI fragment with scoI-flanking DNA plus intervening aphII cassette was excised from each of the two constructs and cloned in pSUP202pol4 (48). The resulting plasmids pRJ2575 and pRJ2576 were mobilized individually by conjugation from E. coli S17-1 into B. japonicum 110spc4 for marker exchange, yielding mutants 2575 and 2676 with aphII in the same and opposite directions as scoI, respectively (Table 2). The correct chromosomal cassette integration was verified by PCR. The deletion end points are at genome coordinates 1,244,627 and 1,245,226.
Plasmid pRJ3563K was used to mutate the coxB gene. Construction of this plasmid started from pRJ3557, which has a 1.1-kb SmaI-XhoI fragment containing coxB. A unique NcoI-NruI fragment from within coxB was excised and replaced by the HindIII-SmaI kanamycin resistance fragment of pUC4-KIXX (Amersham Biosciences), and then the coxB-flanking BamHI-SmaI and EcoRI fragments were added on both sides of the ΔcoxB construct, resulting in pRJ3562K. This DNA construct was cloned in suicide vector pSUP202pol4 (48) to give pRJ3563K, which was then mobilized via E. coli strain S17-1 into B. japonicum 110spc4. Kanamycin-resistant, tetracycline-sensitive exconjugants were selected and checked for double-crossover events by Southern blot analysis. An isolate containing the coxB deletion (with aphII in the same orientation as coxB) was named 3563 (Table 2). The deletion end points are at genome coordinates 1,288,938 and 1,289,556.
To delete the bll4480–81 genes, 665-bp upstream and 843-bp downstream regions were PCR-amplified with appropriate primer pairs, cloned in pGEM-T Easy (Promega), and verified by sequencing. Both amplicons were then cloned tail-to-head in pBluescript II KS(+) (Stratagene). The aphII gene from pBSL86 (47) was inserted in between, using a PstI restriction site, which resulted in plasmids pRJ6532 and pRJ6533. They were mobilized individually by conjugation from E. coli S17-1 into B. japonicum strain 110spc4 for marker exchange, yielding mutants 6532 and 6533 with the aphII orientations given in Table 2. The deletion end points are at genome coordinates 4,961,905 and 4,962,644.
Preparation of Membrane Fraction
B. japonicum cells were disrupted by three passages through a French pressure cell at 9,000 p.s.i. After the removal of cell debris by centrifugation at 28,000 × g for 30 min at 4 °C, membrane pellets were collected by ultracentrifugation at 129,000 × g for 90 min at 4 °C. They were solubilized by slow stirring overnight at 4 °C. If used for Western blots, the membrane pellets were solubilized in 50 mm Tris-HCl, pH 7.5. If used for cytochrome c oxidase measurements, the buffer additionally contained 1% (w/v) dodecylmaltoside (Glycon Biochemicals, Luckenwalde, Germany).
Determination of Protein Concentration
Concentration of solubilized membrane protein was determined with the Bradford method (49), using a Bio-Rad assay with bovine serum albumin as the standard.
Cytochrome Difference Spectra
Dithionite-reduced minus air-oxidized spectra were recorded from 500 to 650 nm in a Hitachi U-3300 spectrophotometer using solubilized membrane proteins prepared from aerobically grown cells at a concentration of 2 mg/ml. Similarly, dithionite-reduced minus APS-oxidized spectra were recorded. For the reduced spectrum, 2 μl of freshly prepared sodium dithionite solution (0.5 m in deionized H2O) was mixed in to give a final concentration of 5 mm. For the oxidized spectrum, 10 μl of APS of 0.1 m solution was added to a final concentration of 5 mm.
Determination of Cytochrome c Oxidase Activity
Cytochrome c oxidase activity of solubilized membrane protein preparations from aerobically grown B. japonicum cells was determined as described (50) with one modification; measurements were performed under continuous stirring in 50 mm HEPES buffer (pH 7.4) containing 1% (w/v) dodecylmaltoside.
Expression and Purification of ScoI and Its Mutant Derivative
The scoI codons for cysteines 74 and 78 were mutated by QuikChange mutagenesis (Stratagene) into serine codons, resulting in pRJ8318. DNA coding for wild-type and mutated versions of the soluble part of ScoI (starting with a glycine at position 30) was cloned into the expression vector pEC425 (51), resulting in pRJ8331 (wild type) and pRJ8339 (C74S/C78S). E. coli BL21 (DE3) was transformed either with pRJ8331 or with pRJ8339. Dense precultures were used to inoculate the main cultures, which had a volume of 200 ml or 1 liter. The medium used was LB with ampicillin. Cultures were grown at 37 °C until they reached an optical density (A600) of 0.5. At this point, expression of the recombinant protein was induced by the addition of arabinose to a final concentration of 0.1% (w/v). After induction, the cultures were transferred to 30 °C, and after 2–4 h, cells were collected by centrifugation and disrupted by three passages through a French press at 9,000 p.s.i. Purification was performed with Strep-Tactin Sepharose columns (IBA GmbH, Göttingen, Germany) according to the supplier's protocol.
UV-visible Spectroscopy of ScoI
The method was used to follow the binding of Cu(II) to reduced ScoI (52). Reduction of ScoIsol was achieved by incubation with 2 mm dithiothreitol for 4 h at 4 °C. Dithiothreitol removal and exchange against 50 mm sodium phosphate buffer (pH 7.0) was done by gel filtration over PD-10 columns (GE Healthcare). Incremental Cu(II) was added in the following way. 5 μl of the solution added contained Cu(II) in amounts that corresponded to 10% of the stoichiometric amount of protein in 1 ml. With a protein concentration of 23 μm, for example, the requested copper solution was 460 μm. For removal of unbound copper by dialysis, a Slide-A-Lyzer® (10,000 molecular weight cut-off, miniunits) from Pierce was used. UV-visible spectra were recorded from 200 to 800 nm on an Agilent diode array photometer (Agilent Technologies, Santa Clara, CA).
Immunological Techniques
Rabbit antibodies specific for the FixO, FixP, and CoxA proteins were available from previous work (8, 24, 53). For the production of antibodies against CoxB, a peptide of the sequence NH2-RVVEDKEFASWVETAKKK-COOH, corresponding to residues 243–260 of the predicted CoxB sequence, was synthesized. Both the peptide and the polyclonal antibodies from rabbits were custom-made by TANA Laboratories, LLC (Houston, TX). Polyclonal antibodies against the soluble part of ScoI were raised in rabbits. Strep-tagII-marked protein was used as antigen, and the immunization was performed by EUROGENTEC S.A. (Liege, Belgium). For Western blot analyses, membrane proteins (30 μg/lane) were separated by SDS-PAGE (54) and blotted as described previously (24). The dilutions of antibodies were 1:2,500 for anti-CoxA, 1:10,000 for anti-CoxB, 1:10,000 for anti-ScoI, 1:1,000 for anti-FixO, and 1:5,000 for anti-FixP. Protein bands with bound immunoglobulins were detected with anti-rabbit IgG-POD (Roche Applied Science) or with anti-rabbit IgG (H+L)-horseradish peroxidase conjugate (Bio-Rad) and chemiluminescence detection kits from Roche Applied Science and Pierce.
RESULTS
The B. japonicum coxG Gene (blr1174) Is Important for the Biogenesis of Cytochrome aa3 but Not for That of Cytochrome cbb3
The genes for the aa3-type cytochrome oxidase (cox) were sequenced in our laboratory (EMBL Nucleotide Sequence Data base, accession number AJ242592) and in the course of the B. japonicum genome sequencing project (34). Three open reading frames (coxEFG) were found to be located between the subunit I (coxA) and subunit III (coxC) genes (Fig. 1). Based on amino acid sequence similarity, coxE codes for a putative protoheme IX farnesyltransferase. The coxF gene encodes a short transmembrane protein that probably corresponds to subunit IV. The coxG gene product shares 49% identity (60% similarity) with CtaG from P. denitrificans and 38% identity (52% similarity) with the Saccharomyces cerevisiae Cox11 protein. CoxG is predicted to have a hydrophobic N-terminal membrane anchor and a hydrophilic periplasmic domain that carries the Cys-X-Cys motif implicated in Cu(I) ligation (55). There is evidence to suggest that Cox11-like proteins transfer copper to the CuB site in subunit I (56, 57). We therefore tested the contribution of B. japonicum coxG to cytochrome oxidase activity.
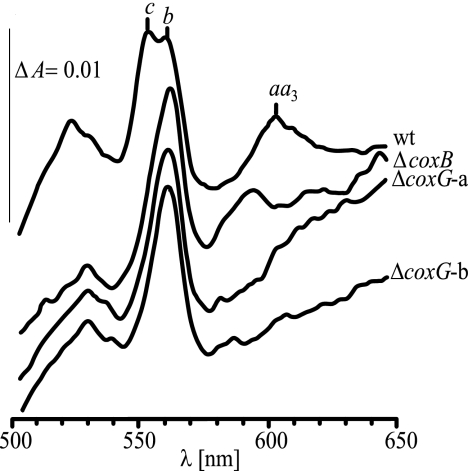
Two coxG-internal deletion mutations were constructed, and the obtained mutant strains 3583 and 3586 carried the inserted aphII cassette in the same or the opposite orientation to coxG, respectively (Table 2 and Fig. 1). Both mutants were NADI-negative, meaning that whole cell cytochrome oxidase activity was defective, and the activity could not be restored by supplementing the growth medium with at least 10 μm CuCl2. The respiratory defect was then quantified in a cytochrome oxidase assay in vitro, using solubilized membrane proteins. A coxB mutant (subunit II gene deletion) (Table 2) and the parental wild type were included for comparison. With the wild-type membrane fraction, an activity of 0.469 μmol of cytochrome c oxidized/min/mg of protein was measured, whereas both the coxB and the two coxG mutants yielded activities of only 0.01 μmol min−1 mg−1, which corresponds to the background oxidation of cytochrome c by air during this assay. A similarly low value was previously reported for a B. japonicum coxA mutant (58). Consistent with this finding was the absence in coxG mutants of the characteristic cytochrome aa3 peak at 603 nm in dithionite-reduced minus air-oxidized difference spectra and a strongly diminished cytochrome c peak at 552 nm (Fig. 2) due to destabilization of the cytochrome bc1-CycM-aa3 supercomplex (58). Curiously, a small peak at 594 nm appeared with the coxB strain (Fig. 2), which might stem from an incompletely formed cytochrome aa3 complex. The spectral defect seen with the coxG mutants could not be restored by the prior addition of 50 μm CuCl2 into the growth medium.
FIGURE 2.
Dithionite-reduced minus air-oxidized difference spectra. The difference spectra were recorded with solubilized membrane protein fractions prepared from aerobically grown B. japonicum cells of the wild type (wt), the coxB deletion mutant 3563 (ΔcoxB), and the two coxG deletion mutants (ΔcoxG) with the aphII cassette inserted in orientation a (strain 3586) and b (strain 3583). The protein concentration was 2 mg/ml. The vertical line on the left spans an absorption difference (ΔA) of 0.01. Peaks characteristic for cytochromes c, b, and aa3 are marked.
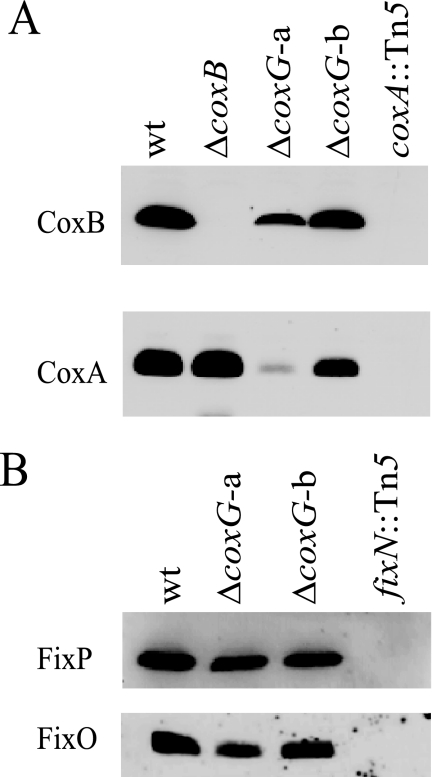
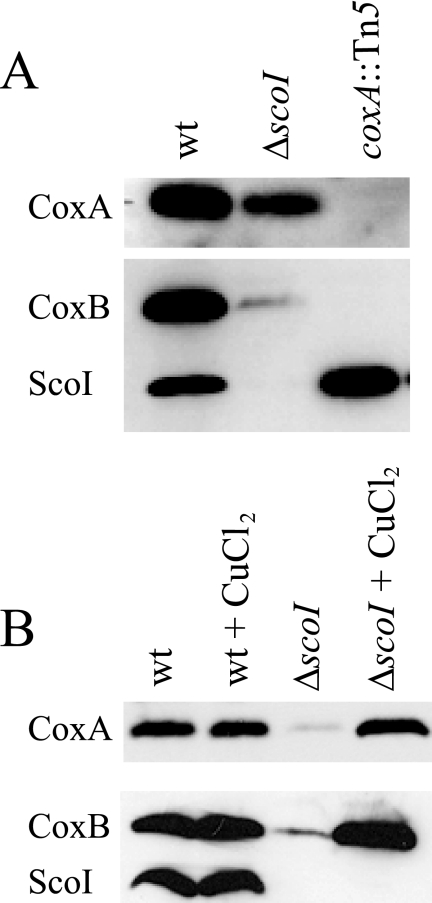
The presence or absence of cytochrome aa3 subunits I and II in coxG mutant membranes was tested by immunoblot analysis, including coxA and coxB mutants for control (Fig. 3A). The controls show that subunits I (CoxA) and II (CoxB) are clearly detectable in the wild type but absent in the coxA mutant. In the coxB mutant, however, only subunit II is missing, as expected, whereas subunit I protein is detectable. We interpret this to mean that subunit II assembly in the membrane depends on the presence of subunit I, whereas subunit I may assemble in the absence of subunit II. In the case of the coxG mutant, the outcome of this experiment is influenced by the orientation of the resistance cassette inserted in the coxG gene (Fig. 3A). Although the ΔcoxG-b mutant (strain 3583) with the cassette in the same orientation as the coxG gene shows only a marginal diminution of the amounts of subunits I and II in its membrane, the ΔcoxG-a mutant (strain 3586) with the cassette in the opposite orientation is strongly depleted for subunit I and, probably as a consequence, exhibits a substantial decrease of subunit II. The divergent behavior finds a likely explanation in the cox operon structure (Fig. 1). We infer that transcription of coxC (subunit III gene) is disturbed in the ΔcoxG-a mutant due to a polar effect of the inserted resistance cassette and that the presence of subunit III is important for the stability of the entire oxidase complex. This polarity effect is largely masked in the ΔcoxG-b mutant in which the out-reading transcription activity originating from the resistance cassette apparently leads to a sufficient expression of coxC. Hence, if coxC is expressed, the deletion in the coxG gene does not significantly interfere with assembly of the oxidase. However, as shown before, the same mutant 3583 had lost cytochrome c oxidase activity, which was also evidenced by the lack of the aa3 peak in the difference spectrum (Fig. 2). The observed phenotype is in line with the proposed role of Cox11-like chaperones in the delivery of copper to the CuB site in subunit I.
FIGURE 3.
Detection of aa3- and cbb3-type oxidase subunits by Western blot analysis. A, antibodies against CoxB and CoxA were used. B, antibodies against FixP and FixO were used. Cells had been cultivated either aerobically (A) or microaerobically (B). Membrane proteins (30 μg/lane) were separated on 14% polyacrylamide gels containing SDS. Proteins from the following strains were analyzed (labeled here by the relevant genotypes): wild type (wt); 3536 (ΔcoxB); 3586 (ΔcoxG-a); 3583 (ΔcoxG-b); COX132 (coxA::Tn5); 3613 (fixN::Tn5).
Next, we addressed the question of whether the biogenesis function of CoxG is also needed for other heme-copper cytochrome oxidases that possess the CuB-containing subunit I. If the cbb3-type cytochrome oxidase, for example, would critically depend on CoxG, the B. japonicum coxG mutants might have a nitrogen fixation defect in symbiosis because this oxidase is essential for bacteroid respiration (cf. Table 1). However, the data in Table 3 show that both coxG strains were able to elicit a fully functional symbiosis (Fix+ phenotype) indistinguishable from that of the B. japonicum wild type. Therefore, it is unlikely that CoxG is involved in the maturation of the cbb3-type oxidase. To substantiate this notion, we confirmed the presence of cbb3 oxidase subunits FixO and FixP in membranes of coxG mutant cells that had been grown under micro-oxic conditions (Fig. 3B). Taken together, it appears as if the role CoxG plays for cytochrome aa3 does not apply to cytochrome cbb3.
TABLE 3.
Symbiotic properties of ΔscoI and ΔcoxG mutants inoculated on soybean
Mutants were tested in parallel with the wild type in two separate series of two (ΔcoxG) and three (ΔscoI) independent experiments. Shown are the data of one representative experiment for each set of strains.
| Strain | Relevant genotype | Number of nodules | Nodule dry weight | Nitrogenase activitya | Relative Fix activityb |
|---|---|---|---|---|---|
| mg | %/min/g | % | |||
| 110spc4 | Wild type | 30.0 ± 3.3 | 0.93 ± 0.03 | 4.15 ± 2.16 | 100 ± 52 |
| 3586 | ΔcoxG-a | 32.7 ± 16.4 | 0.82 ± 0.7 | 4.12 ± 2.11 | 99.3 ± 51.0 |
| 3583 | ΔcoxG-b | 32.0 ± 13.3 | 0.88 ± 0.15 | 4.50 ± 0.93 | 108.4 ± 22.3 |
| 110spc4 | wild type | 35.7 ± 10.3 | 1.12 ± 0.29 | 2.31 ± 0.90 | 100 ± 29 |
| 2575 | ΔscoI | 40.7 ± 7.8 | 0.69 ± 0.15 | 0.82 ± 0.35 | 28.8 ± 11.9 |
a Nitrogenase activity is expressed as percentage of C2H4/min/g.
b Relative nitrogen fixation activity is expressed as a percentage of wild type.
Gene blr1131 Encodes a Sco1-like Protein
The second biogenesis protein that we considered to be relevant for this work is related to Sco1. A copper-chaperoning function specifically in the formation of the CuA site on subunit II has been attributed not only to mitochondrial but also to bacterial Sco1-like proteins (19–23). In many aerobic bacteria, its gene maps immediately adjacent to cytochrome oxidase structural and biogenesis genes (cf. String Data base; available on the EMBL web site). The B. japonicum cox gene cluster shown in Fig. 1, however, does not encode a Sco1-like protein. Instead, based on sequence similarity, the blr1131 gene (34) was identified as a likely candidate to encode a Sco1-like protein. The blr1131 open reading frame codes for a 196-amino acid protein that shows the typical hallmarks of prokaryotic and eukaryotic Sco proteins (i.e. an N-terminal membrane anchor and a C-terminal, membrane-peripheral thioredoxin-like domain with a Cys-X3-Cys motif and a conserved His for copper ligation) (20). For these reasons and further evidence given below and in keeping with the standard nomenclature for bacterial genes (59), blr1131 of B. japonicum was named scoI.
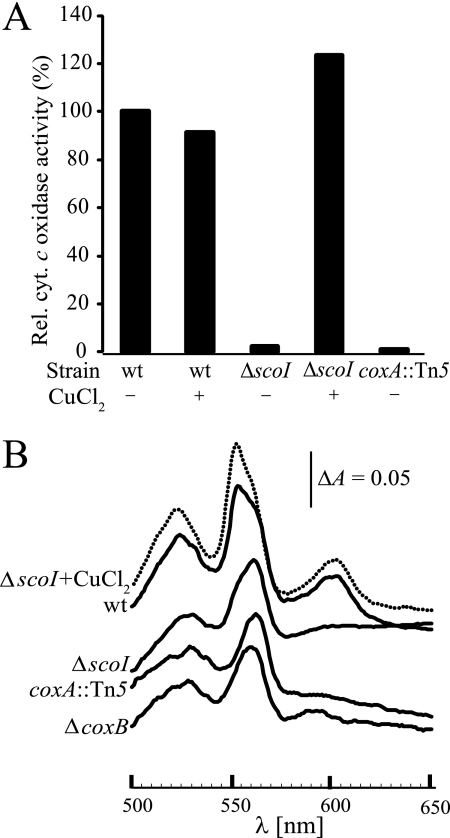
To approach ScoI function, two mutants were constructed (strains 2575 and 2576; Table 2) in which almost the entire scoI gene was deleted and replaced by a kanamycin resistance cassette in either orientation. Because no phenotypic difference was subsequently observed between strains 2575 and 2576, we report here only the results for strain 2575. The ΔscoI mutant exhibited wild type-like growth in complex medium on agar plates and in liquid cultures. A qualitative NADI test revealed a defect in cytochrome c oxidase activity. To further substantiate this defect, quantitative cytochrome c oxidase measurements were done. Mutant COX132 (Table 2) having an insertion in the subunit I gene (coxA::Tn5) was included in these assays for comparison. Fig. 4A shows that the scoI and coxA mutants have negligible cytochrome c oxidase activities below 2% of the wild type, which reflects spontaneous cytochrome c oxidation in air. Cytochrome aa3 formation was analyzed by dithionite-reduced minus APS-oxidized difference spectroscopy of solubilized membranes. Again, scoI and coxA mutants showed the same phenotype (i.e. absence of the diagnostic aa3 peak at 603 nm and a decrease of the cytochrome c peak at 552 nm) (Fig. 4B). In the scoI mutant, the 603 nm peak appeared again when cells had been grown in medium containing 50 μm CuCl2 (Fig. 4B). Likewise, cytochrome c oxidase activity of the scoI mutant was restored to wild-type levels by the same CuCl2 treatment (Fig. 4A). Assembly of cytochrome aa3 in the membrane was then analyzed in Western blots using antibodies specific for CoxA (subunit I) (24) and CoxB (subunit II). Fig. 5(A and B) shows that both subunits are substantially depleted in scoI mutant membranes. When 50 μm CuCl2 had been added to the growth medium, the amounts of both subunits in the scoI mutant were restored to wild-type levels (Fig. 5B).
FIGURE 4.
The scoI mutation affects cytochrome aa3. A, relative cytochrome c oxidase activity of aerobically grown wild type (wt) and mutant strains 2575 (ΔscoI) and COX132 (coxA::Tn5). Wild-type activity (100%) corresponds to ∼0.43 μmol horse heart cytochrome c oxidized/mg of membrane protein/min. Whether or not 50 μm CuCl2 was added to the PSY medium is indicated with a plus or minus sign. B, dithionite-reduced minus APS-oxidized difference spectra of solubilized membrane protein fraction (61 mg/ml) from aerobically grown cells. As labeled, the continuous curves represent the wild type and the ΔscoI, coxA::Tn5, and ΔcoxB mutants (strains 2575, COX132, and 3536, respectively). The dotted curve represents the ΔscoI mutant grown in PSY supplemented with 50 μm CuCl2. The vertical bar indicates a difference in absorption (ΔA) of 0.05. Note that, in contrast to Fig. 2, the cytochrome b is not resolved here as a separate peak but as a shoulder (560 nm).
FIGURE 5.
Western blot analysis of cytochrome aa3 subunits in the ΔscoI mutant. A, comparison of scoI mutant with wild type and coxA mutant. B, restoration of subunit assembly by copper. As marked on the left, antibodies specific against CoxA, CoxB, and ScoI (control) were used. The lower blots in A and B were exposed to a mixture of anti-CoxB and anti-ScoI immunoglobulins. Membrane proteins (30 μg/lane) were separated on 14% polyacrylamide gels containing SDS. They were isolated from the following strains: wild type (wt), 2575 (ΔscoI), and COX132 (coxA::Tn5). For copper supplementation, PSY medium contained 50 μm CuCl2.
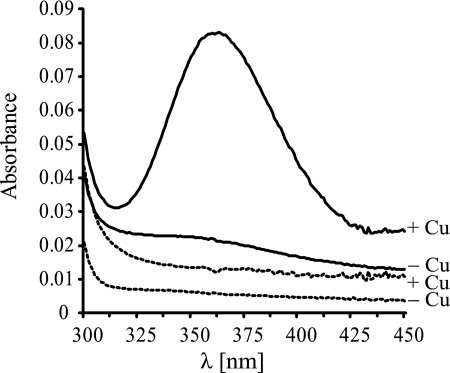
The results suggest that the mutant defect was caused by a lack of copper-dependent maturation of the oxidase complex and that ScoI acts as an aa3-specific copper chaperone. To further support this idea, a soluble ScoI protein derivative (ScoIsol) was purified (see “Experimental Procedures”), and its ability to bind copper was tested by UV-visible absorption spectroscopy. It was essential to reduce ScoIsol with dithiothreitol prior to the experiment. The addition of CuCl2 to the protein resulted in an absorbance peak at ∼360 nm (Fig. 6), reflecting the binding of Cu(II) to the protein (52). Upon incremental copper addition, the peak increased and reached a plateau at a ratio of ∼0.8 stoichiometric equivalents of Cu(II) per ScoIsol molecule. Copper binding did not occur to a ScoIsol mutant derivative in which the two predicted active site cysteines Cys74 and Cys78 had been replaced by serines. By analogy with Sco1-like proteins from other organisms, our results indicate that Cys74 and Cys78 form part of the active site and that they need to be reduced to thiols or thiolate anions to allow complex formation with a copper cation.
FIGURE 6.
Evidence for copper binding to ScoI. UV-visible spectra were taken of 23 μm reduced ScoIsol (solid curves) and ScoI[C74S/C78S]sol (dashed curves), dissolved in 50 mm phosphate buffer, pH 7.0. −, no Cu(II) was added; +, Cu(II) was added to protein in molar equivalents of either 1 (in the case of ScoIsol) or 1.4 (in the case of ScoI[C74S/C78S]sol. The peak at 360 nm corresponds to the absorbance of protein-bound copper.
The B. japonicum scoI Mutant Is Symbiotically Defective
During anoxic growth with nitrate as the terminal electron acceptor, the scoI mutant (strain 2575) exhibited a slight delay in denitrification with a transient accumulation of nitrite, indicating that nitrite reduction or a subsequent N-oxide reduction step is impaired (data not shown). A more striking property of the scoI mutant was its inability to establish a fully functional symbiosis with soybean (G. max). Table 3 shows that it reaches only 29% of the nitrogen fixation (Fix) activity of the wild type, as determined by the acetylene reduction assay. Furthermore, plants inoculated with strain 2575 displayed signs of nitrogen starvation, such as a decreased nodule dry weight (Table 3) and a pale green leaf color. On the one hand, we can safely argue that the symbiotic defect in the scoI mutant was not caused by the described defect in cytochrome aa3 biogenesis because a coxA knock-out mutant has a clear Fix+ phenotype (58). On the other hand, although all of the scoI mutant phenotypes were not as severe as those of a cytochrome cbb3-defective mutant (60), they would plausibly explain the symbiotic defect if ScoI served as a maturation factor for the CuB site of the symbiotically essential cytochrome cbb3.
ScoI Is Not Needed for the Biogenesis of the cbb3-type Oxidase
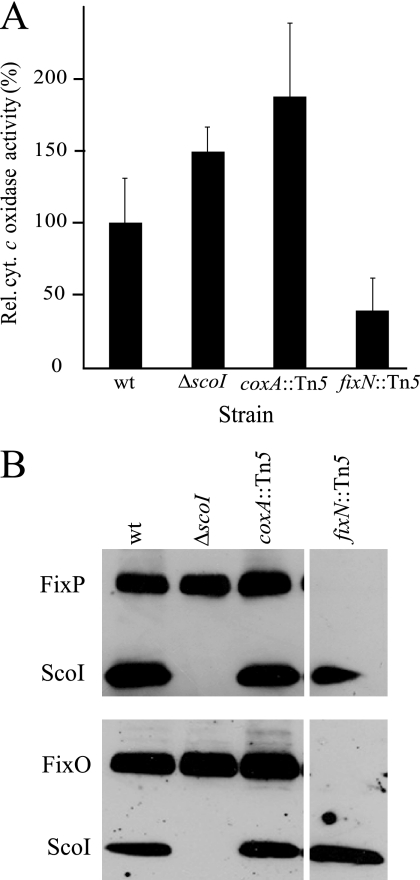
Cytochrome oxidase activity was determined in membranes of the scoI mutant and compared with that of the wild type, the coxA mutant, and a cbb3-defective fixN mutant. All strains were grown under conditions (micro-oxia, or anoxia with nitrate) in which the fixNOQP operon is strongly induced and its product is the predominant cytochrome oxidase expressed (32, 60). Fig. 7A shows the results with membranes isolated from anoxically grown cultures, using reduced horse heart cytochrome c as the electron donor. The fixN mutant had a more than 60% decreased cytochrome c oxidase activity as compared with the wild type, which confirms that the cbb3-type oxidase is the most prominent oxidase under these growth conditions in the wild type. In contrast, neither the scoI nor the coxA mutant was impaired in this assay. Interestingly, they even showed higher activity than the wild type. Perhaps the lack of competing cytochrome aa3 in these mutants allows for a better substrate usage or a facilitated assembly of the cbb3-type oxidase in the membrane. The in vitro experiment of Fig. 7A was corroborated by a N,N,N′,N′-tetramethyl-p-phenylenediamine oxidation assay in vivo with cells that had been cultivated microaerobically. Again, only the fixN mutant exhibited an impaired N,N,N′,N′-tetramethyl-p-phenylenediamine oxidation activity as compared with the other three strains (data not shown). Finally, we examined membranes for the presence of some of the cbb3-type oxidase subunits. For this purpose, membranes were isolated from wild-type and mutant cells that had been cultivated anaerobically or microaerobically and tested by Western blot analysis with antibodies specific for the FixO and FixP proteins. As shown in Fig. 7B, the scoI mutant contains these subunits in undisturbed amounts, just like the wild type and the coxA mutant, but in contrast to the control (fixN mutant), where both subunits are missing. Taken together, none of the phenotypic and biochemical tests performed with the scoI mutant revealed a function of the ScoI protein in the biogenesis of the cbb3-type oxidase.
FIGURE 7.
The scoI mutation does not affect cytochrome cbb3. A, relative cytochrome c oxidase activity of the wild type (wt) and strains 2575 (ΔscoI), COX132 (coxA::Tn5), and 3613 (fixN::Tn5). Cells had been cultivated under conditions of anaerobic nitrate respiration. A total of 11 measurements with three independent biological replicas were done for each strain. Wild type activity (100%) corresponds to 1.2 μmol cytochrome c oxidized/mg of protein/min. B, Western blot analysis of cytochrome cbb3 subunits. As marked on the left, antibodies specific against FixP, FixO, and ScoI (control) were used. The blots in the upper and lower panels were exposed to mixtures of anti-FixP plus anti-ScoI immunoglobulins and anti-FixO plus anti-ScoI immunoglobulins. Membrane proteins (30 μg/lane) were separated on 14% polyacrylamide gels containing SDS. They were isolated from the strains indicated. The separate lane on the right was run on the same gel as the other lanes. Error bars, S.D.
If not cbb3, which protein might then be the symbiotically important target for ScoI? Using bioinformatics, we examined the B. japonicum chromosome for genes encoding potential periplasmic CuA-binding proteins other than CoxB and found three: bll3785 (coxM), bll4481, and blr0315 (nosZ). The first two are in operons for heme-copper cytochrome oxidases, coxMNOPQ, and bll4481–4479 (Table 1). A coxN deletion mutant had previously been shown to be Fix+ in symbiosis (61). Here we constructed and tested bll4481–4480 deletion mutants (strains 6532 and 6533; Table 2) and found that they also had a Fix+ phenotype (Table 1). The nosZ gene codes for a periplasmic CuA-containing nitrous oxide reductase (62, 63). A B. japonicum nosZ mutant (strain GRZ3035 (64)) was reported by Mesa et al. (65) to be Fix+ in symbiosis. We confirmed the phenotype of this mutant with the standard soybean infection test used in our laboratory (data not shown). In conclusion, although the biogenesis of all three proteins may well depend on ScoI, none of them is important for symbiosis. Future work on the identification of other periplasmic ScoI targets may help to explain the symbiotic scoI mutant phenotype.
DISCUSSION
The relative exclusiveness of B. japonicum cytochromes aa3 and cbb3 for aerobic and microaerobic cells, respectively, was of tremendous help in our attempt to assess the contribution of CoxG and ScoI to the biogenesis of either oxidase. It was somewhat surprising that B. japonicum CoxG was found to be essential for the biogenesis of cytochrome aa3 but not for that of cytochrome cbb3. Given that its pro- and eukaryotic homologs (CtaG and Cox11) had been implicated in copper delivery to the CuB site on subunit I (15), one might have expected CoxG to serve as an important chaperone for both oxidases. What are the reasons for the selectivity of CoxG? The question is difficult to answer because, despite the remarkable progress on the structure and function of Cox11-like proteins (55–57, 66), the mechanism of copper insertion into subunit I remains elusive (18, 67). We could think of two possible, mutually not exclusive discriminating factors. The first is subunit III, which is conserved in almost all bacterial heme-copper oxidases except cytochrome cbb3. A hypothetical CoxG-subunit III interaction as a prerequisite for copper insertion into subunit I would not be fulfilled by the completely dissimilar FixO subunit in cytochrome cbb3. A second discriminator might be the reported difference in the heme-CuB active site architecture between the two oxidase classes (68–70). Especially if copper insertion occurs during membrane translocation, as proposed by Khalimonchuk et al. (56), the CoxG protein might be better suited to deliver copper to subunit I of the aa3- rather than cbb3-type oxidase during folding and maturation.
The B. japonicum scoI gene was also shown here to be essential for cytochrome aa3 formation and activity. The soluble ScoI domain devoid of its N-terminal membrane anchor was expressed and purified. It bound Cu(II) with a nearly 1:1 stoichiometry, and the reduced cysteines in a CXXXC motif were demonstrated to be essential for binding. All of these results are consistent with the purported role of the eukaryotic (i.e. mitochondrial) ScoI-like proteins (19, 22, 71, 72) and some prokaryotic ScoI homologs (20, 23, 73) in the biogenesis of the membrane-peripheral, CuA-containing subunit II domain. Akin to Cox11/CtaG/CoxG, however, the direct transfer of copper from the ScoI chaperone to the target subunit remains to be demonstrated experimentally. Moreover, the model of ScoI-like proteins as copper chaperones has been challenged. Because they are thioredoxin-like, some were found to possess protein dithiol:disulfide oxidoreductase activity (26, 74, 75). Also, a Cu2+-reducing activity was reported (76). An interesting case has emerged from work with human mitochondria, which have two ScoI homologs, one acting as a dithiol:disulfide oxidoreductase (Sco2) and the other (Sco1) as a copper chaperone (77). The latter appeared to depend on the activity of the former. We tested the B. japonicum ScoIsol in the standard insulin reduction assay (78) but did not detect any reducing activity. It is an attractive hypothesis, however, that the previously described thioredoxin-like TlpA protein (24, 25) interacts with ScoI in B. japonicum. Incidentally, all of the phenotypes described here for the scoI mutant are similar to those of a tlpA mutant (24). Finally, the biogenesis of the CuA center has experienced a new twist with the recent discovery of the T. thermophilus PCuAC as a copper chaperone that is specific for the formation of the ba3-type oxidase (26). We noticed that the B. japonicum genome harbors two genes for PCuAC-like proteins (bll4880 and blr7088). Future work will tell whether they are involved in copper trafficking.
The ScoI-like protein SenC was shown to be important for the cbb3-type oxidase in Rhodobacter capsulatus (21) and Pseudomonas aeruginosa (79). These discoveries are intriguing in view of the fact that the cbb3-type oxidase does not have a CuA center (3, 7). In our own studies, we have made a considerable effort to either prove or exclude an involvement of ScoI in cytochrome cbb3 biogenesis and function, and the obvious necessity to do so was the partial Fix− phenotype of the scoI mutant. The data provide clear evidence that ScoI is not a copper chaperone for the cbb3-type oxidase in B. japonicum. The reason for the observed deviation between species is not known. The cbb3-type oxidase is required for aerobic growth in R. capsulatus and P. aeruginosa, whereas B. japonicum needs it for respiration at very low oxygen tensions. It is difficult to understand, however, why such a physiological difference should be associated with a selective ScoI function.
Having ascertained that there are no additional coxG and scoI homologs in the genome and having shown that the B. japonicum CoxG and ScoI proteins are involved only in the maturation of the aa3-type cytochrome oxidase, we postulate a copper trafficking pathway for the maturation of cytochrome cbb3 that is uncoupled from these two biogenesis factors. In this context, a previously made observation may now gain momentum. We and others had identified the fixGHIS operon (also called ccoGHIS) immediately downstream of the cytochrome cbb3 structural genes and had shown that it was essential for the assembly of the oxidase (29, 30). One of the genes (fixI/ccoI) codes for a potential metal-transporting P-type ATPase (80, 81), which is perhaps specific for copper. Rather than transporting copper through the cytoplasmic membrane (to which destination?), the role of FixI might be to sequestrate copper on the periplasmic side and deliver it directly to the membrane-embedded CuB site in subunit I (FixN/CcoN).
Acknowledgments
We gratefully acknowledge the expert technical assistance of Olivera Volarevic (deceased, August 2009). The help of Patrizia Rossi in DNA sequencing, Sabine Haueter in mutant construction, and Rainer Follador in phenotypic analysis is greatly appreciated. Socorro Mesa is thanked for providing strain GRZ3035.
This work was supported by grants from ETH Zürich and the Swiss National Foundation for Scientific Research.
- PSY
- peptone salts-yeast extract
- APS
- ammonium persulfate
- Fix
- nitrogen fixation
- NADI
- 1-naphthol plus N,N-dimethyl-1,4-phenylendiamine.
REFERENCES
- 1.García-Horsman J. A., Barquera B., Rumbley J., Ma J., Gennis R. B. (1994) J. Bacteriol. 176, 5587–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castresana J., Lübben M., Saraste M., Higgins D. G. (1994) EMBO J. 13, 2516–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira M. M., Santana M., Teixeira M. (2001) Biochim. Biophys. Acta 1505, 185–208 [DOI] [PubMed] [Google Scholar]
- 4.Iwata S., Ostermeier C., Ludwig B., Michel H. (1995) Nature 376, 660–669 [DOI] [PubMed] [Google Scholar]
- 5.Trumpower B. L., Gennis R. B. (1994) Annu. Rev. Biochem. 63, 675–716 [DOI] [PubMed] [Google Scholar]
- 6.Poole R. K., Cook G. M. (2000) Adv. Microb. Physiol. 43, 165–224 [DOI] [PubMed] [Google Scholar]
- 7.Ducluzeau A. L., Ouchane S., Nitschke W. (2008) Mol. Biol. Evol. 25, 1158–1166 [DOI] [PubMed] [Google Scholar]
- 8.Zufferey R., Preisig O., Hennecke H., Thöny-Meyer L. (1996) J. Biol. Chem. 271, 9114–9119 [DOI] [PubMed] [Google Scholar]
- 9.Kulajta C., Thumfart J. O., Haid S., Daldal F., Koch H. G. (2006) J. Mol. Biol. 355, 989–1004 [DOI] [PubMed] [Google Scholar]
- 10.Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) Science 272, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 11.Carr H. S., Winge D. R. (2003) Acc. Chem. Res. 36, 309–316 [DOI] [PubMed] [Google Scholar]
- 12.Khalimonchuk O., Rödel G. (2005) Mitochondrion 5, 363–388 [DOI] [PubMed] [Google Scholar]
- 13.Bundschuh F. A., Hoffmeier K., Ludwig B. (2008) Biochim. Biophys. Acta 1777, 1336–1343 [DOI] [PubMed] [Google Scholar]
- 14.Bundschuh F. A., Hannappel A., Anderka O., Ludwig B. (2009) J. Biol. Chem. 284, 25735–25741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobine P. A., Pierrel F., Winge D. R. (2006) Biochim. Biophys. Acta 1763, 759–772 [DOI] [PubMed] [Google Scholar]
- 16.van der Oost J., Haltia T., Raitio M., Saraste M. (1991) J. Bioenerg. Biomembr. 23, 257–267 [DOI] [PubMed] [Google Scholar]
- 17.Cao J., Hosler J., Shapleigh J., Revzin A., Ferguson-Miller S. (1992) J. Biol. Chem. 267, 24273–24278 [PubMed] [Google Scholar]
- 18.Greiner P., Hannappel A., Werner C., Ludwig B. (2008) Biochim. Biophys. Acta 1777, 904–911 [DOI] [PubMed] [Google Scholar]
- 19.Nittis T., George G. N., Winge D. R. (2001) J. Biol. Chem. 276, 42520–42526 [DOI] [PubMed] [Google Scholar]
- 20.Balatri E., Banci L., Bertini I., Cantini F., Ciofi-Baffoni S. (2003) Structure 11, 1431–1443 [DOI] [PubMed] [Google Scholar]
- 21.Swem D. L., Swem L. R., Setterdahl A., Bauer C. E. (2005) J. Bacteriol. 187, 8081–8087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abajian C., Rosenzweig A. C. (2006) J. Biol. Inorg. Chem. 11, 459–466 [DOI] [PubMed] [Google Scholar]
- 23.Cawthorn T. R., Poulsen B. E., Davidson D. E., Andrews D., Hill B. C. (2009) Biochemistry 48, 4448–4454 [DOI] [PubMed] [Google Scholar]
- 24.Loferer H., Bott M., Hennecke H. (1993) EMBO J. 12, 3373–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Capitani G., Rossmann R., Sargent D. F., Grütter M. G., Richmond T. J., Hennecke H. (2001) J. Mol. Biol. 311, 1037–1048 [DOI] [PubMed] [Google Scholar]
- 26.Abriata L. A., Banci L., Bertini I., Ciofi-Baffoni S., Gkazonis P., Spyroulias G. A., Vila A. J., Wang S. (2008) Nat. Chem. Biol. 4, 599–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zufferey R., Hennecke H., Thöny-Meyer L. (1997) FEBS Lett. 412, 75–78 [DOI] [PubMed] [Google Scholar]
- 28.Thöny-Meyer L. (2002) Biochem. Soc. Transact. 30, 633–638 [DOI] [PubMed] [Google Scholar]
- 29.Preisig O., Zufferey R., Hennecke H. (1996) Arch. Microbiol. 165, 297–305 [DOI] [PubMed] [Google Scholar]
- 30.Koch H. G., Winterstein C., Saribas A. S., Alben J. O., Daldal F. (2000) J. Mol. Biol. 297, 49–65 [DOI] [PubMed] [Google Scholar]
- 31.Jünemann S. (1997) Biochim. Biophys. Acta 1321, 107–127 [DOI] [PubMed] [Google Scholar]
- 32.Preisig O., Zufferey R., Thöny-Meyer L., Appleby C. A., Hennecke H. (1996) J. Bacteriol. 178, 1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arslan E., Kannt A., Thöny-Meyer L., Hennecke H. (2000) FEBS Lett. 470, 7–10 [DOI] [PubMed] [Google Scholar]
- 34.Kaneko T., Nakamura Y., Sato S., Minamisawa K., Uchiumi T., Sasamoto S., Watanabe A., Idesawa K., Iriguchi M., Kawashima K., Kohara M., Matsumoto M., Shimpo S., Tsuruoka H., Wada T., Yamada M., Tabata S. (2002) DNA Res. 9, 225–256 [DOI] [PubMed] [Google Scholar]
- 35.Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 36.Regensburger B., Hennecke H. (1983) Arch. Microbiol. 135, 103–109 [DOI] [PubMed] [Google Scholar]
- 37.Mesa S., Hauser F., Friberg M., Malaguti E., Fischer H. M., Hennecke H. (2008) J. Bacteriol. 190, 6568–6579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hauser F., Lindemann A., Vuilleumier S., Patrignani A., Schlapbach R., Fischer H. M., Hennecke H. (2006) Mol. Genet. Genomics 275, 55–67 [DOI] [PubMed] [Google Scholar]
- 39.Hauser F., Pessi G., Friberg M., Weber C., Rusca N., Lindemann A., Fischer H. M., Hennecke H. (2007) Mol. Genet. Genomics 278, 255–271 [DOI] [PubMed] [Google Scholar]
- 40.Göttfert M., Hitz S., Hennecke H. (1990) Mol. Plant-Microbe Interact. 3, 308–316 [DOI] [PubMed] [Google Scholar]
- 41.Göttfert M., Grob P., Hennecke H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2680–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn M., Hennecke H. (1984) Mol. Gen. Genet. 193, 46–52 [Google Scholar]
- 43.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 44.Studier F. W., Moffatt B. A. (1986) J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 45.Rossmann R., Stern D., Loferer H., Jacobi A., Glockshuber R., Hennecke H. (1997) FEBS Lett. 406, 249–254 [DOI] [PubMed] [Google Scholar]
- 46.Simon R., Priefer U., Pühler A. (1983) in Molecular Genetics of the Bacteria-Plant Interaction (Pühler A. ed) pp. 98–106, Springer Verlag, Heidelberg, Germany [Google Scholar]
- 47.Alexeyev M. F. (1995) BioTechniques 18, 52–56 [PubMed] [Google Scholar]
- 48.Fischer H. M., Babst M., Kaspar T., Acuña G., Arigoni F., Hennecke H. (1993) EMBO J. 12, 2901–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 50.Gerhus E., Steinrücke P., Ludwig B. (1990) J. Bacteriol. 172, 2392–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulz H., Hennecke H., Thöny-Meyer L. (1998) Science 281, 1197–1200 [DOI] [PubMed] [Google Scholar]
- 52.Imriskova-Sosova I., Andrews D., Yam K., Davidson D., Yachnin B., Hill B. C. (2005) Biochemistry 44, 16949–16956 [DOI] [PubMed] [Google Scholar]
- 53.Loferer H., Hennecke H. (1994) Eur. J. Biochem. 223, 339–344 [DOI] [PubMed] [Google Scholar]
- 54.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 55.Banci L., Bertini I., Cantini F., Ciofi-Baffoni S., Gonnelli L., Mangani S. (2004) J. Biol. Chem. 279, 34833–34839 [DOI] [PubMed] [Google Scholar]
- 56.Khalimonchuk O., Ostermann K., Rödel G. (2005) Curr. Genet. 47, 223–233 [DOI] [PubMed] [Google Scholar]
- 57.Banting G. S., Glerum D. M. (2006) Eukaryot. Cell 5, 568–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bott M., Bolliger M., Hennecke H. (1990) Mol. Microbiol. 4, 2147–2157 [DOI] [PubMed] [Google Scholar]
- 59.Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. (1968) J. Gen. Microbiol. 50, 1–14 [DOI] [PubMed] [Google Scholar]
- 60.Preisig O., Anthamatten D., Hennecke H. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bott M., Preisig O., Hennecke H. (1992) Arch. Microbiol. 158, 335–343 [DOI] [PubMed] [Google Scholar]
- 62.Haltia T., Brown K., Tegoni M., Cambillau C., Saraste M., Mattila K., Djinovic-Carugo K. (2003) Biochem. J. 369, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zumft W. G. (2005) J. Mol. Microbiol. Biotechnol. 10, 154–166 [DOI] [PubMed] [Google Scholar]
- 64.Velasco L., Mesa S., Xu C. A., Delgado M. J., Bedmar E. J. (2004) Antonie van Leeuwenhoek 85, 229–235 [DOI] [PubMed] [Google Scholar]
- 65.Mesa S., Alché J., Bedmar E., Delgado M. J. (2004) Physiol. Plant. 120, 205–211 [DOI] [PubMed] [Google Scholar]
- 66.Carr H. S., Maxfield A. B., Horng Y. C., Winge D. R. (2005) J. Biol. Chem. 280, 22664–22669 [DOI] [PubMed] [Google Scholar]
- 67.Hiser L., Di Valentin M., Hamer A. G., Hosler J. P. (2000) J. Biol. Chem. 275, 619–623 [DOI] [PubMed] [Google Scholar]
- 68.Buse G., Soulimane T., Dewor M., Meyer H. E., Blüggel M. (1999) Protein Sci. 8, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hemp J., Christian C., Barquera B., Gennis R. B., Martínez T. J. (2005) Biochemistry 44, 10766–10775 [DOI] [PubMed] [Google Scholar]
- 70.Rauhamäki V., Bloch D. A., Verkhovsky M. I., Wikström M. (2009) J. Biol. Chem. 284, 11301–11308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horng Y. C., Leary S. C., Cobine P. A., Young F. B., George G. N., Shoubridge E. A., Winge D. R. (2005) J. Biol. Chem. 280, 34113–34122 [DOI] [PubMed] [Google Scholar]
- 72.Banci L., Bertini I., Ciofi-Baffoni S., Gerothanassis I. P., Leontari I., Martinelli M., Wang S. (2007) Structure 15, 1132–1140 [DOI] [PubMed] [Google Scholar]
- 73.Saenkham P., Vattanaviboon P., Mongkolsuk S. (2009) FEMS Microbiol. Lett. 293, 122–129 [DOI] [PubMed] [Google Scholar]
- 74.Ye Q., Imriskova-Sosova I., Hill B. C., Jia Z. (2005) Biochemistry 44, 2934–2942 [DOI] [PubMed] [Google Scholar]
- 75.Williams J. C., Sue C., Banting G. S., Yang H., Glerum D. M., Hendrickson W. A., Schon E. A. (2005) J. Biol. Chem. 280, 15202–15211 [DOI] [PubMed] [Google Scholar]
- 76.Badrick A. C., Hamilton A. J., Bernhardt P. V., Jones C. E., Kappler U., Jennings M. P., McEwan A. G. (2007) FEBS Lett. 581, 4663–4667 [DOI] [PubMed] [Google Scholar]
- 77.Leary S. C., Sasarman F., Nishimura T., Shoubridge E. A. (2009) Hum. Mol. Genet. 18, 2230–2240 [DOI] [PubMed] [Google Scholar]
- 78.Holmgren A. (1979) J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 79.Frangipani E., Haas D. (2009) FEMS Microbiol. Lett. 298, 234–240 [DOI] [PubMed] [Google Scholar]
- 80.Solioz M., Vulpe C. (1996) Trends Biochem. Sci. 21, 237–241 [PubMed] [Google Scholar]
- 81.Lu Z. H., Solioz M. (2002) Adv. Protein Chem. 60, 93–121 [DOI] [PubMed] [Google Scholar]
- 82.Surpin M. A., Lübben M., Maier R. J. (1996) Gene 183, 201–206 [DOI] [PubMed] [Google Scholar]
- 83.Surpin M. A., Maier R. J. (1999) Appl. Environ. Microbiol. 65, 339–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Göttfert M., Hennecke H., Tabata S. (2004) in Genomes and Genomics of Nitrogen-fixing Organisms (Palacios R., Newton W. E. eds) pp. 99–112, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 85.Arslan E. (2001) The cbb3- and bd-type Oxidases of the Soybean Symbiont Bradyrhizobium japonicum, Ph.D. dissertation, Federal Institute of Technology, Zurich, Switzerland [Google Scholar]
- 86.Pessi G., Ahrens C. H., Rehrauer H., Lindemann A., Hauser F., Fischer H. M., Hennecke H. (2007) Mol. Plant-Microbe Interact. 20, 1353–1363 [DOI] [PubMed] [Google Scholar]