Abstract
Triacylglycerol (TAG) hydrolysis, membrane lipid biosynthesis, and lipid turnover are largely interlinked processes. In yeast, TAG is mobilized by three TAG lipases named Tgl3p, Tgl4p, and Tgl5p, which are localized to lipid particles/droplets. These TAG lipases posses a conserved GXSXG motif that is characteristic of hydrolytic enzymes. Here, we demonstrated that the yeast TAG lipase Tgl4p, the functional ortholog of the adipose TAG lipase, ATGL, catalyzes multiple functions in lipid metabolism. An extended domain and motif search analysis revealed that Tgl4p bears not only a lipase consensus domain but also a conserved motif for calcium-independent phospholipase A2. We show that Tgl4p exhibits TAG lipase, steryl ester hydrolase, and phospholipase A2 activities, but surprisingly it also catalyzed the acyl-CoA-dependent acylation of lysophosphatidic acid to phosphatidic acid (PA). Heterologous overexpression of Tgl4p in Pichia pastoris increased total phospholipid and specifically PA synthesis. Moreover, deletion of TGL4 in Saccharomyces cerevisiae showed an altered pattern of phosphatidylcholine and PA molecular species. Altogether, our data suggest that yeast Tgl4p functions as a hydrolytic enzyme in lipid degradation but also contributes to fatty acid channeling and phospholipid remodeling.
Keywords: Enzymes/Lipid, Lipid, Lipid/Lipase, Lipid/Phospholipases, Lipid/Phospholipid/Metabolism, Lipid/Triacylglycerol, Membrane/Lipids, Organisms/Yeast
Introduction
Emerging evidence indicates that membrane phospholipids not only form passive hydrophobic barriers for cellular compartmentalization but also play important roles in other processes, e.g. as potent lipid second messengers and in receptor-mediated signaling, enzyme activation, and solute transport (1–3). Moreover, phospholipids serve as molecular chaperons (4, 5) and precursors for the synthesis of macromolecules (6–10). Many of these processes involve specific acyltransferases, which form phospholipids from lysophospholipids and fatty acids and thus play key roles in fatty acid metabolism and lipid biosynthesis (11).
Lysophospholipid acyltransferases and transacylases are also involved in the remodeling of phospholipids. In general, phospholipids are metabolically active and are subjected to deacylation-reacylation reactions, which are crucial to the maintenance of proper membrane fluidity, the formation of specific lipid domains, vesicle flux, and other important cellular processes (12–15). Lands (16) proposes that phospholipids contain saturated fatty acids in the sn-1 position, whereas polyunsaturated fatty acids are present in the sn-2 position. In mammalian cells, fatty acyl chains in the sn-2 position of the glycerol backbone can be removed by specific phospholipases A2 (PLA2),2 which leads to the formation of 1-acyl lysophospholipids. These intermediates are then reacylated by lysophospholipid acyltransferases in the course of remodeling processes (17, 18).
Among the phospholipids, phosphatidic acid (PA) plays a key role in lipid metabolism, because major phospholipids such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), and phosphatidylserine (PS) are mostly synthesized from PA via the CDP-DAG pathway (19–21). PA can be formed through different reactions involving lyso-PA acyltransferase, DAG kinase, and phospholipase D. In plants, PA is implicated in seed germination and the response to stress induced by drought, salinity, and low temperature. Also, in mammalian cells, PA plays key roles as an activator of cell growth and proliferation and in vesicular trafficking, secretion, and endocytosis (22–24). In Saccharomyces cerevisiae, the formation of PA by phospholipase D Pld1p/Spo14p is essential for the fusion of vesicles with the prospore membrane during sporogenesis (25, 26). In addition, PA is important for cytokinesis in budding yeast (27) and for microvesicle formation from the plasma membrane. Also, proper membrane curvature properties depend mostly on lyso-PA acyltransferases (28).
Another important phospholipid is PC, which contributes with its cylindrical shape to the structural integrity of membrane bilayers. Therefore, the remodeling of PC is also highly important in maintaining membrane properties. In mammalian cells, PLA2 plays a key role in generating lyso-PC and is involved in maintaining a balanced pattern of PC molecular species (17, 18). In the yeast, remodeling also determines the molecular species of PC and its function as a major constituent of membranes. In most cases, an unsaturated fatty acid is present in the sn-2 position of PC, whereas palmitate predominates in the sn-1 position (29). Two biosynthetic routes are involved in PC remodeling and maintaining PC molecular species with specific acyl chains at different ratios (30, 31).
This far, three yeast phospholipases B have been identified and characterized. These enzymes hydrolyze PC in both the sn-1 and sn-2 positions and thus produce mainly glycerophosphocholine and fatty acids (32–34). An sn-2 position-specific PLA2 has not yet been identified in S. cerevisiae. Recent investigations in our laboratory and by others (35–37), however, suggest that fatty acids from TAG may be channeled to phospholipid synthesis. Moreover, TAG lipases and/or related enzymes may be involved in phospholipid remodeling. In mammalian cells, a TAG lipase named ATGL (38), desnutrin (39), or calcium-independent phospholipase A2ζ (iPLA2ζ) has been identified independently by three different groups. This protein possesses two signature motifs, namely the iPLA2 ((G/A)XGXXG) and lipase (GXSXG) consensus sequences, and has been identified as a novel member of the iPLA2/lipase family (40). Later on, ATGL orthologs were identified and characterized in yeast (Tgl4p), plants (sugar-dependent-1), flies (Brummer lipase-4), and other vertebrates (41–46). In addition, two patatin domain-containing lipolytic enzymes from S. cerevisiae lipid particles, Tgl3p and Tgl5p, have been identified and partially characterized (47). Surprisingly, the function of these polypeptides in TAG hydrolysis in vivo and maintenance of TAG homeostasis appears to be conserved. However, the possible function of these enzymes as PLA2 has never been addressed.
TAG hydrolysis appears to be a major source of fatty acid and DAG, which is a precursor of membrane lipid biosynthesis, specifically for the generation of PA. It has been proposed that DAG formed by TAG lipases might be channeled to PE and PC synthesis via the Kennedy pathway (48–50). In the present work, biochemical studies of the yeast TAG lipase Tgl4p led us to more detailed in silico and enzyme analytical investigations of this protein. We show that Tgl4p harbors a PLA2-specific protein motif, (G/A)XGXXG. Moreover, we demonstrate that Tgl4p is not only a TAG/steryl ester (SE)- and phospholipid-hydrolyzing enzyme but also functions as a lysophosphatidic acid (LPA) acyltransferase. These surprising findings will be discussed in the context of the possible role of Tgl4p in lipid degradation, remodeling, and phospholipid homeostasis.
MATERIALS AND METHODS
Strains and Culture Conditions
Yeast strains used throughout this study are listed in Table 1. Cells were grown in either YPD (1% yeast extract, 2% bacto peptone, 2% glucose) or in synthetic minimal medium containing 0.67% yeast nitrogen base (Difco) supplemented with the appropriate amino acids and 2% glucose or 2% galactose. For heterologous expression of His6-tagged Tgl4p, Pichia pastoris cells were grown on buffered minimal methanol medium (BMM10) containing 1.34% yeast nitrogen base, 4 × 10−5 % biotin (Sigma), 5% methanol (Merck), and 200 mm potassium phosphate, pH 6.5.
TABLE 1.
Strains used in this study
| Genus/species | Strain | Genotype | Source |
|---|---|---|---|
| S. cerevisiae | BY4741, wild type | Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | EUROSCARF collection, Frankfurt, Germany |
| S. cerevisiae | tgl4Δ | MATa, ura3–52, trp1Δ63, leu2Δ1, his3Δ200, Δtgl4::kanMX4 | EUROSCARF collection, Frankfurt, Germany |
| P. pastoris | GS115, wild type | his4 | Invitrogen |
| P. pastoris | Overexpressing Tgl4p-His6 | GS115, TGL4-His6 tag at the N terminus | Ref. 41 |
| P. pastoris | Overexpressing mutant Tgl4p-His6 | GS115, TGL4T675A, S890A-His6 tag at the N terminus | This study |
Bioinformatics Analysis
The conserved protein domain of Tgl4p and its motifs were examined using the Conserved Domain Database at NCBI (http://www.ncbi.nih.gov/Structure/cdd) and the pfam database as described (51).
In Vivo Labeling of Phospholipids and Neutral Lipids
Tgl4p was expressed heterologously in P. pastoris under the AOX1 promoter as described previously (41). The Tgl4p-His6-overexpressing strain and its corresponding wild type, GS115, were precultured in 5 ml of YPD containing 2% glucose. For in vivo labeling, cells at an OD of 0.2 were inoculated in BMM10 medium and cultivated until growth reached the midlog phase. Then, cells were shifted to 1% methanol as a carbon source to induce the AOX promoter. At the time of induction, 0.5 μCi/ml [14C]acetate (specific activity, 51 mCi/mmol) was added, and cells were grown for an additional 12 h. Then, cells (OD = 10) were harvested by centrifugation, and lipids were extracted using chloroform/methanol (2:1 v/v). Individual phospholipids were separated by two-dimensional TLC (silica gel 60) using chloroform/methanol/25% ammonia (65:35:5 v/v) as a first developing solvent, and chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5 v/v) as solvent for the second dimension. Neutral lipids were separated by one-dimensional TLC using light petroleum:diethyl ether:acetic acid (70:30:1 v/v) as the solvent system.
Overexpression and Purification of Tgl4p Using Affinity Chromatography
For the isolation of His-tagged Tgl4p (41), cytosol was prepared from P. pastoris cells overexpressing Tgl4p. Cells were suspended in 50 mm Tris buffer, pH 7.5, 0.3 m sucrose, 1 mm 2-mercaptoethanol containing protease inhibitors, 0.1 mm phenylmethylsulfonyl fluoride, and 1 μg ml−1 leupeptin and disintegrated using glass beads. Unbroken cells and debris were removed by centrifugation at 3,000 × g for 10 min. The cell-free extract was centrifuged at 100,000 × g for 90 min to obtain cytosolic and total membrane fractions. The cytosolic fraction was subjected to nickel-nitrilotriacetic acid affinity column chromatography following the manufacturer's instructions, yielding Tgl4p preparation for further analyses.
SDS-PAGE and Western Blot Analysis
Protein samples were quantified by the method of Lowry et al. (52) using bovine serum albumin as a standard. SDS-PAGE was carried out by the method of Laemmli (53). For Western blot analysis proteins were separated by 12% SDS-PAGE, transferred to a Hybond ECL nitrocellulose membrane (Amersham Biosciences), and analyzed using standard procedures (54).
Enzyme Analyses
The affinity chromatography-purified Tgl4p was assayed for calcium-independent PLA2, lipase, and transacylase activities as described by Jenkins et al. (40). The PLA2 assay was carried out with purified fractions in 100 mm Tris-HCl, pH 7.5, containing 4 mm EGTA in a final volume of 200 μl for 30 min at 30 °C in the presence of 100 μm (1 μCi/assay) dipalmitoyl-2-[9,10-3H]glycero-3-phosphocholine (92.3 Ci/mmol) or [14C]phosphatidic acid (54.8 mCi/mmol) or 40,000 dpm of 32P-labeled PE or PS (produced by labeling the Escherichia coli cells and extracted lipids). Purified fractions were also assayed for TAG lipase and SE hydrolase activities using 100 mm Tris-HCl, pH 7.5, containing 2 mm EDTA and 1 mm dithiothreitol in the presence of 100 μm [9,10-3H]triolein (53 Ci/mmol) or [3H]cholesteryl-oleate (43 Ci/mmol), respectively, for 30 min at 30 °C as described previously (55, 56). All reactions were terminated by lipid extraction with 200 μl of butanol. Radiolabeled products were analyzed by TLC using chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5 v/v) for the PLA2 assay and light petroleum/diethyl ether/acetic acid (70:30:1 v/v) as a solvent system for the lipase and SE hydrolase assays. Spots were visualized by iodine staining, identified by standards, scraped off, and quantified by liquid scintillation counting using a Packard Tri-Carb 2900TR scintillation counter and LSC Safety Cocktail (Baker, Deventer, The Netherlands) with 5% water as a scintillation mixture.
Lysophospholipid acyltransferase activity was determined by measuring the incorporation of [14C]oleoyl-CoA into phospholipids in the presence of lysophospholipids as acyl acceptors. The reactions were performed in the presence of 100 mm Tris-HCl, pH 7.5, the respective lysophospholipid at 50 μm, and 25 μm [14C]oleoyl-CoA (58 mCi/mmol) at 30 °C with 0.2–1 μg of purified Tgl4p in a total volume of 100 μl for 15 min. The reaction was terminated by extracting lipids with 800 μl of chloroform/methanol/2% phosphoric acid (1:2:1 v/v). Subsequently, lipids were separated by one-dimensional TLC using chloroform/methanol/acetone/acetic acid/water (50:10:20:15:5 v/v) as the solvent system. Individual radioactive lipid spots were analyzed as described above.
Phospholipid Quantification
Lipids were extracted from yeast cells (57), and phospholipids were separated by two-dimensional thin-layer chromatography as described above. Lipids were stained with iodine vapor, spots were scraped off the plate, and lipids were quantified as described by Broekhuyse (58).
Electrospray Ionization Tandem Mass Spectrometry (ESI-MS/MS)
Yeast cells were grown in synthetic complete medium until A600 reached 1.0–2.0, harvested by centrifugation, and washed with sterile water. Lipids were extracted as described previously (59), and the chloroform-soluble fraction was dried, suspended in high pressure liquid chromatography-grade methanol, and subjected to ESI-MS/MS (Bruker Esquire 3000 Plus electrospray ion trap instrument; Bremen, Germany). Lipid samples (10 μl) were applied directly to the ESI source through a polytetrafluoroethylene line at a rate of 4 μl/min. The following ESI-MS/MS settings were used. The turboelectrospray ionization source was maintained at 260 °C, and data were collected in the positive and negative ion mode from 400 to 1200 m/z. The efficiency of lipid extraction was calculated to be 83–87%, and the procedure was monitored in every experiment. The efficiency of ionization was highly reproducible. Experiments were repeated three times yielding almost identical spectra.
Site-directed Mutagenesis
Plasmid pPIC 3.5K-TGL4 wild type was obtained as described previously by Athenstaedt and Daum (41). Plasmid pPIC 3.5K-T675A,S890A was constructed using the QuikChange polymerase chain reaction-based mutagenesis procedure (Stratagene) with the pPIC 3.5K-TGL4 wild type plasmid as template following the manufacturer's instructions. Primers used to construct site-directed mutants were as described previously (35).
RESULTS
Dual Signature Motifs in Tgl4p
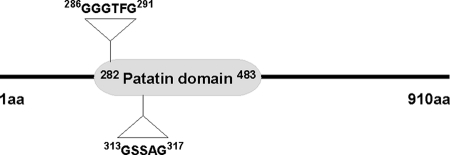
Previously the yeast protein Tgl4p had been reported as TAG lipase and an ortholog of the mammalian ATGL (see the Introduction). Those findings indicate that the TAG lipase activity of the two related enzymes was conserved from yeast to mammals. Tgl4p is a core patatin domain-containing protein and is categorized as a member of the α/β-hydrolase family. A more detailed search using the S. cerevisiae genome database (SGD), however, revealed that this polypeptide contains not only the typical lipase consensus sequence, GXSXG, but also a sequence motif, (G/A)XGXXG, that is characteristic of calcium-independent PLA2 proteins (Fig. 1). These in silico data tempted us to speculate that Tgl4p might also catalyze calcium-independent PLA2 activity and that this function might be conserved similarly to the lipase activity of this protein.
FIGURE 1.
Dual signature motifs in Tgl4p. The positions of conserved GXSXG lipase and GXGXXG calcium-independent phospholipase motifs in Tgl4p are indicated by triangles with the respective amino acid (aa) residues.
Tgl4p Catalyzes TAG Lipase, Steryl Ester Hydrolase, and Phospholipase A2 Reactions
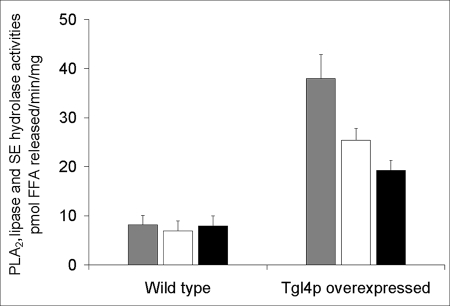
To test for hydrolytic activities of Tgl4p, we overexpressed N-terminal His6-tagged Tgl4p heterologously in P. pastoris under an AOX1 promoter. As described previously (41), the protein was localized to the cytosolic fraction and detected by Western blot analysis using an anti-His6 monoclonal antibody (supplemental Fig. S1). Using cytosolic fractions from the wild type and the Tgl4p overexpressed strain, enzymatic assays for TAG lipase, SE hydrolase, and calcium-independent PLA2 were carried out with [9,10-3H]triolein, [3H]cholesteryl oleate, and dipalmitoyl-2-[9,10-3H]glycero-3-phosphocholine as substrates. As shown in Fig. 2, TAG lipase, SE hydrolase, and PLA2 activities were detected in the cytosol from the Tgl4p-overexpressing strain, whereas the release of radiolabeled fatty acid from the respective substrates was marginal in wild type cytosol. The presence of Ca2+ and/or ATP, a known stabilizer and activator of calcium-independent PLA2, in the assay mixture did not affect the enzyme activity (data not shown).
FIGURE 2.
Tgl4p catalyzes hydrolysis of TAG, SE, and PC. TAG lipase, SE hydrolase, and PLA2 activities were measured in cytosol prepared from P. pastoris wild type and a TGL4-overexpressing strain. Gray bar, PLA2 activity; white bar, TAG lipase activity; black bar, SE hydrolase activity. Mean values from four independent measurements ± S.D. are shown.
For further investigation of PLA2 activity, the cytosolic fraction of the Tgl4p-overexpressing P. pastoris strain was subjected to affinity column chromatography, and Tgl4p was purified close to homogeneity as described previously (41). Enzyme activity assays performed with radiolabeled PC revealed that purified Tgl4p exhibited a PLA2 activity of 152.5 ± 8.5 pmol/min/mg protein. The release of [3H]palmitic acid was linear within 40 min and was protein-dependent to a maximum of 0.8 μg of protein under the given assay conditions. To ascertain the substrate preference of Tgl4p for various phospholipids, PLA2 activity was also measured using [14C]PA, [14C]PE, and [14C]PS. We found that Tgl4p hydrolyzed PE at an activity rate of 28.6 pmol/min/mg protein. PA and PS as substrates were not hydrolyzed. Because hydrolysis of the acyl bond in the sn-1 position may follow the release of the fatty acid from the sn-2 position, we also tested Tgl4p for lysophospholipase activity using 1-[9,10-3H]lysophosphatidic acid as a substrate. However, no such activity of Tgl4p was detectable (data not shown).
Tgl4p Preferentially Mediates Acyl-CoA-dependent Acylation of Lysophosphatidic Acid
Previous reports described the patatin motif of hydrolases as also catalyzing ATP- and acyl-CoA-independent transacylation reactions (60–62). For this reason, we also tested Tgl4p for possible ATP- and/or acyl-CoA-independent transfer of acyl moieties among TAG, diacylglycerol, and monoacylglycerol by sequential hydrolysis and transacylation reactions. However, Tgl4p did not catalyze any of these reactions (data not shown). But to our surprise, we found that Tgl4p acted as an acyl-CoA-dependent lysophospholipid acyltransferase. The substrate specificity of Tgl4p was tested with [1-14C]oleoyl-CoA as the acyl donor and various lysophospholipids such as lysophosphatidylcholine, lysophosphatidylethanolamine, lysophosphatidylinositol, lysophosphatidylserine, and lysophosphatidic acid as acyl acceptors. As shown in Fig. 3A, Tgl4p acylated LPA with high preference and formed PA as a product, whereas lysophosphatidylethanolamine, lysophosphatidylcholine, lysophosphatidylinositol, and lysophosphatidylserine were utilized with only marginal efficiency. The identity of PA as a product of the acylation reaction was confirmed by two-dimensional TLC (data not shown).
FIGURE 3.
Tgl4p mediates oleoyl-CoA-dependent acylation of lysophosphatidic acid. The acyl acceptor and donor specificity of Tgl4p were analyzed with the purified protein. A, lysophospholipid acyltransferase assays were carried out in the presence of 50 μm lysophospholipids (LPA, lysophosphatidic acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; LPS, lysophosphatidylserine; and LPI, lysophosphatidylinositol) and 25 μm [1-14C]oleoyl-CoA. B, acyl chain preference of Tgl4p. Acyl-CoAs with different chain length as indicated were used. NE, no enzyme; NS, no substrate. The data are derived from at least three independent experiments and are shown as mean values ± S.D. (error bars).
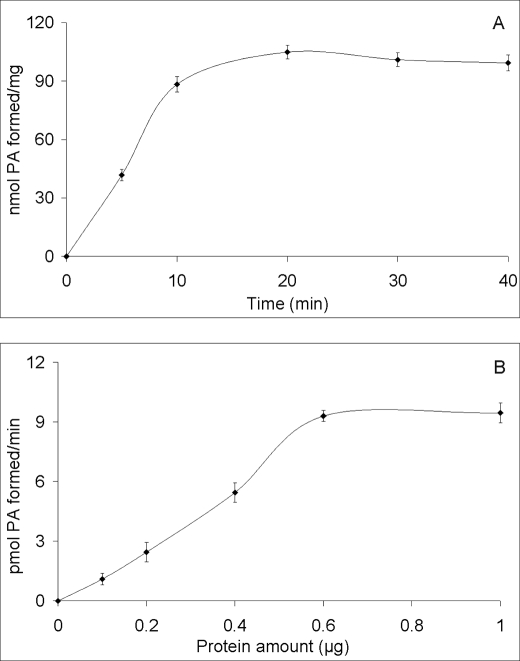
We then extended our studies to investigate the acyl donor preference for Tgl4p using [9,10-3H]lysophosphatidic acid (47 Ci/mmol) and different unlabeled acyl-CoAs as substrates (Fig. 3B). The highest activity was observed with oleoyl-CoA, whereas palmitoyl-CoA, stearoyl-CoA, arachidyl-CoA, and myristoyl-CoA were less efficient acyl donors under standard conditions. The Tgl4p-LPAAT reaction was linear within 10 min and reaching as high as 600 ng of protein in the assay mixture (Fig. 4).
FIGURE 4.
Tgl4p mediates time- and protein-dependent LPA acyltransferase activity. A, acyltransferase assays with 50 μm LPA and 25 μm [14C]oleoyl-CoA were performed for the indicated time periods. B, protein-dependent LPAAT activity of Tgl4p. The data represent the mean values of triplicate measurements ± S.D. (error bars).
Finally, we examined the kinetic properties of the LPA acyltransferase reaction with isolated Tgl4p. The apparent Km and Vmax values for oleoyl-CoA and LPA were calculated from Lineweaver-Burk plots. The Km and Vmax values of Tgl4p for oleoyl-CoA were 15.1 μm and 11.98 nmol/min/mg, respectively. The Km and Vmax values for 1-oleoyl-LPA were 14.3 μm and 11.4 nmol/min/mg, respectively. Moreover, we estimated the pH optimum for the LPAAT reaction at between 7.0 and 9.0. The addition of Mg2+ or Ca2+ did not affect the enzymatic activity. The Zwitterionic detergent CHAPS and the non-ionic detergent Triton X-100 inhibited Tgl4p-derived LPAAT activity at concentrations of 12 and 4 mm, respectively.
Phosphorylation of Tgl4p Is Dispensable for Acyltransferase Function
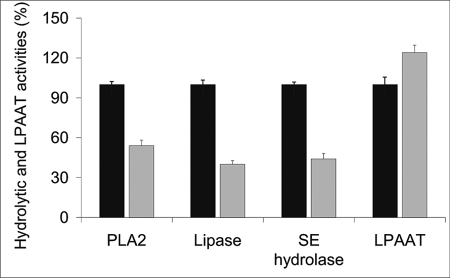
Recently, Kurat et al. (35) demonstrated that Tgl4p is phosphorylated and activated by the cyclin-dependent kinase 1 (Cdk1/Cdc28) in S. cerevisiae. The phosphorylated form of Tgl4p catalyzed lipolysis during cell cycle progression. To determine the effect of phosphorylation at Thr-675 and Ser-890 shown by these authors on hydrolytic and acyltransferase activities of Tgl4p, we constructed variants of Tgl4p by site-directed mutagenesis. To this end, Thr-675 and Ser-890 residues were replaced by alanine. Both the wild type and the mutated form (TGL4-T675A,S890A) of Tgl4p were overexpressed in P. pastoris and confirmed by Western blot analysis (supplemental Fig. S1). Biochemical analyses clearly indicated that substitution of amino acids as described above had a strong impact on the functions of Tgl4p (Fig. 5). We found that mutations of Thr-675 and Ser-890 resulted in a marked reduction of hydrolytic activities but did not affect the LPA acyltransferase activity of Tgl4p in vitro. These data clearly demonstrate that the hydrolytic and acyltransferase activities of Tgl4p act independently of each other.
FIGURE 5.
Evaluation of the mutation of Thr-675 and Ser-890 to alanine. The wild type and mutated form of Tgl4p (TGL4-T675A,S890A) were overexpressed in P. pastoris and analyzed for multiple functions of Tgl4p. Black bar, wild type Tgl4p; gray bar, mutated form of Tgl4p (TGL4-T675A,S890A). The mean values of three independent experiments ± S.D. are shown.
Impact of TGL4 Overexpression or Deletion on Phospholipid and Neutral Lipid Metabolism
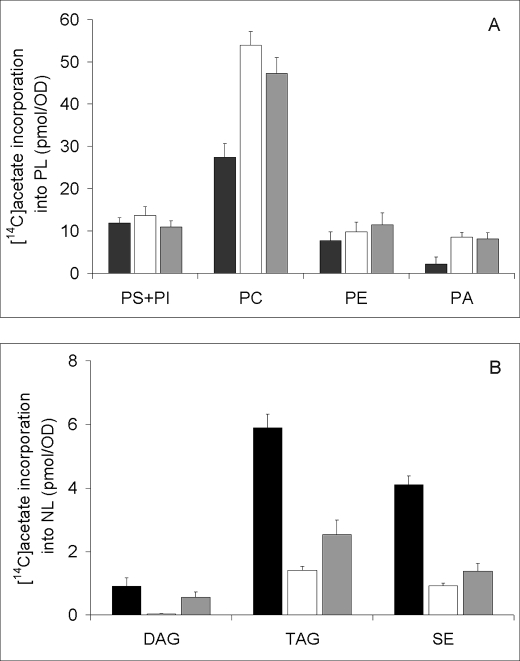
The different enzymatic functions of Tgl4p in vitro such as TAG lipase, SE hydrolase, PLA2, and LPA acyltransferase intrigued us so that we investigated the multifunctional role of this protein in lipid metabolism in greater detail. To this purpose, we analyzed the phospholipid profiles of wild type TGL4-His6 and His-tagged TGL4-T675A,S890A overexpressed in P. pastoris and incubated for 24 h in the presence of [14C]acetate. As shown in Fig. 6A, overexpression of both forms of TGL4 increased the total cellular phospholipid levels, especially the amounts of PC and PA. A slight increase in PE was also observed in these strains. The same results were obtained when phospholipids from these overexpressing strains were quantified chemically (data not shown). In contrast, overexpression of TGL4-His6 drastically decreased the amounts of TAG, DAG, and SE (Fig. 6B). Cells overexpressing His-tagged TGL4-T675A,S890A also showed a marked decrease of the three neutral lipids although not quite as pronounced as TGL4-His6. This finding is in line with the view that the mutated form of Tgl4p was less active than the wild type. We concluded from this experiment that enhanced TAG and SE hydrolysis led to an increased availability of fatty acids and an enhanced phospholipid production in the overexpressing strains. Acylation of LPA may specifically stimulate phospholipid biosynthesis.
FIGURE 6.
Heterologous expression of Tgl4p in P. pastoris affects the lipid composition. TGL4 and TGL4-T675A,S890A were overexpressed in P. pastoris as described under “Materials and Methods.” Lipids were labeled with [14C]acetate in an induction medium for 12 h. A, incorporation of [14C]acetate into individual phospholipids. black bar, wild type; white bar, TGL4-overexpressing strain; gray bar, mutated form of TGL4 (TGL4-T675A,S890A)-overexpressing strain. PI, phosphatidylinositol. B, neutral lipid profile of P. pastoris. Black bar, wild type GS115; white bar, TGL4-overexpressing strain; gray bar, mutated form of TGL4-overexpressing strain.
As Tgl4p functions both as PLA2 and LPAAT, and overexpression in P. pastoris enhanced the total phospholipid content, we speculated that Tgl4p might also affect the pattern(s) of phospholipid molecular species as a result of the remodeling process that appears to take place. To address this question, we employed ESI-MS/MS and compared the profile of phospholipid species in S. cerevisiae wild type and tgl4Δ. As shown in Table 2, the composition of fatty acid species in PC and PA from the tgl4Δ deletion mutant was different from the wild type. The tgl4Δ strain showed an increase of 32:2 species in PC, which was compensated by decreased levels of 32:1 and 34:1 species. In addition, the tgl4Δ mutant showed a strong increase of single unsaturated fatty acid (32:1) PA species. Interestingly, the molecular species of phosphatidylinositol and PE did not vary significantly between the wild type and tgl4Δ (data not shown), indicating that Tgl4p affects the fatty acid species composition of PC and PA in a rather specific way.
TABLE 2.
Molecular species of phosphatidylcholine and phosphatidic acid in S. cerevisiae wild type and tgl4Δ
| Phosphatidylcholine |
Phosphatidic acid |
||||
|---|---|---|---|---|---|
| Molecular species | Wild type | tgl4Δ | Molecular species | Wild type | tgl4Δ |
| 28:1 | 3.34 ± 0.6 | 1.60 ± 0.5 | 32:0 | 6.46 ± 1.2 | 6.68 ± 0.7 |
| 30:1 | 4.87 ± 0.54 | 3.80 ± 0.8 | 32:1 | 9.37 ± 2.3 | 17.53 ± 1.2 |
| 30:2 | 4.30 ± 0.8 | 1.90 ± 0.5 | 32:2 | 12.9 ± 2.1 | 11.52 ± 1.8 |
| 32:1 | 18.1 ± 2.2 | 15.3 ± 1.8 | 34:0 | 4.2 ± 0.85 | 5.01 ± 0.9 |
| 32:2 | 26.7 ± 2.5 | 39.9 ± 2.9 | 34:1 | 11.8 ± 1.5 | 10.02 ± 1.4 |
| 34:1 | 9.06 ± 1.4 | 4.75 ± 0.7 | 34:2 | 19.4 ± 2.4 | 13.35 ± 1.3 |
| 34:2 | 29.6 ± 2.4 | 30.4 ± 3.2 | 36:0 | 9.20 ± 1.6 | 11.68 ± 1.7 |
| 36:1 | 1.90 ± 0.62 | 1.33 ± 0.4 | 36:1 | 13.7 ± 2.1 | 14.22 ± 0.85 |
| 36:2 | 2.11 ± 0.63 | 1.01 ± 0.8 | 36:2 | 12.9 ± 2.3 | 10.01 ± 1.34 |
DISCUSSION
In yeast cells, three different TAG lipases, named Tgl3p, Tgl4p, and Tgl5p, have been identified thus far and partially characterized at the molecular level. Among these TAG lipases, Tgl4p has been identified as the functional ortholog of mammalian adipose triglyceride lipase (42). The yeast lipases were shown to catalyze the degradation of TAG with different levels of efficiency and to induce lipolysis, especially when cells enter the vegetative growth from the stationary phase. It has also been reported that these enzymes channel the released metabolites, fatty acids and DAG, toward membrane lipids synthesis (41, 63). More recently, Kurat et al. (35) showed that Tgl4p is phosphorylated and activated by the cyclin-dependent kinase 1 Cdk1p/Cdc28p. The phosphorylated form of Tgl4p activates lipolysis and supports early bud formation and cell cycle progression. Our results presented here are in line with these findings. The dephosphorylated mutant form of Tgl4p was strongly inhibited in its hydrolytic capacity but was unaffected as LPA acyltransferase in vitro and in vivo (Figs. 5 and 6). In contrast, the wild type form of Tgl4p exhibited both hydrolytic and LPA acyltransferase functions. Thus, it appears that the phosphorylated form of Tgl4p stimulates lipolysis and the dephosphorylated form of the enzyme to a certain extent promotes membrane lipid synthesis.
Although the role of Tgl4p as a TAG lipase has been reasonably well characterized, it appears that this protein acts rather as a multifunctional enzyme. It came as a surprise that Tgl4p also exhibited functions different from TAG lipase that had not been realized previously or even anticipated. In the present study, we have demonstrated that Tgl4p can also function as SE hydrolase, sn-2-specific PLA2, and acyl-CoA-dependent LPA acyltransferase. The discrepancy with previous reports (41, 47) appears to be because of the modified assay conditions used in this study (see “Materials and Methods”), which specifically improved substrate presentation to the hydrolytic enzyme. Notably, the additional enzyme activities may be critical for membrane lipid metabolism with TAG lipase, SE hydrolase, and PLA2 acting in catabolic processes, whereas LPA acyltransferase contributes to the anabolic branch of lipid biosynthesis. Similar to Tgl4p, the plant-soluble LPA acyltransferase (At4g24160) possess four motifs that are conserved across the plant species: a GXSXG lipase motif; a GXGXXG calcium-independent PLA2 motif; an HX4D motif; and V(X)3HGF, a probable lipid-binding motif (37). At4g24160 was shown to hydrolyze both TAG and phosphatidylcholine.
Recently, we also demonstrated that the yeast lipases Tgl3p and Tgl5p mediate acyl-CoA-dependent acylation of lysophosphatidylethanolamine and LPA, respectively. We showed that the lysophosphatidylethanolamine acyltransferase function of Tgl3p was essential for efficient yeast sporulation (55). It has to be noted, however, that Tgl3p and Tgl5p exhibited neither phospholipase nor steryl ester hydrolase activity. Thus, these two enzymes are clearly different from Tgl4p with respect to their enzymatic properties and most likely contribute differently to lipid metabolism.
The multiple enzymatic functions of Tgl4p raise a question as to the physiological role of this enzyme. An obvious explanation could be that Tgl4p (i) catalyzes TAG and SE turnover and provides a continuous pool of fatty acids by its hydrolytic activities; (ii) at the same time is involved in de novo phospholipid biosynthesis by its activity as LPA acyltransferase; and (iii) finally contributes to membrane lipid remodeling by PLA2 activity. Such roles may be in line with our findings that Tgl4p overexpression in P. pastoris resulted in: (i) an increased FFA content because of enhanced TAG, DAG, and SE hydrolysis (see Fig. 6); and (ii) an increased phospholipid level due to channeling of the released FFA toward membrane lipid biosynthesis or acylation of LPA to PA. In mammalian cells, it was shown that adiponutrin/ATGL was up-regulated when oleate was fed, indicating that it may also function as transacylase to channel FFA to TAG, thereby effectively preventing the cell from lipotoxicity (40). Likewise, yeast Tgl4p may mediate acyl-CoA-dependent acylation to deviate released or excess FFA toward phospholipid synthesis. This channeling, however, does not occur by transacylation, because Tgl4p preferentially acylates LPA in an acyl-CoA-dependent reaction to produce PA as a general precursor for phospholipids. Finally, our ESI-MS/MS analyses (see Table 2) support the view that Tgl4p may also contribute to phospholipid remodeling. Both the PLA2 and LPA acyltransferase functions of Tgl4p appear to be important for the maintenance of the appropriate ratios of different molecular phospholipid species in vivo, especially that of PC and PA.
The hypothesis that Tgl4p acts as a multifunctional enzyme in general yeast lipid metabolism as outlined above, does not, however, take into account the subcellular localization of this polypeptide in lipid particles/droplets. As most other lipid particle proteins, Tgl4p lacks transmembrane-spanning domains and may thus be assumed to reside in the phospholipid monolayer of the lipid particle surface (64). This topology raises the question how this lipase gets access to the substrate TAG that is a component of the highly hydrophobic core of lipid particles. Recently, we were able to present a more detailed model of the lipid particle interior (65). We showed that the SE of lipid particles form a number of ordered shells below the surface phospholipid monolayer surrounding a random inner core of TAG. Although the protrusion of TAG molecules through the SE shells may occur, it is difficult to understand how TAG lipases get access to their substrates through the phospholipid monolayer membrane. The multiple functions of Tgl4p may support such a process by poking holes into the phospholipid monolayer through the PLA2 activity and softening SE shells by its SE hydrolase activity. Once the TAG substrate has been utilized by the lipase function, the acyltransferase function of the enzyme may remodel phospholipids and contribute to the “healing” of the surface monolayer. This hypothesis, however, does not exclude the possibility that substantial amounts of PA or other phospholipids formed through the recycling and channeling functions of Tgl4p may be transported to other subcellular sites.
In summary, our work presented here demonstrates the multifunctional role of Tgl4p as TAG lipase, SE hydrolase, PLA2, and LPA acyltransferase. Consequently, this enzyme serves both the catabolic and anabolic pathways of lipid metabolism in the yeast. As usual, some questions remain open. One of these questions is whether Tgl4p occurs exclusively in lipid particles. It has been shown previously that other lipid particle proteins are also found in the endoplasmic reticulum, although in some cases only in marginal amounts. Nevertheless, it cannot be excluded that lipid particles are not the only subcellular site at which Tgl4p is active and contributes to lipid metabolism. Another open question is how the multiple functions of Tgl4p are regulated and linked to neutral lipid/phospholipid-synthesizing and -degrading routes. Finally, we have to take into account that the multiple functions of Tgl4p may also affect other cellular processes that have escaped our attention thus far. Identifying the genetic and molecular interaction partners of TGL4 and its gene product may contribute to a better understanding of these open questions.
Supplementary Material
Acknowledgments
We thank Dr. Karin Athenstaedt and Prof. Ram Rajasekharan for providing yeast strains and antibodies for this study and for helpful discussions.
This work was supported by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich, Projects 18857 and W901-B05 (to G. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PLA2
- phospholipase A2
- iPLA2
- independent PLA2
- DAG
- diacylglycerol
- LPA
- lysophosphatidic acid
- LPAAT
- lysophosphatidic acid acyltransferase
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- SE
- steryl ester
- TAG
- triacylglycerol
- ATGL
- adipose triacylglycerol lipase
- ESI
- electrospray ionization
- MS/MS
- tandem mass spectrometry
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Munnik T. (2001) Trends Plant Sci. 6, 227–233 [DOI] [PubMed] [Google Scholar]
- 2.Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., Ohsumi Y. (2000) Nature 408, 488–492 [DOI] [PubMed] [Google Scholar]
- 3.Iwanyshyn W. M., Han G. S., Carman G. M. (2004) J. Biol. Chem. 279, 21976–21983 [DOI] [PubMed] [Google Scholar]
- 4.Bogdanov M., Dowhan W. (1999) J. Biol. Chem. 274, 36827–36830 [DOI] [PubMed] [Google Scholar]
- 5.Bogdanov M., Sun J., Kaback H. R., Dowhan W. (1996) J. Biol. Chem. 271, 11615–11618 [DOI] [PubMed] [Google Scholar]
- 6.Becker G. W., Lester R. L. (1980) J. Bacteriol. 142, 747–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menon A. K., Stevens V. L. (1992) J. Biol. Chem. 267, 15277–15280 [PubMed] [Google Scholar]
- 8.Lester R. L., Dickson R. C. (1993) Adv. Lipid Res. 26, 253–274 [PubMed] [Google Scholar]
- 9.Fankhauser C., Homans S. W., Thomas-Oates J. E., McConville M. J., Desponds C., Conzelmann A., Ferguson M. A. (1993) J. Biol. Chem. 268, 26365–26374 [PubMed] [Google Scholar]
- 10.Bishop R. E., Gibbons H. S., Guina T., Trent M. S., Miller S. I., Raetz C. R. (2000) EMBO J. 19, 5071–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stymne S., Stobart A. K. (1987) The Biochemistry of Plants: A Comprehensive Treatise (Stumpf P. K. ed) Vol. 10, Chapter 8, pp. 175–214, Academic Press, New York [Google Scholar]
- 12.Lands W. E. (1958) J. Biol. Chem. 231, 883–888 [PubMed] [Google Scholar]
- 13.Holub B. J., Kuksis A. (1978) Lipid Res. 16, 1–125 [DOI] [PubMed] [Google Scholar]
- 14.Irvine R. F., Dawson R. M. (1979) Biochem. Biophys. Res. Commun. 91, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 15.Inoue M., Murase S., Okuyama H. (1984) Arch. Biochem. Biophys. 231, 29–37 [DOI] [PubMed] [Google Scholar]
- 16.Lands W. E. (1960) J. Biol. Chem. 235, 2233–2237 [PubMed] [Google Scholar]
- 17.Lands W. E., Merkl I. (1963) J. Biol. Chem. 238, 898–904 [PubMed] [Google Scholar]
- 18.Lands W. E., Inoue M., Sugiura Y., Okuyama H. (1982) J. Biol. Chem. 257, 14968–14972 [PubMed] [Google Scholar]
- 19.Carman G. M., Henry S. A. (1999) Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 20.Gaspar M. L., Aregullin M. A., Jesch S. A., Nunez L. R., Villa-García M., Henry S. A. (2007) Biochim. Biophys. Acta 1771, 241–254 [DOI] [PubMed] [Google Scholar]
- 21.Carman G. M., Han G. S. (2009) J. Lipid Res. 50, S69–S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Testerink C., Munnik T. (2005) Trends Plant Sci. 10, 368–375 [DOI] [PubMed] [Google Scholar]
- 23.Wang X., Devaiah S. P., Zhang W., Welti R. (2006) Prog. Lipid Res. 45, 250–278 [DOI] [PubMed] [Google Scholar]
- 24.Carman G. M., Henry S. A. (2007) J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudge S. A., Sciorra V. A., Iwamoto M., Zhou C., Strahl T., Morris A. J., Thorner J., Engebrecht J. (2004) Mol. Biol. Cell 15, 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi H., Morishita M., Schwartz C. L., Coluccio A., Engebrecht J., Neiman A. M. (2006) J. Cell Sci. 119, 1406–1415 [DOI] [PubMed] [Google Scholar]
- 27.Xie Z., Fang M., Bankaitis V. A. (2001) Mol. Biol. Cell 12, 1117–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt A., Wolde M., Thiele C., Fest W., Kratzin H., Podtelejnikov A. V., Witke W., Huttner W. B., Söling H. D. (1999) Nature 401, 133–141 [DOI] [PubMed] [Google Scholar]
- 29.Wagner S., Paltauf F. (1994) Yeast 10, 1429–1437 [DOI] [PubMed] [Google Scholar]
- 30.Boumann H. A., Gubbens J., Koorengevel M. C., Oh C. S., Martin C. E., Heck A. J., Patton-Vogt J., Henry S. A., de Kruijff B., de Kroon A. I. (2006) Mol. Biol. Cell 17, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Kroon A. I. (2007) Biochim. Biophys. Acta 1771, 343–352 [DOI] [PubMed] [Google Scholar]
- 32.Lee K. S., Patton J. L., Fido M., Hines L. K., Kohlwein S. D., Paltauf F., Henry S. A., Levin D. E. (1994) J. Biol. Chem. 269, 19725–19730 [PubMed] [Google Scholar]
- 33.Merkel O., Fido M., Mayr J. A., Prüger H., Raab F., Zandonella G., Kohlwein S. D., Paltauf F. (1999) J. Biol. Chem. 274, 28121–28127 [DOI] [PubMed] [Google Scholar]
- 34.Fyrst H., Oskouian B., Kuypers F. A., Saba J. D. (1999) Biochemistry 38, 5864–5871 [DOI] [PubMed] [Google Scholar]
- 35.Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., Kohlwein S. D. (2009) Mol. Cell. 33, 53–63 [DOI] [PubMed] [Google Scholar]
- 36.Wagner A., Grillitsch K., Leitner E., Daum G. (2009) Biochim. Biophys. Acta 1791, 118–124 [DOI] [PubMed] [Google Scholar]
- 37.Ghosh A. K., Chauhan N., Rajakumari S., Daum G., Rajasekharan R. (2009) Plant Physiol. 151, 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zechner R., Kienesberger P. C., Haemmerle G., Zimmermann R., Lass A. (2009) J. Lipid Res. 50, 3–21 [DOI] [PubMed] [Google Scholar]
- 39.Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. (2004) J. Biol. Chem. 279, 47066–47075 [DOI] [PubMed] [Google Scholar]
- 40.Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. (2004) J. Biol. Chem. 279, 48968–48975 [DOI] [PubMed] [Google Scholar]
- 41.Athenstaedt K., Daum G. (2005) J. Biol. Chem. 280, 37301–37309 [DOI] [PubMed] [Google Scholar]
- 42.Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S. D. (2006) J. Biol. Chem. 281, 491–500 [DOI] [PubMed] [Google Scholar]
- 43.Eastmond P. J. (2006) Plant Cell 18, 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grönke S., Mildner A., Fellert S., Tennagels N., Petry S., Müller G., Jäckle H., Kühnlein R. P. (2005) Cell Metab. 1, 323–330 [DOI] [PubMed] [Google Scholar]
- 45.Shan T., Wang Y., Wu T., Guo J., Liu J., Feng J., Xu Z. (2008) J. Anim. Sci. 86, 1781–1788 [DOI] [PubMed] [Google Scholar]
- 46.Saarela J., Jung G., Hermann M., Nimpf J., Schneider W. J. (2008) BMC Genomics 9, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Athenstaedt K., Daum G. (2003) J. Biol. Chem. 278, 23317–23323 [DOI] [PubMed] [Google Scholar]
- 48.Igal R. A., Coleman R. A. (1996) J. Biol. Chem. 271, 16644–16651 [DOI] [PubMed] [Google Scholar]
- 49.Cook H. W., Spence M. W. (1985) Can. J. Biochem. Cell Biol. 63, 919–926 [DOI] [PubMed] [Google Scholar]
- 50.Caviglia J. M., de Gómez Dumm I. N., Coleman R. A., Igal R. A. (2004) J. Lipid Res. 45, 1500–1509 [DOI] [PubMed] [Google Scholar]
- 51.Bateman A., Birney E., Durbin R., Eddy S. R., Howe K. L., Sonnhammer E. L. (2000) Nucleic Acids Res. 28, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 53.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 54.Haid A., Suissa M. (1983) Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 55.Rajakumari S., Daum G. (2010) Mol. Biol. Cell 21, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Müllner H., Deutsch G., Leitner E., Ingolic E., Daum G. (2005). J. Biol. Chem. 280, 13321–133218 [DOI] [PubMed] [Google Scholar]
- 57.Folch J., Lees M., Sloane-Stanley G. H. (1957) J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 58.Broekhuyse R. M. (1968) Biochim. Biophys. Acta 152, 307–315 [DOI] [PubMed] [Google Scholar]
- 59.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh M., Tucker D. E., Burchett S. A., Leslie C. C. (2006) Prog. Lipid Res. 45, 487–510 [DOI] [PubMed] [Google Scholar]
- 61.Gao J. G., Shih A., Gruber R., Schmuth M., Simon M. (2009) Mol. Genet. Metab. 96, 253–260 [DOI] [PubMed] [Google Scholar]
- 62.Pinsirodom P., Parkin K. L. (2000) J. Agric. Food Chem. 48, 155–160 [DOI] [PubMed] [Google Scholar]
- 63.Daum G., Paltauf F. (1980) Monatsh. Chem. 111, 355–363 [Google Scholar]
- 64.Rajakumari S., Grillitsch K., Daum G. (2008) Prog. Lipid Res. 47, 157–171 [DOI] [PubMed] [Google Scholar]
- 65.Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., Daum G. (2008) J. Biol. Chem. 283, 17065–17074 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.