Abstract
Recent studies have implicated Epac2, a guanine-nucleotide exchange factor for the Rap subfamily of monomeric G proteins, as an important regulator of insulin secretion from pancreatic β-cells. Although the Epac proteins were originally identified as cAMP-responsive activators of Rap1 GTPases, the role of Rap1 in β-cell biology has not yet been defined. In this study, we examined the direct effects of Rap1 signaling on β-cell biology. Using the Ins-1 rat insulinoma line, we demonstrate that activated Rap1A, but not related monomeric G proteins, promotes ribosomal protein S6 phosphorylation. Using isolated rat islets, we show that this signaling event is rapamycin-sensitive, indicating that it is mediated by the mammalian target of rapamycin complex 1-p70 S6 kinase pathway, a known growth regulatory pathway. This newly defined β-cell signaling pathway acts downstream of cAMP, in parallel with the stimulation of cAMP-dependent protein kinase, to drive ribosomal protein S6 phosphorylation. Activated Rap1A promotes glucose-stimulated insulin secretion, islet cell hypertrophy, and islet cell proliferation, the latter exclusively through mammalian target of rapamycin complex 1, suggesting that Rap1 is an important regulator of β-cell function. This newly defined signaling pathway may yield unique targets for the treatment of β-cell dysfunction in diabetes.
Keywords: Diseases/Diabetes, G Proteins/Low Molecular Weight, Phosphorylation/Serine/Threonine, Signal Transduction/Cyclic Nucleotides/Cyclic AMP, Signal Transduction/G proteins, Tissue/Organ Systems/Pancreatic Islet, Rap1, mTOR Complex 1
Introduction
Both types of diabetes mellitus (1 and 2) involve a failure of the β-cells of the islets of Langerhans to produce the appropriate amount of insulin to properly regulate blood glucose levels. Type 1 diabetes is an autoimmune disease that results from the destruction of the β-cells. Islet transplantation has been studied as a long term treatment for type 1 diabetes, but the results thus far have been disappointing because of a high graft failure rate, necessitating retransplants with an already scarce resource (1). Type 2 diabetes is a multifactorial disease that is characterized by varying degrees of peripheral insulin resistance and insufficient insulin production. Early in the pathogenesis of the disease, the β-cells of the pancreas are able to compensate for peripheral insulin resistance via increased insulin secretion from individual β-cells and an overall increase in β-cell size and number (i.e. β-cell mass). It is these compensatory measures that slow the development of type 2 diabetes, and it is their failure that leads to hyperglycemia (2). As such, the pathways regulating these compensatory mechanisms have been widely studied and have become targets for the treatment of type 2 diabetes (reviewed in Refs. 3 and 4).
At the center of many of these pathways is the second messenger, cAMP (5). It was long thought that cAMP acted exclusively through the cAMP-dependent protein kinase (PKA)6; however, it is now clear that there exist other important intracellular targets for cAMP. Principal among these are the Epac proteins, cAMP-regulated guanine nucleotide exchange factors (6, 7) whose primary function is believed to be the cAMP-induced activation of the Ras-like monomeric G protein Rap1. However, recent studies have suggested that Epac proteins can engage other downstream targets directly to act independently of Rap1 (8, 9).
The role of Epac proteins in β-cell biology has been studied extensively. Early experiments demonstrated that Epac proteins are required for cAMP-stimulated intracellular Ca2+ release (10, 11) and insulin secretion (12, 13). More recently, studies have demonstrated roles for Epac proteins in promoting resistance to fatty acid-induced apoptosis (14) and as the target for antidiabetic sulfonylurea drugs (15). Interestingly, the direct role of Rap in these Epac-mediated processes has not been examined. Moreover, in some of these reports, the effects of Epac were attributed to Rap-independent pathways (12, 13). Thus, the current study was initiated to elucidate the role of Rap signaling in pancreatic β-cells by examining the Rap signaling pathway directly. We confirm that activated Rap1A regulates insulin secretion but also demonstrate that it increases β-cell size and proliferation, the latter exclusively through a stimulation of mTORC1. This newly described pathway might reveal interesting targets for the treatment or prevention of β-cell dysfunction in islet transplantation and type 2 diabetes.
EXPERIMENTAL PROCEDURES
Cell Lines, Reagents, and Antibodies
The Ins-1 cell line (16) was maintained in complete RPMI 1640 medium (11.1 mm glucose) supplemented with 10% fetal bovine serum, 10 mm HEPES, 2 mm glutamine, 1 mm sodium pyruvate, and 50 μm β-mercaptoethanol at 37 °C in a humidified atmosphere containing 95% air and 5% CO2. The cells were passaged 1:4 every 3 days. For immunoblot analysis, the antibodies employed were to Rap1 (Santa Cruz Biotechnology, Santa Cruz, CA; and Cell Signaling Technology, Beverly, MA), to Rap1GAP (EMD/Merck KGaA, San Diego, CA), and to β-actin (Sigma-Aldrich). The monoclonal anti-hemagglutinin and anti-Myc antibodies were obtained from Zymed Laboratories Inc. (San Francisco, CA). Rheb, Phospho-Akt (Ser473), phospho-RPS6 (Ser235/236), and total RPS6 antibodies were from Cell Signaling Technology, and the total Akt antibody was from BD Pharmingen. LI-COR Odyssey®-compatible secondary antibodies were from Rockland (Gilbertsville, PA) and Molecular Probes (Eugene, OR). 8-CPT-cAMP was from Biomol (Plymouth Meeting, PA). H89, epidermal growth factor, LY294002, myristoylated 14-22 amide, and rapamycin were from Calbiochem (La Jolla, CA).
Plasmids and Constructs
pCDNA3.1 plasmids containing human cDNAs for Rap1A, Rac1-G12V, and Myc-CDC42-G12V were obtained from the UMR cDNA Resource (Rolla, MO). The Rap1A-63E and Rap1GAP constructs were gifts of Lawrence Quilliam (Indiana University School of Medicine). The pAdtrack-CMV vector was a gift of Bert Vogelstein (Johns Hopkins University Medical Center). Epac1 and Epac2 adenoviruses were purchased from Vector Biolabs (Philadelphia, PA). siRNA specific for Rap1A and a scrambled control were purchased from Integrated DNA Technologies (Coralville, IA) according to published sequences (17). siRNA specific for Rheb was purchased from Ambion (Austin, TX), and the luciferase control siRNA sequence was purchased from Dharmacon (Layfayette, CO).
Construction of Recombinant Adenoviruses
Recombinant adenoviruses were constructed by subcloning the cDNAs for Rap1A, Rac-G12V, Myc-CDC42-G12V, Rap1A-63E, and Rap1GAP into the pAdtrack-CMV vector and then recombining them with pAdEasy-1 in BJ5183 Escherichia coli (Stratagene). The resulting DNA was transfected into HEK 293 cells with Lipofectamine (Invitrogen). Next, the viruses were serially amplified and purified using Adeno-XTM virus purification kits (BD Biosciences). Purified adenoviral stocks were tested by reverse transcription-PCR assay against the Ad5 E1A gene to ensure that they were free of contamination by wild-type adenovirus. Ins-1 cells were treated at a multiplicity of infection of 10 for 2 h at 37 °C.
Immunoblot Analysis
Ins-1 cells, grown to ∼80–90% confluence, were washed once with cold PBS and lysed with cold lysis buffer (described under “Rap1 Activity Assays”). The lysates were cleared by centrifugation at 14,000 × g for 10 min at 4 °C. The supernatants were assayed for protein concentration (Bio-Rad), and then equal amounts of total protein from lysates were separated by SDS-PAGE and subjected to immunoblot analysis according to standard methods. Western blotting was performed using the Odyssey System (LI-COR, Lincoln, NE) according to the manufacturer's instructions.
Phospho-protein Immunoblot Analysis
One million Ins-1 cells were plated in 6-well plates and allowed to grow for 2 days. The cells were then infected with the indicated adenovirus as described above and then starved for 18 h in starvation medium (RPMI 1640 minus glucose supplemented with 0.1% fatty acid-free bovine serum albumin, 5.6 mm glucose, 10 mm HEPES, 10 mm sodium pyruvate, and 50 μm β-mercaptoethanol). The cells were then stimulated with the indicated agonists, washed once with ice-cold PBS, and lysed with ice-cold buffer (50 mm Tris, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mm EDTA, 50 mm NaF, 150 mm NaCl, 2 mm dithiothreitol, 0.2 mm activated sodium vanadate, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 3 μg/ml leupeptin, 4 μg/ml aprotinin, 30 μm tosylphenylalanyl chloromethyl ketone, 29 μm Nα-p-tosyl-l-lysine chloromethyl ketone, and 133 mm phenylmethylsulfonyl fluoride). The lysates were cleared by centrifugation at 16,000 × g for 10 min at 4 °C. Supernatants were assayed for protein concentration, and then equal amounts of total protein from lysates were separated by SDS-PAGE and subjected to immunoblot analysis according to standard methods.
For siRNA transfection experiments, 2 million Ins-1 cells were plated in 6-well plates and allowed to adhere overnight. The cells were transfected with siRNA specific for Rap1A (R1A) or a scrambled control (17) using Dharmafect 1 transfection reagent according to the manufacturer's protocol (Dharmacon). After 48 h in transfection medium, the cells were infected with Epac1 and 2 viruses (multiplicity of infection of 5 each) or Rap1(63E) (multiplicity of infection of 10) for 2 h. The medium was then changed to starvation medium for 18 h, and the cells were lysed and subjected to immunoblotting. The immunoblot shown is representative of three independent experiments.
Preparation of GST Fusion Protein
A bacterial expression plasmid encoding a GST-tagged version of the RalGDS protein (GST-RalGDS) was transformed into BL21(DE3) E. coli. The cells were grown in Luria broth at 37 °C for 2–3 h until the optical density reached 0.5–0.6 and induced with 0.5 mm isopropyl-d-thiogalactopyranoside (Teknova). The cultures were grown for an additional 2.5 h at 37 °C. The cells were harvested by centrifugation for 15 min at 6,000 × g at 4 °C, and the resulting pellet resuspended in 2.5 ml of buffer A (2.3 m sucrose, 50 mm Tris-HCl, pH 7.7, 1 mm EDTA, and 1:500 protease inhibitor mix: 23 μg/ml phenylmethylsulfonyl fluoride, 11 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone, 11 μg/ml 1-chloro-3-tosylamido-7-amino-2-heptanone, 2 μg/ml leupeptin, and 2 μg/ml aprotinin) followed by dilution with 10 ml of buffer B (50 mm Tris-HCl 7.7, 10 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, and 1:500 protease inhibitor mix). The cells were then passed three times at 10,000 p.s.i. through a microfluidizer (Microfluidics Corporation, Newton, MA). The lysates were cleared by centrifugation at 30,000 × g for 30 min, and the resulting supernatant was incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) equilibrated in buffer B for 2 h at 4 °C with continuous rocking. Protein concentration was determined by Bradford assay (Bio-Rad).
Rap1 Activity Assays
Ins-1 cells were seeded at a density of 4 × 106 in 10-cm dishes and allowed to grow for ∼48 h. The cells were starved for 18 h in starvation medium. The cells were then stimulated with 8CPT-cAMP (100 μm) for the indicated times, washed once with ice-cold PBS, and lysed in a buffer containing 50 mm Tris, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 10 mm NaF, 1 mm EDTA, 10 mm MgCl2, 5% glycerol, 10 μg/ml leupeptin, and 10 μg/ml aprotinin. Rap1 activity assays were performed essentially as in Ref. 18.
Rat Islet Isolation and in Vitro Culture of Rat Islets
As described previously (19), pancreatic islets were harvested from male Sprague-Dawley rats weighing ∼250 g under a protocol approved by the Duke University Institutional Animal Care and Use Committee. Approximately 200 rat islets/condition were cultured in 2 ml of RPMI medium (containing 10% fetal calf serum and 8 mm glucose) and treated with adenoviruses at a concentration of 5 × 109 particles/ml of medium for 18 h. Virus-containing medium was replaced with fresh culture medium, and islets were cultured for various times after treatment, as indicated in the figure legends, with fresh medium daily.
Islet Insulin Secretion Assays
The day before the assay, pools of ∼200 islets were treated with various recombinant adenoviruses as described above and then incubated in complete RPMI medium with and without rapamycin (25 nm) overnight. The islets were preincubated with 2.8 mm glucose-containing Krebs-Ringer bicarbonate buffer (4.38 mm KCl, 1.2 mm MgSO4, 1.5 mm KH2PO4, 129 mm NaCl, 5 mm NaHCO3, 10 mm HEPES, 3.11 mm CaCl2, 0.25% bovine serum albumin, pH 7.4) for 2 × 30 min. Next, 25 islets/replicate were picked into Krebs-Ringer bicarbonate buffer containing the desired concentration of glucose for 1.5 h. Following stimulation, the secretion medium was removed and assayed for insulin levels (Coat-A-Count® [125I]insulin radioimmunoassay; Siemens Medical Solutions, Malvern, PA). The remaining islets or cells were washed once in PBS and lysed in radioimmune precipitation assay buffer (Sigma), and the amount of secreted insulin was normalized to the total protein of the extract as measured by BCA protein assay (Thermo Scientific, Rockford, IL).
Microscopy
Groups of ∼200 rat islets were treated with various recombinant adenoviruses and treated with and without rapamycin, and 5–10 islets were hand-picked per replicate. The islets were fixed with 2% paraformaldehyde and placed in warm agarose on tissue culture dishes. Fixed islet cell nuclei were Hoechst-stained. The entire islet was analyzed, and z-stack images were captured using an SP5 inverted confocal microscope (Leica) at the Duke University Light Microscopy Core Facility. Three nonsequential slices (z-stack)/islet were chosen for analysis. The number of nuclei in a measured area was counted using Metamorph software. The data represent the average number of nuclei counted from four areas/image slice with three image slices/islet (n = 4–6 islets/condition).
Proliferation Assays
DNA synthesis rates were measured as described previously (20). Briefly, [methyl-3H]thymidine was added at a final concentration of 1 μCi/ml to pools of ∼200 islets during the last 18 h of cell culture. Groups of 25 islets were picked in triplicate, washed, and centrifuged twice at 300 × g for 3 min at 4 °C. DNA was precipitated with 500 μl of cold 10% trichloroacetic acid for 30 min and then solubilized and resuspended by the addition of 80–100 μl of 0.3 n NaOH. The amount of [3H]thymidine incorporated into DNA was measured by liquid scintillation counting and normalized to total cellular protein (BCA protein assay; Thermo Scientific).
Statistical Analyses
The data were analyzed using GraphPad Prism v4 (GraphPad Software Inc., San Diego, CA). The data are given as the mean ± S.E. and compared by t test or by two-way analysis of variance followed by Bonferroni's post-test, as appropriate, to determine p values. A probability of p < 0.05 was considered significant.
RESULTS
Rap1 Stimulates the Phosphorylation of RPS6 in an Activation-dependent Manner through mTORC1
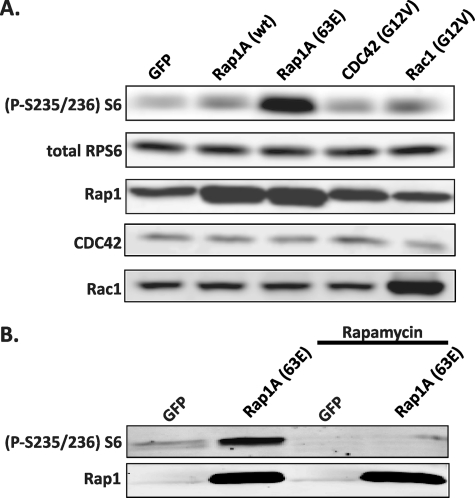
To gain insight into the function of Rap1 in the pancreatic β-cell, we used recombinant adenovirus to express a dominant-active mutant of Rap1A (Rap1A-63E) in the Ins-1 rat insulinoma cell line. After treatment with recombinant adenovirus and starvation, the cells were lysed, and the lysates immunoblotted against a panel of phosphoproteins involved in growth regulatory pathways (Alliance for Cellular Signaling solution protocols PS00000560 and PS00000561; data not shown); a striking change was consistently seen in the phosphorylation status of RPS6 with Rap1A-63E expression (Fig. 1A). This change in RPS6 phosphorylation was dependent on Rap1 activation, because expression of the wild-type form of Rap1A resulted in only a modest increase in RPS6 phosphorylation (Fig. 1A). Moreover, this effect was specific to Rap1A over other GTPases linked previously to RPS6 phosphorylation. To elaborate, both the related Rac1 and CDC42 proteins have been shown to promote phosphorylation of RPS6 (21), but the activated forms of these proteins (Rac1-G12V and CDC42-G12V, respectively) induced little to no increase in the phosphorylation of RPS6 in the Ins-1 cell line (Fig. 1A).
FIGURE 1.
Activated Rap1A stimulates the phosphorylation of RPS6 in an activation-dependent manner through mTORC1. A, Ins-1 cells were transduced with adenovirus expressing the indicated proteins and then serum- and glucose-starved into quiescence. The cells were then lysed, and equal amounts of protein were analyzed by immunoblotting for phosphorylated RPS6. The figure is representative of four independent experiments. B, rat pancreatic islets were transduced with adenovirus expressing the indicated proteins and then treated with or without rapamycin. Islets were then lysed, and equal amounts of protein were analyzed by immunoblotting for phosphorylated RPS6 using a specific antibody. The figure is representative of three independent experiments.
Because expression of activated Rap1A induced an increase in RPS6 phosphorylation in a β-cell-derived line, we sought to determine whether activated Rap1A also promoted RPS6 phosphorylation in primary pancreatic islets. Expression of Rap1A-63E in isolated rat islets induced a strong phosphorylation of RPS6, and this activation was completely blocked by rapamycin treatment (Fig. 1B). Rapamycin binds to its intracellular receptor, FKBP12, which interacts directly with mTOR, specifically blocking signaling through mTORC1, which is a known regulator of cell growth and proliferation. Thus, in isolated primary islets, Rap1A likely promotes the phosphorylation of RPS6 by specifically promoting mTORC1 signaling downstream to RPS6.
cAMP Can Act through Endogenous Rap1 to Stimulate RPS6 Phosphorylation
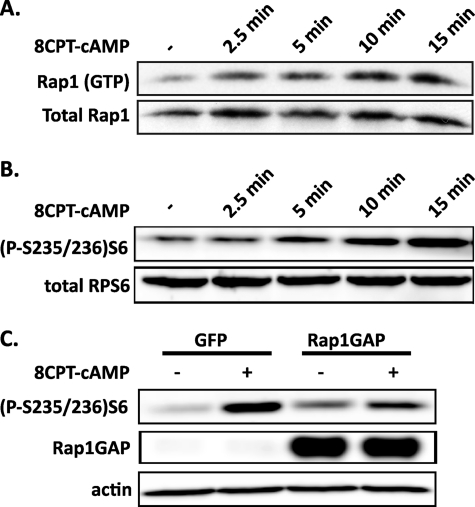
In isolated rat islets maintained at low glucose concentrations, cAMP is able to stimulate S6K1 activity (22). Because Rap1 is a known downstream effector of cAMP, we investigated the role of Rap1 in mediating cAMP-dependent signaling processes in the β-cell. First, we confirmed that cAMP activates Rap1 in the Ins-1 cell line. Ins-1 cells were starved for 18 h in low glucose medium and then treated for various time periods with the cell-permeable cAMP analog, 8CPT-cAMP. Treatment of Ins-1 cells with this compound resulted in a significant increase in the levels of active, GTP-bound Rap1 over time (Fig. 2A). Next, we confirmed that cAMP is able to stimulate RPS6 activity in the Ins-1 cells. Treatment of the Ins-1 cells with 8CPT-cAMP resulted in a significant increase in the phosphorylation levels of RPS6 over time (Fig. 2B), suggesting that both the cAMP-to-Rap1 and the cAMP-to-RPS6 pathways are present and active in Ins-1 cells.
FIGURE 2.
cAMP stimulation of RPS6 phosphorylation is partially dependent on signaling through Rap1. A, cAMP induces Rap1 activation in the Ins-1 cell line. Ins-1 cells were serum- and glucose-starved into quiescence and then incubated with 100 μm 8CPT-cAMP. The cell lysates were used in pull-down assays using a GST fusion of the activated Rap1-binding domain of RalGDS. The levels of precipitated Rap1 were determined by immunoblot analysis using an anti-Rap1 antibody. The levels of total Rap1 in the lysates were also determined. B, cAMP induces RPS6 phosphorylation in the Ins-1 cell line. Ins-1 cells were treated as in A and then stimulated for various times with 100 mm 8CPT-cAMP. The cells were then lysed, and equal amounts of protein were analyzed for phosphorylation of the RPS6 protein by immunoblot using an anti-phospho-RPS6-antibody. The levels of total RPS6 were also determined as a loading control. C, expression of Rap1GAP partially inhibits cAMP-stimulated RPS6 phosphorylation. Ins-1 cell were transduced with adenoviruses expressing either GFP or Rap1GAP and then starved as in A. The cells were the stimulated for 15 min with 100 μm 8CPT-cAMP, lysed, and analyzed for RPS6 phosphorylation status as in B.
To determine whether the activation of Rap1 is required for cAMP stimulation of RPS6 in the Ins-1 cells, we used a recombinant adenovirus to overexpress Rap1GAP, thus abolishing Rap1 signaling (18) (supplemental Fig. S1). Expression of this construct in Ins-1 cells resulted in a reduction, but not elimination, of 8CPT-cAMP-stimulated RPS6 phosphorylation (Fig. 2C). Taken together, these results suggest that cAMP stimulation of RPS6 phosphorylation is at least partially dependent on Rap1 activity in the Ins-1 cell line.
PKA Cooperates with Rap1 in Promoting cAMP-stimulated RPS6 Activation, but the Process Is Independent of PI3K/Akt
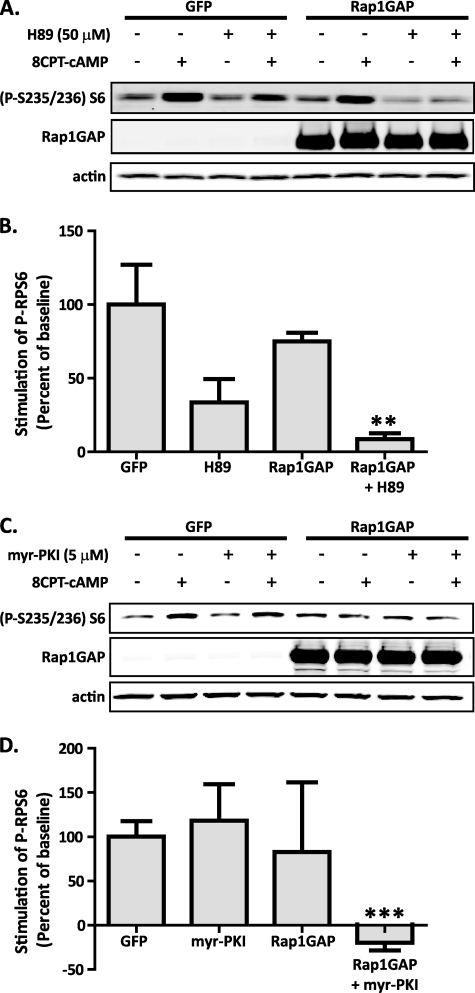
Studies using isolated rat islets have suggested that some of the effects of cAMP on S6K1 are mediated by PKA (22). A contributing effect of PKA would be consistent with our finding noted above that only a portion of the 8CPT-cAMP-stimulated RPS6 phosphorylation was sensitive to inhibition of Rap1 signaling. Therefore, we tested whether PKA was involved in cAMP-stimulated S6K1 activity in the Ins-1 cell line. Preincubation of the Ins-1 cells with the PKA inhibitor H89 resulted in a partial inhibition of 8CPT-cAMP-induced RPS6 phosphorylation (Fig. 3A). Moreover, when Ins-1 cells overexpressing Rap1GAP were treated with H89, 8CPT-cAMP-stimulated phosphorylation of RPS6 was almost completely abrogated (Fig. 3A). Quantification of band intensities from five independent experiments normalized to the actin levels confirms that blockade of both PKA and Rap1 activity is required to completely abrogate cAMP stimulation of RPS6 phosphorylation (Fig. 3B and supplemental Fig. S2). Interestingly, H89 treatment alone had a trend toward a more dramatic effect on the blockade of RPS6 phosphorylation than overexpression of Rap1GAP. In further researching the selectivity of H89, we found that it is as good of an inhibitor of S6K1, which is responsible for phosphorylation of RPS6, as it is of PKA (23, 24). Therefore, these experiments were repeated with a different, highly specific PKA inhibitor, 14-22 amide, myristoylated to make it cell permeable (myr-PKI) (24, 25). In these experiments, myr-PKI alone had no impact on the stimulation of RPS6 phosphorylation by 8CPT-cAMP (Fig. 3C). Although the impact of Rap1GAP treatment alone was more variable than in the previous experiments (Fig. 3B), quantification of band intensities from four independent experiments confirmed that inhibition of both PKA and Rap1 activity is required to completely block RPS6 phosphorylation (Fig. 3D).
FIGURE 3.
cAMP-stimulated RPS6 phosphorylation requires signaling through both PKA and Rap1 and is independent of PI3K signaling. A, Ins-1 cells were transduced with adenovirus encoding either GFP or Rap1GAP and then glucose- and serum-starved into quiescence. The cells were treated with or without 50 μm H89 for 30 min and then stimulated for 15 min with 100 μm 8CPT-cAMP. The cells were then lysed, and equal amounts of protein were analyzed for RPS6 phosphorylation by immunoblot using an anti-phospho-RPS6-antibody. The levels of actin were determined as a loading control. The figure is representative of five separate experiments. B, RPS6 band intensities were normalized to total protein (i.e. the actin band intensities), and then to the percent stimulation of RPS6 by 8CPT-cAMP in the GFP control cells. **, p < 0.01. C, Ins-1 cells were transduced with the indicated adenoviruses as in A, then treated with or without 5 μm myr-PKI for 45 min, and then stimulated for 15 min with 100 μm 8CPT-cAMP. The cells were lysed and subjected to immunoblot analysis as in A. The figure is representative of four separate experiments. D, quantification of stimulation of RPS6 phosphorylation with or without myr-PKI was performed as in B. ***, p < 0.001.
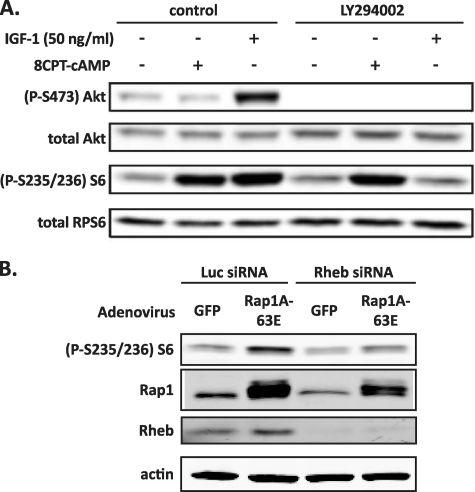
The canonical stimulating event upstream of mTORC1 and S6K1 in many cell types is PI3K-mediated activation of Akt (26, 27). Because Rap1 signaling is able to activate PI3K in some cell types (28), we explored the potential role of this pathway in Rap1-stimulated S6K1 activity in Ins-1 cells. Stimulation of Ins-1 cells with either 8CPT-cAMP or IGF-1 (the latter signals exclusively through the PI3K → Akt pathway) (29) resulted in increased RPS6 phosphorylation. However, only stimulation with IGF-1 resulted in enhanced phosphorylation of Akt (Fig. 4A, first through third lanes). Moreover, treatment of cells with a specific inhibitor of PI3K, LY294002, completely blocked IGF-1-stimulated RPS6 phosphorylation, as expected, but had no effect on 8CPT-cAMP-induced RPS6 phosphorylation (Fig. 4A, fourth through sixth lanes). Taken together, these data suggest that the ability of cAMP to stimulate mTORC1 signaling is independent of the PI3K/Akt signaling pathway in β-cells.
FIGURE 4.
cAMP-stimulated RPS6 phosphorylation is independent of PI3K signaling and dependent on Rheb. A, Ins-1 cells were transduced with adenovirus encoding either GFP or Rap1GAP and then glucose- and serum-starved into quiescence. The cells were treated with or without with 30 μm LY294002 for 30 min and then stimulated for 15 min with 100 μm 8CPT-cAMP or 10 ng/ml IGF-1. The cells were then lysed, and equal amounts of protein were analyzed for RPS6 phosphorylation by immunoblot using an anti-phospho-RPS6-antibody. The levels of total RPS6 were also determined as a control. The figure is representative of three separate experiments. B, Ins-1 cells were transfected with the indicated siRNA sequences (Rheb, siRheb; or a luciferase control sequence, siLuc) for 48 h and then transduced with the indicated adenoviral constructs and starved as in A. The cells were lysed, and equal amounts of protein were analyzed by immunoblotting for phosphorylated RPS6. The figure is representative of four independent experiments.
Although it is possible that Rap1 directly activates mTORC1, another related monomeric G protein, Rheb, is known to be immediately upstream of mTORC1 and S6K1 (30). Rheb is usually negatively regulated by tuberous sclerosis complex protein 2 (TSC2; tuberin) (30). In the canonical PI3K → RPS6 signaling pathway, Akt-mediated inhibition of TSC2 relieves its inhibition on Rheb, allowing Rheb to activate mTORC1 (31, 32). Thus, we aimed to determine whether Rap1-mediated RPS6 phosphorylation was also dependent on Rheb. Indeed, we found that Rap1A-63E was no longer able to promote RPS6 phosphorylation when Rheb expression was blocked by a specific siRNA oligonucleotide (Fig. 4B). These results demonstrate that Rap1 signals through the mTORC1 pathway, and indicate that it acts upstream of Rheb, independent of PI3K activation of Akt.
Activation of Rap1 Promotes GSIS, but the mTOR/S6K1 Pathway Is Not Required for the Impact on GSIS
After establishing the cAMP → Rap1 → mTORC1 → RPS6 signaling pathway in pancreatic β-cell-derived lines and islets, we next sought to analyze the roles of this pathway in pancreatic islet biology. The compensatory response to systemic insulin resistance of type 2 diabetes involves an increase in both β-cell number and size (i.e. β-cell mass) (33) and increased insulin secretion from individual cells. Previous studies have demonstrated that signaling through Epac potentiates glucose-stimulated insulin secretion (GSIS) and insulin granule dynamics (7, 34, 35).
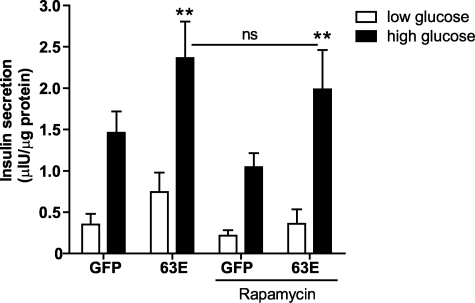
Consistent with these findings, expression of Rap1A-63E in isolated rat and human pancreatic islets enhanced GSIS (Fig. 5 and supplemental Fig. S4, respectively). Rap1–63E expression phenocopies signaling by endogenously activated Rap1A, as has been observed in other cellular systems (17). These results, coupled with previous reports showing Epac potentiation of GSIS (35), suggest that this function of Epac is mediated by Rap1. Additionally, to examine whether the newly identified Rap1 → RPS6 pathway was involved in the potentiation of GSIS by Rap1A-63E, rapamycin treatment was used to block the signaling downstream of mTORC1. Interestingly, we observed that rapamycin had no impact on the ability of activated Rap1 to increase GSIS from rat islets (Fig. 5). These results demonstrate that activation of Rap1 potentiates GSIS, but this activity of Rap1 is not mediated exclusively by the newly described Rap1 → RPS6 pathway.
FIGURE 5.
Activation of Rap1 promotes GSIS in a rapamycin-insensitive manner. Rat pancreatic islets were isolated and treated with adenoviruses expressing GFP or Rap1A-63E the day before the day of the assay and treated with dimethyl sulfoxide alone or rapamycin. Insulin secretion was measured from islets expressing the indicated proteins after a 2 h incubation in low glucose (2.5 mm) or high glucose (15 mm) buffer. The data represent the means ± S.E. of the average values from four independent experiments. **, p > 0.01 as compared with the dimethyl sulfoxide-treated GFP control islets.
Activation of Rap1 β-Cell Proliferation through the mTORC1 Pathway
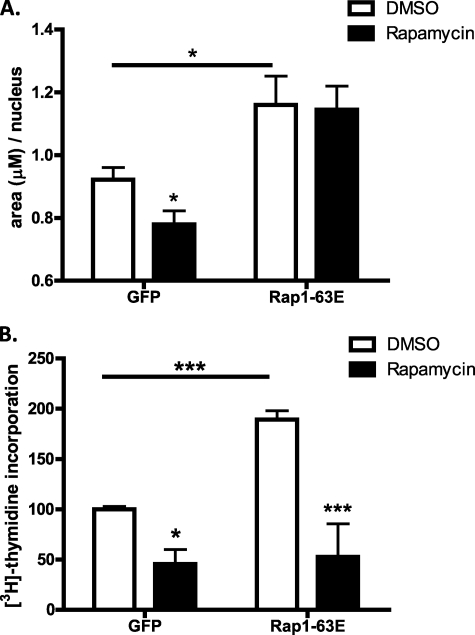
S6K1 activity has been linked to increased β-cell number and size (33), suggesting that Rap1 may play a role in these physiologies. To explore this hypothesis, we first determined whether Rap1 signaling was able to increase islet cell size through activation of RPS6. Rap1A-63E was expressed in isolated rat pancreatic islets, and the average size of islet cells was determined using confocal microscopic images of islet cross-sections. In the control islets, rapamycin pretreatment led to an approximate 20% decrease in islet cell size, as expected from previous reports (36) (Fig. 6A). Interestingly, Rap1A-63E expression led to an approximate 25% increase in islet cell size. Assuming a perfect sphere and using μm/nucleus as the diameter, Rap1A-63E expression can be estimated to dramatically increase the β-cell volume (from 0.4 μm3 with GFP to 0.8 μm3 with Rap1A-63E). Surprisingly, this increase was insensitive to pretreatment with rapamycin, suggesting that Rap1A-63E can also act through another growth regulatory pathway to impact on islet cell size (Fig. 6A).
FIGURE 6.
Activation of Rap1 promotes increases in islet cell size and proliferation, the latter through mTORC1. Rat pancreatic islets were isolated and infected with adenoviruses expressing vector or Rap1A-63E the day before the day of the assay and treated with dimethyl sulfoxide (DMSO) alone or rapamycin. A, fixed islet cell nuclei were Hoechst-stained and analyzed by confocal microscopy. The number of nuclei in a defined area was counted using Metamorph software. The data represent the average numbers of nuclei counted from four areas/image slice, with three image slices/islet (n = 4–6 islets/condition). B, islets were incubated with methyl-[3H]thymidine for 24 h before assay. The amount of [3H]thymidine uptake was then determined as measure of cell proliferation. The data represent the means ± S.E. of the average values from three (A) or four (B) independent experiments. *, p > 0.05; ***, p < 0.001 as compared with the indicated condition.
The potential impact of Rap1 signaling on β-cell proliferation was also examined. Activation of Rap1 signaling, again by expression of Rap1A-63E, enhanced islet proliferation ∼2-fold as determined by [3H]thymidine uptake (Fig. 6B). Importantly, we observed that rapamycin blockade of mTORC1 signaling completely abrogated the Rap1-mediated enhancement of islet proliferation (Fig. 6B). Thus, Rap1-potentiated β-cell proliferation appears to be mediated exclusively by the Rap1 → RPS6 signaling pathway but is not required for Rap1-mediated enhancement of GSIS or islet cell size. Together, these results indicate that Rap1 is involved in at least two distinct signaling pathways impacting on various β-cell functions.
DISCUSSION
In this study we define a Rap1 → RPS6 signaling axis, acting through mTORC1, that is important in β-cell function. The demonstration that cAMP stimulation of Rap1 activates RPS6 phosphorylation and that this signaling process requires the cooperation of Rap1 and PKA reveals a new aspect of cAMP signaling in β-cells. The finding that this mode of Rap1 activation is independent of PI3K and Akt, the canonical upstream activators of mTORC1, is unique, to our knowledge. Finally, the discovery that Rap1 activation not only potentiates islet cell GSIS but also stimulates increases in β-cell size and proliferation suggests that this pathway could be exploited for expansion of functional β-cell mass in diabetes.
A recent study by Xie et al. (37) demonstrated the impact of cAMP signaling on β-cell functions with a β-cell-specific Gαs-deficient mouse. As expected from the loss of this stimulator of cAMP production, cAMP levels in the islets of these mice were significantly reduced. The major defect was not in insulin secretion but rather a decrease in β-cell mass that was explained by a decrease in β-cell size and proliferation. This report correlates well with our data implicating Rap1 as a mediator for β-cell cAMP signaling and the impact of Rap1 on β-cell size and proliferation.
Previous studies have reported that cAMP is able to activate RPS6 phosphorylation in several cell types, including isolated rat islets, pancreatic acinar cells (38), Sertoli cells (39), Swiss 3T3 cells (40), and thyroid cells (41). Interestingly, whereas cAMP appears to stimulate S6K1 activity through a PI3K-independent pathway in both Sertoli (39) and thyroid cells (42), cAMP stimulation of S6K1 appears to be completely PKA-dependent in both cell types (28, 39, 42). Furthermore, to our knowledge, only three studies have directly examined the effects of Rap1 on S6K1 activity (28, 43, 44), and in none of these studies did activation of Rap1 stimulate S6K1. Thus, the involvement of Rap1 in cAMP-stimulated RPS6 phosphorylation appears to be dependent on cell type, a mode of regulation that is becoming increasingly appreciated for other signal transduction cascades (45).
One of the best characterized downstream targets of mTORC1 is p70 S6K1, which phosphorylates and activates RPS6 (26). The significance of S6K1 signaling in the biology of the mouse pancreatic β-cell has been demonstrated by work involving the deletion of the S6K1 gene. The phenotype of the S6K1-null mouse is predominantly characterized by a marked reduction in β-cell mass, resulting in a syndrome that mimics human type 2 diabetes (46). Interestingly, this defect appeared to be specific to the pancreatic β-cells, because the other tissues examined were mostly unaffected. In addition, a recent study found that phosphorylation-deficient RPS6 knock-in mice have a growth defect that is selective for embryonic fibroblasts and β-cells, and these mice have compromised glucose homeostasis (33). Both of these studies correlate well with our findings that Rap1 signaling through mTORC1 and phosphorylation of RPS6 play an important role in regulating β-cell mass and function.
In light of recent reports indicating a role for Epac and Rap1 in secretion from pancreatic β-cells and acinar cells (12, 13, 35, 47), it was not unexpected that expression of the activated form of Rap1 potentiated GSIS. The most thorough characterization of Epac/Rap1 function in GSIS has been performed by Shibasaki et al. (35). Shibasaki et al. inhibited Rap1 function in insulinoma cells and islets by overexpression of Rap1GAP and selective siRNA-mediated reduction in Rap1 expression and demonstrated reduced cAMP-potentiated GSIS (35). They also demonstrated that cAMP activation of Rap1 was mediated through Epac2 and was independent of PKA (35). In an elegant TIRF microscopic characterization, the authors showed that cAMP potentiated insulin granule exocytosis by selectively up-regulating the levels of “restless newcomer” granules, those which dock, prime, and are immediately released. Islets isolated from Epac2-null mice were defective in cAMP-potentiated GSIS; specifically, in the exocytosis events by “restless newcomer” granules, primarily during the first phase of insulin secretion. Shibasaki et al. propose that cAMP-mediated Rap1 signaling regulates the density of the readily releasable pool of insulin granules near the plasma membrane, which has been shown to be critical in the first phase of insulin secretion. Although the results of Shibasaki and colleagues outline a general mechanism for the promotion of GSIS by Rap1A, the precise downstream signaling mechanisms remain undefined.
Our results place Rap1A upstream of Rheb and mTORC1 in the cAMP → RPS6 pathway. Interestingly, the negative regulator of Rheb, TSC2, was originally identified as a GAP for Rap1 (48), until the more recent finding that Rheb is its true biological target (49). Because activation of Akt is the typical mode for TSC2 inhibition, the PI3K/Akt independence of the Rap1 effect on RPS6 phosphorylation (Fig. 4) suggests that Rap1 can independently relieve the inhibition of Rheb by TSC2. Our data using selective siRNA against Rap1A and Rheb support this hypothesis. First, selectively reducing the levels of Rap1A blocks the activated Rap1 effect on S6 phosphorylation (supplemental Fig. S1B). Second, adding back constitutively active Rap1A (Rap1A-63E) restores RPS6 phosphorylation in the absence of endogenous Rap1 activation (supplemental Fig. S1B). These experiments confirm that Rap1A is the active isoform and that Rap1A-63E phenocopies the signaling activity of activated endogenous Rap1. Finally, the fact that Rap1A-63E does not dramatically increase RPS6 phosphorylation above that promoted by endogenous Rap1 suggests that Rap1 does not activate mTORC1 directly and that there is another rate-limiting step in the pathway, perhaps, as we suggest, the Rheb GTPase. Taken together with what is already known about Rap1 and TSC2, our findings support a model in which activated Rap1 recruits TSC2 away from Rheb, allowing Rheb signaling to mTORC1 to remain active, although alternative mechanisms cannot be excluded.
Several notable recent publications support the hypothesis that this Rap1 pathway might yield targets for the treatment of β-cell dysfunction in diabetes. First, there now exists a set of literature on the impact of rapamycin (sirolimus), which blocks mTORC1, on β-cell function. Whereas rapamycin and the related compound FK-506 (tacrolimus) are commonly used nonglucocorticoid immunosuppressive drugs and have been found to delay the onset of insulin dependence in a rodent model of type 1 diabetes (50), rapamycin treatment actually hastened the development of fulminant diabetes in a rodent model of type 2 diabetes (Psammomys obesus) through a loss in both β-cell mass and function (51). The Edmonton protocol for human islet transplantation calls for the use of sirolimus and tacrolimus, which, combined with our results, possibly explains the relatively disappointing results 1–2 years after transplant (52, 53). In fact, Nir et al. (54) demonstrated that the sirolimus/tacrolimus regimen prevented β-cell regeneration in a rodent model of islet transplantation. Furthermore, in isolated human islets and human islet grafts into NOD.scid mice, Johnson et al. (55) demonstrated that tacrolimus impaired GSIS and islet graft function and that these defects could be ameliorated by treatment with a GLP-1 agonist, exenatide. The second set of literature relevant to our work is related to Rap1 signaling in diabetes therapeutics. There are now several papers that indirectly implicate Rap1 in mediating incretin (e.g. GLP-1, GIP) action (12, 13, 35, 47). Interestingly, however, an activator of Rap1, Epac2, has been found to be a direct target of sulfonylurea drugs that stimulate insulin secretion (15). Combined, these recent findings lend credence to the idea that Rap1 or other proteins in the newly defined cAMP-stimulated signaling pathway could serve as targets for diabetes therapeutics.
The results of our study show that the previously undefined signaling axis, cAMP → Rap1 → mTORC1 → RPS6, is responsible for some of the physiologic effects of cAMP elevation on β-cell function. Our findings represent a new link between cAMP signaling and the pathways controlling β-cell proliferation and suggest that directly targeting this pathway may have beneficial therapeutic effects for patients with type 2 diabetes. The ability of Rap1 to potentiate increases in β-cell proliferation and size indicates that it might mediate both islet hyperplasia and hypertrophy, respectively, positively impacting β-cell mass, whereas the Rap1-mediated potentiation of GSIS has interesting implications for the preservation of β-cell function. Finally, the rapamycin independence of the impact on cell size highlights the potential utility of Rap1 pathway agonists in situations where rapamycin-related compounds are used for immunosuppression, such as in islet transplantation.
Supplementary Material
Acknowledgments
We thank Missy Infante for technical assistance, Dr. Danhong Lu and Helena Winfield for providing the isolated rat islets, Dr. Hans Hohmeier for assistance with the human islet experiments, and Dr. Michelle Arlotto and Jon Haldeman for testing the adenoviral constructs.
Note Added in Proof
Although a recent study by Moore et al. (Moore, C. E., Xie, J., Gomez, E., and Herbert, T. P. (2009) J. Mol. Biol. 389, 480–494) primarily implicates PKA as a mediating glucagon-like peptide 1 (GLP-1, a cAMP-raising G protein-coupled receptor agonist) signaling to ribosomal protein S6 in a mouse insulinoma cell line, their work also supports the mTORC1 dependence of cAMP-stimulated ribosomal protein S6 phosphorylation that we see in our experiments while explaining the partial PKA dependence by implicating PKA as a novel ribosomal protein S6 kinase.
This work was supported, in whole or in part, by National Institutes of Health Grants DK078732 (to P. T. F.), DK058398 (to C. B. N.), DK076488 (to P. J. C.), and DK080845 (to M. E. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PKA
- cAMP-dependent protein kinase
- GFP
- green fluorescent protein
- GSIS
- glucose-stimulated insulin secretion
- GST
- glutathione S-transferase
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- RPS6
- ribosomal protein S6
- TSC2
- tuberous sclerosis complex protein 2
- siRNA
- small interfering RNA
- PBS
- phosphate-buffered saline
- PI3K
- phosphatidylinositol 3-kinase
- myr-PKI
- myristoylated 14-22 amide
- S6K1
- S6 kinase 1
- GAP
- GTPase-activating protein.
REFERENCES
- 1.Hohmeier H. E., Newgard C. B. (2005) Nat. Biotechnol. 23, 1231–1232 [DOI] [PubMed] [Google Scholar]
- 2.Kahn S. E. (2003) Diabetologia 46, 3–19 [DOI] [PubMed] [Google Scholar]
- 3.Butler P. C., Meier J. J., Butler A. E., Bhushan A. (2007) Nat. Clin. Pract. Endocrinol. Metab. 3, 758–768 [DOI] [PubMed] [Google Scholar]
- 4.Muoio D. M., Newgard C. B. (2008) Nat. Rev. Mol. Cell Biol. 9, 193–205 [DOI] [PubMed] [Google Scholar]
- 5.Furman B., Pyne N., Flatt P., O'Harte F. (2004) J. Pharm. Pharmacol. 56, 1477–1492 [DOI] [PubMed] [Google Scholar]
- 6.de Rooij J., Zwartkruis F. J., Verheijen M. H., Cool R. H., Nijman S. M., Wittinghofer A., Bos J. L. (1998) Nature 396, 474–477 [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H., Springett G. M., Mochizuki N., Toki S., Nakaya M., Matsuda M., Housman D. E., Graybiel A. M. (1998) Science 282, 2275–2279 [DOI] [PubMed] [Google Scholar]
- 8.López De Jesús M., Stope M. B., Oude Weernink P. A., Mahlke Y., Börgermann C., Ananaba V. N., Rimmbach C., Rosskopf D., Michel M. C., Jakobs K. H., Schmidt M. (2006) J. Biol. Chem. 281, 21837–21847 [DOI] [PubMed] [Google Scholar]
- 9.Shi G. X., Rehmann H., Andres D. A. (2006) Mol. Cell. Biol. 26, 9136–9147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang G., Chepurny O. G., Holz G. G. (2001) J. Physiol. 536, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuboi T., da Silva Xavier G., Holz G. G., Jouaville L. S., Thomas A. P., Rutter G. A. (2003) Biochem. J. 369, 287–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kashima Y., Miki T., Shibasaki T., Ozaki N., Miyazaki M., Yano H., Seino S. (2001) J. Biol. Chem. 276, 46046–46053 [DOI] [PubMed] [Google Scholar]
- 13.Fujimoto K., Shibasaki T., Yokoi N., Kashima Y., Matsumoto M., Sasaki T., Tajima N., Iwanaga T., Seino S. (2002) J. Biol. Chem. 277, 50497–50502 [DOI] [PubMed] [Google Scholar]
- 14.Kwon G., Pappan K. L., Marshall C. A., Schaffer J. E., McDaniel M. L. (2004) J. Biol. Chem. 279, 8938–8945 [DOI] [PubMed] [Google Scholar]
- 15.Zhang C. L., Katoh M., Shibasaki T., Minami K., Sunaga Y., Takahashi H., Yokoi N., Iwasaki M., Miki T., Seino S. (2009) Science 325, 607–610 [DOI] [PubMed] [Google Scholar]
- 16.Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. (1992) Endocrinology 130, 167–178 [DOI] [PubMed] [Google Scholar]
- 17.Bailey C. L., Kelly P., Casey P. J. (2009) Cancer Res. 69, 4962–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wittchen E. S., Worthylake R. A., Kelly P., Casey P. J., Quilliam L. A., Burridge K. (2005) J. Biol. Chem. 280, 11675–11682 [DOI] [PubMed] [Google Scholar]
- 19.Schisler J. C., Fueger P. T., Babu D. A., Hohmeier H. E., Tessem J. S., Lu D., Becker T. C., Naziruddin B., Levy M., Mirmira R. G., Newgard C. B. (2008) Mol. Cell. Biol. 28, 3465–3476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fueger P. T., Schisler J. C., Lu D., Babu D. A., Mirmira R. G., Newgard C. B., Hohmeier H. E. (2008) Mol. Endocrinol. 22, 1251–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou M. M., Blenis J. (1996) Cell 85, 573–583 [DOI] [PubMed] [Google Scholar]
- 22.Kwon G., Marshall C. A., Pappan K. L., Remedi M. S., McDaniel M. L. (2004) Diabetes 53, (Suppl. 3) S225–S232 [DOI] [PubMed] [Google Scholar]
- 23.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray A. J. (2008) Sci. Signal 1, re4. [DOI] [PubMed] [Google Scholar]
- 25.Dalton G. D., Dewey W. L. (2006) Neuropeptides 40, 23–34 [DOI] [PubMed] [Google Scholar]
- 26.Fingar D. C., Blenis J. (2004) Oncogene 23, 3151–3171 [DOI] [PubMed] [Google Scholar]
- 27.Wang L., Gout I., Proud C. G. (2001) J. Biol. Chem. 276, 32670–32677 [DOI] [PubMed] [Google Scholar]
- 28.Tsygankova O. M., Saavedra A., Rebhun J. F., Quilliam L. A., Meinkoth J. L. (2001) Mol. Cell. Biol. 21, 1921–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine A. J., Feng Z., Mak T. W., You H., Jin S. (2006) Genes Dev. 20, 267–275 [DOI] [PubMed] [Google Scholar]
- 30.Inoki K., Li Y., Xu T., Guan K. L. (2003) Genes Dev. 17, 1829–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai S. L., Tee A. R., Short J. D., Bergeron J. M., Kim J., Shen J., Guo R., Johnson C. L., Kiguchi K., Walker C. L. (2006) J. Cell Biol. 173, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tee A. R., Anjum R., Blenis J. (2003) J. Biol. Chem. 278, 37288–37296 [DOI] [PubMed] [Google Scholar]
- 33.Ruvinsky I., Sharon N., Lerer T., Cohen H., Stolovich-Rain M., Nir T., Dor Y., Zisman P., Meyuhas O. (2005) Genes Dev. 19, 2199–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Rooij J., Rehmann H., van Triest M., Cool R. H., Wittinghofer A., Bos J. L. (2000) J. Biol. Chem. 275, 20829–20836 [DOI] [PubMed] [Google Scholar]
- 35.Shibasaki T., Takahashi H., Miki T., Sunaga Y., Matsumura K., Yamanaka M., Zhang C., Tamamoto A., Satoh T., Miyazaki J., Seino S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19333–19338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volarevic S., Thomas G. (2001) Prog. Nucleic Acids Res. Mol. Biol. 65, 101–127 [DOI] [PubMed] [Google Scholar]
- 37.Xie T., Chen M., Zhang Q. H., Ma Z., Weinstein L. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19601–19606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bragado M. J., Groblewski G. E., Williams J. A. (1997) Am. J. Physiol. 273, C101–C109 [DOI] [PubMed] [Google Scholar]
- 39.Lécureuil C., Tesseraud S., Kara E., Martinat N., Sow A., Fontaine I., Gauthier C., Reiter E., Guillou F., Crépieux P. (2005) Mol. Endocrinol. 19, 1812–1820 [DOI] [PubMed] [Google Scholar]
- 40.Withers D. J., Bloom S. R., Rozengurt E. (1995) J. Biol. Chem. 270, 21411–21419 [DOI] [PubMed] [Google Scholar]
- 41.Cass L. A., Meinkoth J. L. (1998) Endocrinology 139, 1991–1998 [DOI] [PubMed] [Google Scholar]
- 42.Cass L. A., Summers S. A., Prendergast G. V., Backer J. M., Birnbaum M. J., Meinkoth J. L. (1999) Mol. Cell. Biol. 19, 5882–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castro A. F., Rebhun J. F., Clark G. J., Quilliam L. A. (2003) J. Biol. Chem. 278, 32493–32496 [DOI] [PubMed] [Google Scholar]
- 44.Shelly C., Herrera R. (2002) J. Cell Sci. 115, 1985–1993 [DOI] [PubMed] [Google Scholar]
- 45.Kimple M. E., Nixon A. B., Kelly P., Bailey C. L., Young K. H., Fields T. A., Casey P. J. (2005) J. Biol. Chem. 280, 31708–31713 [DOI] [PubMed] [Google Scholar]
- 46.Pende M., Kozma S. C., Jaquet M., Oorschot V., Burcelin R., Le Marchand-Brustel Y., Klumperman J., Thorens B., Thomas G. (2000) Nature 408, 994–997 [DOI] [PubMed] [Google Scholar]
- 47.Sabbatini M. E., Chen X., Ernst S. A., Williams J. A. (2008) J. Biol. Chem. 283, 23884–23894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wienecke R., König A., DeClue J. E. (1995) J. Biol. Chem. 270, 16409–16414 [DOI] [PubMed] [Google Scholar]
- 49.Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J. (2003) Curr. Biol. 13, 1259–1268 [DOI] [PubMed] [Google Scholar]
- 50.Baeder W. L., Sredy J., Sehgal S. N., Chang J. Y., Adams L. M. (1992) Clin. Exp. Immunol. 89, 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fraenkel M., Ketzinel-Gilad M., Ariav Y., Pappo O., Karaca M., Castel J., Berthault M. F., Magnan C., Cerasi E., Kaiser N., Leibowitz G. (2008) Diabetes 57, 945–957 [DOI] [PubMed] [Google Scholar]
- 52.Shapiro A. M., Lakey J. R., Ryan E. A., Korbutt G. S., Toth E., Warnock G. L., Kneteman N. M., Rajotte R. V. (2000) N. Engl. J. Med. 343, 230–238 [DOI] [PubMed] [Google Scholar]
- 53.Shapiro A. M., Ricordi C., Hering B. J., Auchincloss H., Lindblad R., Robertson R. P., Secchi A., Brendel M. D., Berney T., Brennan D. C., Cagliero E., Alejandro R., Ryan E. A., DiMercurio B., Morel P., Polonsky K. S., Reems J. A., Bretzel R. G., Bertuzzi F., Froud T., Kandaswamy R., Sutherland D. E., Eisenbarth G., Segal M., Preiksaitis J., Korbutt G. S., Barton F. B., Viviano L., Seyfert-Margolis V., Bluestone J., Lakey J. R. (2006) N. Engl. J. Med. 355, 1318–1330 [DOI] [PubMed] [Google Scholar]
- 54.Nir T., Melton D. A., Dor Y. (2007) J. Clin. Invest. 117, 2553–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson J. D., Ao Z., Ao P., Li H., Dai L., He Z., Tee M., Potter K. J., Klimek A. M., Meloche R. M., Thompson D. M., Verchere C. B., Warnock G. L. (2009) Cell Transplant. 18, 833–845 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.