Abstract
Efficient proliferation of Mycobacterium tuberculosis (Mtb) inside macrophage requires that the essential response regulator MtrA be optimally phosphorylated. However, the genomic targets of MtrA have not been identified. We show by chromatin immunoprecipitation and DNase I footprinting that the chromosomal origin of replication, oriC, and the promoter for the major secreted immunodominant antigen Ag85B, encoded by fbpB, are MtrA targets. DNase I footprinting analysis revealed that MtrA recognizes two direct repeats of GTCACAgcg-like sequences and that MtrA∼P, the phosphorylated form of MtrA, binds preferentially to these targets. The oriC contains several MtrA motifs, and replacement of all motifs or of a single select motif with TATATA compromises the ability of oriC plasmids to maintain stable autonomous replication in wild type and MtrA-overproducing strains, indicating that the integrity of the MtrA motif is necessary for oriC replication. The expression of the fbpB gene is found to be down-regulated in Mtb cells upon infection when these cells overproduce wild type MtrA but not when they overproduce a nonphosphorylated MtrA, indicating that MtrA∼P regulates fbpB expression. We propose that MtrA is a regulator of oriC replication and that the ability of MtrA to affect apparently unrelated targets, i.e. oriC and fbpB, reflects its main role as a coordinator between the proliferative and pathogenic functions of Mtb.
Keywords: Bacteria, Bacterial Signal Transduction, Chromatin Immunoprecipitation (ChIP), DNA-binding Protein, DNA Replication, Autonomous Replication, Footprinting, Histidine-Aspartate-response Regulator, Mycobacteria, Two-component Signal Transduction
Introduction
Approximately one-third of the world population is latently infected with Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis. Studies with the mouse model of aerosol infection suggest that Mtb multiplication in the lung often continues uninterrupted until the mounting of host-adaptive immunity (reviewed in Ref. 1). This process results in pathogen growth arrest in the infected macrophages (MΦ)2 and leads to the formation of granulomas. The bacteria in the granulomatous lesions are believed to be in a “latent or persistent state” and maintain a limited bacterial turnover but to resume active multiplication and cause infection when the host immune system is compromised (reviewed in Refs. 1–3). It is intriguing and largely unknown how Mtb multiplication is regulated upon infection or during survival in the persistent state. Mining the Mtb genome sequence data revealed that the pathogen operates a host of regulatory networks, including the paired two-component regulatory signal transduction systems (2CRS), several of which are believed to be required for virulence (4). The roles of several key players involved in these regulatory networks are largely unknown.
Paired 2CRS are the major means by which bacteria sense and respond to different stimuli and regulate gene expression through a phosphorylation relay (reviewed in Ref. 5). The Mtb genome encodes 11 paired 2CRS, among which MtrAB is the only essential 2CRS (4, 6, 7). This system consists of MtrB, a membrane-bound sensor kinase, which is nonessential for growth, and MtrA, a cytosolic response regulator (RR), which is essential, and knock-out strains of mtrA could not grow in broth unless a second copy of mtrA was present (7). The mtrA promoter activity is differentially expressed in virulent Mtb and in the vaccine strain Mycobacterium bovis BCG. mtrA transcription is sharply up-regulated in M. bovis BCG upon its entry into MΦ. In contrast, mtrA transcription is unchanged in virulent Mtb upon its infection of murine or human monocyte-derived MΦ (6, 7). These initial insights suggested to us that it is the activity of the MtrAB system, along with a few other key regulators, that is necessary for dictating the outcome of proliferation and persistence upon infection.
Because mtrA is an essential gene and the conditional knock-out strains of mtrA are unavailable, to address the roles of the mtrA gene product in Mtb proliferation upon infection, we created and characterized the mtrA merodiploid strains producing elevated levels of MtrA (8). These studies revealed that the merodiploid strain overproducing WT MtrA, designated as Rv78 Mtb(MtrA+), is attenuated in MΦ and in mouse lungs, but it is otherwise as proficient as the WT strain for growth in nutrient broth. In contrast, the merodiploid overproducing phosphorylation-defective MtrA, due to a mutation at the phosphate accepting amino acid Asp-53 (8) and designated as Rv129 Mtb(D53N MtrA+), is not severely attenuated in MΦ (8). We also found that the merodiploid overproducing both the WT MtrA RR and the MtrB kinase is as proficient as the WT strain for growth in MΦ (8). These studies lead to a hypothesis that optimal proliferation of Mtb in MΦ and in murine lungs depends, in part, on the ratio of phosphorylated MtrA (MtrA∼P) and nonphosphorylated MtrA and that MtrB kinase regulates the MtrA∼P state (8).
It is reasonable to assume that the essential RR MtrA or its MtrA∼P form regulates the activities of several essential targets and their associated pathways. A consequence of this regulation could determine whether the bacterium maintains a persistent state or assumes active multiplication and causes disease. A first step in understanding how MtrA RR might control Mtb proliferation upon infection is to identify the targets and define the DNA-binding motifs recognized by the MtrA RR. This study addresses these issues, and we demonstrate that oriC, the site where chromosomal DNA replication initiates, and PfbpB, the promoter for a major secreted antigen 85B, are physiologically significant MtrA targets. Our work also identifies the MtrA DNA recognition motifs, and it is thereby an important step toward understanding the whole MtrA regulon and potentially how Mtb physiology is regulated between acute and latent disease states.
MATERIALS AND METHODS
Bacterial Strains
M. tuberculosis strain H37Rv was grown in Middlebrook 7H9 media. Mtb mtrA merodiploid strains were created by plasmid transformation, and transformants were selected on Middlebrook 7H10 agar supplemented with hygromycin (50 μg/ml) as described previously (8). Escherichia coli Top-10 were used to propagate mycobacterial plasmids.
Chromatin Immunoprecipitation Experiments
Our basic protocol as applied to Mtb cells was described previously (8). Mtb(MtrA+) cultures (Mtb merodiploid overproducing WT MtrA protein) actively growing in Middlebrook 7H9 broth were exposed to 1% formaldehyde for 20 min and processed to prepare cellular lysates, and the genomic DNA was then sheared to an average size of 500 to 1000 bp, as described. Cleared supernatant was obtained and incubated with anti-MtrA or mock antibodies followed by immunopure protein G-agarose beads to collect immunoprecipitates. DNA samples were then purified using DNAzol, and protein-DNA cross-links were reversed by heating, and an aliquot of DNA was directly used to amplify select DNA fragments by PCR. The oligonucleotide primers that were used are listed in Table 1. PCR products were resolved in agarose gels, stained with SYBR Green, and photographed as needed. Typically, 30 cycles of amplification with serially diluted DNA of mock, immunoprecipitation-treated and total input samples were carried out. DNA band intensities were quantified by densitometry scanning and analyzed by Molecular Imager Fx. Typically, DNA samples were diluted 100- and 200-fold and used in PCR. The band intensities for FtsZ are the same with anti-MtrA and mock antibodies, i.e. not enriched with anti-MtrA antibody (8). We considered this ratio as 1 and normalized the same ratio for each of the other targets to this value. For all initial experiments, values 2 and above were considered as potential MtrA targets.
TABLE 1.
Oligonucleotide primers used for ChIP assays
| Primers | Gene/DNA | Remarks |
|---|---|---|
| 1. Q21: gaggatcgcgagccgttgcc | dnaA promoter- | Forward primer, 180-bp |
| 2. MVM97: cggggtcatcggtcaacgacg | dnaA promoter | Reverser primer |
| 3. Mty51: ggggaattcttaaaaaaacttctc | oriC | Forward, 182-bp |
| 4. MTBOriTF189RC: gtcggagttgtggatg | oriC | Reverse |
| 5. P85b.F: ccaagctcgaatctttcggctcacgtctgtc | fbpB promoter | Forward, 202-bp |
| 6. P85bR: ccgcgatagatccataccgccataccgtttg | fbpB promoter | Reverse |
| 7. P85A.F: gtgacggcgccacgaaccctgtcaa | fbpA promoter | Forward, 209-bp |
| 8. P85A.R: ttggccgtgaacgaccgccggataagggtt | fbpA promoter | Reverse primer |
| 9. mtp210F: ctgtgtcgcggctacgacgtg | mtrA promoter | Forward, 223-bp |
| 10 mtp415R: gggtcacgctgggcacggcc | mtrA promoter | Reverse |
| 11. mce1AF: tgcacgatccgcaacttct | mce1 promoter | Forward, 220-bp |
| 12. mce1AR: tcctcagcgagtagccgttac | mce1 promoter | Reverse, 220-bp |
| 13. MVM238 gcggatccgcttcctccctggtggggc | ftsZ promoter | Reverse, 160-bp |
| 14. MVM508FZPE: tgcccgccgcgtatcggcgc | ftsZ promoter | Forward, |
| 15. pfbpCF: ttgcgccgctcgggagccagcc | fbpC promoter, | forward, 200-bp |
| 16. p85CR: actctcatctgccgcacgacgcggtcgaat | fbpC promoter, | reverse |
| 17. YneApSpeI: tggcaacactagtccggctgtccgcaccagcggc | chiZ promoter | forward, 198-bp |
| 18. YneA2HIIIR: gtctcaagcttctcagacggtaatcgctcgcgtg | chiZ promoter | Reverse |
| Oligonucleotide primers used for qRT-PCR | ||
| 1. FbpA | Forward primer: 85AF: | 5′-GGATCTGGGTGGCAACAACCT-3′ |
| Reverse primer: 85AR: | 5′-CAGCTGTGCGTACCGCTGTC-3′ | |
| RT primer: 85ART: | 5′-GTTGCAGGTCGGGCTTCATAG-3′ | |
| Taqman Probe: 85A.TP: | 5′-TGCTGGTCCGCACGAAGCCCTCGA | |
| 2. FbpB: | Forward primer: MVM54685RTY1 | 5′-TCAGGGGATGGGGCCTAGCC-3′ |
| Reverse pimer:RTAg85A2 | 5′-GCTTGGGGATCTGCTGCGTA-3′ | |
| RT primer:MVM545A85BRTY3 | 5′-GCCGGCGCCTAACGAACTCTGC-3′ | |
| Taqman Probe: 85B-TP | 5′-FAM-TCGAGTGACCCGGCATGGGAGCGT-BHQ-1 3′ | |
| 3. 16S rRNA: | Forward primer: RT16S1: | 5′-GAGTGGCGAACGGGTGAGTAACA-3′ |
| Reverse primer: RT16S2 | 5′-CACCCCACCAACAAGCTGATAGG-3′ | |
| RT primer: RT16S3 | 5′-CCGCACGCTCACAG-3′ | |
| Taqman Probe: | 5′-FAM-d(TCCACCACAAGACATGCATCCCGTG)-BHQ-3′ | |
RNA Extraction and Quantitative Real Time-PCR Analysis
Extraction of total Mtb RNA from Middlebrook 7H9 broth-grown and intracellular macrophage-grown cells was done essentially as described previously (8). DNA contamination was removed by treatment with DNase I (Ambion). Approximately 50–100 ng of total RNA was reverse-transcribed to make cDNA specific to fbpB, fbpA, and 16 S rRNA genes using Superscript II reverse transcriptase (Invitrogen). Target cDNA from control and experimental sets were amplified in separate reaction tubes. Real time PCR (TaqMan® chemistry) was carried out in a Bio-Rad I-Cycler using the TaqDNA polymerase (New England Biolabs), TaqMan® probes (Biosearch Technologies), and reverse and forward primers (see Table 1). The calculated threshold cycle (Ct) value for each gene of interest was normalized to the Ct value for 16 S. and the fold expression was calculated using the following formula: fold change = 2−Δ·(ΔCt). No reverse transcription reactions were included as negative controls. Expression data are the average from three independent RNA preparations, each reverse-transcribed and quantified by real time PCR in triplicate.
oriC Plasmid Mutagenesis and Autonomous Replication Analysis
pMQ219 is the Mtb oriC plasmid that has an 814-bp DNA fragment containing the 150-bp 3′-end of the dnaA, the 115-bp 5′-end of the dnaN, and the 553-bp their intergenic region (9). Mutations in the MtrA-boxes F2, F3, F4, and F5 were created by PCR mutagenesis following the previously described protocols (8, 10–12). The “GTCACA” nucleotides of the selected MtrA-boxes in all cases were replaced with a “TATATA” sequence. Plasmid pMMR87 carries mutations in MtrA-box F2, whereas pMMR88 contains mutations in F2, F3, F4, and F5. The integrity of the cloned insert and the mutated residues was confirmed by DNA sequencing. Approximately 250 and 500 ng of oriC plasmid DNA was used to electrotransform Mtb H37Rv (WT), Rv19 (Mtb carrying integrated plasmid lacking the mtrA insert), Rv78 (Mtb MtrA+), and Rv129 (Mtb MtrAD53N) strains, and transformants were selected on Middlebrook 7H10 agar plates containing 10 μg/ml kanamycin (kan) for WT or 10 μg/ml kan and 50 μg/ml hygromycin for Rv19, Rv78, and Rv129. Both Rv78 and Rv129 strains showed poor transformation efficiency. The pZErO 2.1 plasmid was always used as a control. Measurement of transformation frequency, recovery of oriC plasmids, and Southern analysis were performed essentially as described previously (9, 13).
oriC Plasmid Stability Experiments
Actively growing cultures of Mtb merodiploid strains producing normal levels of MtrA (Rv19), elevated levels of phosphorylation-competent Mtb(MtrA+), and phosphorylation-defective Mtb(D53N, MtrA+) MtrA proteins in Middlebrook 7H9 broth containing kan were seeded into fresh growth media lacking kan and grown for different days. At indicated time intervals, aliquots of cultures were removed, diluted, spread on agar plates containing hygromycin, but with and without kan, and incubated at 37 °C for 3 weeks. Colonies from both series of plates were counted, and the proportion of kan-resistant colonies was determined.
DNase I Footprinting Analysis
DNase I protection footprint assays were done as described previously (14). The 32P-5′-end-labeled DNA fragments containing the Mtb oriC and the fbpB promoter were prepared as described previously (14). The oriC end-labeled DNA fragments were prepared from plasmid pMQ219 (9), which contained the same 814 bp of Mtb oriC DNA used for the autonomous replication experiments. The 32P-5′-end label was placed at the polylinker EcoRI (dnaA 3′) site or at the HindIII (dnaN 5′) site (Fig. 3) and at the natural oriC SalI or HpaII sites (footprints not shown). The fbpB promoter end-labeled DNA fragments were prepared from plasmid pGEMT-fbpB and were created by ligating PCR-amplified DNA (a 202-bp fragment amplified by P85bF and P85bR, see primers in Table 1) into the TA overhang plasmid pGEMT-easy (Promega). The 32P-5′-end label was placed at the polylinker NcoI site (Fig. 2) or at the polylinker NdeI site (footprints not shown). Both plasmids pMQ219 and pGEMT-fbpB were sequenced prior to footprint analysis.
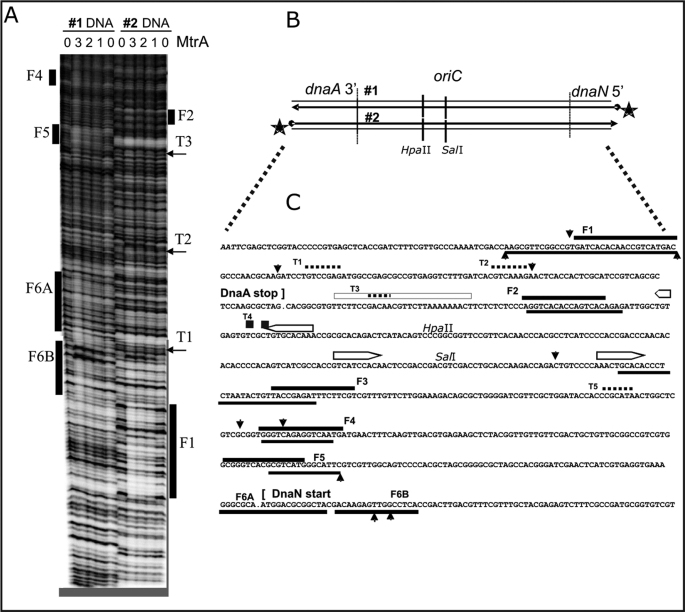
FIGURE 3.
MtrA footprint analysis in the M. tuberculosis oriC region, between 3′ dnaA and 5′ dnaN. A, two representative autoradiograms (#1 and #2) using 32P-5′-end-labeled DNA, diagrammed in B, at the 5′-end of dnaN (#1) and the 3′-end of dnaA (#2). Purified His-tagged MtrA protein was phosphorylated as described in Fig. 2, prior to the DNase I protection footprint analysis, also detailed under “Materials and Methods.” The numbers (0, 1, 2, 3) above the lanes indicate that either 0, 10, 20, or 30 μl of the MtrA (2 mg/ml protein) was added to the standard 200-μl binding reactions. Similarly aligned bars and arrows mark the footprints (F1, F2, … F6A, F6B) and shorter toe prints (T1, T2 …) that are described in the text and aligned with the DNA sequence in C. B, schematic diagram of the M. tuberculosis oriC region is aligned with the 32P-end-labeled DNA (#1 and #2) used in A. The indicated HpaII and SalI sites also served as positions for 32P-end-labeled DNA used in additional footprint experiments not shown. C, summary of MtrA footprint analysis in the M. tuberculosis oriC region. These DNA sequences correspond with the above oriC schematic in B and with the footprints in A. It also summarizes footprint experiments (using internally labeled HpaII and SalI sites) that are not shown. The open bars with pointed ends mark established DnaA-boxes (9). Solid bars mark the footprints (F1, F2, … F6A, F6B), and dotted lines mark the shorter toe prints (T1, T2 …). Perpendicular arrowheads mark the prominent DNase I cut sites that are enhanced by MtrA. When positioned below the presented DNA sequences, these lines and arrows indicate the sequences protected or cleaved on the corresponding bottom strand.
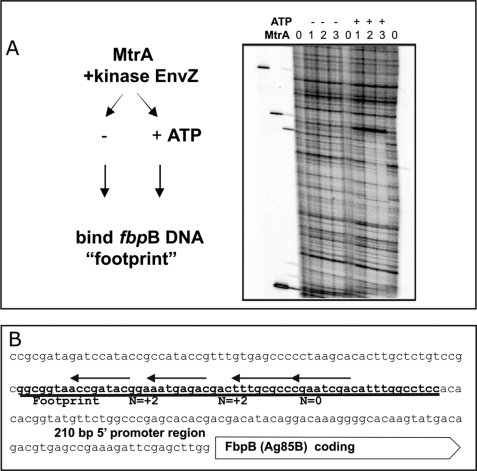
FIGURE 2.
MtrA footprint analysis in the 5′ region of fbpB (Ag85B). A, MtrA binding requires ATP in the protein kinase reaction. Purified His-tagged MtrA protein was phosphorylated by incubation with soluble maltose-binding protein-tagged E. coli EnvZ kinase and 1.0 mm ATP for 30 min prior to the DNase I protection “footprint” analysis using the 32P- 5′-end-labeled DNA fragment, as detailed under “Materials and Methods,” which spans the upstream region of fbpB. The numbers (0, 1, 2, 3) above the lanes indicate that either 0, 10, 20, or 30 μl of the phosphorylated MtrA (2 mg/ml MtrA protein) was added to the standard 200-μl binding reactions. The − or + signs indicate that ATP was either absent or present in otherwise identical reactions. B, DNA sequences bound by MtrA. The DNA sequences immediately upstream to the annotated M. tuberculosis FbpB coding DNA are shown. The zone of MtrA-specific DNase I protection, presented above in footprints A and B, is underlined. The arrows above this footprint align with the best matches to the direct repeats (with variable n = 2 or n = 0 bp spacing). These are the proposed MtrA recognition sequences that are described in the text and in Fig. 5.
In Vitro MtrA Protein Purification and Phosphorylation
The pET plasmid overexpressing His-tagged MtrA protein from a T7 promoter was described previously (8). This plasmid was transformed into the ArcticExpress (DE3) RIL strain (Stratagene, catalog no. 230193), and MtrA overproduction was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside to actively growing cultures at 30 °C and grown for 20 h at 10 °C. The recombinant His-MtrA protein was purified on nickel affinity columns as described previously (8). MalE-EnvZ kinase purification and MtrA phosphorylation by EnvZ protein were essentially performed as described previously (8).
RESULTS
MtrA Phosphorylation Levels and Intramacrophage Growth
Our approach for identifying MtrA RR targets is first to define growth conditions where MtrA phosphorylation-dependent phenotypes are evident and then to infer potential targets and to test them by biochemical and in vivo approaches. Earlier experiments to evaluate the consequences of MtrA overproduction used THP-1 cell lines, which may not have all of the properties of immunoactive MΦ (8). To confirm these findings in a more native system, we evaluated the growth kinetics of Mtb merodiploids producing elevated levels of WT phosphorylation-competent WT MtrA (Rv78) and Mtb merodiploids producing elevated levels of phosphorylation-defective D53N MtrA (Rv129) in human peripheral blood monocyte-derived macrophages. Similar to the results reported with the THP-1 cell lines, the Rv129 merodiploids replicated proficiently in monocyte-derived cells, whereas the Rv78 merodiploids did not (supplemental Fig. S1). These results further support our working hypothesis that, during intracellular growth, the elevated pools of MtrA increase the levels of MtrA∼P and this in turn mis-regulates the targets of MtrA. Such targets could include genes for promoting intramacrophage growth and genes involved in the replication and cell division of Mtb.
ChIP Indicates That Chromosomal Replication Origin, oriC, and the Promoter for Antigen 85B Are MtrA Targets
Based on the above hypothesis, we focused the initial studies on two types of potential targets and analyzed them by ChIP. The first targets included the secreted antigen 85 complex pathway responsible for cell wall biogenesis, and the second targets included the DnaA-mediated oriC replication pathway. The antigen 85 family includes a family of proteins that bind fibronectin and are designated as fbpA, fbpB, and fbpC (15–17). These proteins catalyze mycolyl transferase activity (18), which is critical for generating α-α-trehalose dimycolate or “cord factor”, a major virulence factor (19, 20). Also, infection of monocyte-derived macrophages with Mtb is shown to be associated with a sharp increase in the expression of fbpB, and such a rapid increase is believed to be necessary for establishing infection and subsequent proliferation in macrophages (21). The expression of fbpA is differentially modulated during the acute and chronic stages of infection (22–24). Another critical point for regulating cell proliferation is oriC because it is the site where chromosomal DNA replication begins upon the binding of the initiator protein DnaA as well as several other global regulators that must coordinate replication with cell division (25–27). As additional potential MtrA targets, we selected the promoters of dnaA, the initiator of DNA replication; ftsZ, the initiator of cell division; Rv2719, a cell wall hydrolase regulating Z-ring assembly; mce1, the gene product required for Mtb invasion and possibly virulence (8, 28–30); and finally, mtrA, whose expression, like other RR genes, is expected to be autoregulatory (6, 7).
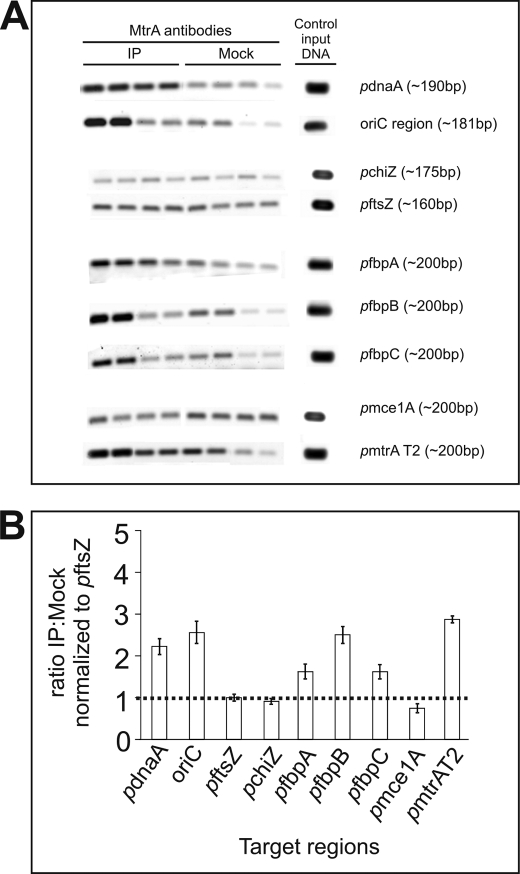
Fig. 1A shows representative ChIP data obtained with anti-MtrA antibodies. As shown, the PCR product signals of oriC and those of the PdnaA and PfbpB were conspicuously increased by binding with anti-MtrA antibodies relative to the no antibody (mock) controls (Fig. 1A). The PCR signals for PRv2719c, PftsZ, and Pmce1 remained unchanged, whereas those of the PfbpA and PfbpC increased modestly relative to the controls (Fig. 1A). Finally, the signal for the PmtrA region was also enriched (Fig. 1A), consistent with the hypothesis that expression of mtrA, like other RR, is autoregulated. To obtain consistent and statistically significant results, we measured the anti-MtrA to mock ratio by densitometric scanning and normalized the data to that of a negative control, the PftsZ ratio. From four independent experiments, we obtained the ratios for oriC (2.6), PfbpB (2.5), PdnaA (2.0), and PmtrA (2.9), whereas those of the PfbpA (1.6) and PfbpC (1.6) were below 2.0 (see Fig. 1B). For initial characterization, we focused on PfbpB and oriC, whose ratios were significantly higher than 2.0.
FIGURE 1.
ChIP experiments. A, agarose gel analysis of ChIP samples. Mtb cell lysates following formaldehyde cross-linking, immunoprecipitation (IP) with anti-MtrA or mock antibodies, and heat reversal of cross-links were used in PCR with select targets. Lysates were diluted 50- and 200-fold for oriC and the promoters of ftsZ, dnaA, chiZ, fbpA, fbpB, and fbpC promoters. For mtrA promoters, 100- and 300-fold diluted lysates were used. PCRs were done in duplicate, and products were resolved by agarose gel electrophoresis, stained with SYBR Green dye, and visualized by scanning in Molecular Imager Fx. Product lanes corresponding to anti-MtrA and Mock antibodies and the target in question are marked. Control input DNA refers to PCR products wherein an aliquot of suitably diluted genomic DNA was used with appropriate primers in amplification reactions. Representative data for each target are shown. B, normalization of ChIP data. PCR products obtained were scanned by densitometry and analyzed by Quantity One software (Bio-Rad Molecular Imager Fx). The ratio of anti-MtrA to mock immunoprecipitation signals was determined for each primer pair and normalized against that of the FtsZ promoter value. The y axis shows the normalized anti-MtrA to mock ratio, and the x axis shows the individual targets.
We supported our ChIP results with electrophoretic mobility shift assays, and we evaluated MtrA binding to oriC with both MtrA and MtrA∼P (phosphorylated MtrA). The MtrA∼P was produced by incubating MtrA with heterologous kinase EnvZ and ATP (8). Incubation of these proteins with the fluorescein isothiocyanate-labeled oriC DNA fragment led to a protein concentration-dependent retardation in the mobility (supplemental Fig. S2). Less MtrA∼P than MtrA was required to shift the oriC DNA, demonstrating that phosphorylated MtrA acquires a higher affinity for oriC-binding sites. Similar results were obtained with the fbpB promoter (data not shown). Together, these in vitro electrophoretic mobility shift assays and in vivo ChIP data demonstrate that both oriC and the PfbpB are significant MtrA targets.
DNase I Footprinting Reveals MtrA-binding Sites
Next, we performed DNase I protein footprinting experiments to identify the MtrA-binding sites in fbpB and oriC. MtrA or MtrA∼P was added to binding reactions with 32P-5′-end-labeled DNA targets. Figs. 2 and 3 provide examples of DNase I protection footprinting experiments in the PfbpB 5′ and the oriC regions. As can be seen, MtrA produces one long footprint (a span of DNase I protection) from 91 to 149 bp upstream of fbpB. This binding requires ATP (Fig. 2A), implying that phosphorylated MtrA∼P gains significant affinity for fbpB target DNA. We did not observe this footprint when ATP was omitted. Assuming 100% MtrA phosphorylation, then MtrA∼P binds fbpB with an apparent Kd ∼10 nm. However, this Kd is a large overestimate. Our unpublished phosphate transfer experiments3 estimated that only ∼1% of MtrA was phosphorylated in these and similar binding assays. Therefore, MtrA∼P affinity is very high, with apparent Kd values around 0.1 nm.
The MtrA protected region is marked with bold line in the ag85B sequence (Fig. 2B). Our MtrA DNA-binding sequence analysis (see below) predicts four direct repeat binding motifs under this long footprint.
Similar footprint experiments in the oriC region produced multiple MtrA footprints (Fig. 3). These experiments used 32P-5′-end-labeled DNA on either side of oriC (number 1, inside the 5′ start of dnaN, and number 2 inside the 3′-end of dnaA). In similar footprint experiments, we also used 32P-end-labeled DNA inside oriC at the HpaII and SalI sites (Fig. 3B) (data not shown). The positions of these footprints are summarized in Fig. 3C. Although the single fbpB 5′ footprint was 60 bp long (Fig. 3C), the oriC footprints were less than half this length, and we distinguished two lengths. Spans of 20–30 bp were labeled as “Footprints” (F1, F2, F3, F4, F5, F6A, and F6B), which are marked as solid lines in Fig. 3C. Spans of less than 10 bp were labeled as “Toe prints” (T1, T2, T3, T4, and T5), and these are marked as dotted lines in Fig. 3C.
MtrA-binding Sequences
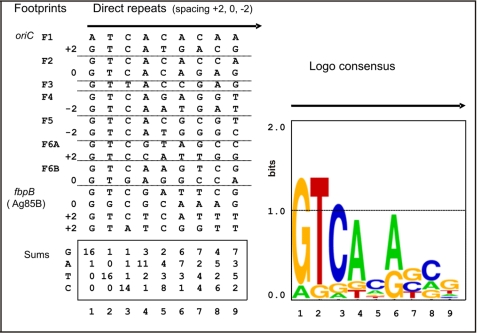
Next, we performed multiple sequence alignments of the MtrA footprinted DNA to identify the common DNA sequences recognized by MtrA. This analysis suggested that MtrA-binding sites were organized as 9-bp direct repeats (Fig. 4), and six of the oriC footprints span two 9-bp direct repeats with either an n = +2, n = 0, or an n = −2 spacing. A base pair frequency analysis of these aligned direct repeats suggested a consensus GTCACAgcg, and a Logo analysis also argued that most of the recognition information lies in the first six positions GTCACA (Fig. 4). The MtrA-binding sites under the fbpB (Ag85B) footprint can also be viewed as four of the same direct repeats with n = 0, +2, +2 spacing, but these direct repeats show more differences from the consensus (compare Fig. 2B with Fig. 4). If one MtrA molecule binds one DNA direct repeat, then four MtrA molecules bind the fbpB (Ag85B). This assumption agrees with the extra length of this footprint. Also, the strict requirement of ATP (MtrA∼P) for binding fbpB (Fig. 2A) suggests an extra cooperative mode of binding by MtrA molecules at fbpB. This view is further supported by the electrophoretic mobility shift assay results in supplemental Fig. S3, where MtrA forms two shifted DNA complexes with fbpB DNA but only one shifted-DNA complex with an oriC-binding site, for example.
FIGURE 4.
Motif log analysis. MtrA binds by apparently recognizing two or more direct repeats similar to GTCACA. The DNA sequences under the footprints in Figs. 2 and 3 were aligned to assume optimum similarity. For 6 out of the 7 oriC footprints, shown in Fig. 3C, the best aligned DNA could be organized as two direct repeats, but a variable spacing of +2, 0 or −2 bp was required for alignment. The exceptional footprint F3 apparently contained only one direct repeat. Frequency and Logo analysis suggests that most of the sequence conservation and presumably most of the information that specifies selective DNA binding lies in the first six positions (GTCACA). Four similar but less exact direct repeats with n = 0, n = +2, and n = +2-bp spacing are also present under the MtrA footprint 5′ to fbpB (Fig. 3C). The supplemental Fig. S3 demonstrates that the n = +2-bp spacing provides a higher affinity than n = 0.
Next, we evaluated the importance of the spacing between the direct repeats (supplemental Fig. S4). For example, the oriC footprint F2 with n = 0 spacing has the closest matching direct repeats (two perfect GTCACAnnn motifs), but F2 occupancy is barely detectable in Fig. 3A (#2). At the same MtrA∼P concentrations, F1 with n = +2 bp spacing is almost fully occupied. Accordingly, we asked if MtrA affinity for F2 would increase with an alternative n = +2 spacing by using electrophoretic mobility shift assays (supplemental Fig. S4). Double-stranded DNA oligonucleotides were modeled after the oriC F2 DNA. Two GTCACAnnn direct repeats with n = 0 (WT) spacing and with n = +2 bp spacing were incubated with serial 1:3 dilutions of an MtrA kinase reaction. Although selective MtrA binding to the WT oriC F2 (n = 0) DNA was barely detectable (supplemental Fig. S4), MtrA affinity for oriC F2 + 2 DNA was substantial, and only these binding reactions could produce reliable Kd measurements (supplemental Fig. S3). MtrA affinity requires both optimal spacing and correct sequence motifs because selective binding is lost with mutant F2 + 2 DNA that has one altered direct repeat motif (supplemental Fig. S4).
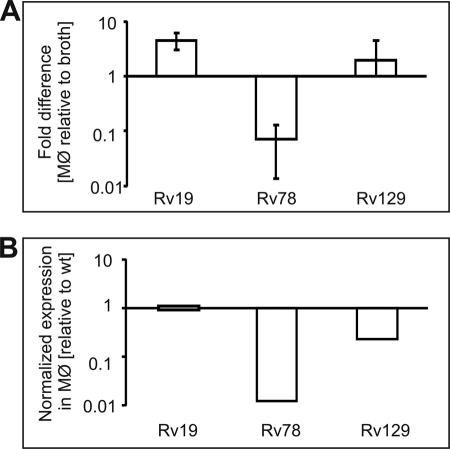
Transcription of fbpB Is Sharply Decreased during Intramacrophage Growth in mtrA Merodiploid Overproducing WT MtrA
As reviewed, the transcription of mtrA in virulent Mtb remains unchanged in broth and during intra-MΦ growth, whereas the fbpB expression is up-regulated upon infection (6, 7, 31). DNase I footprinting data (Fig. 2) imply that fbpB transcription is regulated by MtrA. To test this possibility, we examined the quantitative real time-PCR expression profile of fbpB from intramacrophage and broth-grown bacteria relative to the housekeeping gene, 16 S rRNA. The fold-differences in the fbpB expression in MΦ relative to broth condition are presented (Fig. 5A). The fbpB expression relative to 16 S rRNA was up-regulated upon infection in WT cells producing normal levels of MtrA and MtrB, confirming the published data of Wilkinson et al. (21). In contrast, the fbpB expression was down-regulated in Rv78 Mtb(MtrA+) merodiploids overproducing MtrA (in cells producing normal levels of MtrB) upon infection, although it was restored to near WT levels in the Rv129 Mtb(D53N MtrA+) merodiploids (Fig. 5A). When normalized to WT, the fbpB expression in Rv78 was found to be reduced nearly 70-fold (see Fig. 5B). These results suggest that a consequence of MtrA overproduction leading to an imbalance between MtrA and MtrA∼P levels is reduction of fbpB expression.
FIGURE 5.
Quantitative real time-PCR analysis of fbpB gene expression. The cDNA specific to fbpB and 16 S rRNA genes was synthesized from the RNA samples of Rv19 Mtb (WT), Rv78 Mtb(MtrA+), and Rv129 Mtb (D53N MtrA+) strains grown in broth and macrophages and used to evaluate fbpB expression relative to 16 S rRNA. Data are presented as fold-differences in the expression of macrophage-grown bacteria relative to broth (A). The fbpB RNA expression data of Rv78 and Rv129 were normalized against the wild type strain and are also presented (B).
Mutations in MtrA-boxes Compromise oriC Plasmid Replication
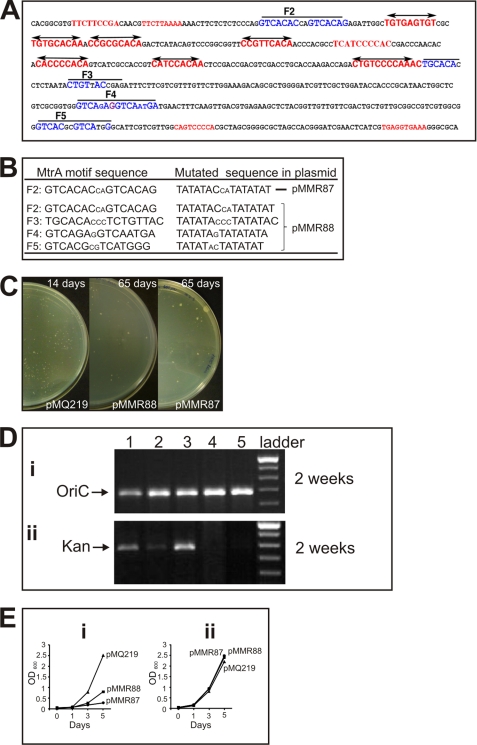
To test how MtrA-boxes (or direct repeats, Fig. 4) might influence autonomous replication, oriC plasmid replication assays were performed with both WT and altered oriC DNA. Previous studies showed that E. coli plasmids do not replicate in Mtb and other mycobacterial species (9, 13, 32). However, such plasmids with an 814-bp DNA Mtb oriC fragment containing the 3′-end of dnaA, the 5′-end of dnaN, and their intergenic region (pMQ219), maintain stable autonomous replication (9, 13). Previous studies also showed that mutations in individual DnaA-boxes, but not in areas located outside the DnaA-boxes, decrease transformation efficiency and autonomous replication activity, indicating that DnaA-boxes are critical for oriC plasmid replication (9, 13, 33). To test if MtrA-boxes are similarly important, we created two plasmids, one containing mutations in the MtrA-box F2 (pMMR87, see Fig. 6, A and B) and the other containing mutations in MtrA-boxes F2, F3, F4, and F5 (pMMR88, see Fig. 6, A and B). The F2-box, although it showed weak affinity to MtrA, is highly conserved in all mycobacterial species, including Mycobacterium leprae (see supplemental Fig. S5). The GTCACA sequence of the MtrA motifs was replaced with TATATA in both plasmids (see Fig. 6B).
FIGURE 6.
oriC sequence, MtrA direct repeat mutations, and oriC plasmid transformation assays. A, oriC sequence (the dnaA-dnaN intergenic region). DnaA-boxes are marked in red, and MtrA-boxes are marked in blue. Mutated residues in the MtrA-boxes F2, F3, F4, and F5 are shown. B, mutated MtrA-box sequences. The presumptive MtrA motifs and the sequences of the mutated boxes are shown for clarity. The plasmid pMMR87 contains only the first F2 mutation and pMMR88 contains all four mutations. C, oriC transformants on agar plates. Approximately 250 ng of WT (pMQ219) and mutant (pMMR87 and pMMR88) plasmid DNA was electrotransformed into Mtb H37Rv WT, and plates were incubated at 37 °C. The pMQ219 transformants in WT background were photographed after 14 days, and the pMMR87 and pMMR88 transformants were photographed after 65 days. D, agarose gel showing oriC and kan cassette PCR products. Genomic DNA of the transformants of pMQ219, pMMR87, pMMR88, and control pZErO 2.1 were extracted and used as templates to amplify oriC (panel i) and the kan cassette (panel ii). The PCR products were resolved on 0.8% agarose gels, stained with ethidium bromide, and photographed. Lane 1, pMQ219; lane 2, pMMR87; lane 3, pMMR88; lane 4, Rv WT; lane 5, pZErO 2.1; lane 6, 1-kb DNA ladder. E, growth curves of Mtb merodiploid strains. The frozen stocks of pooled transformants of pMMR87 and pMMR88 were diluted in Middlebrook 7H9 broth supplemented with oleic acid/dextrose/albumin/sodium chloride with 10 μg/ml kan (in panel i) and without antibiotic (in panel ii) and grown for 10 days to a final A600 between 0.25 and 0.3. The cultures were then diluted to A600 of 0.05, and the growth was monitored daily, as shown. For pMQ219, the freezer stocks were diluted and grown for 3 days to A600 of 0.6 prior to diluting to a final A600 of 0.025. Note that the growth rates of these cultures are very similar when grown in the absence of antibiotic. Similar results were also obtained when antibiotic concentration was increased to 25 μg/ml (data not shown).
Consistent with the published reports, transformation of WT Mtb with the pMQ219 plasmid gave several kan-resistant transformants with a transformation frequency of 104 colonies/μg input DNA (9, 33). The pMQ219 input plasmid was recovered in E. coli when the total DNA preparation of the pMQ219 transformants was used to transform the E. coli Top-10 strain, indicating that this plasmid replicates autonomously in Mtb. In contrast, transformation of Mtb with the pMMR87 and pMMR88 plasmids produced tiny pin-headed colonies, indicating a growth defect from an inability to maintain these oriC plasmids. Transformation of E. coli with the total DNA extracted from the primary transformants collected from several plates failed to produce kan-resistant colonies, indicating that the mutant MtrA plasmids could not be recovered and were therefore not replicating autonomously in Mtb (data not shown). PCR analysis with oligonucleotide primers specific to the oriC or to the kan cassette yielded positive signals (Fig. 6D, panels i and ii), like the WT oriC plasmid, pMQ219, indicating that the mutant oriC plasmids were also present but presumably integrated into the chromosome. Incubation of transformation plates up to 65 days led to a modest increase in the size of the mutant oriC plasmid colonies (Fig. 6C, representative plate is shown). Consistent with the slow colony growth, the liquid broth growth rates of the pMMR87 and pMMR88 transformants were similarly decreased as compared with WT oriC pMQ219, but only when grown in the presence of kan for plasmid selection. No differences in the growth rates were observed when these same cultures were grown without kan (Fig. 6E). A similar PCR analysis of pMMR87 and pMMR88 transformants failed to detect plasmid, i.e. kan cassette after 65 days of incubation indicating plasmid loss (data not shown). Presumably, the presence of the integrated plasmid in these transformants is deleterious in itself. The inability to recover oriC plasmids with mutations in the MtrA-boxes combined with the slow growth of transformants suggests that the mutant plasmids are severely compromised for replication. In particular, pMMR87 demonstrates that mutations in only the one MtrA-box at F2 are sufficient to compromise replication.
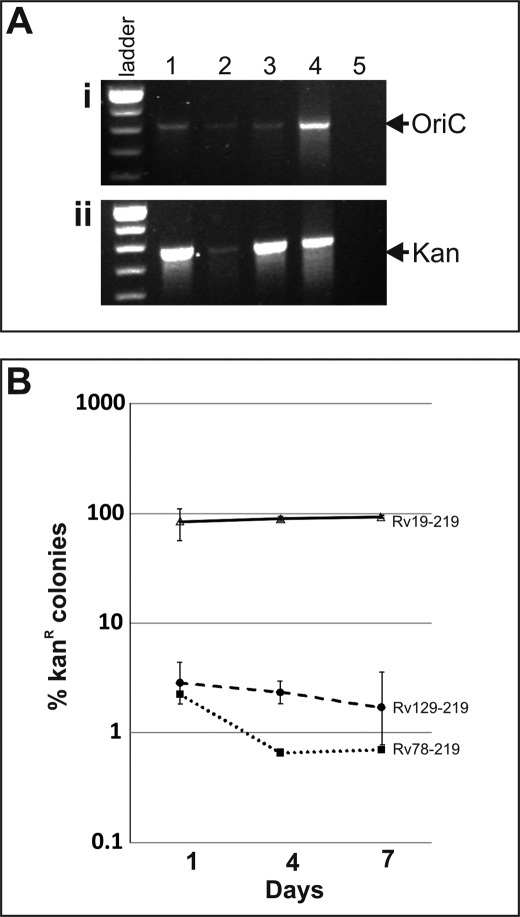
Next, we examined the consequences of MtrA overproduction on oriC plasmid replication and its maintenance by transforming pMQ219 and pMMR88 plasmids into Rv78 and Rv129, which are Mtb merodiploid strains overproducing WT Mtb(MtrA+) and phosphorylation-defective Mtb(D53N, MtrA+) MtrA proteins (8). As a control, these plasmids were also transformed into Rv19, which is WT Mtb carrying an integrated plasmid vector lacking the mtrA insert. Transformation of pMQ219 into Rv78 and Rv129 produced a similar number of transformants (∼104 per μg of DNA) as compared with that into WT (data not shown) indicating that transformation efficiency of pMQ219 plasmid was not affected by MtrA overproduction. PCR analysis of merodiploid transformant genomic DNA confirmed the presence of oriC and kan cassettes of pMQ219 plasmid (Fig. 7A). Efforts to recover pMQ219 plasmid from all three strains were, however, unsuccessful indicating that the pMQ219 plasmid is not replicating extrachromosomally but rather is integrated on the chromosome. Southern analysis supported our interpretation that integration events may have occurred, because we identified the presence of bands corresponding to chromosomal oriC (∼3 kb) and the integrated plasmid (∼5.5 kb) (see supplemental Fig. S6, A and B). DNA band corresponding to ∼4.2 kb would be detected if the pMQ219 plasmid were to be replicating extrachromosomally, and this is what we see in the Mtb WT background (supplemental Fig. S6C, also legend for more details). Because replication potential of the pMQ219 plasmid in Rv WT and Rv19 backgrounds appears to be different, for the sake of clarity, pMQ219 in WT background is referred to as free plasmid and that in Rv19, Rv78, and Rv128 is referred to as integrated plasmid. It is not readily evident why pMQ219 plasmid is excluded in the Rv19 background, and further studies are required to address this issue (see “Discussion”).
FIGURE 7.
Effect of MtrA overproduction on pMQ219 plasmid stability. A, agarose gel showing oriC and kan cassettes. Genomic DNA preparations of Mtb merodiploid strains overproducing WT Mtb(MtrA+) and phosphorylation-defective Mtb(D53N, MtrA+) MtrA proteins, along with the control (RV19), were analyzed by PCR for oriC (panel i) and kan (panel ii). Lane 1, Rv19; lane 2, Rv78; lane 3, Rv129, lane 4, pMQ219 plasmid; and lane 5, Rv WT. B, stability experiments are as follows: Mtb merodiploids actively growing in broth in the absence of kan were diluted at indicated time periods, spread on agar plates with and without kan, and viable colonies appearing after 3 weeks incubation were determined. The ratio of kan-resistant colonies among total cells were presented on semi-log scale.
Assuming that replication is initiated at both origins, i.e. native and integrated oriC, then such activated origins could compete for replication components and pose origin incompatibility issue (36). This could in turn have consequences on cell cycle progression and the stability of integrated plasmid sequences. To evaluate if integrated plasmids bearing oriC sequences are stably maintained, we carried out stability experiments. In these experiments, the pMQ219 merodiploid strains cultured in the absence of kan antibiotic for different days were spread on agar plates without and with kan. As can be seen, less than 5% Rv78 and Rv129 cells were kan-resistant, whereas nearly 100% Rv19 cells were kan-resistant (Fig. 7B). Although differences between the Rv78 and Rv129 strains were minor, plasmid loss in Mtb(MtrA+) was relatively more than that in the Mtb(D53N, MtrA+). These results indicate that the pMQ219 plasmid remained relatively intact in cells producing normal levels of MtrA (Fig. 7B), but was lost significantly under MtrA overproduction conditions, and that MtrA phosphorylation has no effect on plasmid maintenance. Taken together, these results clearly reveal that optimal levels of MtrA are required for maintenance of plasmid bearing oriC sequences and that oriC is the MtrA target.
We also attempted to transform pMMR88 plasmid into Rv19, Rv78, and Rv129. Like the situation seen in the WT background, transformation of pMMR88 plasmid into Rv19 produced several tiny pinhead colonies that grew slowly, thus indicating a growth defect (data not shown). In the case of Rv78, we obtained in two independent experiments 12 and 90 transformants/μg of DNA, respectively, whereas with Rv129 we obtained after repeated attempts only 3 transformants/μg of DNA (data not shown). These results indicated that transformation efficiency of mutant oriC plasmids is severely compromised under MtrA overproduction conditions. The primary transformants grew slowly in liquid broth, but after repeated culturing began to grow like their WT counterparts. Presumably, the newly acquired growth advantage of these strains is due to accumulation of spontaneous mutations. Further studies are required to characterize these strains. Consequently, the pMMR88 transformants were not processed further.
DISCUSSION
In this study, we have identified and validated two important, but rather unrelated, sequences as MtrA targets. These are oriC, the DNA sequence critical for the initiation of chromosomal DNA replication and the duplication of genome, and the promoter for the fbpB gene, which codes for an immunodominant major secreted antigen 85B. Initial in vivo ChIP experiments (Fig. 1) were supported by in vitro DNase I footprinting (Figs. 2 and 3), and their sequence analysis revealed that MtrA recognizes direct repeat motifs resembling GTCACAgcg (Fig. 4) and that binding affinity is increased by MtrA phosphorylation (Fig. 2 and supplemental Figs. S2–S4).
The accuracy of our proposed MtrA recognition sequences is further supported by comparison with the recognition sequences of related RR proteins. MtrA was originally identified by a strong hybridization with cloned PhoB DNA (6), and amino acids sequence alignments confirm a strong affiliation with the PhoB RR family (data not shown). E. coli PhoB RR plays a critical role in regulating the genes involved in the phosphate utilization pathway (5, 37, 38). Alignment of the published PhoB recognition sequences of E. coli, Sinorhizobium meliloti (39), and Streptomyces coelicolor (40) with our proposed MtrA recognition sequences, whose direct repeats are spaced for optimal MtrA binding (+2 spacing, see supplemental Fig. S5), revealed that all of these sequences share the common core GTCA direct repeats with an 11-bp helical repeat. This would place the recognition motifs on the same face of the B-form DNA helix, like other typical DNA-binding proteins.
Overall, most of our MtrA binding data is consistent with the standard model for PhoB and related response regulators, where phosphorylation stimulates dimer protein binding to two direct DNA repeats. This is how we interpreted MtrA binding at the individual oriC footprints, and this interpretation is easily extended to the fbpB promoter, where we propose that phosphorylation stimulates the binding of two MtrA dimers to four direct DNA repeats. It should be noted that the fbpB promoter direct repeats only partially match our proposed MtrA recognition sequences. However, this defect is presumably compensated by the optimal +2 spacing and perhaps by cooperative binding between four MtrA molecules (one per direct repeat). It is pertinent to recognize that although the fbpB genes of Mtb and other mycobacterial members are identified, detailed promoter analyses, including the identification of −35 and −10 elements, have not been reported. Nonetheless, analysis of the 175 bp upstream of different fbpB regions clearly shows significant sequence conservation under the MtrA footprint (see supplemental Fig. S7 for further details). Conservation of the sequences in the footprinted region of different mycobacterial species further supports our conclusion that the fbpB promoter is an MtrA target.
Although our results argue that MtrA recognizes GTCACA motifs with this typical 11-bp helical repeat, our results also unexpectedly argue for the usage of atypical 9- and 7-bp helical repeats (described as +2, 0, and −2 spacing between 9-bp direct repeats in Fig. 4). Presumably, MtrA binding to DNA is best when the 6-bp motif is on the same face of the helix. Using the F2 oriC as an example, we showed that MtrA binding to F2 requires the GTCACA motif, and it is barely detectable unless the natural spacing is increased from 0 to +2 or +3 (supplemental Fig. S4). Despite this intrinsically weak affinity for MtrA, three points argue that F2 is physiologically important and perhaps critical for replication control. First, changing only the GTCACA repeats in F2 abolished the autonomous replication activity of the Mtb oriC plasmid in Mtb (pMMR87, Fig. 6). Previous studies showed that mutations in DnaA-boxes comparably impaired the autonomous replication activity of oriC plasmids (9, 13, 33). Second, the atypically spaced GTCACA repeats in F2 are conserved among diverse Mycobacteria species (supplemental Fig. S4). Only DnaA-boxes show a comparable degree of conservation among these oriC DNA sequences. F2 is perfectly conserved among Mtb, M. leprae, and Mycobacterium avium (supplemental Fig. S6). Interestingly, in the more distant species, where base pair changes are seen in the corresponding GTCACA repeats, additional and perhaps compensatory GTCACA repeats are present. For example, Mycobacterium smegmatis and Mycobacterium flavescens have a third perfect GTCACA motif with the same atypical spacing. This observation further suggests a selective pressure for creating MtrA-binding sites and not simply the lack of genetic drift.
Third, F2 is located inside oriC, between the DnaA-boxes and the AT-rich unwinding site (supplemental Fig. S5) (41). Although detailed molecular details on how the initiation of DNA replication in Mtb occurs are unknown, recent studies reveal that the initial interactions of DnaA with DnaA-boxes in the presence of ATP are necessary for a rapid oligomerization of DnaA at oriC and the formation of the DnaA-oriC initiation complex competent for initiation (42). This initial step is followed by unwinding of oriC at the AT-rich sequences, which can occur in the absence of helicases and other replication proteins (41). Thus, the strategic location of F2 in oriC suggests that MtrA bound at F2 either aids or interferes with these key replication steps (see below). Although detailed studies are required to understand the mechanism of action, results presented in Fig. 6 suggest that MtrA activity is necessary for maintaining stable autonomous replication of oriC plasmids. The relative weakness of the F2 MtrA-binding site argues that it is bound in vivo when the ratio of MtrA∼P to MtrA is high.
The MtrA-oriC footprinting data show that MtrA at the 814-bp oriC region produces seven shorter (∼30 bp) footprints (F1, F2, F3, F4, F5, F6A, and F6B) and several even shorter (∼10 bp) points of DNase I protection that we called toe prints (Fig. 3). It should however be noted that the dnaA-dnaN intergenic region has just four MtrA-binding sites (see Fig. 3C: F2, F3, F4, and F5). Such dispersed contacts across all of oriC, including the dnaA 3′- and dnaN 5′-coding DNA, are more typical of DnaA protein binding to multiple DnaA-boxes that span bacterial oriCs, including Mtb oriC (42). It is possible that the toe prints are not true MtrA-binding sites but rather could be due to the consequence of the formation of large MtrA-oriC nucleoprotein complexes. Presumably, these could be similar to the observed weaker DnaA-binding sites of E. coli oriC, which can be mutated without any effect (43). Further studies are required to address this issue. It is pertinent to note that in E. coli cells, DnaA persistently binds to the high affinity DnaA-boxes in oriC, but the key low affinity DnaA-boxes are unoccupied until they are required during the initiation of chromosome replication. This high to low affinity DnaA-box hierarchy is an important part of the E. coli oriC regulatory system (44). The Caulobacter crescentus response regulator CtrA binds five sites spread across its replication origin (45). Perhaps like the multiple DnaA-boxes, the multiple oriC MtrA-binding sites may form a hierarchy of persistently bound (high affinity) sites and transiently bound (low affinity such as F2) sites that are similarly required for Mtb oriC replication control. Clearly, further studies are required to address how MtrA controls oriC replication.
How might MtrA activity function to regulate oriC replication? The MtrA contact sites in oriC are distinctly different from the DnaA-boxes (42). This makes it unlikely that MtrA binding to oriC affects the first step of initiation, namely the binding of the DnaA protein to DnaA-boxes and vice versa. However, MtrA binding to oriC could either aid or hinder the ability of the DnaA protein to oligomerize at oriC and could thereby control the replication initiation process. Because MtrA∼P preferentially binds to these targets, we propose that, under normal growth conditions where signals for MtrA phosphorylation are limiting and tightly regulated by the cognate MtrB sensor kinase, the MtrA-oriC interactions lead to regulated replication resulting in stable autonomous replication. Thus, the absence of MtrA-boxes as in pMMR87 and pMMR88 (see Fig. 6) could impair oriC plasmid replication possibly leading to plasmid loss. However, for growth conditions that promote an elevated MtrA∼P state, i.e. during intracellular growth in cells overproducing MtrA in the absence of MtrB, it is expected that the elevated pools of MtrA∼P can access all of its recognition sequences in oriC. This situation could lead to an interference with the DnaA protein binding to oriC and the formation of the oriC-DnaA initiation complex. This line of thinking leads us to speculate that MtrA can play a positive or a negative role depending upon its phosphorylation potential. Perhaps the elevated pools of MtrA∼P limits the ability of the DnaA protein to initiate replication during nonreplicative persistence or the chronic stage of infection.
One unexpected finding is that the pMQ219 plasmid, which replicates extrachromosomally in the WT background, is excluded in WT cells carrying integrated plasmid vector, i.e. Rv19. This result is independent of MtrA overproduction phenotypes (see supplemental Fig. S6 and legend). The mycobacterial shuttle vectors based on pAL5000 replication origin (34) generally replicate as free plasmids in the integrated plasmid background. The pMQ219, which is a derivative of pZErO 2.1 plasmid (9), and the integration plasmid pJFr-19, which is used to affect MtrA levels (8), share more than the 800-bp region of homology. It is possible that some factors or components involved in Mtb oriC replication function in recombination; these mechanisms are not yet fully understood. Although speculative, such factors, if any, could preferentially promote recombination between pMQ219 and the resident plasmid at the region of homology resulting in integration events when both oriC sequences are primed for initiation. Further studies, including the construction and characterization of minichromosomes lacking such homologous sequences, are required to understand why the pMQ219 plasmid is excluded in the Rv19 background.
Although detailed studies are required to understand the roles of MtrA on oriC replication, stability experiments revealed that optimal levels of MtrA, irrespective of its phosphorylation state, are required for the maintenance of integrated oriC plasmid sequences in Rv78 and Rv129 backgrounds (Fig. 7). We envision that MtrA influences oriC replication, and hence under overproduction conditions promote activation of oriC at the integrated locus. This could then potentially create oriC incompatibility issues as the presence of activated oriC at two different regions on the chromosome, i.e. native as well as integrated locus, could be detrimental for normal cell cycle progression (36). A consequence is the loss of plasmid-bearing oriC sequences. One intriguing finding of our data is that, unlike the WT Rv19 cells, the integrated oriC plasmids are not stably maintained in merodiploids overproducing phosphorylation-defective Mtb(D53N, MtrA+) MtrA protein. It should be noted that our experiments were carried out with MtrA merodiploid strains but not with the strains producing phosphorylation-defective Mtb(D53N, MtrA+) as the sole source. Thus, it is likely the Mtb(D53N, MtrA+) will have both homo- and heterodimers of MtrA, and both species could be competent in affecting oriC replication. Future studies with Mtb mtrA mutants producing MtrA(D53N) as the sole source for MtrA will be required to address these issues.
In contrast to oriC, MtrA at the fbpB promoter produced one discrete long (∼60 bp) footprint (Fig. 2), consistent with four MtrA molecules binding together. This binding clearly shows a preference for MtrA∼P (Fig. 2A). Evaluation of the binding characteristics of MtrA to the fbpB promoter and oriC may reflect different regulatory functions. For example, MtrA makes one focused contact inside the fbpB promoter, as is typical of transcriptional regulators. This argues that MtrA∼P acts as a transcriptional repressor for the fbpB gene. Such transcriptional repression could occur under the conditions that elevate the MtrA phosphorylation state. Earlier studies revealed that the phosphorylation potential of MtrA is elevated during intramacrophage growth in cells overproducing phosphorylation-competent MtrA in the absence of the MtrB sensor kinase (8). Mtb secretes copious amounts of highly immunogenic Ag85B protein (46), whose processed peptides contain epitopes for T-cell recognition (35). Thus, the reduced expression of fbpB upon infection under elevated MtrA∼P conditions could limit the available pools of secreted antigen 85B for its subsequent processing by antigen presentation pathways. It is pertinent to note that the MtrA footprinted region of fbpB promoter is well conserved in other mycobacterial species as well (see supplemental Fig. S7).
Interestingly, analysis of fbpA expression under MtrA overproduction conditions, however, revealed a different expression profile from that of fbpB (supplemental Fig. S8). The expression of fbpA was reduced in Rv19 Mtb WT cells upon macrophage infection, although it was elevated in both Rv78 Mtb(MtrA+) and Rv129 mtrA merodiploids (supplemental Fig. S8). The phosphorylation-independent increase in the fbpA expression in Rv129 Mtb (D53N MtrA+) merodiploid suggests that the regulation of fbpA is different and not directly dependent on a balanced amount of MtrA phosphorylation. Also, because the ChIP signal for PfbpA is weaker than for pfbpB (Fig. 1), it is possible that the observed changes in the fbpA expression are due to secondary effects or to more distant binding sites. It is unknown if expression of genes responsible for other secreted antigens and other immunodominant proteins are also affected by MtrA. Nonetheless, MtrA∼P-dependent reduction of the expression of fbpB and possibly other unrecognized genes responsible for antigenic proteins could be one of the many immune evasion mechanisms the pathogen uses for its long term persistent growth during chronic infection and in granuloma.
Supplementary Material
Acknowledgments
We thank Naveen Nair for technical assistance, Dr. A. Chauhan and Erin Maloney for RNA preparation and Southern experiments, Meredith Moomey for help with the construction of oriC plasmids, and Dr. Julia Grimwade for helpful advice and carrying out preliminary footprinting experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants AI 084734 and AI 73966 (to M. V. M.) and AI 48417 (to M. R.). This work was also supported by Canadian Institutes for Health Research Operating Grant MT-13453 (to M.-C. O., D. P. B., and G. T. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Methods” and Figs. S1–S8.
M. Rajagopalan and M. V. Madiraju, unpublished data.
- MΦ
- macrophages
- 2CRS
- two-component regulatory systems
- RR
- response regulator
- ChIP
- chromatin immunoprecipitation
- WT
- wild type
- kan
- kanamycin.
REFERENCES
- 1.North R. J., Jung Y. J. (2004) Annu. Rev. Immunol. 22, 599–623 [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann S. H., Cole S. T., Mizrahi V., Rubin E., Nathan C. (2005) J. Exp. Med. 201, 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith I. (2003) Clin. Microbiol. Rev. 16, 463–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 5.Hoch J. A. (2000) Curr. Opin. Microbiol. 3, 165–170 [DOI] [PubMed] [Google Scholar]
- 6.Via L. E., Curcic R., Mudd M. H., Dhandayuthapani S., Ulmer R. J., Deretic V. (1996) J. Bacteriol. 178, 3314–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahrt T. C., Deretic V. (2000) J. Bacteriol. 182, 3832–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fol M., Chauhan A., Nair N. K., Maloney E., Moomey M., Jagannath C., Madiraju M. V., Rajagopalan M. (2006) Mol. Microbiol. 60, 643–657 [DOI] [PubMed] [Google Scholar]
- 9.Qin M. H., Madiraju M. V., Rajagopalan M. (1999) Gene 233, 121–130 [DOI] [PubMed] [Google Scholar]
- 10.Chauhan A., Madiraju M. V., Fol M., Lofton H., Maloney E., Reynolds R., Rajagopalan M. (2006) J. Bacteriol. 188, 1856–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan M., Atkinson M. A., Lofton H., Chauhan A., Madiraju M. V. (2005) Biochem. Biophys. Res. Commun. 331, 1171–1177 [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto K., Low B., Rutherford S. A., Rajagopalan M., Madiraju M. V. (2001) Biochem. Biophys. Res. Commun. 280, 898–903 [DOI] [PubMed] [Google Scholar]
- 13.Qin M. H., Madiraju M. V., Zachariah S., Rajagopalan M. (1997) J. Bacteriol. 179, 6311–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siam R., Brassinga A. K., Marczynski G. T. (2003) J. Bacteriol. 185, 5563–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abou-Zeid C., Garbe T., Lathigra R., Wiker H. G., Harboe M., Rook G. A., Young D. B. (1991) Infect. Immun. 59, 2712–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Content J., de la Cuvellerie A., De Wit L., Vincent-Levy-Frébault V., Ooms J., De Bruyn J. (1991) Infect. Immun. 59, 3205–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiker H. G., Harboe M. (1992) Microbiol. Rev. 56, 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., Besra G. S. (1997) Science 276, 1420–1422 [DOI] [PubMed] [Google Scholar]
- 19.Brennan P. J., Nikaido H. (1995) Annu. Rev. Biochem. 64, 29–63 [DOI] [PubMed] [Google Scholar]
- 20.Goren M. B. (1972) Bacteriol. Rev. 36, 33–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson R. J., DesJardin L. E., Islam N., Gibson B. M., Kanost R. A., Wilkinson K. A., Poelman D., Eisenach K. D., Toossi Z. (2001) Mol. Microbiol. 39, 813–821 [DOI] [PubMed] [Google Scholar]
- 22.Pai S. R., Actor J. K., Sepulveda E., Hunter R. L., Jr., Jagannath C. (2000) Microb. Pathog. 28, 335–342 [DOI] [PubMed] [Google Scholar]
- 23.Shi L., Jung Y. J., Tyagi S., Gennaro M. L., North R. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi L., North R., Gennaro M. L. (2004) Infect. Immun. 72, 2420–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bramhill D., Kornberg A. (1988) Cell 54, 915–918 [DOI] [PubMed] [Google Scholar]
- 26.Bramhill D., Kornberg A. (1988) Cell 52, 743–755 [DOI] [PubMed] [Google Scholar]
- 27.Zyskind J. W., Smith D. W. (1986) Cell 46, 489–490 [DOI] [PubMed] [Google Scholar]
- 28.Casali N., White A. M., Riley L. W. (2006) J. Bacteriol. 188, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chauhan A., Lofton H., Maloney E., Moore J., Fol M., Madiraju M. V., Rajagopalan M. (2006) Mol. Microbiol. 62, 132–147 [DOI] [PubMed] [Google Scholar]
- 30.Shimono N., Morici L., Casali N., Cantrell S., Sidders B., Ehrt S., Riley L. W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 15918–15923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haydel S. E., Clark-Curtiss J. E. (2004) FEMS Microbiol. Lett. 236, 341–347 [DOI] [PubMed] [Google Scholar]
- 32.Rajagopalan M., Qin M. H., Nash D. R., Madiraju M. V. (1995) J. Bacteriol. 177, 6527–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dziadek J., Rajagopalan M., Parish T., Kurepina N., Greendyke R., Kreiswirth B. N., Madiraju M. V. (2002) J. Bacteriol. 184, 3848–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kana B. D., Mizrahi V. (2004) Tuberculosis 84, 63–75 [DOI] [PubMed] [Google Scholar]
- 35.Mustafa A. S., Shaban F. A., Abal A. T., Al-Attiyah R., Wiker H. G., Lundin K. E., Oftung F., Huygen K. (2000) Infect. Immun. 68, 3933–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dasgupta S., Løbner-Olesen A. (2004) Plasmid 52, 151–168 [DOI] [PubMed] [Google Scholar]
- 37.Blanco A. G., Sola M., Gomis-Rüth F. X., Coll M. (2002) Structure 10, 701–713 [DOI] [PubMed] [Google Scholar]
- 38.Zundel C. J., Capener D. C., McCleary W. R. (1998) FEBS Lett. 441, 242–246 [DOI] [PubMed] [Google Scholar]
- 39.Yuan Z. C., Zaheer R., Morton R., Finan T. M. (2006) Nucleic Acids Res. 34, 2686–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sola-Landa A., Rodríguez-García A., Apel A. K., Martín J. F. (2008) Nucleic Acids Res. 36, 1358–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Farhana A., Kumar S., Rathore S. S., Ghosh P. C., Ehtesham N. Z., Tyagi A. K., Hasnain S. E. (2008) PLoS ONE 3, e2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madiraju M. V., Moomey M., Neuenschwander P. F., Muniruzzaman S., Yamamoto K., Grimwade J. E., Rajagopalan M. (2006) Mol. Microbiol. 59, 1876–1890 [DOI] [PubMed] [Google Scholar]
- 43.Riber L., Fujimitsu K., Katayama T., Løbner-Olesen A. (2009) Mol. Microbiol. 71, 107–122 [DOI] [PubMed] [Google Scholar]
- 44.Leonard A. C., Grimwade J. E. (2005) Mol. Microbiol. 55, 978–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siam R., Marczynski G. T. (2000) EMBO J. 19, 1138–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harth G., Horwitz M. A., Tabatadze D., Zamecnik P. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 15614–15619 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.