Abstract
Modification of the lipid A moiety of bacterial lipopolysaccharide influences cell wall properties, endotoxic activity, and bacterial resistance to antimicrobial peptides. Known modifications are variation in the number or length of acyl chains and/or attached phosphoryl groups. Here we identified two genes (gnnA and gnnB) in the major foodborne pathogen Campylobacter jejuni that enable the synthesis of a GlcN3N precursor UDP 2-acetamido-3-amino-2,3-dideoxy-α-d-glucopyranose (UDP-GlcNAc3N) in the lipid A backbone. Mass spectrometry of purified lipooligosaccharide verified that the gene products facilitate the formation of a 2,3-diamino-2,3-dideoxy-d-glucose (GlcN3N) disaccharide lipid A backbone when compared with the β-1′-6-linked d-glucosamine (GlcN) disaccharide observed in Escherichia coli lipid A. Functional assays showed that inactivation of the gnnA or gnnB gene enhanced the TLR4-MD2-mediated NF-κB activation. The mutants also displayed increased susceptibility to killing by the antimicrobial peptides polymyxin B, colistin and the chicken cathelicidin-1. The gnnA and gnnB genes are organized in one operon with hemH, encoding a ferrochelatase catalyzing the last step in heme biosynthesis. These results indicate that lipid A modification resulting in amide-linked acyl chains in the lipid A is an effective mechanism to evade activation of the innate host defense and killing by antimicrobial peptides.
Keywords: Antimicrobial Peptides, Bacterial Genetics, Innate Immunity, Lipopolysaccharide (LPS), Toll-like Receptors (TLR), Campylobacter jejuni, Lipid A
Introduction
Lipopolysaccharide (LPS)2 is an amphipathic molecule that is an essential component of the outer membrane of most Gram-negative bacteria. LPS generally consists of three distinct structural domains: the lipid A moiety that anchors the molecule in the membrane, the core oligosaccharide, and a variable number of repeating oligosaccharide units, the O-antigen (1). LPS is not only critical for cell wall integrity, it is also one of the most potent activators of the innate immune system. LPS can activate the Toll-like receptor 4 (TLR4)-MD2 receptor complex, which triggers the production of proinflammatory mediators and antimicrobial peptides and indirectly steers the adaptive immune response (2–4).
To escape recognition by the innate immune system, Gram-negative bacteria have evolved several mechanisms to modify the structure of lipid A. Most lipid A-modifying enzymes are known to be regulated in response to changes in environmental conditions (5). The modifications can alter recognition by the TLR4 complex and/or promote resistance of the bacterial cell wall to host cationic antimicrobial peptides. The addition of polar groups, such as phosphoethanolamine, 4-amino-4-deoxy-l-arabinose, and/or palmitate, to the lipid A reduces its negative charge and limits its interaction with the cationic antibiotic polymyxin B or other antimicrobial peptides (AMPs) (6–9). Another mechanism of lipid A modification is to change the number of acyl chains attached to the disaccharide backbone. Hereto, Salmonella has the outer membrane enzymes PagP and PagL. The altered number of acyl chains increases the resistance to AMPs and decreases the cellular signaling through TLR4 (10, 11). This indicates that lipid A modifications can have immune-evasive as well as immune-modulating effects.
Biologically potent lipid A usually consists of a conserved β-1′-6-linked glucosamine (GlcN) disaccharide backbone with two ester- and two amide-linked acyl chains (12). Some bacterial species, including Leptospira interrogans and Acidithiobacillus ferrooxidans, can synthesize lipid A with only amide-linked acyl chains (GlcN3N). In A. ferrooxidans, the dehydrogenase (GnnA) and the transaminase (GnnB) convert UDP-GlcNAc to the analogue UDP-GlcNAc3N, in which the GlcNAc 3-OH group is replaced with an amine (13). Recombinant GnnA and GnnB from A. ferrooxidans, expressed in Escherichia coli, also yield large quantities of UDP-GlcNAc3N lipid A molecules. The possible immune-modulating or immune-evasive role of this type of modification of lipid A has thus far not been investigated (14).
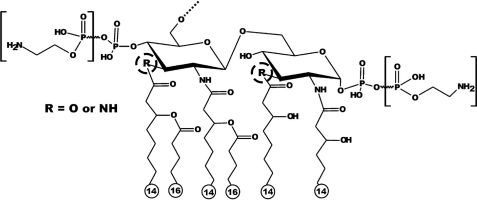
Campylobacter jejuni is a Gram-negative spiral-shaped bacterium that causes gastrointestinal illness in humans. The LPS of C. jejuni lacks the O-antigen and therefore is often referred to as lipooligosaccharide (LOS). The structure of the Campylobacter LOS is highly variable, and this may contribute to modulation of the host innate immune response. Structural analysis of C. jejuni lipid A indicates that this bacterium displays heterogeneity in its lipid A (Fig. 1). The major structure of the C. jejuni lipid A contains a hybrid backbone of a β-1′-6-linked GlcN3N-GlcN disaccharide with two phosphate groups and six saturated fatty acids (15). C. jejuni has mixed lipid A with mainly three amide-linked acyl chains and only one ester-linked acyl chain instead of two ester- and two amide-linked acyl chains, present in most Enterobacteriaceae (16). It is proposed that lipid A with more hydroxyl-bound acyl chains is more biologically active toward TLR4 than lipid A with more amide-bound acyl chains (12, 16, 17). This might explain the reported difference in biological activity between E. coli and C. jejuni lipid A (12). In this study, we identified the C. jejuni genes responsible for adding amide-linked acyl chains in the lipid A and demonstrated that this modification influences the biological activity of lipid A by altering the TLR4 response and the bacterial resistance to antimicrobial peptides.
FIGURE 1.
Schematic structure of the lipid A of C. jejuni wild-type 11168. The variable acyl linkages are indicated by R (encircled by a dotted line), which can be either oxygen (O) or NH. Variable phosphoethanolamine group(s) are indicated between brackets (43).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The strains and plasmids used in this study are listed in Table 1. C. jejuni strains were routinely grown on plates containing blood agar base II (Oxoid, London, UK) supplemented with 5% horse blood (Biotrading Benelux b.v., Mijdrecht, The Netherlands) lysed with 0.5% saponin (Sigma) or in heart infusion broth (HI) (Oxoid) at 37 or 42 °C under microaerophilic conditions (5% O2, 10% CO2, 85% N2). E. coli strains were grown in Luria-Bertani medium (Biotrading) at 37 °C. When appropriate, medium was supplemented with chloramphenicol (20 μg/ml), kanamycin (50 μg/ml), and/or ampicillin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristics | Source or reference |
|---|---|---|
| C. jejuni strains | ||
| 11168 (H1) | Wild-type | (44, 45) |
| 11168 gnnA::Cm | 11168 derivative gnnA::Cm | This study |
| 11168 gnnB::Cm | 11168 derivative gnnB::Cm | This study |
| 11168 gnnA::CmR | 11168 derivative gnnA::Cm (reverse) | This study |
| 11168 gnnB::CmR | 11168 derivative gnnB::Cm (reverse) | This study |
| 11168 gnnB::Cm + pMA1–504-505 | 11168 derivative gnnB::Cm + pMA1–504-505 | This study |
| E. coli strains | ||
| PC2955 | relA1 Φ80dlacZΔM15 phoA8 hsdR17 recA1 endAl gyrA96 thi-1 relA1 luxS glnV44 | Netherlands Culture Collection of Bacteria (NCCB) |
| Plasmids | ||
| pGEM-T easy | PCR cloning vector, AmpR | Promega Corp. |
| pGEMhemH-504–505 | pGEM-T easy containing hemH, gnnA and gnnB | This study |
| pGEM504 | pGEM-T easy containing gnnA | This study |
| pGEM505 | pGEM-T easy containing gnnB | This study |
| pGEM504::Cm | pGEM-T easy containing gnnA::Cm | This study |
| pGEM505::Cm | pGEM-T easy containing gnnB::Cm | This study |
| pGEM504–505 | pGEM-T easy containing gnnA and gnnB | This study |
| pMA1 | Suitable for conjugation | (19) |
| pMA1–504-505 | pMA1 containing C. jejuni gnnA and gnnB | This study |
| pAV35 | pBluescript II SK containing E. coli Cmr cassette | (18) |
Construction of C. jejuni gnnA::Cm and gnnB::Cm Mutants
The Campylobacter genes hemH, Cj0504c (gnnA), and Cj0505c (gnnB) were amplified by PCR using the primers HemHRev and 505Fwd (Table 2). The resulting fragment of 1912 bp was cloned into the pGEM-T easy vector (Promega Corp., Madison, WI) to form plasmid pGEMhemH-504-505. Inverse PCR using the primers Cj504BamHIR and Cj504BamHIF was performed on pGEMhemH-504-505 to replace 811 bp of the Cj0504 gene by a unique BamHI site. The inverse PCR product was self-ligated to form plasmid pGEMΔ504. The primers Cj505BamHIR and Cj505BamHIF were used in a similar procedure to replace 441 bp of the Cj0505 gene by a unique BamHI site, yielding pGEMΔ505. The pGEMΔ504 and pGEMΔ505 plasmids were digested with BamHI to insert a 0.7-kb BamHI chloramphenicol cassette (Cm) isolated from plasmid pAV35 (18). Both Cm orientations were selected, resulting in the plasmids pGEMΔ504::Cm, pGEMΔ504::CmR, pGEMΔ505::Cm, and pGEMΔ505::CmR. The plasmids isolated from E. coli DH5α were introduced into C. jejuni 11168H1 via electroporation. Homologous recombination resulting in double-crossover events was verified by PCR.
TABLE 2.
Primers used in this study
| Primer name | DNA sequence (5′ → 3′) |
|---|---|
| 505Fwd | GCAACAGCTGAAGTTGTGG |
| HemHRev | CACAGGCTTTGAGTAAGTTC |
| Cj504stopXbaI | AGATCTTTCACGCAAACCTTTCTAAAATTT |
| Cj505startSacI | GAGCTCATTTCTCTAAGGTTTTTTATGAATTT |
| 21Fwd (CmFwd) | GGAGGATAAATGATGCAATTCAC |
| 22Rev (CmRev) | CTGGGATTTTATTTATTCAGCAAG |
| HemHR | CTTAGTTAAGATCAGATAAATAC |
| Cj505F | ATGAATTTTATCAATCTTCAAG |
| Cj504BamHIR | AGGATCCGACCTATGATACCTATTTTC |
| Cj504BamHIF | TGGATCCACAAAGTTCAAGAAATTTTA |
| Cj505BamHIR | TGGATCCATCATCATGGCAAAAAATAG |
| Cj505BamHiF | AGGATCCGCAAGAGCAAGTTATTTGTATTTTT |
| pmetKsphI | GCATGCAGTTGATTTTAACTAACTTTTGCT |
| GFPrev | ACAAGTGTTGGCCATGGAACA |
Complementation of C. jejuni gnnA::Cm and gnnB::Cm Mutants
To complement the C. jejuni gnnA:: Cm and gnnB::Cm mutants, a shuttle plasmid was constructed with intact copies of Cj0504c and Cj0505c as insert. Therefore, the genes were amplified from the chromosome of C. jejuni strain 11168H1 by PCR with the primers Cj504stopXbaI and Cj505startSacI and the proofreading enzyme platinum Pfx DNA polymerase (Invitrogen). After the addition of a 5′-A overhang to the PCR product with Taq polymerase (Invitrogen), the product was ligated into pGEM-T easy to obtain plasmid pGEM504-505. Finally, pGEM504-505 and pMA1 (19) were digested with SacI and SacII, and the resulting 1986-bp fragment of pGEM504-505 was ligated into plasmid pMA1 behind the C. jejuni metK promoter to yield plasmid pMAI-504-505. E. coli S17 was used to conjugate plasmid pMAI-504-505 into C. jejuni gnnB::Cm, following described procedures (20).
Protoporphyrin IX Detection
For the detection of protoporphyrin IX, Campylobacter grown overnight in HI were harvested by centrifugation and resuspended in TEN buffer (40 mm Tris/HCl, pH 7.5, 1 mm EDTA, and 150 mm NaCl). Fluorescence of bacterial cells from 2 × 109 colony-forming units/ml was measured using an LS50B luminescence spectrometer (PerkinElmer Life Sciences) (excitation at 405 nm, emission at 630 nm) (21).
Isolation of LOS
C. jejuni LOS was isolated by the hot phenol extraction method as described (22). In brief, bacteria grown on blood agar plates for 20 h at 42 °C under microaerophilic conditions were harvested in 5 ml of water, pelleted by centrifugation (5500 × g, 30 min, 4 °C), and resuspended in 10 volumes of bacterial wet weight in distilled water and an equal volume of hot phenol (70 °C). After 2 h of incubation (70 °C) with intermittently vortexing and centrifugation (18,000 × g, 20 min, 4 °C), the water phase containing the LOS was collected. To increase the yield of LOS, a second extraction was done by adding an equal volume of distilled water to the phenol phase. LOS was precipitated from the water phase by adding sodium acetate (0.1 g/g of wet weight of bacteria) and 2 volumes of cold acetone (−20 °C). After incubation for 16 h at −20 °C, LOS was collected by centrifugation (18,000 × g, 30 min, 4 °C), washed in cold acetone, and dissolved in 3.5 ml of distilled water. Centrifugation (100,000 × g, 2 h, 4 °C) was performed to remove DNA. For further purification, the pellet was resuspended in 250 μl of water and treated overnight with 1 unit of DNase and 0.25 mg of proteinase K. Finally, the LOS was collected by centrifugation (100,000 × g, 2 h, 4 °C) and dissolved in distilled water up to a final concentration of 1 mg/ml. Purified LOS of C. jejuni was quantified by the Purpald assay (23).
Tricine-SDS-PAGE
C. jejuni LOS samples were analyzed by Tricine-SDS-PAGE (24). Each lane was loaded with 100 ng of purified C. jejuni LOS. The gels were run at a constant current of 20 mA for about 4 h and silver-stained to visualize the LOS (25).
Mass Spectrometry Analysis
C. jejuni lipid A analyses were done as described by Geurtsen et al. (26) with a modification in the LOS hydrolysis. The LOS sample was dissolved in 500 μl of 10 mm sodium acetate, pH 4.5, containing 1% SDS and placed in an ultrasonic bath until the sample was completely dissolved. After 1 h of incubation (100 °C), the sample was dried by vacuum centrifugation. To remove the SDS, the sample was washed with 100 μl of distilled water and 500 μl of acidified ethanol (100 μl of 4 m HCl with 20 ml of 95% ethanol) followed by centrifugation (2000 × g, 10 min). To remove the acid, the sample was washed twice with non-acidified 95% ethanol and centrifuged (2000 × g, 10 min). The sample was lyophilized to yield solid lipid A. Just prior to the analysis, the sample was reconstituted in 200 μl of a mixture of chloroform/methanol/water (3:1.5:0.25, v/v), and a 10-μl aliquot of the clear supernatant was pipetted into a gold-coated borosilicate glass nanospray needle (manufactured in-house, essentially as described by Wilm and Mann (27)). A static nanoelectrospray source (built in-house) was mounted onto an LCQ Classic mass spectrometer (Thermo Scientific), operated in negative ion mode at mass unit resolution. The heated capillary was set at 200 °C, and the atmospheric pressure ionization source collision-induced dissociation energy (SID) was enabled at 15 V. Automatic gain control was used to control the filling of the ion trap at a full mass spectrometry target of 1 × 107 ions. Scans were acquired from 400–2000 Da with 3 microscans and a maximum inject time of 150 ms.
Cell Culture, Transfection, and TLR Activation
The HeLa 57A cell line stably transfected with an NF-κβ luciferase reporter construct (28) was routinely cultured in 25-cm2 tissue culture flasks (Corning) in Dulbecco's modified Eagle's medium (Invitrogen) with 5% fetal calf serum in a humidified incubator at 37 °C and 10% CO2. Transfection of the cells with plasmids encoding TLR4, MD2, and CD14 was essentially performed as described (29). After 48 h of incubation, the medium was replaced with fresh medium, and cells were stimulated (5 h) with the indicated concentrations of purified LOS. After stimulation with LOS, the cells were rinsed twice with 500 μl of Dulbecco's phosphate-buffered saline (pH 7.4) and lysed in 100 μl of reporter lysis buffer (Promega) for 30 min at −80 °C. Firefly luciferase activity was measured with the luciferase assay system (Promega) using a luminometer (TD-20/20, Turner Designs, Sunnyvale, CA). The transfection efficiency was normalized using the β-galactosidase assay (Promega) as described (29).
Antimicrobial Peptide Susceptibility
Log phase bacteria grown at 42 °C in HI were resuspended to a concentration of 106 colony-forming units/ml in 10 mm sodium phosphate buffer, pH 7.0, supplemented with 1/100 volume HI medium (minimal HI medium) in 96-well polypropylene microtiter plates. After the addition of the indicated concentrations of polymyxin B, colistin E, or cathelicidin-1 and 1 h of incubation (37 °C, microaerophilic conditions), serial dilutions were plated and incubated 24 h at 42 °C under microaerophilic conditions on charcoal plates to enable counting of colony-forming units.
Statistical Analyses
Data were analyzed with a two-tailed paired t test (Graphpad prism). A p value of <0.05 was considered to be significant.
RESULTS
Identification of the Putative C. jejuni gnnA and gnnB Homologs
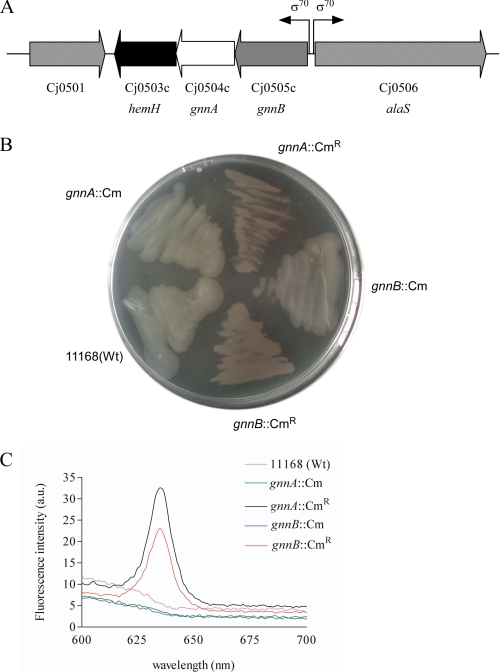
C. jejuni lipid A contains both GlcN saccharides and GlcN3N saccharides in contrast to E. coli LPS (12, 17). In A. ferrooxidans, the genes gnnA and gnnB are responsible for the synthesis of GlcN3N-containing lipid A (14). Bioinformatics revealed that the genome of C. jejuni strain 11168 contains two genes, Cj0504c and Cj0505c, which are respectively 40 and 55% similar at the amino acid level to the GnnA and GnnB proteins of A. ferrooxidans. The C. jejuni gnnA and gnnB gene and the downstream gene hemH, putatively involved in the protoheme biosynthesis, have a similar gene orientation and overlapping start and stop codons, suggesting that they may form an operon (Fig. 2A). The upstream gene alaS coding for an alanyl-tRNA synthetase has an opposite orientation, indicating that the promoter of the putative gnnB-gnnA-hemH operon may be located directly in front of gnnB. Genomic analysis of all C. jejuni genome sequences available in the public databases revealed a similar organization of the gnnB-gnnA-hemH genes in all strains.
FIGURE 2.
The gnnA and gnnB genes are organized in one operon with hemH. A, chromosomal arrangement of the hemH-Cj0504(gnnA)-Cj0505(gnnB) operon and its flanking regions in wild-type C. jejuni 11168. The position of the presumed sigma 70 (σ70) promoter is indicated. B, thioglycollate agar plate inoculated with wild type strain 11168 and the mutants gnnA::Cm, gnnA::CmR, gnnB::Cm, and gnnB::CmR illustrating the red colonies caused by the accumulation of protoporphyrin. C, spectrofluorometry of the strains mentioned in B after resuspension in TEN buffer to verify the presence of protoporphyrin. a.u., arbitrary units; Wt, wild type.
Fluorescent Phenotype of C. jejuni gnnA and gnnB Mutants
To investigate the function of the putative gnnA and gnnB homologs in C. jejuni strain 11168, both genes were disrupted by insertion of a chloramphenicol resistance (CmR) cassette in either orientation. PCR analysis verified that the desired four types of transformants (gnnA::Cm and gnnB::Cm with CmR in two orientations) were obtained. Unexpectedly, the pellets of the mutants that contained the CmR cassette in the opposite direction as the gnnA or gnnB genes appeared red. This red color was also visible when these mutants were grown on a transparent thioglycollate plate (Fig. 2B). Fluorescence microscopy revealed that these bacteria were intensely red fluorescent (data not shown). In other bacterial species, the HemH protein is involved in the protoheme biosynthesis and catalyzes the insertion of a ferrous ion into protoporphyrin IX to synthesize protoheme. Disruption of hemH in Pseudomonas fluorescens causes the accumulation of protoporphyrin, resulting in typical reddish colonies, which are highly fluorescent under UV light (21). To investigate whether the red fluorescence observed for the C. jejuni gnnA::CmR and gnnB::CmR mutants is due to the accumulation of protoporphyrin, we performed fluorescence spectrum analysis (Fig. 2C). This yielded a spectrum with a peak at 630 nm only for the mutants with the CmR cassette in a polar orientation, which corresponds to that of protoporphyrin (21).
We hypothesized that the fluorescent bacterial phenotype may have been caused by a dampening effect of the CmR promoter on the transcription of the hemH gene. Real-time reverse transcriptase-PCR analysis confirmed the strong reduction of hemH transcript in these gnnA and gnnB mutants. These results confirm that the genes gnnA, gnnB, and hemH are transcribed from a promotor in front of gnnB. To investigate whether the transcription of the gnnB-gnnA-hemH operon is altered in response to heme or oxidative stress, real-time reverse transcriptase-PCR analysis was performed on RNA isolated from bacteria grown in the presence of the heme precursor d-aminolevulinic acid, iron, dipyridyl, or H2O2. None of these conditions significantly changed the gnnB-gnnA-hemH transcript levels.
Further analysis of the transformants demonstrated that the gnnA and gnnB mutants that carried the CmR cassette in the non-polar orientation were not fluorescent and showed parental growth rates and motility in semi-solid agar (data not shown). These mutants were used for further structural analysis of lipid A and in the biological assays described below.
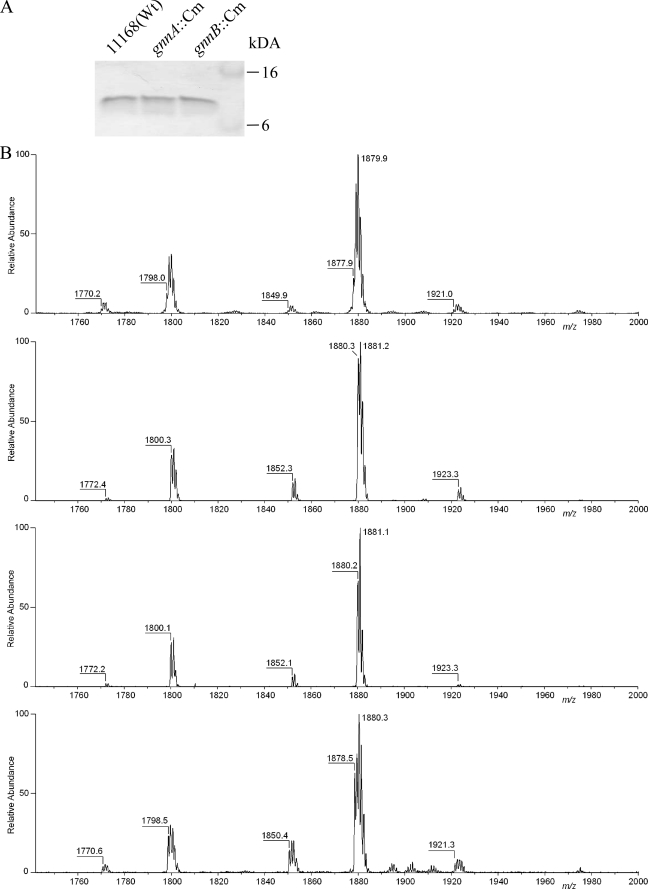
LOS Structural Analysis
The main function of GnnA and GnnB in A. ferrooxidans is in the biosynthesis of GlcN3N-substituted lipid A (13, 14). To analyze the linkage of the acyl chains in the lipid A of the C. jejuni gnnA::Cm or gnnB::Cm mutants, LOS was isolated from the mutants and parental strain after growth at the optimal C. jejuni growth temperature of 42 °C. Tricine-SDS-PAGE of these samples demonstrated similar electrophoretic mobility and staining of the LOS (Fig. 3A), indicating no large structural differences. To enable more detailed analysis of the lipid A, the different LOS samples were hydrolyzed and analyzed by static nanoelectrospray ionization mass spectrometry in the negative ion mode. This showed similar relative abundances of peaks for the lipid A of the wild-type and the mutant strains (Fig. 3B), indicating a similar distribution of lipid A isoforms. The lipid A of the wild type (Fig. 3B, upper trace) showed an abundant peak at a monoisotopic molecular ion of m/z 1877.9 Da, representing the hexa-acylated diphosphate containing lipid A, whereas the peak at m/z 1798.0 Da corresponds to a hexa-acylated monophosphate species (30). Because of the heterogeneity of the constituents, we have confirmed these monoisotopic m/z values with higher mass resolution data acquisitions (data not shown). The remaining minor peaks at m/z 1770.2, 1849.9, and 1921.0 Da represent respectively hexa-acylated monophosphate species with a C14 instead of a C16 acyl chain, hexa-acylated diphosphate species with a C14 instead of a C16 acyl chain, and hexa-acylated monophosphate species with a phosphoethanolamine. As expected, all peaks observed for the lipid A of the parent strain had shifted by two mass units in the gnnA::Cm and gnnB::Cm mutants, indicating substitution of one N-linked acyl chain by an O-linked acyl chain. To verify that this difference was solely due to disruption of the gnnA or gnnB gene, we analyzed the LOS of the gnnB mutant complemented in trans with plasmid pMA1-504-505. This plasmid contains intact copies of the gnnA and gnnB gene placed under the control of a constitutive (metK) promoter. Mass spectrometry demonstrated a decrease in lipid A masses to wild-type levels for the complemented gnnB mutant strain, although some heterogeneity in mass was observed (Fig. 3B, bottom panel). Overall, these results demonstrate that inactivation of either gnnA or gnnB results in replacement of one N-linked by an O-linked acyl chain in the lipid A.
FIGURE 3.
LOS structural analysis. A, silver-stained Tricine-SDS-PAGE gel loaded with equal amounts of LOS isolated from C. jejuni strain 11168 and its gnnA::Cm and gnnB::Cm derivatives grown at 42 °C. Wt, wild type. B, negative ion electrospray mass spectra of purified lipid A obtained from the wild-type strain, the gnnA::Cm mutant, the gnnB::Cm mutant, and the gnnB::Cm mutant strain containing the complementation plasmid pMA1-504-505 (top to bottom). Monoisotopic peaks at m/z 1798.0 and 1877.9 Da represent a hexa-acylated monophosphate species and the characteristic hexa-acylated biphosphate species, respectively. Three additional minor peaks were observed in all strains, the monoisotopic molecular ions corresponding to penta-acylated monophosphate (m/z 1770.2 Da), a hexa-acylated biphosphate species substituted with a C14 instead of a C16 acyl chain (m/z 1849.9 Da), and hexa-acylated lipid A with one phosphate molecule and one phosphoethanolamine group attached to the lipid A (m/z 1921.0 Da).
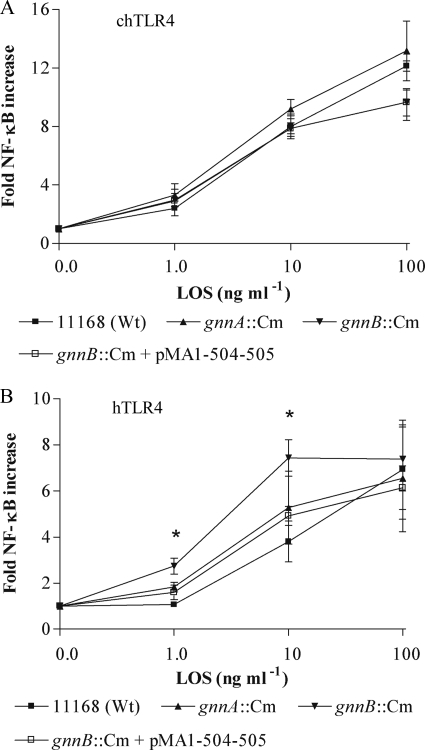
Biological Activity of the gnnA::Cm and gnnB::Cm LOS
To investigate whether the type of linkage of the acyl chain is important for activation of the innate immune response by lipid A, we tested the effect of isolated gnnA and gnnB LOS on the activation of HeLa 57A cells transfected with plasmids containing TLR4, MD2, and CD14. Cellular activation was measured using the NF-κB luciferase reporter construct that is stably expressed by HeLa 57A cells (29). Incubation of the cells expressing the human TLR4-MD2 complex with the different purified LOS samples showed significant enhanced cellular activation for the mutant strains when compared with the parent strain at 1 ng/ml purified LOS and for the gnnB::Cm mutant also at 10 ng/ml (p < 0.05) (Fig. 4A). As TLR4 ligand specificity varies between species and chickens are a natural habitat for C. jejuni, we also evaluated the effect of the different LOS types on the activation of the chicken TLR4-MD2 complex. HeLa 57A cells transfected with chTLR4-chMD2, stimulated with the different C. jejuni LOS, showed no difference in NF-κB activation (Fig. 4B), verifying that the effect of the lipid A modification varies between species. The increased activation of the human TLR4-MD2 complex by the mutant LOS was partially restored to wild-type levels after complementation of the gnnB mutant with pMA1-504-505. Overall, our results indicate that the substitution of an N-linked with an O-linked acyl chain in the lipid A results in a more biologically active LOS, at least in humans.
FIGURE 4.
Species-specific recognition of C. jejuni GnnA and GnnB LPS. NF-κB-luciferase activity in HeLa 57A cells transfected with (A) chTLR4-chMD2-hCD14 or (B) hTLR4-hMD2-hCD14 after 5 h of stimulation with LOS purified from wild-type (Wt) strain 11168, the gnnA::Cm mutant, gnnB::Cm mutant, and the gnnB::Cm mutant complemented with pMA1-504-505. Values are indicated as -fold NF-κB increase and are the mean ± S.D. of four independent experiments. *, p < 0.05.
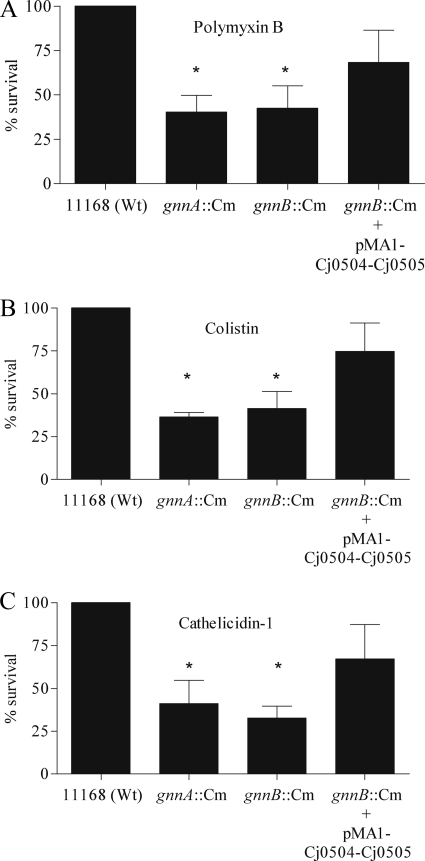
Modification of Lipid A with GlcN3N Influences Resistance to Antimicrobial Peptides
Modification of lipid A can alter the resistance to AMPs (31). To investigate whether the lipid A modification caused by C. jejuni GnnA and GnnB influenced the susceptibility to AMPs, we incubated log phase-grown wild-type strain 11168, the gnnA and gnnB mutants, and the complemented gnnB mutant strain with increasing concentrations of polymyxin B, colistin E, or chicken cathelicidin-1. After 1 h of incubation, bacterial viability was determined by colony-forming unit counting. The gnnA and gnnB mutants showed at least 50% less survival to all three antimicrobial peptides when compared with the parental strain (Fig. 5). Equal survival percentages for the wild type and mutants were seen when antibiotics with a different bacterial target were used such as kanamycin (data not shown). The increased sensitivity of the gnnB mutant to AMPs was partially abolished in the complemented gnnB mutant strain. These data clearly demonstrate that the lipid A modification not only inhibits the activation of the innate immunity but also decreases the susceptibility to AMPs.
FIGURE 5.
Susceptibility of gnnA::Cm and gnnB::Cm mutant bacteria to AMPs. Resistance to antimicrobial peptides of the C. jejuni strain 11168 wild type (Wt), the gnnA::Cm and gnnB::Cm mutants, and the gnnB::Cm mutant containing the complementation plasmid pMA1-504-505 is shown. Logarithmic phase bacteria were incubated for 1 h with 5 μg/ml polymyxin B (A), 5 μg/ml colistin E (B), and 0.1 μm chicken cathelicidin-1 (C). Bacterial survival was determined 24 h after plating of the suspensions on charcoal plates. Results are expressed as the percentage of the inoculum that survives the exposure to the antimicrobial agents and are the mean ± S.D. of three independent experiments. *, p < 0.05.
DISCUSSION
Lipid A is the main constituent of the outer membrane of Gram-negative microorganisms and is the principal ligand of the innate immune receptor TLR4. During evolution, many bacteria have evolved enzymes that modify the lipid A structure and thus alter the physico-chemical and biological properties of the molecule. Here we identified two C. jejuni genes, gnnA and gnnB, that enable the addition of an N-linked acyl chain instead of an O-linked acyl chain to the disaccharide lipid A backbone. This modification decreases the potency of lipid A as a TLR4 agonist as demonstrated by the enhanced activation of the human TLR4-MD2 complex by purified LOS from the mutant microbes. In addition, these mutants are more susceptible to the antimicrobial peptides polymyxin B, colistin E, and chicken cathelicidin-1. The altered linkage of the acyl chain in the lipid A conferred by GnnA and GnnB may thus have evolved to enable the bacterium to resist the innate host defense.
The C. jejuni genes gnnA and gnnB are located in a single operon with hemH. This gene organization is conserved in all sequenced C. jejuni strains and Campylobacter lari but not in other Campylobacter strains or different bacterial species (13). The potential significance of the coordinated transcription of the gnnA/B and hemH genes is unknown. HemH is a protein that catalyzes the insertion of a ferrous ion into protoporphyrin IX to synthesize protoheme. In E. coli, hemH transcription is modestly increased in response to heme limitation (32). Furthermore, high heme concentrations cause lipid A modifications in the oral pathogen Porphyromonas gingivalis (33). The hemH gene in E. coli and Salmonella enterica is also activated in response to hydrogen peroxide (34, 35). We investigated whether the transcription of the gnnB-gnnA-hemH operon is altered in response to heme, the heme precursor d-aminolevulinic acid, iron, or H2O2, but none of these conditions significantly changed the gnnB-gnnA-hemH transcript levels.
The function of C. jejuni gnnA and gnnB in lipid A modification was established by mass spectrometry analyses of the isolated LOS of the wild-type and mutant strains. The observed two mass units increase of the lipid A isoforms for the mutants, which was reversed after introduction of an intact copy of the defective gene, indicates that inactivation of either gene caused the addition of an ester-linked acyl chain instead of an amide-linked acyl chain. This is fully consistent with the assumed function of GnnA and GnnB as lipid A-modifying enzymes as determined for A. ferrooxidans GnnA and GnnB (13, 14).
Our primary goal was to investigate the impact of variation in the number of amide-bound acyl chains on the biological properties of the lipid A. This was tested using the activation of NF-κB in HeLa cells transfected with genes encoding human or chicken TLR4-MD2 as a read-out system. This assay has previously been shown to be instrumental in demonstrating differences in TLR4 recognition of LOS species (29). Our finding that in this assay the purified LOS of the gnnA::Cm and gnnB::Cm mutants is more potent than wild-type LOS (Fig. 4A) provides for the first time an experimental basis for the hypothesis that lipid A containing GlcN3N-GlcN is less biologically active than lipid A that has a GlcN disaccharide backbone (16). The altered potency of the LOS was only detected toward the human TLR4-MD2 complex and not for cells expressing the chicken TLR4-MD2 receptor complex (Fig. 4B). These results are in agreement with previous work that indicates differences in ligand specificity of human, murine, and chicken TLR4 (36, 37). The altered potency of the LOS with variable acyl chain linkages extends the known repertoire of immune-modulating lipid A modifications such as variation in the length and number of acyl chains, the addition of polar groups, and variation in the number of phosphate molecules attached to the lipid A (5, 38).
The molecular basis for the variable potency of lipid A may be related to a decreased flexibility of the N-linked acyl chains when compared with the O-linked acyl chains (16). Crystal structures of the interaction between LPS and the TLR4-MD2 complex (PDB code 3FXI) have revealed that the number and the flexibility of the acyl chains might influence the interaction between lipid A and the TLR4-MD2 complex (39). We speculate that because the gnnA::Cm and gnnB::Cm mutants contain more flexible acyl chains in their LOS, the lipid A may fit more easily into the grove of the TLR4-MD2 complex, resulting in enhanced hTLR4 activation.
The composition of LPS may also have major effects on the bacterial resistance to antimicrobial agents. C. jejuni is very resistant to the AMP polymyxin B, a compound often added to medium to select for Campylobacter species (40). We observed that wild-type C. jejuni strain 11168 has a lower susceptibility to polymyxin B, colistin E, and cathelicidin-1 when compared with the gnnA::Cm and gnnB::Cm mutants. This suggests that the reduced number of N-linked acyl chains in the lipid A of the mutants causes increased sensitivity to AMPs. Bacterial killing by AMPs supposedly involves interaction of the small amphipathic peptides with the negatively charged LPS. A number of Gram-negative pathogens resist AMPs by modifying their LPS, especially the lipid A part (41). For example, the incorporation of phosphoethanolamine in the lipid A of Salmonella strongly reduces the susceptibility toward polymyxin B, whereas phosphate molecules attached to the lipid A backbone appear important for cathelicidin-1 binding (42). Our mass spectrometry results indicate that apart from adding phosphate or phosphoethanolamine (15), C. jejuni exploits the gnnA and gnnB modification to increase its resistance to polymyxin B, colistin E, and cathelicidin-1 (Fig. 5). The altered resistance may be attributed to increased membrane stability and permeability caused by the amide linkage when compared with ester-linked acyl chains.
In conclusion, our results indicate a novel mechanism of lipid A modification in C. jejuni that involves GnnA- and GnnB-mediated replacement of an ester-linked by an amide-linked acyl chain in the lipid A. This results in a lipid A species with reduced endotoxin activity but increased bacterial resistance to antimicrobial peptides.
Acknowledgment
We thank Dr. A. van Dijk for providing AMP cathelicidin-1.
This work was supported by NWO-VIDI Grant 917.66.330 (to M. M. S. M. W.) and ZonMW Grant 9120-6150 (to J. P. M. v. P.).
- LPS
- lipopolysaccharide
- LOS
- lipooligosaccharide
- AMP
- antimicrobial peptide
- TLR
- Toll-like receptor
- Cm
- chloramphenicol cassette
- HI
- heart infusion broth
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- h
- human
- ch
- chicken
- GlcN3N
- 2, 3-diamino-2,3-dideoxy-d-glucose
- UDP-GlcNAc3N
- UDP 2-acetamido-3-amino-2,3-dideoxy-α-d-glucopyranose.
REFERENCES
- 1.Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pålsson-McDermott E. M., O'Neill L. A. (2004) Immunology 113, 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H. M., Park B. S., Kim J. I., Kim S. E., Lee J., Oh S. C., Enkhbayar P., Matsushima N., Lee H., Yoo O. J., Lee J. O. (2007) Cell 130, 906–917 [DOI] [PubMed] [Google Scholar]
- 4.Miller S. I., Ernst R. K., Bader M. W. (2005) Nat. Rev. Microbiol. 3, 36–46 [DOI] [PubMed] [Google Scholar]
- 5.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo L., Lim K. B., Poduje C. M., Daniel M., Gunn J. S., Hackett M., Miller S. I. (1998) Cell 95, 189–198 [DOI] [PubMed] [Google Scholar]
- 7.Trent M. S., Ribeiro A. A., Lin S., Cotter R. J., Raetz C. R. (2001) J. Biol. Chem. 276, 43122–43131 [DOI] [PubMed] [Google Scholar]
- 8.Lee H., Hsu F. F., Turk J., Groisman E. A. (2004) J. Bacteriol. 186, 4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Z., Ribeiro A. A., Lin S., Cotter R. J., Miller S. I., Raetz C. R. (2001) J. Biol. Chem. 276, 43111–43121 [DOI] [PubMed] [Google Scholar]
- 10.Bishop R. E., Gibbons H. S., Guina T., Trent M. S., Miller S. I., Raetz C. R. (2000) EMBO J. 19, 5071–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trent M. S., Pabich W., Raetz C. R., Miller S. I. (2001) J. Biol. Chem. 276, 9083–9092 [DOI] [PubMed] [Google Scholar]
- 12.Schromm A. B., Brandenburg K., Loppnow H., Moran A. P., Koch M. H., Rietschel E. T., Seydel U. (2000) Eur. J. Biochem. 267, 2008–2013 [DOI] [PubMed] [Google Scholar]
- 13.Sweet C. R., Ribeiro A. A., Raetz C. R. (2004) J. Biol. Chem. 279, 25400–25410 [DOI] [PubMed] [Google Scholar]
- 14.Sweet C. R., Williams A. H., Karbarz M. J., Werts C., Kalb S. R., Cotter R. J., Raetz C. R. (2004) J. Biol. Chem. 279, 25411–25419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran A. P., Zähringer U., Seydel U., Scholz D., Stütz P., Rietschel E. T. (1991) Eur. J. Biochem. 198, 459–469 [DOI] [PubMed] [Google Scholar]
- 16.Moran A. P. (1997) J. Infect. Dis. 176, Suppl. 2, S115–S121 [DOI] [PubMed] [Google Scholar]
- 17.Rietschel E. T., Brade H., Holst O., Brade L., Müller-Loennies S., Mamat U., Zähringer U., Beckmann F., Seydel U., Brandenburg K., Ulmer A. J., Mattern T., Heine H., Schletter J., Loppnow H., Schönbeck U., Flad H. D., Hauschildt S., Schade U. F., Di Padova F., Kusumoto S., Schumann R. R. (1996) Curr. Top. Microbiol. Immunol. 216, 39–81 [DOI] [PubMed] [Google Scholar]
- 18.van Vliet A. H., Wooldridge K. G., Ketley J. M. (1998) J. Bacteriol. 180, 5291–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Mourik A., Bleumink-Pluym N. M., van Dijk L., van Putten J. P., Wösten M. M. (2008) Microbiology 154, 584–592 [DOI] [PubMed] [Google Scholar]
- 20.Labigne-Roussel A., Harel J., Tompkins L. (1987) J. Bacteriol. 169, 5320–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baysse C., Matthijs S., Pattery T., Cornelis P. (2001) FEMS Microbiol. Lett. 205, 57–63 [DOI] [PubMed] [Google Scholar]
- 22.Keestra A. M., van Putten J. P. (2008) J. Immunol. 181, 4354–4362 [DOI] [PubMed] [Google Scholar]
- 23.Lee C. H., Tsai C. M. (1999) Anal. Biochem. 267, 161–168 [DOI] [PubMed] [Google Scholar]
- 24.Lesse A. J., Campagnari A. A., Bittner W. E., Apicella M. A. (1990) J. Immunol. Methods 126, 109–117 [DOI] [PubMed] [Google Scholar]
- 25.Tsai C. M., Frasch C. E. (1982) Anal. Biochem. 119, 115–119 [DOI] [PubMed] [Google Scholar]
- 26.Geurtsen J., Angevaare E., Janssen M., Hamstra H. J., ten Hove J., de Haan A., Kuipers B., Tommassen J., van der Ley P. (2007) J. Biol. Chem. 282, 37875–37884 [DOI] [PubMed] [Google Scholar]
- 27.Wilm M. S., Mann M. (1994) Int. J. Mass Spectrom. Ion Processes 136, 167–180 [Google Scholar]
- 28.Rodriguez M. S., Thompson J., Hay R. T., Dargemont C. (1999) J. Biol. Chem. 274, 9108–9115 [DOI] [PubMed] [Google Scholar]
- 29.van Aubel R. A., Keestra A. M., Krooshoop D. J., van Eden W., van Putten J. P. (2007) Mol. Immunol. 44, 3702–3714 [DOI] [PubMed] [Google Scholar]
- 30.Marsden G. L., Li J., Everest P. H., Lawson A. J., Ketley J. M. (2009) J. Bacteriol. 191, 2392–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peschel A. (2002) Trends Microbiol. 10, 179–186 [DOI] [PubMed] [Google Scholar]
- 32.McNicholas P. M., Javor G., Darie S., Gunsalus R. P. (1997) FEMS Microbiol. Lett. 146, 143–148 [DOI] [PubMed] [Google Scholar]
- 33.Coats S. R., Jones J. W., Do C. T., Braham P. H., Bainbridge B. W., To T. T., Goodlett D. R., Ernst R. K., Darveau R. P. (2009) Cell Microbiol. 11, 1587–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng M., Wang X., Templeton L. J., Smulski D. R., LaRossa R. A., Storz G. (2001) J. Bacteriol. 183, 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elgrably-Weiss M., Park S., Schlosser-Silverman E., Rosenshine I., Imlay J., Altuvia S. (2002) J. Bacteriol. 184, 3774–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steeghs L., Keestra A. M., van Mourik A., Uronen-Hansson H., van der Ley P., Callard R., Klein N., van Putten J. P. (2008) Infect. Immun. 76, 3801–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hajjar A. M., Ernst R. K., Tsai J. H., Wilson C. B., Miller S. I. (2002) Nat. Immunol. 3, 354–359 [DOI] [PubMed] [Google Scholar]
- 38.Raetz C. R., Guan Z., Ingram B. O., Six D. A., Song F., Wang X., Zhao J. (2009) J. Lipid Res. 50, S103–S108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) Nature 458, 1191–1195 [DOI] [PubMed] [Google Scholar]
- 40.Skirrow M. B. (1977) Br. Med. J. 2, 9–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trent M. S., Stead C. M., Tran A. X., Hankins J. V. (2006) J. Endotoxin Res. 12, 205–223 [DOI] [PubMed] [Google Scholar]
- 42.Bhunia A., Mohanram H., Bhattacharjya S. (2009) Biopolymers 92, 9–22 [DOI] [PubMed] [Google Scholar]
- 43.Szymanski C. M., Michael F. S., Jarrell H. C., Li J., Gilbert M., Larocque S., Vinogradov E., Brisson J. R. (2003) J. Biol. Chem. 278, 24509–24520 [DOI] [PubMed] [Google Scholar]
- 44.Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., Chillingworth T., Davies R. M., Feltwell T., Holroyd S., Jagels K., Karlyshev A. V., Moule S., Pallen M. J., Penn C. W., Quail M. A., Rajandream M. A., Rutherford K. M., van Vliet A. H., Whitehead S., Barrell B. G. (2000) Nature 403, 665–668 [DOI] [PubMed] [Google Scholar]
- 45.Gundogdu O., Bentley S. D., Holden M. T., Parkhill J., Dorrell N., Wren B. W. (2007) BMC Genomics 8, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]