Abstract
Phosphatidic acid (PA) is a pleiotropic lipid second messenger in mammalian cells. We report here that extracellular PA acts as a leukocyte chemoattractant, as membrane-soluble dioleoyl-PA (DOPA) elicits actin polymerization and chemotaxis of human neutrophils and differentiated proleukemic HL-60 cells. We show that the mechanism for this involves the S6 kinase (S6K) signaling enzyme. Chemotaxis was inhibited >90% by the S6K inhibitors rapamycin and bisindolylmaleimide and by S6K1 silencing using double-stranded RNA. However, it was only moderately (∼30%) inhibited by mTOR siRNA, indicating the presence of an mTOR-independent mechanism for S6K. Exogenous PA led to robust time- and dose-dependent increases in S6K enzymatic activity and Thr421/Ser424 phosphorylation, further supporting a PA/S6K connection. We also investigated whether intracellular PA production affects cell migration. Overexpression of phospholipase D2 (PLD2) and, to a lesser extent, PLD1, resulted in elevation of both S6K activity and chemokinesis, whereas PLD silencing was inhibitory. Because the lipase-inactive PLD2 mutants K444R and K758R neither activated S6K nor induced chemotaxis, intracellular PA is needed for this form of cell migration. Lastly, we demonstrated a connection between extracellular and intracellular PA. Using an enhanced green fluorescent protein-derived PA sensor (pEGFP-Spo20PABD), we showed that exogenous PA or PA generated in situ by bacterial (Streptomyces chromofuscus) PLD enters the cell and accumulates in vesicle-like cytoplasmic structures. In summary, we report the discovery of PA as a leukocyte chemoattractant via cell entry and activation of S6K to mediate the cytoskeletal actin polymerization and leukocyte chemotaxis required for the immune function of these cells.
Keywords: Cell/Chemotaxis, Cell/Phagocytosis, Immunology, Immunology/Innate Immunity, Lipid, Lipid/Phospholipid, Phosphorylation, Signal Transduction
Introduction
Chemotaxis is the migratory response of an organism or a cell to a chemical stimulus in the environment. Specifically, during inflammation chemicals released by pathogens cause a migration of phagocytes (neutrophils and macrophages) toward the infected area. The migration of leukocytes is a crucial component of the primary or “innate” immune response during inflammation, other components being phagocytosis and the release of oxygen radicals. Neutrophils are attracted to the chemical markers released by damaged cells during the invasion of healthy tissues by microorganisms.
The neutrophils then proceed to engage in chemotaxis toward pathogens, engulfing and destroying them. In addition to signals from microorganisms (such as the peptide formyl-Met-Leu-Phe peptide) and from dying cells (like tumor necrosis factor), neutrophils are attracted by chemokines and cytokines (such as GM-CSF)2 that alone or in combination with interleukine-8 are powerful chemoattractants (1, 2). Other chemotactic agents are the lipids leukotriene B4 and platelet activating factor.
In the process of chemotaxis, monomeric actin is polymerized to form F-actin (filamentous actin). The polymerization of actin filaments is tightly regulated through a variety of signaling pathways. One such pathway is the phosphatidylinositol 3-kinase pathway.
Phosphorylation of phosphatidylinositol diphosphate to phosphatidylinositol triphosphate by phosphatidylinositol 3-kinase enables proteins with pleckstrin homology domains to associate to phosphatidylinositol triphosphate in the plasma membrane. One such pleckstrin homology-bearing protein is AKT, which once recruited to the membrane becomes phosphorylated by phosphoinositide-dependent protein kinase 1 and TorC2 (3). Akt is then able to associate with Rac and cdc42 proteins involved with actin polymerization. The mTOR/S6K pathway is another phosphatidylinositol 3-kinase effector, and mounting evidence indicates that it is involved in controlling motility in several cell types. Both molecules are known to bind to phosphatidic acid (4–6). Phosphatidic acid (PA) is a pleiotropic lipid second messenger in yeast, plants, and mammalian cells. In the latter it has been shown that vesicular trafficking and exocytosis are increased through promotion of membrane fusion by PA in vivo (7). PA also causes the formation of transformed colonies in soft agar or tumors in xenografted nude mice of H-RasV12 oncogene-transfected fibroblasts (8).
Proteins that are recruited or activated by PA have been identified in mammalian cells (for review, see Ref. 9). Examples include Rac1, Fer kinase, the Ras exchange factor SOS, PI4P5 kinase, mTOR, phosphoinositide-dependent protein kinase, S6K, Raf, Fgr, protein kinase Cζ, SHP, PP1, Arf kinesin, and the Phox-47 component of the NADPH oxidase of phagocytes. PA is generated in the cell by either the combined action of phospholipase C and diacylglycerol kinase (10) or directly by the action of phospholipase D (PLD). PLD is a key signaling enzyme in stimulated platelets, neutrophils, mast cells, adipocytes, and certain cancer cells. In turn, PA can be metabolized to lysophosphatidic acid and diacylglycerol, further enhancing the second messenger prowess of PA in these mammalian cells (11). Two isoforms of PLD exist in mammalian cells, PLD1 and PLD2. Both enzymes are involved in the process of leukocyte cell polarization (12) and adhesion (13) and in angiogenesis in Zebrafish (14). Recently, Nishikimi et al. (15) demonstrated that PA enriches the localization of the atypical guanine exchange factor DOCK2 at the leading edge of chemoattractant-stimulated neutrophils as they begin to polarize and migrate. DOCK2-deficient neutrophils are unable to activate Rac2 in a polarized orientation and migrate in a directed manner. Thus, localized PA accumulation within the cell is crucial for the production of movement.
We began this study by reasoning that the accumulation of PA in or near the inner leaflet of the cell membrane of budding lamellipodia could mimic a gradient of chemoattractant (or PA itself) outside the cell. We report for the first time that extracellular PA serves as a neutrophil chemoattractant. For the mechanism of how this would occur, we propose and show that PA enters the cell and activates S6K, leading to cytoskeletal actin polymerization and chemotaxis. Similarly, we also demonstrate that cell-derived PA activates S6K and chemotaxis.
EXPERIMENTAL PROCEDURES
Materials
HL-60 and COS-7 cells were purchased from American Type Culture Collection (ATCC) (Rockville, MD). Dulbecco's modification of Eagle's medium was purchased from Cellgro (Herndon, VA). Lipids were from Avanti Polar Lipids (Alabaster, AL). Enhanced chemiluminescence (ECL) Western blotting detection reagents were purchased from GE Healthcare. Various primers were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Lipofectamine and PLUS reagents, Opti-MEM reduced serum medium, and dithiothreitol were purchased from Invitrogen. UltraClean Plasmid Prep kit was purchased from MOBIO Laboratories, Inc. (Carlsbad, CA). Quick Ligation kit, TaqDNA polymerase, ThermoPol buffer, restriction enzymes and their buffers, Lambda DNA-HindIII digest, and Antarctic phosphatase were purchased from New England Biolabs Inc. (Ipswich, MA). [γ-32P]ATP (500 μCi) was purchased from PerkinElmer Life Sciences. Vectashield mounting media was purchased from Vector Laboratories (Burlingame, CA). Ion exchange chromatography cellulose phosphate paper was purchased from Whatman (Hillsboro, OR).
Peripheral Blood Neutrophils and HL-60 Cell Differentiation (“dHL-60”)
Neutrophils were isolated from peripheral blood of volunteer donors (who signed an Institutional Review Board-approved consent form) using a Ficoll-Histopaque discontinuous gradient and were resuspended in Hanks' balanced salt solution at 5 × 106 cells/ml. In some experiments lymphocytes and monocytes were isolated from peripheral blood after a second gradient centrifugation in Percoll. Purity was >91 ± 4%, as confirmed by Wright-stained cytopreparations and cytological examination. Promyelocytic leukemic HL-60 cells were grown at 37 °C in a 5% CO2 incubator in Iscove's modified Eagle's medium + 20% (v/v) heat-inactivated fetal bovine serum and 2 μm l-glutamine. Cell density was maintained between 0.1 and 1.0 × 106/ml. HL-60 cells were induced to differentiate to the neutrophilic phenotype (dHL-60) by the addition to the culture medium of 1.25% (v/v) DMSO for 4 days. Viability assays were conducted using 0.4% trypan blue stain in cell preparation samples before analyses. Viability was uniformly >95%. The assessment of cell differentiation was performed by flow cytometric analysis of surface expression of differentiation-related antigens as indicated in Lehman et al. (16).
Plasmid Transfection (Nucleofection) into Neutrophilic HL-60 Cells (dHL-60)
Two days after induction of differentiation, cells were transfected with myc-PLD2 plasmid constructs using nucleofection as per the manufacturer's protocol (Amaxa, Gaithersburg, MD). Fresh DMSO (1.25% (v/v) was added to the media after nucleofection, and cells were cultured for an additional 48-h period. In selected experiments, cells were overexpressed with a Grb2 plasmid (pcDNA-XGrb2) (17). The expression construct containing the full cDNA of human Grb2 variant 1 (pCMV6-Grb2, OriGene Technologies) was the template for PCR subcloning. The Grb2 open reading frame was PCR-amplified using the sense (EcoRI) primer (5′-CAC TGA GCA GAA TTC AGA ATG GAA GCC ATG GCC-3′) and the antisense (PspOM I) primer (5′-TTA AAT CCA ACG GGC CCT CCC ACC CCC TAA-3′). The PCR product was EcoRI/PspOMI-digested, purified, and C-terminal subcloned into the EcoRI/NotI-digested Xpress-tagged pcDNA3.1/HisC expression vector (Invitrogen).
Gene Silencing
For silencing of gene RPS6KB1, we used two siRNAs (at 100 nm) from Ambion (Austin, TX) (“Select validated”), one of which targeted exon 1 (sequence 5′-CAUGGAACAUUGUGAGAAAtt-3′) and another that targeted exon 3. For mTOR (FRAP1) we used two siRNAs; one that targeted exon 3 (locus ID 2475; sense, 5′-GGAGUCUACUCGCUUCUAUtt-3′, and another that targeted exon 6. For efficient PLD silencing, as our laboratory has previously shown (12), two different sets of siRNAs (at 100 nm) were used; one from Ambion (“Silencer Pre-designed”) that targets PLD1 on exon 10 (sense sequence GGC AAA UGA AGA GAU UUU UTT) and the other from Dharmacon (La Fayette, CO) (“SMARTselection”) that targets exon 21 on PLD1 (sense sequence UAA CUG AGC UUA UCU AUG UUU); siRNA targeting PLD2 (exon 15) was from Ambion (sense sequence GGA CUA CAG CAA UCU UAU CTT). A negative control for all silencing was 100 nm siRNA, purchased from Ambion. Neg-siRNA#2 is a 19-bp scrambled sequence with 3′-dT overhangs (the sequence is not disclosed by Ambion) certified to lack significant homology to any known gene sequences from mouse, rat, or human and causes no significant changes in gene expression of transfected cells after 48 h at the same concentration as the siRNA in tests. The protocol for gene silencing comprised induction of differentiation of HL-60 cells with 1.25% DMSO on day 0. On day 1, the cells were then transfected with siRNA. Aliquots of 1 μl of siRNA (resuspended in nuclease-free H2O and added at 100 nm final concentration to cells) were mixed with 300 μl of Opti-MEM and incubated at room temperature for 5 min. siRNAs were added to an Amaxa (Gaithersburg, MD) electroporation cuvette with 2 × 106 HL-60 cells/100 μl in nucleofection solution V. Cells were electroporated using program T-019 (as per the manufacturer's instructions) after which they were resuspended in 1 ml of Iscove's modified Eagle's medium + 20% fetal bovine serum + 1.25% DMSO (to allow cells to continue differentiation). Cells were immediately plated into 6-well plates and incubated at 37 °C for 48 h to allow for maximum gene expression silencing. Lastly, the cells were harvested on day 4 and used for the experiments described herein.
PLD Enzyme Activity
Measurement of lipase activity began with the addition of the following reagents (final concentrations): 3.5 mm PC8 phospholipid, 45 mm HEPES, pH 7.8, and 1 μCi of n-[3H]butanol in a liposome form. Samples were incubated for 20 min at 30 °C with continuous shaking. The addition of 0.3 ml of ice-cold chloroform/methanol (1:2) stopped the reactions. Lipids were then isolated and resolved by thin layer chromatography. TLC plates were developed in the upper phase of a ethyl acetate/isooctane/acetic acid/water (4.3:0.6:1:3.3 v/v) mixture. Lipids were visualized with iodine vapors. The amount of [3H]phosphatidylbutanol that co-migrated with phosphatidylbutanol standards was measured by scintillation spectrometry. Control reactions lacking PC8 were used to remove background counts.
S6K Enzyme Activity
Neutrophil or dHL-60 lysates were obtained after centrifugation (7000 × g, 1 min, 4 °C), and supernatants were mixed with anti-p70S6K antibody conjugated to agarose beads. Immune complex beads were resuspended in a final volume of 40 μl with ice-cold lysis buffer. The in vitro kinase assay contained 75 μm S6 kinase substrate peptide KKRNRTLTK prepared in kinase buffer (13.4 mm HEPES, pH 7.3, 2.5 mm MgCl2, 30 μm Na2VO3, 5 μm p-nitrophenyl phosphate, 2 mm EGTA, 2 μm cAMP-dependent kinase inhibitor TTYADFIASGRTGRRNAIHD, 0.420 mCi of [γ-32P]ATP (7 nm), and 68 μm unlabeled ATP). Aliquots (20 μl) of kinase buffer containing the appropriate substrates were mixed 1:3 (v/v) with immunocomplex beads. The reaction was carried out at 37 °C for 20 min with rotation and terminated by blotting 40 μl of the reaction mixture onto P81 ion exchange chromatography cellulose phosphate papers. Filter squares were washed, dried, and counted for radioactivity. Controls were run in parallel without S6 kinase peptide.
Immunoprecipitation and Western Blot Analyses
Neutrophils or dHL-60 cells were resuspended at 3 × 106 cells/ml. After cell stimulation, aliquots (1 ml) were taken and sedimented (14,000 × g, 15 s), and the pellets were resuspended in 0.2 ml of boiling SDS solution (1% SDS in 10 mm Tris-HCl, pH 7.4), placed on ice, and mixed with 0.3 ml of cold distilled H2O and 0.4 ml of cold Triton X-100-based lysis buffer (12 mm Tris-HCl, pH 7.2, 0.75 mm NaCl, 100 μm Na2VO3, 10 μm phenylmethylsulfonyl fluoride, 0.2 μm β-glycerophosphate, 5 mg/ml each of aprotinin, pepstatin A and leupeptin, and 0.12% Triton X-100). The resulting 1-ml total cell lysates were spun down (14,000 × g, 1 min, 4 °C) to remove insoluble material, and the supernatants then used for immunoprecipitation with the appropriate antibody (2 μg/ml). Pellets were washed twice with lysis buffer, twice with buffer A (100 mm Tris-HCl, pH 7.4, 400 mm LiCl), and twice with buffer B (10 mm Tris-HCl, pH 7.4 100 mm NaCl, 1 mm EDTA). Immune complex beads were resuspended in a final volume of 60 ml with lysis buffer and mixed with 2× SDS sample buffer (1:1 v/v) for subsequent protein gel electrophoresis/immunoblotting with either phosphotyrosine or phospho-Thr421/Ser424-p70S6K antibodies at a 1:1000 dilution. Signals from secondary antibodies conjugated to horseradish peroxidase were detected on x-ray films.
Preparation of Lipids
dHL-60 or neutrophils were resuspended at 5 × 105 and 5 × 106 cells/ml, respectively, in chemotaxis buffer (Hanks' balanced salt solution + 0.5% bovine serum albumin). 200 μl were placed in the upper chambers (or “inserts”) of Transwell plates separated from the lower wells by a 6.5-mm diameter, 5-μm pore Nucleopore polycarbonate membrane. For the study of chemotaxis, DOPA (dioleoyl-PA, 1,2-dioleoyl-sn-glycero-3-phosphate, from Avanti Polar Lipids) was prepared fresh the day of the experiment in 1× phosphate-buffered saline + 0.5% bovine serum albumin, pH 7.2, at a stock concentration of 1 μm. Lipid solutions were sonicated on ice twice for 5 s each (medium setting). Selected experiments used PC (1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)) or PS (1,2-dioleoyl-DOPS-glycero-3-[phospho-l-serine] (DOPS)) prepared similarly to the protocol for PA.
Cell Migration Assay
When ready for chemotaxis, PA was diluted to a working concentration in 500 μl of chemotaxis buffer and placed in the lower wells of Transwell plates. Transwell plates were incubated for 1 h at 37 °C under a 5% CO2 atmosphere. The number of cells that migrated to the lower wells was calculated by placing 10-μl aliquots on a hemocytometer and counting 4 fields in duplicate. It should be also indicated that the chemotaxis assay collects leukocyte cells in the lower well after migration through a Transwell filter. Even though we have titrated the concentration of bovine serum albumin in our chemotaxis buffer (0.5%) to minimize adherence to the underside of the filter, there is a small percentage (8–10%). However, control experiments have determined that the level of adherent cells remains constant for both controls and PA-stimulated cells.
Actin Polymerization Measurements by Flow Cytometry
F-actin polymerization was initiated in vivo by the addition of PA to a neutrophil cell suspension (5 × 106 cells/ml) in Hanks' balanced salt solution (phenol red-free) for 10 min at 37 °C. Then 0.2-ml aliquots were taken and mixed with 1 ml of prechilled fixing solution (2 parts 2× phosphate-buffered saline, pH 7.4, 1 part 20% formaldehyde, and 1 part 75% glycerol in water) and centrifuged. Pellets were resuspended in freshly prepared F-actin staining solution (35 μl of a 3.3 μg/ml methanol FITC-phalloidin stock plus 315 μl of H20) and stained in the dark for 30 min at room temperature. Samples were centrifuged as above, and pellets were resuspended in 1 ml of FACSFlow. The cells were then analyzed by flow cytometry on a FACScan BD Biosciences flow cytometer at a 488-nm excitation wavelength. Data were analyzed using Cell Quest software. The relative F-actin content in cells was expressed in comparison with resting cells that received no treatment (and normalized to 100%).
Generation of Lipase-inactive PLD2 Mutants
The construct pcDNA-mycPLD2 (18) was used as a template to create two Lys → Arg point substitutions (K444R and K758R) and one double substitution (K444R/K758R), as first reported in Sung et al. (19), after the QuikChange XL site-directed mutagenesis protocol (Stratagene, La Jolla, CA) using the following sets of primers: K444R forward (5′-GCC CAT CAT GAG AGG CTC CTG GTG GTG-3′) and K444R reverse (5′-CAC CAC CAG GAG CCT CTC ATG ATG GGC-3′); K758R forward (5′-TCT ACA TCC ACA GCA GGG TGC TCA TCG CAG-3′ and K758R reverse (5′-CTG CGA TGA GCA CCC TGC TGT GGA TGT AGA-3′). All oligonucleotides and their reverse complements were PAGE/high performance liquid chromatography-purified (Integrated DNA Technologies). Molecular identity of the three pcDNA-mycPLD2 mutants was confirmed by direct sequence analysis (Agencourt Bioscience Corp., Beverly MA).
Detection of Intracellular PA Using a GFP-based Sensor
The yeast (Spo20p) soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) PA binding domain (wtPABD) was cloned in a pEGFPC1 vector (Clontech) (20) and denoted “pEGFP-Spo20PABD-WT.” The origin of the coding sequence of Spo20p amino acids 51–91 was reported in Nakanishi et al. (21), who amplified it by PCR using BBO5 and PDO2 as primers and pRS306-SPO20pr-SPO20 as a template and introduced XhoI and XbaI sites. This construct is cut by ApaLRI at the pUC origin (in addition, AgeI and DraII are one-cut, PstI is a two-cut, and NcoI is a four-cut restriction enzyme). The pEGFP-Spo20PABD-WT construct was used as template to generate a PABD mutated domain (MutPABD) by site-directed mutagenesis using a QuikChange mutagenesis kit (Stratagene). This new construct had a leucine-to-proline mutation at residue 67, and it was named “pEGFP-Spo20PABD-L67P.” The wild type and the L67P mutant PA binding sensors have been characterized (22).
COS-7 or HL-60 cells were transfected with either the wild type or the L67P mutant PA sensor. Cells were grown on glass coverslips (coated with collagen for HL-60 cells) inside 6-well tissue cultures plates. Two days posttransfection, fresh media containing no serum was added to the dishes, and cells were incubated with exogenous PA or with bacterial PLD (to generate in situ PA from the cell membrane PC). In either case after 7 min of stimulation, the medium was aspirated, and cells were washed three times with phosphate-buffered saline to remove any traces of extracellular PA. Cells were fixed with 4% paraformaldehyde and imaged using the FITC channel (DAPI was also included to visualize the nucleus). Cells were examined on a Nikon Eclipse 50i fluorescent microscope and photographed (SPOT camera; Diagnostic Instruments) by consecutive exposure using green and blue filters with a 100× (numerical aperture 0.9) objective.
Confocal Microscopy
Confocal imaging of HL-60 cells was performed using an Olympus FV 1000 confocal microscope (DAPI excitation: argon laser, 488 nm; FITC krypton, 568 nm). Double color immunofluorescence preparations were imaged using a 60× oil objective (numerical aperture 1.35) magnified ×2.7 and a series of confocal optical sections (z-step, 0.5 μm). Integration was performed with the Kalman filter (applied twice). Images were acquired with the FV1000 software Version 1.7.3.0.
Statistical Analysis
Data are presented as the mean ± S.E. The difference between means was assessed by the single factor analysis of variance (ANOVA) test. Probability of p < 0.05 was considered to indicate a significant difference.
RESULTS
Exogenous PA Enhances Leukocyte Chemotaxis
The initial finding that prompted this study was an unexpected observation that PA stimulated chemotaxis and actin polymerization, as shown in Fig. 1. The number of cells that migrated increased with concentrations of PA up to ∼100 nm, after which the numbers decreased, forming a bell-shaped curve (Fig. 1A) that is consistent with the behavior of other chemoattractants. It should be pointed out that throughout this study we have utilized a membrane-soluble species of PA (DOPA) known to enter the cells readily and activate intracellular signaling pathways such as NADPH in leukocytes and that this form of PA is more effective than the shorter varieties with saturated sn-2 acyl chains (23). Also, the PA used in this study does not contain any traces of lyso-PA or any other contaminant phospholipid (supplemental Fig. S1).
FIGURE 1.
Exogenously provided PA stimulates neutrophil chemotaxis and actin polymerization. A, freshly isolated human neutrophil suspensions were resuspended in Hanks' balanced salt solution-based chemotaxis buffer at a density of 3 × 106 cells/ml. Aliquots containing 6 × 105 cells were placed in the upper insert portion of Transwell plates. The bottom wells received the indicated concentrations of PA, lyso-PA, GM-CSF, PC, or PS. B, neutrophil suspensions were incubated at 37 °C in the absence or presence of PA (20, 100, and 300 nm), 100 nm lyso-PA, 100 nm PC, 100 nm PS, 7 nm GM-CSF, or 50 ng/ml PMA for 5 min or 100 nm formyl-Met-Leu-Phe peptide for 30 s. The incubations were stopped at the end of the indicated lengths of time by formaldehyde fixation. 200-μl samples of 1 × 104 cells were incubated with phalloidin-FITC and analyzed by flow cytometry. Fluorescence on gated neutrophil populations was measured and plotted in the y axis as relative fluorescence with respect to the control. Results are the mean ± S.E. from three independent experiments conducted in duplicate. The asterisks denote the difference between means that were statistically significant (p < 0.05) by ANOVA.
As a positive control, Fig. 1 also shows chemotaxis induced by the human cytokine GM-CSF, a known neutrophil chemoattractant (1) to which PA compared favorably. Quantitatively, PA elicited ∼130% more chemotaxis than GM-CSF, albeit with a 10-fold higher concentration needed to exert the peak effect. In contrast, lysophosphatidic acid (lyso-PA), PC, and PS failed to elicit chemotaxis.
We next assessed F-actin polymerization, a hallmark of cell migration for chemoattractant-stimulated neutrophils, using flow cytometry to quantify the changes in cytoskeletal reorganization. Changes in actin polymerization occurred quickly (∼2 min), remained high for at least 10 min after the addition of PA to the cells, and were significant at 20–300 nm PA (Fig. 1B). In contrast, lyso-PA, PC, and PS failed to induce changes in F-actin. Quantitatively, PA elicited changes in F-actin that were comparable in potency to the GM-CSF and 55–60% of those observed with formyl-Met-Leu-Phe peptide, a known strong neutrophil inducer of actin polymerization. In conclusion, PA causes an increase in F-actin polymerization and a 3-fold increase in chemotaxis, which are relevant to the processes directed migration.
Chemotaxis Is Inhibited by S6K Inhibitors and by Silencing with Double-stranded RNA Targeting S6K
We next explored the molecular mechanism underlying the effect of PA on chemotaxis. We previously reported that ribosomal S6K is involved in GM-CSF-induced chemotaxis (24), and we hypothesized that this pathway would connect to the PA-signaling pathway. The S6K inhibitors rapamycin and bisindolylmaleimide Ro31-8220 (“Ro31”) were very effective in reducing the migration elicited by PA for primary neutrophils (Fig. 2A) and dHL-60 cells (Fig. 2B). Rapamycin exhibited an IC50 of ∼0.7 nm, and Ro31-8220 exhibited an IC50 of ∼1.3 nm, with >90% inhibition at 10 nm. Viability of cells (assessed by trypan blue exclusion) after exposure to rapamycin and Ro31-8220 was ∼95%, thus excluding cytotoxicity as an explanation for the inhibition of migration in these assays. Rapamycin and Ro31-8220 both inhibited neutrophil adhesion to glass and F-actin polymerization (not shown).
FIGURE 2.
Inhibition of S6K blocks PA-induced chemotaxis. A, freshly isolated human neutrophil suspensions were incubated with the S6K inhibitors rapamycin and Ro31-8220 (Ro31) at 3 nm for 20 min before performing chemotaxis assays as described in Fig. 1. # and *, p < 0.05. B, dHL-60 cells preincubated with S6K inhibitors were assayed for chemotaxis. 100% represents 6.3 ± 0.5 × 104 cells/ml for cells stimulated with PA but no inhibitor.
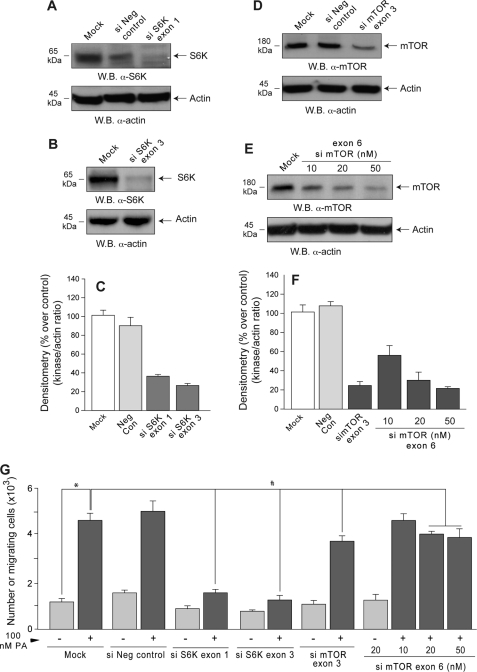
To confirm that S6K plays a functional role in PA-induced leukocyte motility, siRNAs targeting two different exons were used to knockdown S6K gene expression in dHL-60 cells. Western blotting (Fig. 3, A and B) and band densitometry (Fig. 3C) indicated that ∼70–75% depletion was achieved. Loading controls, lack of knockdown of S6K by an irrelevant (negative control) siRNA, and effective knockdown (∼80%) of mTOR by mTOR siRNAs (Fig. 3, D–F) are also shown.
FIGURE 3.
Silencing S6K but not mTOR substantially decreases cell migration. S6K- and mTOR-siRNAs and a negative control siRNA (si Neg control) were transfected into dHL-60s. Half of each cell culture was subsequently probed by Western blotting (W.B.) to determine the extent of silencing using anti-S6K (A and B) or anti-mTOR (D and E) antibodies (as well as with anti-actin to demonstrate equal loading of all gel lanes). C and F, shown is densitometry of the bands for S6K and mTOR as a ratio of the densitometry of actin in each gel lane. The remainder of the silenced cells was assayed for chemotaxis as before (G). Results are the mean ± S.E. from three independent experiments conducted in duplicate. The symbols * and # denote difference between statistically significant (p < 0.05) means by ANOVA between control and PA and between control and silenced samples, respectively.
Both siRNAs targeting S6K, but not the irrelevant siRNA, suppressed dHL-60 chemotaxis toward PA by ∼85% (Fig. 3G). As shown previously (4, 5, 26), PLD-derived PA binds to and activates mTOR. Because mTOR is one of the upstream activators of p70S6K, it could be proposed that the observed results of PA activation in vivo are mediated through PA stimulation of mTOR. However, mTOR siRNA decreased PA-stimulated chemotaxis only to a modest extent (∼30%, Fig. 3G). Accordingly, this PA-stimulated S6K signaling pathway appears to operate largely independently of mTOR.
PA Induces S6K Enzymatic Activation and Phosphorylation
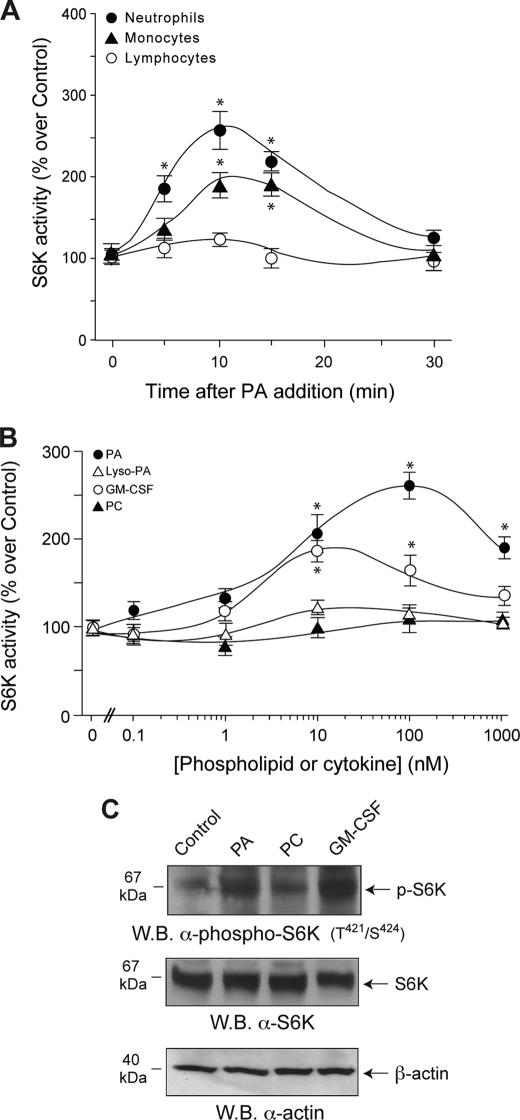
We next examined whether PA directly activates S6K in several different primary cell types. PA activated S6K 2.5-fold with the peak response at 10 min after stimulation (Fig. 4A). A similar but weaker response was observed for monocytes, whereas lymphocytes did not respond at all. PA and GM-CSF stimulated S6K activation (Fig. 4B) with dose-response curves similar to those observed for chemotaxis in Fig. 1A, whereas lyso-PA and PC failed to do so, again consistent with the findings for chemotaxis.
FIGURE 4.
PA activates S6K in neutrophils and monocytes but not lymphocytes. A, time course is shown. Neutrophils, monocytes, and lymphocytes isolated from human peripheral blood and cells were incubated with 100 nm PA, and then S6K activity was measured. B, dose response is shown. Freshly isolated neutrophils were incubated with the indicated concentrations of PA, lyso-PA, GM-CSF, or PC before S6K activity assays. 100% values represent 4973 ± 398 dpm/mg of protein. Results are the mean ± S.E. from three independent experiments conducted in duplicate. The asterisks denote difference between means that were statistically significant (p < 0.05) by ANOVA. C, the effect of PA on S6K phosphorylation is shown. Neutrophils were incubated with either 10 nm GM-CSF or 100 nm PC or PA. Cell lysates were immunoprecipitated with anti-p70S6K and immunoblotted (W.B.) with anti-phospho-Thr421/Ser424-p70S6K antibodies. Shown is a representative Western blot of three experiments with similar results.
S6K activation proceeds through phosphorylation. As shown in Fig. 4C, PA and GM-CSF, but not PC, induced phosphorylation of S6K on residues Thr421/Ser424, two residues key in the enzyme activation. The PA-induced phosphorylation occurred rapidly (∼4 min after the addition of PA) and dose-dependently, becoming detectable at 10 nm PA (not shown).
Endogenous PA, Generated through PLD, Increases Chemokinesis and S6K Activity
We next asked if endogenous PA would enhance basal cell migration (“chemokinesis”) in the same manner that provision of exogenous PA stimulated chemotaxis. We overexpressed the two classic PLD isoforms, PLD1 and PLD2, in dHL-60 cells and observed enhanced cell migration, in particular for PLD2 (Fig. 5A). To assess whether PLD1 or PLD2 are required for chemokinesis, we employed PLD1 and PLD2 siRNAs (Fig. 5B). As indicated in Fig. 5C, the enhancing effect previously observed due to PA stimulation was reduced and more so in the setting of PLD2 depletion than for PLD1 depletion, indicating that at least in dHL-60 cells, the former contributes more than the latter to the overall induction of cell migration.
FIGURE 5.
PLD2 stimulates and is required for chemokinesis. A, dHL-60 cells were transfected with HA-PLD1 or Myc-PLD2 and assayed for Transwell cell migration in the absence of chemoattractants (chemokinesis). B, silencing of PLD1 and PLD2 was accomplished by transfecting dHL-60 with double-stranded RNA and culturing for 4 days. W.B., Western blot. C, silenced cells were used for Transwell cell migration with PA as a chemoattractant.
We also generated PLD2 isoforms with mutations in the HKD domains (Fig. 6A) that are essential for activity (19). PLD2-K444R and PLD2-K758R failed to exhibit significant lipase activity (Fig. 6B) and did not stimulate S6K activity (Fig. 6C) or cell migration (Fig. 6D). Taken together, these findings demonstrate that intracellular PA generated by the action of PLD2 activates S6K and enhances basal cell migration (chemokinesis).
FIGURE 6.
Dependence of S6K activation on the state of PLD activity. A, scheme of PLD2 structure. PX, pleckstrin homology domain; PH, Phox homology domain; HKD1 and HKD2, phospholipase D catalytic half-sites, include Lys-444 and Lys-758, which are essential for activity. B–D, transfected dHL-60 cells were divided into three equal sets. One was used to measure PLD activity (B), a second to measure S6K activity (C), and a third to measure Transwell chemotaxis (D). 100% values represent 3031 ± 289 dpm/mg for PLD activity, 4515 ± 380 dpm/mg of protein for S6K activity, and 6.0 × 103± 0.5 cells in bottom wells. Results represent the mean ± S.E. of three independent experiments in duplicate. * and # denote differences between means that were statistically significant (p < 0.05) by ANOVA between control and WT and between control and mutants, respectively.
Extracellular PA Accumulates within the Cell
Our experiments posed the question of how extracellular PA acts as a chemoattractant, as the mTOR findings suggested that it does not require receptor activation, and no receptor for PA has been identified to date. We, thus, hypothesized that the PA crosses the plasma membrane and accumulates inside the cell to activate S6K in a manner similar to PA generated intracellularly by PLD2.
To explore this possibility, we first used a means of inducing the cells to generate PA that could be encountered physiologically by neutrophils. In nature, PA can be generated on the surface of host cells by extracellular PLD secreted by microorganisms such as Streptomyces chromofuscus. Streptomyces PLDs are among the smallest PLD enzymes and, like the mammalian PLDs, also have the two catalytic HKD motifs, a signature of the PLD superfamily (28). S. chromofuscus PLD was added to neutrophils that were subsequently lysed and analyzed for S6K activity. Bacterial PLD increased leukocyte S6K in a dose- and activity-dependent fashion (Fig. 7).
FIGURE 7.
Bacterial PLD stimulates S6K activity. Neutrophils were incubated with the indicated concentrations of S. chromofuscus PLD (2 units/μg of solid capable of generating 200 μmol/ml PA in vitro) or with same concentrations of heat-inactivated PLD (boiled at 100 °C for 10 min). Cell lysates were immunoprecipitated with anti-S6K antibodies, and S6K activity was measured. 100% values represent 4173 ± 334 dpm/mg of protein. The asterisks denote differences between means that were statistically significant (p < 0.05) by ANOVA and above controls. The number symbol (#) denotes significant differences in the samples with inhibited activity.
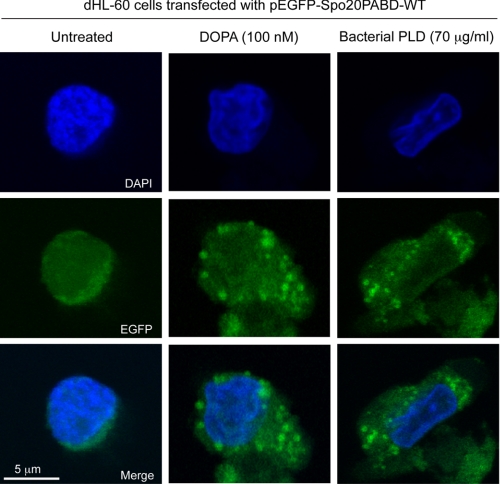
We then attempted to directly demonstrate that PA accumulates inside the cell after exogenous provision or generation using a fluorescent PA sensor consisting of EGFP fused to a 40-amino acid PA binding domain from the yeast Spo20 protein (20–22). We first tested this sensor in COS-7 cells, as they are larger than dHL-60, and details of subcellular structures are more readily distinguished by fluorescence microcopy. In resting cells, as previously described (4), the sensor localizes primarily to the nucleus and/or nucleoli (Fig. 8), due either to the propensity of EGFP to weakly localize there or to local sources of PA. After exogenous provision of PA, translocation of the sensor to punctate cytoplasmic structures is observed (Fig. 8). In contrast, no such translocation for a mutated sensor unable to bind PA (20–22) is observed (supplemental Fig. S2). Exposure of the COS-7 cells to S. chromofuscus PLD similarly caused a translocation of the wild type PA sensor (Fig. 8), but not the mutant sensor, to cytoplasmic vesicular-like structures (supplemental Fig. S2).
FIGURE 8.
Demonstration of the PA into COS-7 cells. COS-7 cells were transfected with the pEGFP-Spo20PABD PA sensor and cultured for 2 days. They were stimulated with PA (100 nm) or bacterial (S. chromofuscus) PLD for 7 min, then washed extensively, fixed, stained with DAPI, and examined by fluorescence using FITC or DAPI filters. Shown are representative fields of images among 20 that were visualized.
Similar results were obtained for neutrophilic HL-60 cells for a redistribution to vesicle-like structures in the cytoplasm upon PA addition or S. chromofuscus PLD treatment in cells overexpressing the wild type PA sensor (Fig. 9) but not in cells overexpressing the mutant sensor (supplemental Fig. S3). Taken together, these results provide evidence that PA, whether exogenously provided as DOPA or generated in situ on the outer surface of the cell, crosses the plasma membrane and accumulates on internal membrane structures from which signaling pathways that lead to S6K activation can be triggered.
FIGURE 9.
Demonstration of the PA into neutrophilic HL-60 cells. HL-60 cells were induced to differentiate and then they were transfected with the pEGFP-Spo20PABD PA sensor and cultured for 2 days in collagen-coated glass slides. The cells were stimulated with PA (100 nm) or bacterial (S. chromofuscus) PLD for 7 min, then washed extensively, fixed, stained with DAPI, and examined by confocal microscopy. Shown are representative fields of images among 20 that were visualized.
DISCUSSION
We report here that PA is a powerful chemoattractant for neutrophils and differentiated monomyelocytic leukemic cells and that the mechanism by which this is accomplished involves activation of S6K, which in turn elicits actin polymerization, cell spreading, pseudopodia formation, and chemotaxis. PA appears comparable in potency to other known neutrophil chemoattractants. As presented here, the PA that stimulates these intracellular pathways can originate from (a) exogenous PA added to the cells, (b) PC on the outer leaflet of the plasma membrane, cleaved by the action of PLD secreted by microorganisms that a phagocyte might encounter (modeled here by the use of purified PLD from S. chromofuscus), and (c) intracellular PLD-derived PA generated by PLD2. All this is summarized in the scheme of Fig. 10. PA can also be produced by the combined action of phospholipase C and diacylglycerol kinases, but this route was not examined here.
FIGURE 10.
A model for the mechanism of action of extracellular PA. Exogenous PA or PA produced after cleavage of cell membrane PC by the action of bacterial PLD (Bact PLD), such as that from S. chromofuscus, stimulates S6K (this report) and mTOR (3). PA is able to induce its own synthesis as the addition of exogenous PA leads to activation of PLD (29) in a positive feedback mechanism. PLD-produced PA from activated cells (Host PLD) or from cells overexpressing PLD2 constructs can add to the transient pool of PA as a second messenger that also binds and activates S6K. It is the activation of S6K rather than the activation of PLD that induces cell spreading, actin polymerization, and chemotaxis. Dashed arrows indicate lipid-protein binding. Continuous blue arrows indicate enzymatic reactions. Continuous black arrows indicate intracellular signaling pathways connections.
It has been reported that extracellular PA can stimulate intracellular PLD to generate intracellular PA, thus amplifying the original signal (29). It is entirely possible then that a network of PA → PLD → PA exists, connecting intercellular and intracellular communication in a similar fashion to that described in Dictyostelium (30) where group migration proceeds as a consequence of cAMP signal relay.
Utilizing a PA sensor, pEGFP-Spo20PABD (20–22), we visualized PA inside the cell. Exposure of cells to PA resulted in the accumulation of PA inside the cell clustered into discrete patches/vesicles where it can lead to the activation of S6K. Lehman et al. (6) have shown immunofluorescent staining of S6K shifting away from perinuclear regions and colocalizing with PLD2 in the cytosol. It is also possible that PA could be on early endosomes during triggered endocytosis and subsequently move into recycling endosomes. There is a precedent for this in that Rizzo et al. (31) have shown that insulin stimulation of fibroblasts causes such redistribution of the PA binding domain of Raf1.
As for the mechanism of action of the PA, there are hypothetically several possibilities. Zhou et al. (32) reported that haptotactic migration of monocytes induced by PA was decreased by pertussis toxin. Breast cancer cells MDA-MB-231 increased their invasive potential in response to PA, and that, too, was attenuated by pretreatment of cells with Clostridium difficile toxin B and pertussis toxin (33). These reports imply a role for a Gi-dependent mechanism, although this was not firmly established. By using the tyrosine kinase inhibitor herbimycin A, Siddiqui and English (34) demonstrated that PA-elicited calcium mobilization and actin polymerization are mediated by tyrosine kinase-based signaling pathways.
However, we report here that the participation of S6K is necessary for the migration-induced effects of PA for neutrophils. S6K was originally described as a kinase involved in the regulation of protein synthesis and ribosomal biogenesis by its effect on the ribosomal 40 S subunit and its protein S6. Phosphorylation of S6 on multiple Ser/Thr residues by S6K is observed quickly after cell stimulation with growth factors such as epidermal growth factor (35). S6K is a physiologic substrate for phosphoinositide-dependent protein kinase 1 through phosphorylation of Thr229, and it is also phosphorylated by the mTOR complex 1. As for its relation to cell migration, our laboratory showed earlier that S6K is a signaling kinase activated during chemotaxis elicited by GM-CSF (24, 36).
PA has been shown to activate mTOR directly or indirectly by binding to Rheb (3, 4), and it also binds and activates S6K in the absence of mTOR activation (6). Lipopolysaccharide-induced PA accumulation in macrophages was reported to initiate an Akt-dependent activation of mTOR-S6K1 that was required for the elevated biosynthesis of inflammatory mediators (37). mTOR is a critical target for survival signals generated by PLD (38). Growth factors and hormones such as insulin regulate mTOR-complex 1 through the generic class I phosphatidylinositol 3-kinase signaling pathway. GTP-bound Rheb enables mTOR complex 1 signaling to downstream substrates, such as S6K1 (39). Even though S6K and mTOR both are part of the same signaling pathway, to observe chemotaxis we find that S6K needs to be active. Chemotaxis can be elicited in the absence of mTOR but not in the absence of S6K.
The association of S6K with the cytoskeletal machinery has been hinted at before as rapamycin inhibits calcium-associated stress fiber formation (40). Actin dynamics localized in the actin arc suggested that the actin arc functions to concentrate proteins actively involved in cell migration and to maintain S6K at the leading edge (41). However, the signaling mechanisms that lead to this are not well understood. Overexpression of cdc42 and Rac (both small GTPases involved in the regulation of membrane ruffling, migration, and actin polymerization) increase S6K levels in cells. It has not been proven that Rho GTPases activate S6K; however, Rac may activate S6K through activation of mixed lineage kinase (MLK3).
S6K colocalizes with those stress fibers in the lamellipodia of moving leukocytes (42), and its phosphorylation induces migration of vascular smooth cells (43, 44) and chick embryo fibroblasts (45). S6K mediates leukocyte chemotaxis induced by the human growth factor GM-CSF, which is inhibited by rapamycin at subnanomolar concentrations. Rhoads et al. (46) have shown that amino acid excess and arginine particularly could promote S6K activation and control the rate of enterocyte migration. Arginine activation of S6K in the enterocyte cell line was determined by wounding the cells and assaying their immunoreactivity (46). Berven et al. (41) have indicated that cytochalasin D and the Rho kinase inhibitor Y-27632, both stress fiber disrupters, increase S6K activity using fibronectin in Swiss 3T3 fibroblasts.
Siddiqui and English (34) have demonstrated that neutrophil chemotaxis occurs in response to PA. The unique physical properties of PA could explain the observed effects on chemotaxis. The cone-shaped geometry of PA gives this molecule the propensity to induce cellular membranes to form hexagonal II phase conformations (47). It has been shown before that PA induces a large fusogenic effect in membranes of several cell systems (48, 49). Vesicular trafficking and exocytosis are all increased after membrane fusion of PA in vivo (7). PA location within the cell should be crucial to the production of movement. Nishikimi et al. (15) have demonstrated that PA provides directionality in the cell. PA is also part of a “PA loop” (25, 27) with PA activating actin, which then activates PLD enzymes, providing more PA that can, in turn, further modulate actin.
In summary, we report the novel discovery that PA derived from PLD action, whether extracellularly (from microbial action) or intracellularly (from leukocytes), induces polymerization of actin, pseudopodia formation, and chemotaxis in a process that is mediated by S6K activation. Phagocyte migration is paramount to the body primary immune response. Neutrophils are attracted to the chemical markers released by damaged cells during the invasion by microorganisms. One of the unintended signals (at least from the point of view of the pathogen) could be the generation of PA in the cell membrane of the leukocyte. In a sense, a gradient of chemoattractants (or PA itself) outside of the cell can be mimicked inside the cell by PA. Some of this PA would further signal through S6K to help initiate membrane polarization and chemotaxis, events that are ultimately crucial to the neutrophil mission of phagocytosis.
Supplementary Material
Acknowledgments
We thank Dr. Francisco Alvarez for help acquiring the confocal images of HL-60 cells at the Microscopy Core Facilities Center of the Boonshoft School of Medicine (Wright State University). We also thank Nicholas Lehman for technical assistance with bacterial PLD activation of S6K experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant HL056653 and Research challenge Grant 667131 (Ohio Board of Regents) (to J. G.-C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- GM-CSF
- granulocyte-macrophage colony-stimulating factor
- PA
- phosphatidic acid
- DOPA
- dioleoyl-PA (1,2-dioleoyl-sn-glycero-3-phosphate)
- PABD
- PA binding domain
- S6K
- ribosomal S6 kinase
- mTOR
- mammalian target of rapamycin
- ANOVA
- single factor analysis of variance
- lyso-PA
- lysophosphatidic acid
- PLD
- phospholipase D
- siRNA
- small interfering RNA
- PC
- phosphatidylcholine
- PS
- phosphatidylserine
- FITC
- fluorescein isothiocyanate
- DAPI
- 4′,6-diamidino-2-phenylindole
- EGFP
- enhanced green fluorescent protein
- WT
- wild type.
REFERENCES
- 1.Gomez-Cambronero J., Horn J., Paul C. C., Baumann M. A. (2003) J. Immunol. 171, 6846–6855 [DOI] [PubMed] [Google Scholar]
- 2.Shen L., Fahey J. V., Hussey S. B., Asin S. N., Wira C. R., Fanger M. W. (2004) Cell. Immunol. 230, 23–32 [DOI] [PubMed] [Google Scholar]
- 3.Kamimura Y., Xiong Y., Iglesias P. A., Hoeller O., Bolourani P., Devreotes P. N. (2008) Curr. Biol. 18, 1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., Chen J. (2001) Science 294, 1942–1945 [DOI] [PubMed] [Google Scholar]
- 5.Sun Y., Fang Y., Yoon M. S., Zhang C., Roccio M., Zwartkruis F. J., Armstrong M., Brown H. A., Chen J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8286–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehman N., Ledford B., Di Fulvio M., Frondorf K., McPhail L. C., Gomez-Cambronero J. (2007) FASEB J. 21, 1075–1087 [DOI] [PubMed] [Google Scholar]
- 7.Corrotte M., Chasserot-Golaz S., Huang P., Du G., Ktistakis N. T., Frohman M. A., Vitale N., Bader M. F., Grant N. J. (2006) Traffic 7, 365–377 [DOI] [PubMed] [Google Scholar]
- 8.Buchanan F. G., McReynolds M., Couvillon A., Kam Y., Holla V. R., Dubois R. N., Exton J. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1638–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghu P., Manifava M., Coadwell J., Ktistakis N. T. (2009) Biochim. Biophys. Acta 1791, 889–897 [DOI] [PubMed] [Google Scholar]
- 10.Topham M. K. (2006) J. Cell. Biochem. 97, 474–484 [DOI] [PubMed] [Google Scholar]
- 11.Cummings R., Parinandi N., Wang L., Usatyuk P., Natarajan V. (2002) Mol. Cell. Biochem. 234, 99–109 [PubMed] [Google Scholar]
- 12.Lehman N., Di Fulvio M., McCray N., Campos I., Tabatabaian F., Gomez-Cambronero J. (2006) Blood 108, 3564–3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powner D. J., Payne R. M., Pettitt T. R., Giudici M. L., Irvine R. F., Wakelam M. J. (2005) J. Cell Sci. 118, 2975–2986 [DOI] [PubMed] [Google Scholar]
- 14.Zeng X. X., Zheng X., Xiang Y., Cho H. P., Jessen J. R., Zhong T. P., Solnica-Krezel L., Brown H. A. (2009) Dev. Biol. 328, 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishikimi A., Fukuhara H., Su W., Hongu T., Takasuga S., Mihara H., Cao Q., Sanematsu F., Kanai M., Hasegawa H., Tanaka Y., Shibasaki M., Kanaho Y., Sasaki T., Frohman M. A., Fukui Y. (2009) Science 324, 384–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehman J. A., Paul C. C., Baumann M. A., Gómez-Cambronero J. (2001) Am. J. Physiol. Cell Physiol. 280, C183–C191 [DOI] [PubMed] [Google Scholar]
- 17.Di Fulvio M., Frondorf K., Henkels K. M., Lehman N., Gomez-Cambronero J. (2007) J. Mol. Biol. 367, 814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez I., Arnold R. S., Lambeth J. D. (1998) J. Biol. Chem. 273, 12846–12852 [DOI] [PubMed] [Google Scholar]
- 19.Sung T. C., Roper R. L., Zhang Y., Rudge S. A., Temel R., Hammond S. M., Morris A. J., Moss B., Engebrecht J., Frohman M. A. (1997) EMBO J. 16, 4519–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeniou-Meyer M., Zabari N., Ashery U., Chasserot-Golaz S., Haeberlé A. M., Demais V., Bailly Y., Gottfried I., Nakanishi H., Neiman A. M., Du G., Frohman M. A., Bader M. F., Vitale N. (2007) J. Biol. Chem. 282, 21746–21757 [DOI] [PubMed] [Google Scholar]
- 21.Nakanishi H., de los Santos P., Neiman A. M. (2004) Mol. Biol. Cell 15, 1802–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Su W., Yeku O., Olepu S., Genna A., Park J. S., Ren H., Du G., Gelb M. H., Morris A. J., Frohman M. A. (2009) Mol. Pharmacol. 75, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor R. M., Foubert T. R., Burritt J. B., Baniulis D., McPhail L. C., Jesaitis A. J. (2004) Biochim. Biophys. Acta 1663, 201–213 [DOI] [PubMed] [Google Scholar]
- 24.Lehman J. A., Calvo V., Gomez-Cambronero J. (2003) J. Biol. Chem. 278, 28130–28138 [DOI] [PubMed] [Google Scholar]
- 25.Lee S., Park J. B., Kim J. H., Kim Y., Kim J. H., Shin K. J., Lee J. S., Ha S. H., Suh P. G., Ryu S. H. (2001) J. Biol. Chem. 276, 28252–28260 [DOI] [PubMed] [Google Scholar]
- 26.Chen J., Fang Y. (2002) Biochem. Pharmacol. 64, 1071–1077 [DOI] [PubMed] [Google Scholar]
- 27.Kusner D. J., Barton J. A., Wen K. K., Wang X., Rubenstein P. A., Iyer S. S. (2002) J. Biol. Chem. 277, 50683–50692 [DOI] [PubMed] [Google Scholar]
- 28.Uesugi Y., Hatanaka T. (2009) Biochim. Biophys. Acta 1791, 962–969 [DOI] [PubMed] [Google Scholar]
- 29.Mazie A. R., Spix J. K., Block E. R., Achebe H. B., Klarlund J. K. (2006) J. Cell Sci. 119, 1645–1654 [DOI] [PubMed] [Google Scholar]
- 30.Kriebel P. W., Parent C. A. (2009) Methods Mol. Biol. 571, 111–124 [DOI] [PubMed] [Google Scholar]
- 31.Rizzo M. A., Shome K., Watkins S. C., Romero G. (2000) J. Biol. Chem. 275, 23911–23918 [DOI] [PubMed] [Google Scholar]
- 32.Zhou D., Luini W., Bernasconi S., Diomede L., Salmona M., Mantovani A., Sozzani S. (1995) J. Biol. Chem. 270, 25549–25556 [DOI] [PubMed] [Google Scholar]
- 33.Sliva D., Mason R., Xiao H., English D. (2000) Biochem. Biophys. Res. Commun. 268, 471–479 [DOI] [PubMed] [Google Scholar]
- 34.Siddiqui R. A., English D. (1997) Biochim. Biophys. Acta 1349, 81–95 [DOI] [PubMed] [Google Scholar]
- 35.Codeluppi S., Svensson C. I., Hefferan M. P., Valencia F., Silldorff M. D., Oshiro M., Marsala M., Pasquale E. B. (2009) J. Neurosci. 29, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomez-Cambronero J. (2003) FEBS Lett. 550, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim H. K., Choi Y. A., Park W., Lee T., Ryu S. H., Kim S. Y., Kim J. R., Kim J. H., Baek S. H. (2003) J. Biol. Chem. 278, 45117–45127 [DOI] [PubMed] [Google Scholar]
- 38.Chen Y., Zheng Y., Foster D. A. (2003) Oncogene 22, 3937–3942 [DOI] [PubMed] [Google Scholar]
- 39.Ali S. M., Sabatini D. M. (2005) J. Biol. Chem. 280, 19445–19448 [DOI] [PubMed] [Google Scholar]
- 40.Crouch M. F. (1997) Biochem. Biophys. Res. Commun. 233, 193–199 [DOI] [PubMed] [Google Scholar]
- 41.Berven L. A., Willard F. S., Crouch M. F. (2004) Exp. Cell Res. 296, 183–195 [DOI] [PubMed] [Google Scholar]
- 42.Berven L. A., Crouch M. F. (2000) Immunol. Cell Biol. 78, 447–451 [DOI] [PubMed] [Google Scholar]
- 43.Tanski W. J., Nicholl S. M., Kim D., Fegley A. J., Roztocil E., Davies M. G. (2005) J. Vasc. Surg. 41, 91–98 [DOI] [PubMed] [Google Scholar]
- 44.Goncharova E. A., Goncharov D. A., Eszterhas A., Hunter D. S., Glassberg M. K., Yeung R. S., Walker C. L., Noonan D., Kwiatkowski D. J., Chou M. M., Panettieri R. A., Jr., Krymskaya V. P. (2002) J. Biol. Chem. 277, 30958–30967 [DOI] [PubMed] [Google Scholar]
- 45.Qian Y., Corum L., Meng Q., Blenis J., Zheng J. Z., Shi X., Flynn D. C., Jiang B. H. (2004) Am. J. Physiol. Cell Physiol. 286, C153–C163 [DOI] [PubMed] [Google Scholar]
- 46.Rhoads J. M., Niu X., Odle J., Graves L. M. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G510–G517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kooijman E. E., Chupin V., Fuller N. L., Kozlov M. M., de Kruijff B., Burger K. N., Rand P. R. (2005) Biochemistry 44, 2097–2102 [DOI] [PubMed] [Google Scholar]
- 48.Blackwood R. A., Smolen J. E., Transue A., Hessler R. J., Harsh D. M., Brower R. C., French S. (1997) Am. J. Physiol. 272, C1279–C1285 [DOI] [PubMed] [Google Scholar]
- 49.Yang L., Huang H. W. (2002) Science 297, 1877–1879 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.