Abstract
In vivo fluorescence microscopy studies of bacterial cells have shown that the bacterial shape-determining protein and actin homolog, MreB, forms cable-like structures that spiral around the periphery of the cell. The molecular structure of these cables has yet to be established. Here we show by electron microscopy that Thermatoga maritime MreB forms complex, several μm long multilayered sheets consisting of diagonally interwoven filaments in the presence of either ATP or GTP. This architecture, in agreement with recent rheological measurements on MreB cables, may have superior mechanical properties and could be an important feature for maintaining bacterial cell shape. MreB polymers within the sheets appear to be single-stranded helical filaments rather than the linear protofilaments found in the MreB crystal structure. Sheet assembly occurs over a wide range of pH, ionic strength, and temperature. Polymerization kinetics are consistent with a cooperative assembly mechanism requiring only two steps: monomer activation followed by elongation. Steady-state TIRF microscopy studies of MreB suggest filament treadmilling while high pressure small angle x-ray scattering measurements indicate that the stability of MreB polymers is similar to that of F-actin filaments. In the presence of ADP or GDP, long, thin cables formed in which MreB was arranged in parallel as linear protofilaments. This suggests that the bacterial cell may exploit various nucleotides to generate different filament structures within cables for specific MreB-based functions.
Keywords: Bacteria, Cell Surface Protein, Electron Microscopy (EM), Kinetics, Microfilaments, Protein Self-assembly, Protein Stability, Actin Homolog, High Pressure SAX, TIRF Microscopy
Introduction
Despite usually being constrained by a cell wall, bacterial shapes are highly diverse, reflecting the large phylogenetic range. For example Escherichia coli, Bacillus subtilis, and Thermatoga maritime are straight rods, Vibrio cholera is a curved rod, Borrelia burgdorferi forms flat waves, whereas Spiroplasma species are helical. One of the main cytoskeletal proteins involved in determining the shapes of bacteria is thought to be MreB an actin homolog whose atomic structure is very similar to G-actin, despite the low sequence homology (1). It can assemble into polymers both in vitro (1) and in vivo (2). Several studies suggest that MreB plays roles in chromosome segregation (3), polar localization of proteins (4), and maintenance of cell shape and resistance to external mechanical stresses (2). Peptidoglycan cell wall synthesis has been linked to the role of the MreB homolog MbI in B. subtilis (5); however, mechanisms by which MreB may provide mechanical support either directly to the cell or indirectly by affecting peptidoglycan wall integrity remain unclear.
In vivo studies of MreB have mainly been limited to visualization under the fluorescence microscope (2, 5). At the low resolution of this technique (∼0.2 μm), MreB was seen to form cable-like structures, which spiral around the periphery of the cell in B. subtilis, presumably just underneath the cytoplasmic membrane. By electron microscopy (EM),2 MreB has been observed in vitro to form straight or curved sheets and bundles (1, 6).
Using cryoelectron tomography of helical-shaped bacteria Spiroplasma melliferum at an intermediate resolution of ∼40 Å, three parallel sheet-like structures underneath the cell membrane were identified. At this low resolution, the molecular structures and organization remained elusive, and it was unclear which of the ribbons may be associated with MreB filaments (7).
Because of these shortcomings, we found it necessary to clarify the molecular structure of MreB in vitro by high resolution electron microscopy. We show that MreB from T. maritime forms novel sheets consisting of interwoven helical-like filaments, which may be related to the cables observed by light microscopy and the ribbons observed by cryoelectron tomography in vivo. This design has appeal as it will be mechanically stronger than a simple sheet constructed of linear filaments and may be of importance for maintaining bacterial cell shape. We have characterized various MreB supramolecular structures under a wide variety of conditions by electron microscopy and studied their polymerization properties, steady state dynamics, and stability of MreB filaments by light scattering, TIRF microscopy, and high pressure small angle x-ray scattering (HP-SAXS).
EXPERIMENTAL PROCEDURES
Chemicals
All chemicals were obtained from WAKO, except nucleotides that were from Sigma; fluorophores from Invitrogen; and EDTA-free protease inhibitor tablets that were supplied by Roche.
Proteins
MreB was engineered into a pET expression vector, which adds an N-terminal thrombin cleavable His tag MD(H)5ENLYFQG-MreB to the original MreB1 plasmid, which was a kind gift from Fusinita van den Ent, MRC. The protein was expressed in E. coli by adding 0.15 mm isopropyl-1-thio-β-d-galactopyranoside to the culture when the absorbance at 600 nm reached ∼0.6. After 4–5 h, cells were centrifuged and resuspended in buffer containing 20 mm imidazole, 300 mm KCl, pH 7.4 (buffer A), and 0.2 mg/ml lysozyme and EDTA-free protease inhibitor. Cells were sonicated and centrifuged at 40,000 rpm for 1.5 h, and the supernatant loaded onto a Qiagen-Ni-nitrilotriacetic acid (NTA) Superflow column. The column was washed with 20 mm imidazole, pH 7.4 and 300 mm KCl. MreB was eluted with 250 mm imidazole, pH 7.4, 300 mm KCl, 1 mm dithiothreitol, and further subjected to gel filtration chromatography on a HiLoad 26/60 Superdex pg column (GE Healthcare). The His tag was cleaved by adding 2 units/mg thrombin at room temperature in buffer A for ∼3 h, and the protein collected as the flow-through fraction after loading onto Qiagen-Ni-NTA Superflow. The protein was concentrated and either used fresh or was frozen in small aliquots in Eppendorf tubes cooled to the temperature of liquid nitrogen. The protein concentration was determined according to Bradford using a Bio-Rad kit. Before experiments, MreB was dialyzed against appropriate buffers. Proteins were briefly centrifuged at high speed before use. Lysines were labeled with Alexa 488 carboxylic acid succinimidyl ester 6-isomer or 5-(and-6) carboxyl tetramethylrhodamine succinimidyl ester at a molar ratio of about 1 dye to 4 MreB overnight at 4 °C as described previously for ParM (8).
TIRF Microscopy
Experiments were conducted in a similar fashion as described earlier in detail for ParM (8). MreB was diluted to the desired concentration, using buffer (300 mm KCl, 2 mm MgCl2, 40 mm Hepes, pH 7.5) in the presence of 10 mm dithiothreitol and crowding agents (1% MC or 8% PVA). ATP or GTP (2 mm final concentration) were added, directly mixed on the glass slide, covered with a glass coverslip and sealed with nail polish. ParM filaments labeled by fluorescent dye were observed with an inverted Nikon microscope as previously described (8). Filament length was analyzed and error estimation determined using the programs Aquacosmos, ImageJ, and KaleidaGraph 3.6 previously described in more detail (9).
Kinetics, Pi Release, and Critical Concentration Studies
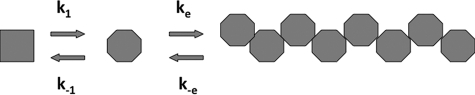
MreB polymerization was initiated by the addition of 2 mm nucleotide in appropriate buffer. The amount of MreB polymer was monitored by 90° perpendicular light scattering at 600 nm using a Hitachi F-4500 fluorescence spectrophotometer at 24 °C. The data were fitted using the program Dynafit (10). k1 and k−1 describe a concentration-independent step of monomer activation when assembly is initiated by the addition of nucleotide (11). All subsequent rate constant pairs are equal to (ke and k-e) and describe the steps of elongation. Adding a dimerization step did not improve the fit. The release of inorganic phosphate upon nucleotide hydrolysis during MreB polymerization was measured at 24 °C using Phosphate Assay Kit (E-6646) from Molecular Probes, based on a method described (12). To determine the critical concentration of ParM for polymerization, various protein concentrations were tested. Reactions were equilibrated at 24 °C for 1 h. 90° perpendicular light scattering experiments were carried out using a Hitachi F-4500 fluorescence spectrophotometer at 300 nm and 24 °C.
Electron Microscopy
Samples were applied to glow discharged, carbon-coated, copper grids, blotted, negatively stained with 1% uranyl acetate, air dried, and examined under a Jeol JEM-2010 HC microscope operated at 100 keV at a nominal magnification of 10,000 to 40,000. Films were digitized with Photo Scan 2000 (Z/I Imaging) at 7-μm steps. Optical transforms, filtered images, and single particle averaging were performed using the EM software package EOS (13). Otherwise electron micrographs were scanned at 1200 DPI on an Epson scanner and displayed by Photoshop.
HP-SAXS
MreB at 4 mg/ml was polymerized by the addition of 2 mm nucleotide and placed into specially designed chambers (14). Temperature was controlled at 24 °C. Hydrostatic pressure was applied in multiple steps. Using beam line BL45-XU (λ, 0.9 Å) and camera lengths of 2.2 m, the small angle scattering patterns of MreB were recorded on a cooled CCD camera, and data were analyzed as described (15).
RESULTS
MreB Sheet Morphology
In vitro MreB formed multilayered sheets in the presence of both ATP and GTP under a large variety of ionic conditions, different pH values, and over a wide temperature range as visualized by EM (Fig. 1). A sheet is a structure that has a considerably larger width (W) than height (H); W ≫ H, opposed to bundles that are roughly round objects in cross section, where W ≈ H. The length L of both sheets and bundles is usually much larger than both width and height L ≫ W, L ≫ H. At physiological salt concentrations around 350 mm KCl found in bacterial cells (16) and at pH values between 7.0 and 7.7 typical for most bacteria, including T. maritime (17), most sheets were about 30–100 nm wide and up to several μm long (Fig. 1, E and H), consistent with values obtained from in vivo light microscopy observations of MreB (2). These sheets had a tendency to roll up or display chirality (Fig. 1, E and I). In contrast, no polymer structures were observed in the absence of nucleotide (Fig. 1A), whereas nonhydrolyzable nucleotides, like AMPPNP or GMPPNP, led only to the formation of amorphous suprastructures (Fig. 1D).
FIGURE 1.
MreB sheets. Typical electron micrographs under various conditions are shown. A, pH 7.0, 70 mm KCl, no nucleotide. B, 70 mm KCl, pH 7.7, 2 mm ADP. C, 70 mm KCl, pH 7.7, 2 mm GDP, arrows mark twisting of the sheet. D, 70 mm KCl, pH 7.7, 2 mm AMPPNP. E, 350 mm KCl, pH 7.7, 2 mm ATP. F, 350 mm KCl, pH 6.5, 2 mm ATP. G, 70 mm KCl, pH 7.7, 2 mm EDTA, 2 mm ATP. H, 350 mm KCl, pH 7.0, 2 mm GTP. I, 350 mm KCl, pH 9.0, 2 mm GTP. J, 70 mm KCl, pH 7.7, 2 mm GTP, scale bars, 200 nm.
Reduction of salt concentrations at constant pH led to the formation of much wider sheets (Fig. 1J). Increasing the pH under a given ionic condition also led to wider sheets (Fig. 1I), while decreasing the pH led to sheets that were smaller in diameter (Fig. 1F). This behavior indicated that sheet formation and dimensions are predominantly a function of the effective electrostatic surface charge of MreB under the conditions studied.
Sheets were formed both in the presence and absence of divalent cations (Fig. 1G) as had been observed in previous studies on MreB suprastructures (1, 18), yet the observed crystalline ordering in the sheets was highest in the presence of millimolar amount of MgCl2 (Fig. 2). Sheets also formed in the presence of millimolar concentrations of CaCl2. These were similar in appearance to those formed in MgCl2; however they tended to curl and twist more easily than sheets formed in the presence of MgCl2 (supplemental Fig. S1A).
FIGURE 2.
Organization and structure in GTP-MreB sheets. A, typical electron micrograph of a GTP-MreB sheet, scale bar 200 nm. B, its Fourier transform showing sharp diffraction spots. Indicated with arrows and numbers: (1) equatorial reflection at ∼ 37 Å (2) first row-line at ∼ 51 Å spacing (3) second row-line at ∼ 25.5 Å spacing (4) third row-line at ∼ 17 Å spacing. C–E, representative filtered and averaged images of GTP-MreB sheets. C, this sheet belonged to group 1, scale bar 100 Å (D) A sheet belonging to group 2, scale bars, 100 Å (E) A very thin sheet allowing us to see the fine structure of a single layer within the sheet, scale bar 200 Å.
Molecular Organization of MreB Sheets and Filament Structures
To learn about the molecular structure of the observed sheets, image analysis of electron micrographs was performed. The larger sheets with high crystalline order that formed at lower salt concentrations (∼70–150 mm KCl) were well suited, as their optical transforms showed sets of discrete reflections (Fig. 2). The Fourier transforms were similar for both ATP and GTP sheets (supplemental Fig. S2A and Fig. 2). However, GTP sheets usually diffracted to higher resolution perhaps because their formation was slower as shown later. The optical diffraction pattern showed discrete spots arranged on row lines. The first meridional reflection was observed at 51 Å, and the first equatorial occurred at 37 Å. Reflections up to the 3rd order on the meridian (∼17 Å) and radial reflections out to spacings of ∼18.5 Å were observed for MreB-GTP (Fig. 2B).
Sheets formed at physiological salt concentrations gave similar optical diffraction patterns, yet the resolution was limited to the first equatorial reflection and the two most inner reflections on the first row line indicating a higher degree of disorder (supplemental Fig. S2B). Filtered images were obtained from electron micrographs, which diffracted to high resolution and further refined by single particle averaging using the EOS electron microscopy analysis package (13) (Fig. 2). The filtered and single particle averaged sheets showed structures that consisted of a more complex arrangement than parallel MreB filaments observed in previous studies (1). Structures from different sheets were not identical but could be separated into two groups (Fig. 2, A and B). Whereas Fourier shell correlation of in-group images agreed up to 21 Å, close to the maximum resolution for negative stain EM, inter-group Fourier shell correlation did not agree at resolutions higher than 28 Å (supplemental Fig. S3). Sheets belonging to group 1 were predominantly formed. These sheets consisted of sets of multiple diagonally interwoven filament-like strands crossing at an angle of about 70 degrees (Fig. 2C). Filaments apparently were not straight protofilaments, rather these polymers adopted a sinusoidal appearance. Sheets of group 2 were similar but more difficult to interpret (Fig. 2D).
A few very thin sheets allowed the investigation of the filamentous structures formed by MreB within the sheets in detail. Filtered and averaged images showed that MreB formed helical 3/1 filaments with a repeat of about 200 Å within the sheets (Fig. 2E). The subunit repeat and the filament radius were about 50 Å, values that could be directly extracted from the filtered and averaged images. Note that every 3rd molecule in Fig. 3C is very bright, indicating that these molecules are close to the carbon-coated EM grid, whereas the molecules inbetween have substantially less stain, indicating that they are not lying in the same plane. These findings apparently differed from a previous study that had used a His-tagged Thermatoga MreB construct, which led solely to linear protofilament formation (1). Direct inspection indicated that the helical filaments observed in Fig. 2E appeared to have the same polarity. If this conclusion is also valid in the mostly observed multilayered sheets is unclear.
FIGURE 3.
Organization and structure in GDP-MreB sheets. A, typical GDP-MreB sheet displaying chirality marked by the arrows. B, Fourier transform of a part of the sheet in A, showing a weak layer line (2) an equatorial reflection (1) both at ∼ 50 Å. C, filtered and averaged image shows GDP-MreB filaments arranged as linear protofilaments.
Opposed to ATP or GTP, the addition of ADP or GDP led to the formation of very long, thin wavy, or twisted ribbons among unstructured aggregates, (Figs. 1, B and C and 3A). The molecular structure of MreB-ADP or MreB-GDP ribbons also differed. The optical transforms of MreB-ADP and MreB-GDP sheets consistently showed two reflections, a wide layer line at about 51 Å arising from the axial subunit repeat of MreB protofilaments and a diffuse equatorial reflection around 50 Å arising from the filament packing (Fig. 3B). No obvious reflection corresponding to twice the interfilament spacing (100 Å) was observed on the equator, which could indicate that filaments may be polar rather than bipolar (19). The filtered images of MreB-ADP polymers apparently show sets of linear filaments, each composed of a string of monomers with an ∼51 Å longitudinal spacing (Fig. 3C). Surprisingly the optical diffraction patterns as well as the filtered images were very similar to those observed from His-tagged MreB-ATP filaments in a previous study (1).
Dynamic Properties
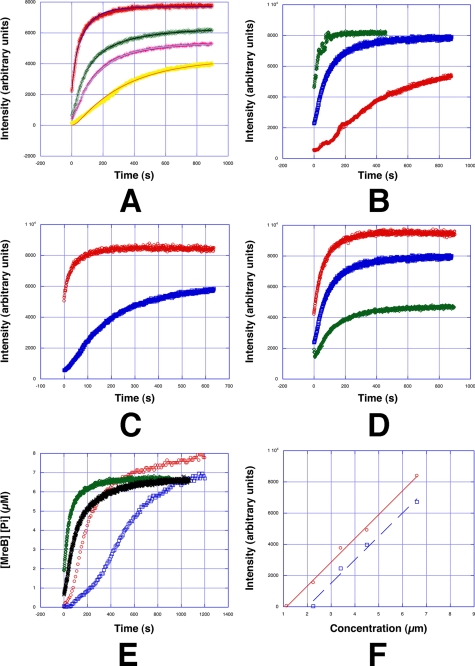
The assembly mechanism of MreB sheets was investigated by light scattering. After adding nucleotide (ATP or GTP is NTP) to physiological concentrations of MreB (∼6 mm) at room temperature (24 °C) and intermediate salt concentrations (150 mm KCl) at pH 7.7, the light scattering signal increased rapidly and reached steady state after about 400 s (Fig. 4A). Assembly was faster with ATP than with GTP (Fig. 4A), and the light scattering intensity of MreB-ATP was stronger than MreB-GTP at steady state, indicating that MreB-ATP formed larger sheets on average than MreB-GTP. Consistent with these findings, the critical concentration for polymerization was about 1 μm for MreB-ATP and 2 μm for MrB-GTP (Fig. 4F). Increasing the salt concentration to physiological values found in the bacterial cytoplasm (∼350 mm), decreased both the polymerization rate and light scattering intensity at steady state by about 25% (Fig. 4A) in agreement with the smaller dimensions measured by EM. Assembly kinetics were substantially faster at 37 °C and about 2.5 times slower at 12 °C as compared with room temperature (24 °C) (Fig. 4B). The rate of polymerization was faster, and more polymer was formed at pH 9 than at the lower pH of 6.5, pH 7.7 gave intermediate values (Fig. 4D). Polymerization was slower, and fewer sheets were formed with NaCl than with KCl (Fig. 4C). This contrasts to previous studies in which different MreB constructs (6) or preparation methods (18) were used. In these reports, MreB either only properly polymerized at high temperatures around 65 °C (6) or favored polymerization at low pH (18). In both previous studies, the formation of filaments was inhibited at physiological salt concentrations (6, 18). In both studies MreB formed suprastructures in the form of bundles, but structural details were not investigated.
FIGURE 4.
A, typical kinetic data obtained by light scattering. MreB concentration was 6.6 μm in all experiments: A, red, 150 mm KCl and ATP; green, 150 mm KCl and GTP; pink, 350 mm KCL and ATP; yellow, 350 mm KCl and GTP. All experiments were in the presence of 2 mm MgCl2, the pH was 7.7 and temperature 24 °C. The thin lines in blue, black, light blue, and orange are the minimal kinetic model fits using DYNAFIT B, temperature dependence of polymerization. Conditions were 150 mm KCl, 2 mm MgCl2, pH 7.7 and ATP. Red, 12 °C; blue, 24 °C; green, 37 °C. C, monovalent cation dependence. Red, 70 mm KCl and ATP; blue, 70 mm NaCl and ATP. Experiments were carried out in the presence of 2 mm MgCl2 at 24 °C, the pH was 7.7 D, pH dependence. Experiments in the presence of 2 mm MgCl2, 150 KCl, and ATP. Red, pH 9.0; blue, pH 7.7; green, pH 6.5. E, relationship between the time course of polymerization and Pi release. The units of the vertical axis for light scattering were converted to the concentration of MreB; green, ATP-MreB light scattering and red, the corresponding Pi release, black, GTP-MreB light scattering and blue the corresponding Pi release. F, steady-state light scattering intensities plotted as a function of MreB concentration. Conditions were 150 mm KCl, 2 mm MgCl2, pH 7.7, 24 °C red, ATP, blue, GTP.
Polymerization of eukaryotic actin requires a nucleation step prior to elongation (20). This was observed by actin polymerization experiments using light scattering; a lag phase prior to the intensity increase of light scattering was characteristic for the period of nucleation. In the present experiments, where the first ∼10 s of MreB polymerization could not be recorded, no apparent lag phase under conditions closer to physiological (at higher temperature or in the presence of KCL) was detected (Fig. 4). Assembly kinetics data could be fit using a minimal kinetics model similar to that described for the linear protofilaments of E. coli FtsZ (11) (Fig. 4A) involving only two steps: monomer activation and elongation (Table 1 and Scheme 1). The rate constants are summarized in Table 1. For MreB, the middle step in forming a dimer was not needed and gave no improvement to the fit. Strong cooperativity within the sheets initial nucleus may not require any additional nucleation mechanism prior to filament elongation, which may be essential for the formation of individual filaments. At low temperature or in the presence of NaCl, the data (Fig. 4) showed a slight lag phase indicating the possible necessity of a nucleation-dependent assembly process under those for T. maritime physiologically unfavorable conditions.
TABLE 1.
The kinetic rate constants
k1 and k−1 describe a concentration-independent step of monomer activation when assembly is initiated by the addition of NTP. All subsequent rate constant pairs are equal to ke and k−e and describe the steps of elongation.
| ATP, 150 mm KCl | GTP, 150 mm KCl | ATP, 350 mm KCl | GTP, 350 mm KCl | |
|---|---|---|---|---|
| k1 (s−1) | 0.021 | 0.008 | 0.005 | 0.003 |
| k−1 (s−1) | 0.164 | 0.093 | 0.077 | 0.028 |
| ke (μm−1s−1) | 0.099 | 0.058 | 0.242 | 0.145 |
| k−e (μm−1 s−1) | 0.012 | 0.016 | 0.054 | 0.033 |
SCHEME 1.
The kinetic scheme or MreB assembly.
Phosphate release lagged behind polymerization, in a similar fashion to that observed for F-actin (21) and the lag was observed to be larger for MreB-GTP than for MreB-ATP. Phosphate release displayed a biphasic behavior (Fig. 4E). The initial phase was fast until the curve intersected the polymerization time course that had already reached steady state. This indicates that each protomer hydrolyzes a single ATP or GTP molecule and does not undergo rapid polymerization dynamics subsequent to its initial incorporation into a polymer. The second phase was slower, yet Pi release continued even after MreB filaments had long reached steady state (Fig. 4E), indicating that the mature polymers are not entirely static. The steady-state dynamics of MreB filaments were observed by TIRF microscopy (Fig. 5A). Electron micrographs of sheets formed from fluorescently labeled MreB after addition of nucleotide were first examined. These appear similar to unlabeled MreB sheets (supplemental Fig. S1B) indicating that labeling does not impair sheet formation. On a microscope slide, flat sheets, such as those formed by MreB, may be expected to adsorb more readily to the glass surface than single filaments or round bundles and therefore appear relatively immobile. However, under the TIRF microscope a large population of observed sheets underwent fluctuations in length at steady state, fluctuations that were larger than the error in the measurement (∼5%) and similar in magnitude to the fluctuations observed for actin filaments (22) (Fig. 5B). The interpretation of these fluctuations is more complicated than for single filaments as here multilayered sheets were being observed. Yet these observations seem consistent with a treadmilling process and in agreement with in vivo observations of fluorescently labeled MreB in Caulabacter cells, which also suggested an F-actin like treadmilling process (23).
FIGURE 5.
TIRF microscopy and HP-SAX. A, TIRF microscopy images of MreB-GTP sheets at steady state, scale bar 5 μm. B, typical examples of length fluctuations of individual sheets over time. C, disintegration of MreB sheets by high pressure as seen by the decrease of the total integrated intensity; red, GTP-MreB; blue, ATP-MreB.
To further quantify the stability and mechanical resiliency of MreB filaments bound to different nucleotides, HP-SAXS studies were performed. Exerting pressure on a filamentous structure, can lead to the destruction of noncovalent interactions, loss of nucleotide and subsequently to filament depolymerization, as shown for actin and ParM filaments (24, 8). MreB filaments were stable unto about 2000 bar after which they disintegrated (Fig. 5C, supplemental Fig. S4). MreB-ATP and MreB-GTP behaved similarly (Fig. 5C). The behavior of MreB filaments under pressure was similar to that of F-actin, which was a stable filament until about 2000 bar (24) but differed from ParM-R1 which displayed microtubule-like dynamic instability and dissociated at much lower pressures of around 400–500 bar (8).
DISCUSSION
Mutations in MreB resulted in a shape conversion from rod to sphere morphology (25), implying a role for MreB in cell shape determination. Disruption of pole-to-pole MreB helically arranged cables by mutational inactivation resulted in failure of lateral murein synthesis and formation of spherical cells (26). Disruption in the two genes immediately downstream of MreB, MreC, and MreD also yielded spherical phenotypes in E. coli and B.subtilis (27, 28). In B. subtilis the synthesis of lateral murein is governed by MreB homologue MbI (5) and the localizations of GFP-MreC and GFP-MreD were observed to be similar to that of MbI (28). Under the light microscope, all of these proteins followed helical patterns underneath the cell membrane.
In a bacterial two-hybrid system, E. coli MreC directly interacted with MreB and MreD (27). In addition, the bitopic membrane protein MreC interacts with several members of the PBP family of bifunctional transglycosylase-transpeptidases (29), which are thought to participate in both lateral and septal murein synthesis. These results suggest that murein synthesis in E. coli is carried out by a helical array of interacting proteins that includes MreB, MreC, MreD, and members of the PBP family. The model predicts that new peptidoglycan is inserted into the lateral cell wall in a helical pattern that reflects the helical pattern of the biosynthesis complex.
Despite this knowledge, the actual role of the MreB cytoskeleton remains unclear, as helical distributions of MreC and MreD in E. coli observed by light microscopy were found to be independent of MreB (26) and in Caulobacter, GFP-PBP2 adopted a helical distribution, which partially overlapped with MreC but not with MreB. In the absence of electron tomography data of T. maritime, the in vivo molecular structure of MreB within the cable-like spirals observed by light microscopy remains elusive. Only one electron tomography study of the helical bacteria Spiroplasma showed ribbon-like structures underneath the cell membrane (7), but the molecular structure and organization within the ribbons could not be unambiguously determined.
Here we have shown that in the presence of NTP Thermatoga MreB formed multilayered sheets of apparently interwoven filaments. The mechanical rigidity of these architectures can be expected to be substantially higher than that of a simple set of parallel linear protofilaments. Indeed, the HP-SAX experiments indicate that MreB filaments have a mechanical resilience similar to F-actin, which is the highest of any cytoskeletal protein known.
Recent rheology experiments have characterized MreB from T. maritime to have an elastic module that is more than 3-fold higher than F-actin (30). Because the elasticity of a polymer network is predicted to depend weakly on the intrinsic rigidity of the filaments, this high elastic modulus can be mostly accounted for by strong interfilament interactions. The elasticity and concentration of MreB are related by a power law relationship between network elasticity (G′) and protein concentration (C): G′ ∼ C0.44 and G' ∼ C0.29 for MreB-ATP and MreB-GTP, respectively (30), which is consistent with that predicted for cross-linked filaments. In contrast, non-cross-linked and entangled semiflexible polymers, such as F-actin, have a predicted power-law relationship G′ ∼ C1.4 (31). The lower concentration dependence index of GTP-MreB indicates that GTP-MreB filaments are more strongly cross-linked than ATP-MreB filaments (30). This is consistent with our electron microscopy observations that Thermatoga NTP-MreB forms sheets consisting of interwoven filaments and that GTP-MreB sheets show a higher degree of order than ATP-MreB sheets. The strong interaction of MreB filaments within the sheets observed here should be able to resist large bending and compression forces and help in imposing a rigid cylindrical architecture to rod-shaped bacterial cells.
Individual MreB filaments within the sheets formed in the presence of GTP or ATP were not linear protofilaments as described earlier (1), which may have to do with the fact that the previous study used a C-terminally His-tagged MreB. Rather untagged MreB assembled into single-stranded sinusoidally varying or helical 3/1 filaments with a repeat of about 200 Å (Fig. 3), whereas in the presence of GDP or ADP linear protofilaments were observed (Fig. 1, B and C). This may indicate that the binding of GTP or ATP to MreB monomers stabilizes a different conformation than ADP or GDP, which in turn leads to the preferential formation of either helical filaments or linear protofilaments. It is unknown whether bacterial cells utilize ATP, GTP, or both nucleotides to assemble MreB in vivo and whether the cell exploits different nucleotides for specific MreB-based functions. In nitrogen-fixing bacteria like T. maritime the rate of ATP regeneration is considered to be growth rate-limiting because of the high ATP requirement for N2 fixation (32), and the ATP/ADP ratio was estimated to be between 1.4 and 2.0. In E. coli, during exponential growth, the ATP and GTP concentrations are reported to be relatively constant at about 3 mm and 900 μm, respectively (33). Assuming similar concentrations in T. maritime, the nucleotide concentrations can be estimated to be about 2 mm ATP, 1 mm ADP, and 1 mm GTP. Should such concentrations lead to interwoven or linear sheet structures at the molecular level or coexistence of both forms in T. maritime is an open question.
It should be noted that opposed to most rod-shaped bacteria described above where MreB is thought to shape the cell wall via MreC, MreC is not present in T. maritime (34). It may be that different bacteria which evolved during evolution and adopted to specific environments have found different ways to elongate their cell wall while maintaining a constant cell width between each division. Other protein factors like Rod Z regulate MreB assembly in rod-shaped bacteria like E. coli, while cell division protein FtsZ is required for the collapse of MreB cables from extended helices into rings near the future division site (34). Whether any of the MreB-associated proteins preferentially binds to MreB-NTP or MreB-NDP cables is unknown.
MreB has been shown to be a determinant of polar protein localization and the translocation of chromosomal origins toward cell poles in both Caulabacter and E. coli (3, 35) leading to the hypothesis that MreB structures possess a uniform polarity that can be interpreted by trafficking factors. The organization of sheets indicates the possibility that both Thermatoga MreB-NTP and MreB-NDP may form polar suprastructures.
The kinetic dynamics of Thermatoga MreB-NTP filament formation was faster than F-actin but significantly slower than ParM (8, 21). The critical concentration of MreB-NTP filament formation was similar to ParM, yet assembly under conditions closer to physiological occurred without a nucleation step, making Thermatoga MreB a very efficient polymerizing machine. Polymerization kinetics and critical concentration differed substantially from a previous study (6) which had used C-terminal His-tagged Thermatoga MreB, but was more similar to the work of Bean and Amann (18), which also employed native MreB, although prepared differently than in our study. The variations in the biochemistry may reflect the differences in molecular structure and organization of Thermatoga MreB suprastructures we have observed here compared with the only previous structural study which used a His-tagged construct (1).
Time-lapse TIRF microscopy of Thermatoga MreB-NTP sheets at steady state support a treadmilling process consistent with in vivo observations of MreB in Caulobacter (23). Yet interpreting of treadmilling in terms of monomer motions is complicated by the fact that we are dealing with complex interwoven sheets rather than single filaments. Second, the site of GTP hydrolysis and what triggers hydrolysis is one of the major unknowns. Nevertheless, our findings define clear predictions on the structures, dynamics, and functions of T. maritime MreB, which can be tested by future in vivo light microscopy and electron tomography studies.
Supplementary Material
Acknowledgments
We thank Dr. Hiroshi Matsuo (Niigata Industrial Creation Organization & Kinki University) and Dr. Ryo Ishiguro (Gifu University) for technical support of HP-SAXS experiments.
This work was supported by the Agency for Science, Technology and Research (A*STAR), Singapore (to R. C. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- EM
- electron microscopy
- HP-SAXS
- high pressure small angle x-ray scattering
- TIRF
- total internal reflection fluorescence microscopy
- AMPPNP
- 5′-adenylyl imidodiphosphate
- GMPPNP
- guanyl-5′-yl imidodiphosphate.
REFERENCES
- 1.van den Ent F., Amos L. A., Löwe J. (2001) Nature 413, 39–44 [DOI] [PubMed] [Google Scholar]
- 2.Jones L. J., Carballido-López R., Errington J. (2001) Cell 104, 913–922 [DOI] [PubMed] [Google Scholar]
- 3.Kruse T., Møller-Jensen J., Løbner-Olesen A., Gerdes K. (2003) EMBO J. 22, 5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gitai Z., Dye N., Shapiro L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel R. A., Errington J. (2003) Cell 113, 767–776 [DOI] [PubMed] [Google Scholar]
- 6.Esue O., Cordero D., Wirtz D., Tseng Y. (2005) J. Biol. Chem. 280, 2628–2635 [DOI] [PubMed] [Google Scholar]
- 7.Kürner J., Frangakis A. S., Baumeister W. (2005) Science 307, 436–438 [DOI] [PubMed] [Google Scholar]
- 8.Popp D., Narita A., Oda T., Fujisawa T., Matsuo H., Nitanai Y., Iwasa M., Maeda K., Onishi H., Maéda Y. (2008) EMBO J. 27, 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popp D., Gov N. S., Iwasa M., Maéda Y. (2008) Biopolymers 89, 711–721 [DOI] [PubMed] [Google Scholar]
- 10.Kuzmic P. (1996) Ann. Biochem. 237, 260–273 [DOI] [PubMed] [Google Scholar]
- 11.Chen Y., Bjornson K., Redick S. D., Erickson H. P. (2005) Biophys. J. 88, 505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Webb M. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasunaga T., Wakabayashi T. (1996) J. Struct. Biol. 116, 155–160 [DOI] [PubMed] [Google Scholar]
- 14.Nishikawa Y., Fujisawa T., Inoko Y., Moritoki M. (2001) Nuclear Instr. Methods A 467, 1384–1387 [Google Scholar]
- 15.Fujisawa T. (2000) J. Crystall. Soc. Japan 42, 97–105 [Google Scholar]
- 16.Cayley S., Lewis B. A., Guttman H. J., Record M. T., Jr. (1991) J. Mol. Biol. 222, 281–300 [DOI] [PubMed] [Google Scholar]
- 17.Krulwich T. A., Agus R., Schneier M., Guffanti A. A. (1985) J. Bacteriol. 162, 768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bean G. J., Amann K. J. (2008) Biochemistry 47, 826–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sukow C., deRosier D. (1998) J. Mol. Biol. 284, 1039–1050 [DOI] [PubMed] [Google Scholar]
- 20.Cooper J. A., Buhle E. L., Jr., Walker S. B., Tsong T. Y., Pollard T. D. (1983) Biochemistry 22, 2193–2202 [DOI] [PubMed] [Google Scholar]
- 21.Iwasa M., Maeda K., Narita A., Maeda Y., Oda T. (2008) J. Biol. Chem. 283, 21045–21053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujiwara I., Takahashi S., Tadakuma H., Funatsu T., Ishiwata S. (2002) Nat. Cell Biol. 4, 666–673 [DOI] [PubMed] [Google Scholar]
- 23.Kim S. Y., Gitai Z., Kinkhabwala A., Shapiro L., Moerner W. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10929–10934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeuchi Y., Suzuki A., Oota T., Hagiwara K., Tatsumi R., Ito T., Balny C. (2002) Eur. J. Biochem. 269, 364–371 [DOI] [PubMed] [Google Scholar]
- 25.Wachi M., Doi M., Tamaki S., Park W., Nakajima-Iijima S., Matsuhashi M. (1987) J. Bacteriol. 169, 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dye N. A., Pincus Z., Theriot J. A., Shapiro L., Gitai Z. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruse T., Bork-Jensen J., Gerdes K. (2005) Mol. Microbiol. 55, 78–89 [DOI] [PubMed] [Google Scholar]
- 28.Leaver M., Errington J. (2005) Mol. Microbiol. 57, 1196–1209 [DOI] [PubMed] [Google Scholar]
- 29.Divakaruni A. V., Loo R. R., Xie Y., Loo J. A., Gober J. W. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18602–18607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esue O., Wirtz D., Tseng Y. (2006) J. Bacteriol. 188, 968–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer A., Xu J., Kuo S. C., Wirtz D. (1999) Biophys J. 76, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Upchurch R. G., Mortenson L. E. (1980) J. Bacteriol. 143, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jewett M. C., Miller M. L., Chen Y., Swartz J. R. (2009) J. Bacteriol. 191, 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margolin W. (2009) Curr. Biol. 19, R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gitai Z., Dye N. A., Reisenauer A., Wachi M., Shapiro L. (2005) Cell 120, 329–341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.