Abstract
Missense mutations represent the most common cause of many genetic diseases including cystathionine β-synthase (CBS) deficiency. Many of these mutations result in misfolded proteins, which lack biological function. The presence of chemical chaperones can sometimes alleviate or even restore protein folding and activity of mutant proteins. We present the purification and characterization of eight CBS mutants expressed in the presence of chemical chaperones such as ethanol, dimethyl sulfoxide, or trimethylamine-N-oxide. Preliminary screening in Escherichia coli crude extracts showed that their presence during protein expression had a significant impact on the amount of recovered CBS protein, formation of tetramers, and catalytic activity. Subsequently, we purified eight CBS mutants to homogeneity (P49L, P78R, A114V, R125Q, E176K, P422L, I435T, and S466L). The tetrameric mutant enzymes fully saturated with heme had the same or higher specific activities than wild type CBS. Thermal stability measurements demonstrated that the purified mutants are equally or more thermostable than wild type CBS. The response to S-adenosyl-l-methionine stimulation or thermal activation varied. The lack of response of R125Q and E176K to both stimuli indicated that their specific conformations were unable to reach the activated state. Increased levels of molecular chaperones in crude extracts, particularly DnaJ, indicated a rather indirect effect of the chemical chaperones on folding of CBS mutants. In conclusion, the chemical chaperones present in the expression medium were able to fully restore the activity of eight CBS mutants by improving their protein folding. This finding could have direct implications for the development of a therapeutical approach to pyridoxine unresponsive homocystinuria.
Keywords: Chaperone Chaperonin, Enzyme Mutation, Heme, Homocysteine, Protein Folding, Pyridoxal Phosphate, S-Adenosylmethionine (SAM), Conformational Disease, Homocystinuria, Misfolding
Introduction
Cystathionine β-synthase (CBS)3-deficient homocystinuria (CBSDH) is an autosomal recessive disorder, which is characterized by extremely elevated levels of plasma homocysteine and methionine (1). The clinical phenotype includes mental retardation, dislocated optic lenses, skeletal abnormalities, and thrombotic vascular complications. About half of CBS-deficient patients respond to treatment with pharmacological doses of pyridoxine (vitamin B6) with a significant lowering of plasma homocysteine levels and an alleviation of clinical phenotypes.
Human CBS has a modular domain structure and a complex regulatory mechanism (Fig. 1) (reviewed in Refs. 2–4). The enzyme contains four identical subunits each having three domains: an N-terminal domain, which binds heme cofactor of unclear function; a highly conserved catalytic core with pyridoxal-5′-phosphate (PLP) cofactor; and a C-terminal regulatory domain, which contains the CBS domain tandem and binds S-adenosyl-l-methionine (AdoMet). The AdoMet serves as an allosteric activator and increases CBS activity up to 8-fold. Removal of the C-terminal region, accompanied with a loss of response to AdoMet and ∼3-fold increase in activity, leads to a change of a homotetramer into a homodimer whose crystal structure was successfully solved (5, 6). In addition to the AdoMet stimulation, a similar increase in activity can be achieved by heating the enzyme close to its melting temperature or by a point mutation in the C-terminal region (7, 8). These forms of CBS activity stimulation are not additive, suggesting that they are acting through a similar mechanism, most likely by displacing the C-terminal autoinhibitory domain from the active site (7, 9, 10).
FIGURE 1.
The modular domain structure of human CBS. The human CBS consists of three domains: the N-terminal heme-binding domain with Cys-52 and His-65 as axial ligands of heme b, the highly conserved catalytic core with the ϵ-amino group of Lys-119 binding the carbonyl group of PLP, and the C-terminal AdoMet-binding regulatory domain with a tandem of CBS domains. The positions of the CBS mutations characterized in this study are also shown.
Missense mutations account for ∼87% of all mutations in CBSDH,4 the most common disorder of sulfur amino acids metabolism (1). Many of these mutations do not target the catalytic residues in CBS but rather result in misfolded proteins, which lack biological function and/or are destined for degradation. Shan and Kruger (11) showed that the activity of I278T, the most prevalent missense mutation found among CBSDH patients, can be partially restored by truncation of the C-terminal AdoMet-binding domain or by a point mutation in the C-terminal region (9). These observations suggested that the I278T mutation affects folding rather than catalytic activity of CBS. Furthermore, a study of 30 Slavic CBSDH patient cell lines revealed that a majority of CBS mutants formed large aggregates devoid of heme (12). These data suggest that the inability of mutants to properly incorporate heme may prevent correct folding, thus altering the tertiary structure and finally leading to a loss of biological function, formation of aggregates, and/or degradation. Thus, abnormal folding is the most common pathogenic mechanism in CBS deficiency as observed previously for other diseases (reviewed in Refs. 13 and 14).
The folding process represents a natural ability of each polypeptide to reach its final destined conformation, which is adversely affected by the presence of missense mutations in CBS. In the living cell, misfolded proteins are refolded by molecular chaperones to their active conformations, form non-functional aggregates, or are degraded by proteases to prevent aggregation. In recent years, small molecules referred to as chemical or pharmacological chaperones have been employed to rescue proteins from such folding defects (reviewed in Ref. 15), utilizing diverse modes of action from a direct effect on protein conformations and folding pathways to a modulation of cellular protein quality control systems. Singh et al. (16) showed that osmolytes such as trimethylamine-N-oxide (TMAO), glycerol, or dimethyl sulfoxide (DMSO) increased enzyme activity of I278T and three other CBS mutants by facilitating the formation of active tetramers through stabilization of the folded protein. Their positive effect on mutant folding was not exclusive to their presence in the yeast growth medium, but they exert their effect also in a coupled in vitro transcription/translation system. Furthermore, recent work of Singh and Kruger (17) suggests that ethanol treatment of Saccharomyces cerevisiae WY35, expressing the I278T human CBS mutant, induces HSP70 and leads to increased CBS activity and steady-state levels of the mutant. Taken together, mutant CBS is amenable and responsive to treatment with chemical chaperones in vivo, and this may represent a new therapeutical approach, especially for pyridoxine non-responsive CBSDH patients.
In this study, we describe the expression of eight CBS mutants in Escherichia coli in the presence of various chemical chaperones. The selected mutations span the entire CBS primary sequence. They include P49L and R125Q as the N-terminal heme pocket mutations, P78R, A114V, and E176K as the dimer interface mutations and P422L, I435T, and S466L as representatives of the C-terminal regulatory domain mutations. The most effective chemical chaperone and its concentration for a particular CBS mutant were chosen based on screening for catalytic activity and formation of tetramers in crude extracts of E. coli expressing the CBS mutant. All CBS mutants were subsequently expressed in E. coli grown in the presence of the selected chemical chaperone, purified to near homogeneity, and characterized in terms of CBS specific activity, formation of tetramers, heme saturation, PLP saturation, AdoMet response, heat activation, and thermal stability.
EXPERIMENTAL PROCEDURES
Chemical Chaperones
Three compounds were tested as chemical chaperones: ethanol, DMSO (Sigma D8418), and TMAO (Sigma T0514). TMAO was dissolved in double distilled H2O to a final concentration of 1 m and filter-sterilized. The following concentrations were used: 1 and 3% (v/v) of ethanol; 1, 3, and 6% (v/v) of DMSO; 1, 10, and 100 mm TMAO.
Mutant CBS Construct Preparation
The mutant CBS cDNA generated from patient samples was initially subcloned into the pKK388 vector and subsequently recloned into the pGEX-6P1 expression vector (18). The desired CBS mutants, i.e. P49L (c.146C > T), P78R (c.233C > G), A114V (c.341C > T), R125Q (c.374G > A), E176K (c.526G > A), P422L (c.1265C > T), I435T (c.1304T > C), and S466L (c.1397C > T), were verified by DNA sequencing. The verified constructs were transformed into E. coli Rosetta2 (DE3) cells (Novagen) for CBS expression studies.
Chemical Chaperone Screening
E. coli cells with the desired construct were grown (30 °C, 275 rpm) in 30 ml of LB medium supplemented with 0.001% thiamine-HCl, 0.0025% pyridoxine-HCl, 0.3 mm δ-aminolevulinic acid, 100 μg/ml ampicillin, and one of the chemical chaperones at a selected final concentration. When the bacterial culture reached A600 of ∼0.8–1, CBS expression was induced with 0.5 mm isopropyl-β-d-1-thiogalactopyranoside and continued overnight. The next day, the cells were harvested by centrifugation (4 °C, 7500 × g, 8 min), and the crude extracts were prepared according to the initial steps of our CBS purification procedure described elsewhere (18). Protein concentrations were determined by the Lowry procedure using bovine serum albumin as a standard (19). The total amount of CBS protein or the amount of tetramers was inspected by SDS-PAGE or native gel Western blots, respectively (see below).
Mutant CBS Protein Expression and Purification
After the initial chemical chaperone screening, the mutant CBS enzymes were expressed on a large scale (6 liters of the medium) in the presence of the most effective concentration of the selected chemical chaperone and purified as described previously (18).
CBS Activity Assay
CBS catalytic activity in the classical reaction was determined by a previously described radioisotope assay using l-[14C]serine as the labeled substrate (20). A crude extract (12 μl) or purified enzyme (420 ng) was assayed in a 100-μl reaction for 30 min or 1 h at 37 °C. The reaction was performed in the presence or absence of AdoMet. The saturation of the purified enzyme with PLP was tested by running the activity assay in the presence or absence of 0.5 mm PLP. One unit of CBS specific activity is defined as the amount of CBS that catalyzes the formation of 1 μmol of cystathionine in 1 h at 37 °C under standard assay conditions.
Protein Gel Electrophoresis and Western Blot
Denatured proteins were separated by SDS-PAGE (21) using a 9% separating gel with a 4% stacking gel. Native samples were separated in 4–15% polyacrylamide gradient precast gels (ReadyGels, Bio-Rad). For visualization, the denatured gels were stained with Simple Blue (Invitrogen).
Western blot analysis of crude cell lysates under denaturing or native conditions was performed as described previously (22). After electrophoresis, the separated proteins were transferred onto a polyvinylidene difluoride membrane (Millipore) and probed with monoclonal anti-CBS antibody (Abnova), monoclonal anti-DnaK antibody, polyclonal anti-DnaJ antibody, and monoclonal anti-GroEL antibody (Assay Designs/Stressgen), respectively. The Precision Plus dual color standards (Bio-Rad) were used as molecular weight marker for the SDS-PAGE gels. The quantitative analysis was performed using Quantity One software (Bio-Rad).
Enzyme Activation by Heating
Purified CBS was diluted to a final concentration of 0.1 mg/ml in Tris-buffered saline, pH 8.6, 100 μm PLP. The enzyme solutions (50 μl) were heated in 200-μl thin-walled PCR tubes in an Eppendorf Mastercycler gradient PCR thermal cycler. The temperature was raised from 37 to 53 °C in 0.5 °C-increments with a 1-min incubation at each temperature. After an additional 1-min incubation at 53 °C, samples were chilled on ice, and CBS activity was determined as described above.
Spectroscopic Characterization
UV-visible spectra were measured on an Agilent diode array model 8453 UV-visible spectrophotometer in 1× Tris-buffered saline, pH 8.6, at 25 °C.
Thermostability Determinations
Thermal stability of WT and mutant CBS purified proteins was compared by using the onset and melting temperatures of the proteins. The optical density at 320 nm, mainly caused by the light scattering of the thermally unfolded and aggregated protein, was monitored between 10 and 90 °C with 2.5 °C-increments and a 3-min equilibrium time at each temperature. Measurements were made on an Agilent diode array model 8453 UV-visible spectrophotometer equipped with a Peltier temperature controller. Two ml of protein samples (0.2 mg/ml) were prepared by diluting the stock protein with the appropriate amount of buffer (20 mm HEPES, pH 7.4, 1 mm tris(2-carboxyethyl)phosphine, 0.01% Tween 20) and then placed in a quartz cuvette with a 1-cm path length. A micro stir bar (100 rpm) was put into the cuvette to reduce the thermal gradient of the sample. The onset and melting temperatures were determined by the first derivative of the plots of A320 versus temperature. The second derivative UV spectra at each temperature were also used to compare the changes of tertiary structure of proteins as a function of temperature.
RESULTS
Screening of Chemical Chaperones in E. coli Yields Optimal Conditions for Mutant CBS Expression
The impact of the three chemical chaperones at various effective concentrations varied with CBS mutations; however, in general, 1 and 3% (v/v) ethanol and 3% (v/v) DMSO showed the best results (Table 1). The presence of a chemical chaperone in the medium during expression in nearly all cases increased the mutant CBS activity in E. coli crude extracts when compared with its basal activity. The least effective chemical chaperone in our hands was TMAO, which in some cases (WT, I435T, and S466L) led to a decrease of CBS activity when compared with an enzyme expressed in the absence of a chemical chaperone.
TABLE 1.
CBS activities of WT and selected mutants in E. coli crude extracts after expression in the absence or presence of three chemical chaperones
CBS activities are expressed in percent relative to CBS activity of the WT expressed in the absence of a chaperone representing 100%. The average activity of WT CBS expressed from pGEX-6P1-hCBS construct in E. coli Rosetta2 (DE3) crude extract was 402 ± 86 milliunits/mg of protein. Values represent average values with S.E. from at least three measurements. The bold values denote the conditions, which were subsequently used for the large-scale expression and protein purification of each mutant.
| CBS protein | CBS activity (% of WT) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No chaperone | 1% ethanol | 3% ethanol | 1 mm TMAO | 10 mm TMAO | 100 mm TMAO | 1% DMSO | 3% DMSO | 6% DMSO | |
| WT | 100 | 130 ± 15 | 171 ± 31 | 89 ± 14 | 94 ± 6 | 82 ± 7 | 106 ± 19 | 160 ± 14 | 166 ± 25 |
| P49L | 91 ± 5 | 161 ± 22 | 241 ± 25 | 114 ± 17 | 123 ± 10 | 177 ± 24 | 115 ± 17 | 154 ± 11 | 254 ± 21 |
| P78R | 51 ± 8 | 89 ± 12 | 60 ± 4 | 56 ± 8 | 60 ± 5 | 55 ± 11 | 60 ± 12 | 77 ± 5 | 58 ± 4 |
| A114V | 43 ± 4 | 115 ± 13 | 137 ± 12 | 68 ± 6 | 77 ± 6 | 77 ± 4 | 72 ± 6 | 110 ± 15 | 62 ± 8 |
| R125Q | 11 ± 5 | 59 ± 7 | 29 ± 3 | 103 ± 4 | 97 ± 6 | 87 ± 12 | 103 ± 10 | 79 ± 7 | 49 ± 5 |
| E176K | 21 ± 2 | 57 ± 8 | 47 ± 8 | 32 ± 6 | 34 ± 4 | 42 ± 3 | 34 ± 5 | 43 ± 11 | 27 ± 4 |
| P422L | 125 ± 13 | 279 ± 45 | 433 ± 65 | 135 ± 11 | 154 ± 17 | 221 ± 35 | 295 ± 18 | 299 ± 36 | 348 ± 52 |
| I435T | 426 ± 51 | 855 ± 71 | 1001 ± 21 | 287 ± 42 | 139 ± 20 | 531 ± 44 | 160 ± 13 | 557 ± 27 | 505 ± 31 |
| S466L | 828 ± 63 | 933 ± 34 | 1063 ± 32 | 760 ± 37 | 755 ± 64 | 990 ± 51 | 816 ± 76 | 976 ± 65 | 1119 ± 79 |
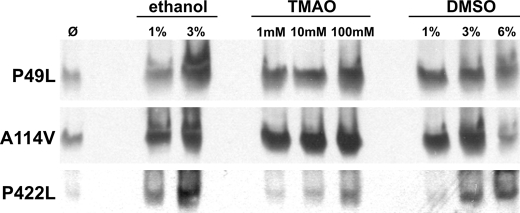
The presence of a chemical chaperone in the medium during expression did not significantly impact the total amount of CBS subunits based on denaturing gel Western blot analysis of the crude extracts (data not shown). There were, however, significant changes in the levels of active CBS tetramers as shown on native gel Western blots for three representative CBS mutants (Fig. 2). The increased abundance of tetramers correlates well with the measured increase in CBS activity. These data suggest that the presence of chemical chaperones resulted in an improved folding and thus formation of active tetramers.
FIGURE 2.
Native gel Western blots of three representative CBS mutants expressed in the absence and presence of chemical chaperones. The crude extracts (soluble fraction, 30 μg) were separated in a 4–15% Tris-HCl native gel, transferred to a polyvinylidene difluoride membrane, and probed with monoclonal anti-CBS antibody (Abnova). For brevity, other studied CBS mutants, which yielded similar results, are not shown.
Purified CBS Mutants Retain an Active Conformation
To determine whether the effect of a given chemical chaperone on mutant CBS is retained, we purified eight mutant CBS enzymes to near homogeneity by our two-step purification procedure. The chemical chaperones, selected on the basis of screening in E. coli crude extracts, were present only in the cultivation medium during cell growth and protein expression. The purification and characterization of each protein were carried out in the complete absence of any chemical chaperones.
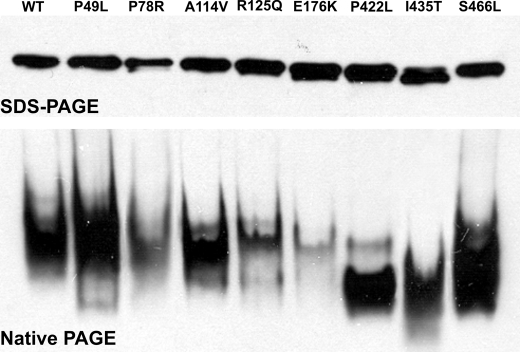
The purified mutant enzymes migrated in a native gel as tetramers similar to WT, suggesting that they retained their active conformations induced by the chaperones (Fig. 3, bottom). Even when the amounts of the mutants applied to the native gel were nearly equal, as shown on the SDS gel Western blot (Fig. 3, top), the intensity of the mutant tetramers varied. These differences may be attributed to the different sensitivity of the employed monoclonal anti-CBS antibody to the native conformations of each particular mutant. No higher oligomers or aggregates were detected on the native gel Western blot. The tetramers of the C-terminal mutants (i.e. P422L, I435T, and S466L) migrated faster than WT or other mutant CBS enzymes as observed previously (8).
FIGURE 3.
SDS-PAGE (top) and native-PAGE (bottom) Western blots of WT and purified CBS mutants. The purified CBS enzymes (100 ng) were separated in a 10% SDS-PAGE gel under denaturing conditions (top) and in a 4–15% PAGE gel under native conditions (bottom). Following electrophoresis, the gels were transferred to polyvinylidene difluoride membranes and probed with a monoclonal anti-CBS antibody (Abnova).
Purified CBS Mutants Are at Least as Active as WT
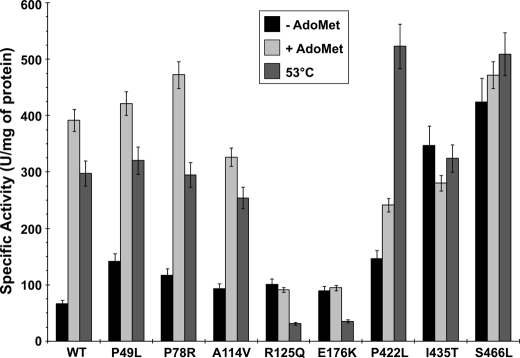
The specific activity measurements of the purified mutants showed interesting results when compared with WT CBS. All the purified mutants had specific activities equal or higher than WT CBS in the presence of exogenous PLP and in the absence of AdoMet (Table 2). On the other hand, several differences were detected in their PLP saturation as well as AdoMet response. The P49L, P78R, and A114V mutants were indistinguishable from WT CBS in terms of specific activity, PLP saturation, and AdoMet response. The significant impact of exogenous PLP on CBS specific activity was detected for the R125Q and E176K mutants, which also did not respond to AdoMet activation. The C-terminal mutants P422L, I435T, and S466L also showed slight response to additional PLP. No response to AdoMet activation was observed for the P422L, I435T, and S466L mutants, which also displayed significantly higher basal activities than WT CBS as we described before (7, 8).
TABLE 2.
Basal activity, PLP saturation, and AdoMet response of the purified CBS enzymes
The purified enzymes were assayed for CBS catalytic activity in the classical reaction. The saturation of the purified enzyme with PLP cofactor was tested by running the activity assay in the presence or absence of 500 μm PLP. The AdoMet response was determined by assaying the enzyme in the presence or absence of 300 μm AdoMet. The + and − signs denote the presence and absence, respectively, of PLP or AdoMet in the reactions. Values represent average values with S.E. from at least three independent assays.
| CBS specific activity | |||
|---|---|---|---|
| units/mg of protein | |||
| CBS protein | |||
| WT | 128 ± 15 | 148 ± 21 | 530 ± 45 |
| P49L | 117 ± 14 | 122 ± 12 | 388 ± 39 |
| P78R | 97 ± 12 | 110 ± 15 | 501 ± 27 |
| A114V | 92 ± 13 | 100 ± 11 | 401 ± 31 |
| R125Q | 57 ± 10 | 139 ± 18 | 106 ± 9 |
| E176K | 66 ± 11 | 138 ± 14 | 111 ± 8 |
| P422L | 196 ± 20 | 226 ± 28 | 238 ± 41 |
| I435T | 322 ± 37 | 564 ± 44 | 582 ± 60 |
| S466L | 497 ± 31 | 626 ± 48 | 639 ± 43 |
| 500 μm PLP | − | + | + |
| 300 μm AdoMet | − | − | + |
Heating or AdoMet Addition Results in CBS Activation
The increase in activity observed in the presence of AdoMet can be mimicked by heating the enzyme to a temperature just below the melting point (55 °C) of WT CBS (7, 23). The response of WT and CBS mutants to AdoMet or to heating is shown in Fig. 4. The WT CBS responded to AdoMet stimulation or heating to 53 °C with a 6- and a 4.5-fold increase in activity, respectively. The P49L, P78R, and A114V mutants behaved similarly to WT. Both R125Q and E176K mutants did not respond to AdoMet stimulation and decreased in activity after heating at 53 °C to 31% of mutant basal activity for R125Q or 40% of mutant basal activity for E176K. The I435T and S466L mutants did not respond significantly to AdoMet stimulation or to the heating. On the other hand, another C-terminal mutant P422L slightly responded to AdoMet stimulation (1.7-fold), but a significant increase in activity was observed after heating it to 53 °C when compared with basal activity (3.6-fold).
FIGURE 4.
Activity changes of WT and CBS mutants in response to AdoMet stimulation or heating. The black bars represent basal activity of WT or mutant CBS without the addition of AdoMet. The light gray bars represent CBS activity assayed in the presence of AdoMet, the allosteric activator of CBS. The dark gray bars show the response of CBS activity to gradual heating of the enzyme solution to 53 °C for 1 min prior to the enzyme assay (see “Experimental Procedures” for details). Error bars denote the S.E. values and are derived from a minimum of three independent measurements.
Purified Mutant CBS Enzymes Are Fully Saturated with Heme
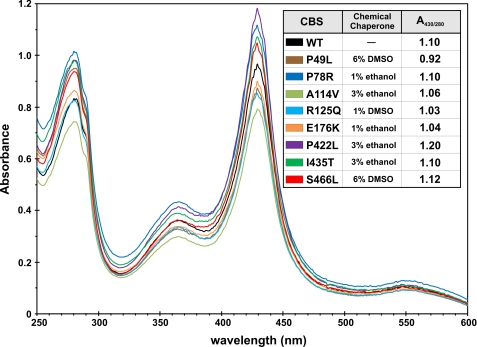
All purified CBS enzymes showed normal heme saturation (Fig. 5). The lowest A430/280 ratio was detected for P49L (0.92). The highest heme content was detected for the P422L mutant with a value of 1.2. All other mutants were comparable with WT CBS in heme saturation with values around 1.05.
FIGURE 5.
UV-visible spectra of purified CBS proteins. The purified CBS enzymes were diluted in 1× Tris-buffered saline, pH 8.6. Spectra were recorded on an Agilent 8453 UV-visible spectrophotometer. The chemical chaperones listed in the inset table were only present in the medium during protein expression.
CBS Mutants Are at Least as Thermostable as WT
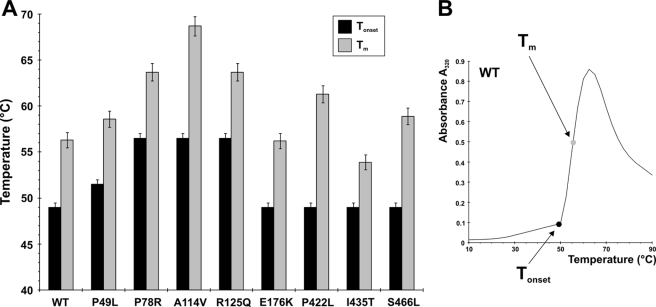
We subjected the purified CBS mutants to thermal denaturation to investigate their thermal stability and changes in the tertiary structure as a function of temperature. The second derivative UV spectra for the mutant proteins at each temperature did not show any significant differences when compared with that for the WT protein, suggesting that the CBS mutants expressed in the presence of chemical chaperone are folded into a similar conformation as WT CBS (data not shown). Furthermore, the same analysis revealed that the purified CBS mutants are equally or more thermostable than WT (Fig. 6). The lowest temperature at which aggregates started to form was 49 °C for WT and for four of the mutants: E176K and all the C-terminal mutants, P422L, I435T, and S466L. For the rest of the CBS mutants, the initial temperature of protein aggregation was 51 °C for P49L or 56 °C for P78R, A114V, and R125Q.
FIGURE 6.
Thermostability of purified WT and CBS mutants. The individual enzymes were gradually heated from 10 to 90 °C as described under “Experimental Procedures.” A, Tonset designates the temperature at which aggregates start to form, and Tm represents melting temperatures. B, an example of the thermostability analysis of WT CBS. Error bars denote the S.E. values and are derived from a minimum of three independent measurements.
Chemical Chaperones May Act Indirectly via Activation of Molecular Chaperones
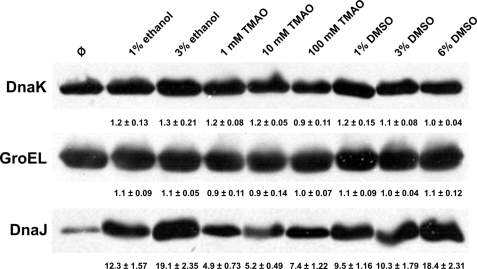
To investigate whether the mode of action of the chemical chaperones is direct by assisting the mutant protein to fold or rather indirect by inducing molecular chaperones, we probed the crude extracts with antibodies against three different molecular chaperones, DnaK, GroEL, and DnaJ (Fig. 7). No significant changes in protein levels of DnaK or GroEL were detected in the presence of chemical chaperones. In contrast, a significant increase of the DnaJ levels was observed for all the tested chemical chaperones. The increase in the amount of DnaJ in the presence of a chemical chaperone when compared with its absence varied from 4.9-fold in the presence of 1 mm TMAO up to 19.1- or 18.4-fold in the presence of 3% ethanol or 6% DMSO, respectively. Our data suggest that chemical chaperones may act indirectly by inducing a specific class of molecular chaperones (particularly DnaJ), which in turn facilitate improved folding of mutant CBS.
FIGURE 7.
Protein levels of three molecular chaperones, DnaK, GroEL, and DnaJ, in response to the absence or presence of chemical chaperones. The crude extracts (50 μg) of mutant CBS enzymes expressed in the presence or in the absence of the chemical chaperones were separated in 9% Tris-HCl SDS-PAGE gel, transferred to polyvinylidene difluoride membrane, and probed with monoclonal anti-DnaK antibody, monoclonal anti-GroEL antibody, or polyclonal anti-DnaJ antibody. Values under the respective Western blot bands represent the -fold of increase of the molecular chaperone protein levels in response to the presence of a given chemical chaperone with S.E. calculated from data for each CBS mutant.
DISCUSSION
To function properly, a newly synthesized polypeptide for an enzyme must fold into its specific native conformation. All the necessary information for correct folding is encoded in the primary amino acid sequence. However, this process is accelerated and assisted in vivo by molecular chaperones (24, 25). Molecular chaperones also prevent the unfolded or partially folded protein from assembling into non-functional structures by initiation of protein refolding or targeting them for degradation (26). Despite tight quality control, misfolding and subsequent protein aggregation and/or degradation occur as a response to various stresses such as aging, temperature fluctuation, or genetic defect and represent the major causes of a diverse group of diseases called protein misfolding or protein conformational diseases (25, 27). In many cases (e.g. Gaucher disease and other lysosomal storage disorders), mutant enzyme misfolding results in degradation of the aberrant protein, the loss of enzymatic function, and the subsequent accumulation of the corresponding enzyme substrate (e.g. glucocerebroside in Gaucher disease). On the other hand, for Alzheimer disease, Parkinson disease, type II diabetes mellitus, and amyloid diseases, impaired protein folding results in the toxic intra- or extracellular accumulation of misfolded, aggregated proteins in the form of amyloid plaques.
Janosik et al. (12) first showed that CBS deficiency may be considered a protein misfolding disease. They tested CBS from a number of patient fibroblast lines by electrophoresis in native PAGE followed by Western blot analysis. The CBS mutants did not form active tetramers and accumulated as inactive aggregates. Moreover, these misfolded CBS aggregates were devoid of heme, which resulted in the hypothesis that heme may be necessary for correct folding and active tetramer formation of CBS. Our recent work suggested that despite the heme being crucial for proper CBS folding and assembly, it is not essential for expression of active CBS and can be substituted with other metalloporphyrins or even completely replaced by including a chemical chaperone such as TMAO in the growth medium of a heme-deficient S. cerevisiae (28). The usefulness of chemical chaperones in restoration of mutant CBS function was first demonstrated by Singh et al. (16). They showed that chemical chaperones, if present during the initial folding process, can facilitate proper folding of several mutant CBS proteins in vivo as well as in vitro. Similar results were observed for a group of 27 CBS mutants assembled in the presence of glycerol, δ-aminolevulinic acid, or betaine.5 The recent work by Singh and Kruger (17) suggests that a chemical chaperone such as 4% ethanol is able to dramatically restore the I278T CBS mutant stability and enzymatic function in S. cerevisiae by manipulation of the cellular levels of molecular chaperones. Ethanol treatment induced the expression of HSP70 and increased activity and steady-state levels of the I278T mutant. On the other hand, overexpression of HSP26 resulted in rapid degradation of I278T protein. Therefore, the HSP70:HSP26 ratio in S. cerevisiae seems to be crucial in the determination of whether misfolded CBS proteins will undergo refolding or will be targeted for degradation via the ubiquitin/proteasome-dependent mechanism. Taken together, these data suggest that treatment with chemical chaperones or manipulation of the levels of molecular chaperones increases or completely restores the function of some mutant CBS enzymes in E. coli, S. cerevisiae, and a mouse transgenic for I278T (29).
In this study, we present the purification of eight CBS mutants from E. coli expressed in the presence of chemical chaperones. Initial screening of various chemical chaperones at several concentrations showed that many CBS mutants responded well to the chaperone treatment by increased formation of tetramers and a corresponding increase in enzymatic activity. These promising results from E. coli crude extracts encouraged us to attempt to purify the mutants to homogeneity. Several previous attempts to purify CBS mutants resulted in low yields or failed completely due to the unstable mutant conformations, their extreme tendency to aggregation, and heme loss (12, 30, 31). The presence of chemical chaperones during CBS expression helped to overcome these problems, resulting in homogeneous mutant CBS enzymes as active as WT. All the purified CBS mutants formed predominantly tetramers with virtually no aggregation or formation of lower or higher order multimers detected. The C-terminal CBS mutants (i.e. P422L, I435T, and S466L) showed a relatively subtle but distinct shift in their migration in native PAGE when compared with WT or other CBS mutants. This faster migration of C-terminal mutants agreed with previous observations and is most likely indicative of conformational changes associated with mutations in the C-terminal domain (7, 8).
All CBS mutants were fully saturated with heme as also noted for WT CBS. Our previous results indicated that heme does not function in redox sensing, ligand binding, or catalysis but rather supported the structural role of heme in proper folding and/or subunit assembly (22, 28, 32–35). With respect to this hypothesis and the presented data, heme saturation could serve as a marker for properly folded CBS and could be indicative of the activity of purified CBS.
Although all the mutants were at least as active as the WT enzymes, they varied in their response to the presence of exogenous PLP as well as to AdoMet stimulation. The properties of the purified C-terminal mutants I435T and S466L were in good agreement with the previous results (7, 23, 36). Their basal activity when compared with WT was increased, and they did not respond to AdoMet stimulation. This behavior has been explained previously by a mutation-induced conformational change, which results in increased basal activity of C-terminal mutants when compared with WT and at the same time a lack of response to AdoMet stimulation. Our previous data on the S466L CBS enzyme showed a systematic decrease in activity with increasing temperature (7). As we detected a small increase in activity for the heated enzyme when compared with its basal activity rather than a decrease, we suppose that the chemical chaperones stabilized the mutant conformation and thus prevented its rapid denaturation during gradual heating.
Two mutants in the catalytic core, R125Q and E176K, were distinctly different from the WT. Both purified mutants were as active as WT CBS, but they did not respond to AdoMet stimulation and were only partially saturated with PLP. This decreased PLP content may be due to a lower affinity of the mutants for the coenzyme. Considering the lack of AdoMet response of these two mutants, we can speculate that a mutation in the catalytic region such as R125Q or E176K causes an undesirable interaction with the C-terminal region, resulting in an inability of AdoMet to further stimulate CBS activity.
Previous observations that specific C-terminal mutations, AdoMet, limited proteolysis, or heating all induce a very similar level of WT CBS activation suggested that these different forms of activation share a common mechanism (7, 23, 37, 38). We showed that the P49L, P78R, and A114V mutants behave similarly as WT in terms of their basal activity, AdoMet response, and thermal activation. In contrast, heating of R125Q and E176K mutants did not result in their activation and thus further supports our hypothesis about their specific conformation. In the absence of AdoMet, they are as active as WT but unable to change their conformation in response to AdoMet stimulation or heating to 53 °C. The observed decrease in specific activity of R125Q and E176K after heating to 53 °C may be accounted for by an irreversible loss of PLP given their decreased ability to bind the cofactor (Table 2).
Finally, very interesting results were observed for another C-terminal CBS mutant, P422L. We showed that the P422L expressed slightly higher basal activity than WT and partially responded to AdoMet stimulation, suggesting that it is not a typical representative of the C-terminal mutants. The heating study separated P422L from the other C-terminal CBS mutants even more when the enzyme increased its activity 3.6-fold when compared with untreated enzyme. This result may indicate that activation of CBS by heating or AdoMet binding is likely acting through different mechanisms although they share a similar consequence, i.e. conformational change. Furthermore, improved folding of CBS mutants induced by the presence of a chemical chaperone surprisingly yielded enzymes with equal or higher thermostability than WT CBS. Similar improvement in thermal stability was found for human phenylalanine hydroxylase mutants expressed in the presence of 1% glycerol or 5 mm TMAO (39).
Whether chemical chaperones induce proper folding of mutant enzymes directly by stabilizing their conformation in vivo or indirectly by increasing levels of molecular chaperones is still unclear. Singh et al. (16) suggested that chemical chaperones such as osmolyte glycerol could act directly because its presence in both the expression medium and the in vitro transcription/translation system resulted in an increase of the I278T mutant activity. However, no stimulation was observed when chaperones were added directly to the purified I278T mutant, indicating that the species amenable for correction via the chemical chaperone are only the newly synthesized polypeptide chains (16, 39). Furthermore, Singh and Kruger (17) have recently reported that another chemical chaperone, ethanol, increased activity and steady-state levels of the I278T mutant in S. cerevisiae by induction of a molecular chaperone, particularly HSP70, which promotes proper folding of I278T polypeptides. In our study, the steady-state levels of DnaK, a microbial analog of HSP70, were found to be comparable in the presence of a chemical chaperone or in its absence. However, steady-state levels of another molecular chaperone, DnaJ, a microbial analog of HSP40 and co-chaperone of DnaK, were found significantly elevated in the presence of all the tested chemical chaperones. The expression levels of another molecular chaperone, GroEL, a microbial analog of HSP60, were not significantly affected in the presence of a chemical chaperone, which agrees with the results of chemical chaperone screening of human phenylalanine hydroxylase mutants (39). Our data indicate that proper folding of CBS mutants expressed in the presence of chemical chaperones could be facilitated by induction of molecular chaperones, particularly DnaJ. DnaJ is necessary for DnaK function; DnaJ stimulates the ATP turnover rate of DnaK and facilitates the interaction of DnaK with polypeptide substrates (40). Therefore, we suggest that increased levels of DnaJ induced by chemical chaperones resulted in higher activity and affinity of DnaK for the polypeptide substrate, which subsequently facilitated proper folding of CBS mutants.
Our findings suggest that the expression of several CBS mutants in the presence of various chemical chaperones resulted in their proper folding and full restoration of catalytic activity. These acquired characteristics seem to have become permanent properties of the purified mutant CBS proteins. Moreover, the chemical chaperone-induced CBS mutant folding resulted in a conformation thermodynamically as or more stable than WT CBS. Chemical chaperones have been shown to be preferentially excluded from the surface of proteins, an effect that causes the compact native conformation to be favored over unfolded states (15). However, this is not the only mode of action of such compounds in cells. These small chemical substances also induced the expression of molecular chaperones, mainly DnaJ, which likely resulted in improved interaction of DnaK with the newly synthesized polypeptide, thus generating properly folded proteins. This study provides a proof of principle, i.e. that these mutant enzymes appear to be misfolded in vivo and that they can be properly folded in the presence of chemical chaperones. Obviously, compounds such as ethanol or DMSO will not be used in patients. A high-throughput screen of available approved substances may yield compounds capable of similar restoration of CBS folding and activity as the chemicals used in this study. Such compounds would be a very attractive alternative for treating patients with CBS deficiency caused by a missense mutation. Even a small rescue of enzyme activity could have substantial impact on the clinical picture or the course of this disease.
Acknowledgments
We thank Kirk Hansen and Lauren Kiemele of the Proteomics Core of the Colorado IDDRC for assistance with proteomic analysis of CBS double bands in SDS-PAGE, Sarah Venezia (Department of Pediatrics, University of Colorado at Denver) for critical reading of the manuscript, and Dr. Viktor Kozich and Eva Rehulkova (Institute of Inherited Metabolic Disorders, Charles University of Prague) for sharing with us some of the CBS mutant constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant HL065217-06A2 (to J. P. K.). This work was also supported by American Heart Association Grant-in-Aid 09GRNT2110159, a grant from the Jerome Lejeune Foundation (to J. P. K.), and a generous gift from Margie McGlynn.
J. P. Kraus, unpublished data.
Kopecka, J., Krijt, J., Rakova, K., and Kozich, V. (2010) J. Inherit. Metab. Dis., in press.
- CBS
- cystathionine β-synthase
- CBSDH
- CBS-deficient homocystinuria
- PLP
- pyridoxal-5′-phosphate
- AdoMet
- S-adenosyl-l-methionine
- WT
- wild type
- TMAO
- trimethylamine-N-oxide
- DMSO
- dimethylsulfoxide.
REFERENCES
- 1.Mudd S. H., Levy H. L., Kraus J. P. (2001) in The Metabolic and Molecular Bases of Inherited Disease (Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K., Vogelstein B. eds) 8th Ed., pp. 2007–2056, McGraw-Hill, New York [Google Scholar]
- 2.Miles E. W., Kraus J. P. (2004) J. Biol. Chem. 279, 29871–29874 [DOI] [PubMed] [Google Scholar]
- 3.Banerjee R., Zou C. G. (2005) Arch. Biochem. Biophys. 433, 144–156 [DOI] [PubMed] [Google Scholar]
- 4.Singh S., Madzelan P., Banerjee R. (2007) Nat. Prod. Rep. 24, 631–639 [DOI] [PubMed] [Google Scholar]
- 5.Meier M., Janosik M., Kery V., Kraus J. P., Burkhard P. (2001) EMBO J. 20, 3910–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taoka S., Lepore B. W., Kabil O., Ojha S., Ringe D., Banerjee R. (2002) Biochemistry 41, 10454–10461 [DOI] [PubMed] [Google Scholar]
- 7.Janosík M., Kery V., Gaustadnes M., Maclean K. N., Kraus J. P. (2001) Biochemistry 40, 10625–10633 [DOI] [PubMed] [Google Scholar]
- 8.Maclean K. N., Gaustadnes M., Oliveriusová J., Janosík M., Kraus E., Kozich V., Kery V., Skovby F., Rüdiger N., Ingerslev J., Stabler S. P., Allen R. H., Kraus J. P. (2002) Hum. Mutat. 19, 641–655 [DOI] [PubMed] [Google Scholar]
- 9.Shan X., Dunbrack R. L., Jr., Christopher S. A., Kruger W. D. (2001) Hum. Mol. Genet. 10, 635–643 [DOI] [PubMed] [Google Scholar]
- 10.Banerjee R., Evande R., Kabil O., Ojha S., Taoka S. (2003) Biochim. Biophys. Acta 1647, 30–35 [DOI] [PubMed] [Google Scholar]
- 11.Shan X., Kruger W. D. (1998) Nat. Genet. 19, 91–93 [DOI] [PubMed] [Google Scholar]
- 12.Janosík M., Oliveriusová J., Janosíková B., Sokolová J., Kraus E., Kraus J. P., Kozich V. (2001) Am. J. Hum. Genet. 68, 1506–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staretz-Chacham O., Lang T. C., LaMarca M. E., Krasnewich D., Sidransky E. (2009) Pediatrics 123, 1191–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sideras K., Gertz M. A. (2009) Adv. Clin. Chem. 47, 1–44 [PubMed] [Google Scholar]
- 15.Leandro P., Gomes C. M. (2008) Mini. Rev. Med. Chem. 8, 901–911 [DOI] [PubMed] [Google Scholar]
- 16.Singh L. R., Chen X., Kozich V., Kruger W. D. (2007) Mol. Genet. Metab. 91, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh L. R., Kruger W. D. (2009) J. Biol. Chem. 284, 4238–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank N., Kent J. O., Meier M., Kraus J. P. (2008) Arch. Biochem. Biophys. 470, 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 20.Kraus J. P. (1987) Methods Enzymol. 143, 388–394 [DOI] [PubMed] [Google Scholar]
- 21.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 22.Oliveriusová J., Kery V., Maclean K. N., Kraus J. P. (2002) J. Biol. Chem. 277, 48386–48394 [DOI] [PubMed] [Google Scholar]
- 23.Frank N., Kery V., Maclean K. N., Kraus J. P. (2006) Biochemistry 45, 11021–11029 [DOI] [PubMed] [Google Scholar]
- 24.Walter S., Buchner J. (2002) Angew. Chem. Int. Ed. Engl. 41, 1098–1113 [DOI] [PubMed] [Google Scholar]
- 25.Hartl F. U., Hayer-Hartl M. (2009) Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 26.Liberek K., Lewandowska A., Zietkiewicz S. (2008) EMBO J. 27, 328–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos C. H., Ferreira S. T. (2005) Protein Pept. Lett. 12, 213–222 [DOI] [PubMed] [Google Scholar]
- 28.Majtan T., Singh L. R., Wang L., Kruger W. D., Kraus J. P. (2008) J. Biol. Chem. 283, 34588–34595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh L. R., Gupta S., Honig N. H., Kraus J. P., Kruger W. D. (2010) PLoS Genet. 6, e1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kraus J. P., Rosenberg L. E. (1983) Arch. Biochem. Biophys. 222, 44–52 [DOI] [PubMed] [Google Scholar]
- 31.Singh S., Madzelan P., Stasser J., Weeks C. L., Becker D., Spiro T. G., Penner-Hahn J., Banerjee R. (2009) J. Inorg. Biochem. 103, 689–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bruno S., Schiaretti F., Burkhard P., Kraus J. P., Janosik M., Mozzarelli A. (2001) J. Biol. Chem. 276, 16–19 [DOI] [PubMed] [Google Scholar]
- 33.Pazicni S., Lukat-Rodgers G. S., Oliveriusová J., Rees K. A., Parks R. B., Clark R. W., Rodgers K. R., Kraus J. P., Burstyn J. N. (2004) Biochemistry 43, 14684–14695 [DOI] [PubMed] [Google Scholar]
- 34.Pazicni S., Cherney M. M., Lukat-Rodgers G. S., Oliveriusová J., Rodgers K. R., Kraus J. P., Burstyn J. N. (2005) Biochemistry 44, 16785–16795 [DOI] [PubMed] [Google Scholar]
- 35.Cherney M. M., Pazicni S., Frank N., Marvin K. A., Kraus J. P., Burstyn J. N. (2007) Biochemistry 46, 13199–13210 [DOI] [PubMed] [Google Scholar]
- 36.Evande R., Blom H., Boers G. H., Banerjee R. (2002) Biochemistry 41, 11832–11837 [DOI] [PubMed] [Google Scholar]
- 37.Kozich V., Kraus J. P. (1992) Hum. Mutat. 1, 113–123 [DOI] [PubMed] [Google Scholar]
- 38.Kery V., Poneleit L., Kraus J. P. (1998) Arch. Biochem. Biophys. 355, 222–232 [DOI] [PubMed] [Google Scholar]
- 39.Nascimento C., Leandro J., Tavares de Almeida I., Leandro P. (2008) Protein J. 27, 392–400 [DOI] [PubMed] [Google Scholar]
- 40.Bukau B., Horwich A. L. (1998) Cell 92, 351–366 [DOI] [PubMed] [Google Scholar]