Abstract
The immunoglobulin (Ig) superfamily is one of the largest families in the vertebrate genome, found most frequently in cell surface molecules. Intercellular adhesion molecule-1 (ICAM-1) contains five extracellular Ig superfamily domains (D1-D5) of which the first domain, D1, is the binding site for the integrin lymphocyte function-associated antigen-1 (LFA-1) and human rhinovirus. Despite the modular nature of many Ig superfamily domains with respect to domain folding and ligand recognition, D1 does not fold on its own due to the loss of its interaction with the second domain. The goal of this study was to engineer ICAM-1 D1 by introducing mutations that would stabilize the Ig superfamily domain fold while retaining its ability to bind to LFA-1 and rhinovirus. First, with a directed evolution approach, we isolated mutations in D1 that showed binding to conformation-specific antibodies and the ligand binding domain of LFA-1 called the inserted, or I, domain. Then, with a rational design approach we introduced mutations that contributed to the stability of ICAM-1 D1 in solution. The mutations that restored native folding of D1 in isolation were those that would convert hydrogen bond networks in buried regions into hydrophobic contacts. Notably, for most mutations, identical or similar types of substitutions were found in ICAM-1 molecules of different species and other ICAM family members. The systematic approach demonstrated in this study to engineer a single Ig superfamily fold in ICAM-1 can be broadly applicable to the engineering of modular Ig superfamily domains in other cell surface molecules.
Keywords: Cell Adhesion, Integrin, Protein Domains, Protein Folding, Receptor Structure-Function, Virus Entry, ICAM-1, Immunoglobulin Superfamily, Intercellular Adhesion Molecule-1, Rhinovirus
Introduction
Intercellular adhesion molecule-1 (ICAM-1)4 is an inducible transmembrane receptor expressed on the surface of several cell types including leukocytes and endothelial cells and is up-regulated by inflammatory mediators such as interleukin-1, interferon-γ, tumor necrosis factor, and lipopolysaccharide (1). ICAM-1 is the counter receptor for the leukocyte integrins, lymphocyte function-associated antigen-1 (LFA-1, αLβ2), Mac-1 (αMβ2), and p150,95 (αxβ2), and promotes cellular interactions important in immunity and inflammation (2–7). ICAM-1 is also used as a sequestration receptor for Plasmodium falciparum-infected erythrocytes (8) and has been subverted as a receptor for human rhinovirus (9, 10) and Coxsackievirus A21 (11). Structurally, ICAM-1 consists of five extracellular domains (D1-D5), a transmembrane spanning region, and a short cytoplasmic domain. The second, third, and fourth domains of ICAM-1 contain a number of glycan chains that appear to contribute to ICAM-1 stability and function (12–14). All five extracellular domains of ICAM-1 belong to the immunoglobulin (Ig) superfamily, a family that is the largest in the vertebrate genome (15) and found in many cell surface receptors (16). Ig superfamily domains frequently exist in tandem arrays in surface receptors, and each domain in one molecule may pair with different ligands.
The Ig superfamily fold is characterized by a β-sandwich structure consisting of seven to nine β strands labeled A-G along the polypeptide chain. The two sides of the sandwich are formed by two β sheets, one β sheet containing ABED and the other containing the A′GFCC' β-strands. The two β-sheets surround a hydrophobic interior with a conserved disulfide bond between β-strands B and F. The N-terminal domain of ICAM-1 D1 has an additional disulfide bond near its N-terminal face. Among the five Ig superfamily domains in ICAM-1, the integrins LFA-1 and Mac-1 bind via their inserted or I domains to domains D1 and D3, respectively (17, 18). This domain-specific binding is not confined to the host immune molecules. Of the 102 HRV (human rhinovirus) serotypes known to date, ∼90% use ICAM-1 as a receptor. For all the ICAM-1 binding HRV serotypes that have been examined, the binding site in ICAM-1 is limited to D1. This was shown by mutational analyses of specific residues (10, 19) as well as by a cryoelectron microscopy reconstruction of a complex of HRV16 with the two N-terminal domains of ICAM-1 (20). There do not appear to be any residue contacts between domain D2 of ICAM-1 and the viral surface (21). Additionally, Coxsackievirus A21 has been shown to have contacts with D1 of ICAM-1 (22). The subversion of Ig superfamily host cell receptors by a wide variety of pathogens is well documented. One example is poliovirus, another member of the picornavirus family, which binds D1 of the poliovirus receptor (PVR or CD155) (23, 24). Another example is human immunodeficiency virus, whose gp120 protein binds to CD4 (another Ig superfamily receptor), with all of the contacts limited to the first Ig superfamily domain (25).
Many Ig superfamily domains in cell surface receptors are modular, i.e. domain folding and function is retained in isolation from neighboring domains. Although many Ig superfamily domains appear to be independently folded units, not all Ig superfamily domains in cell surface receptors retain native fold and function on their own. However, previous attempts to produce D1 alone from either a mammalian or a bacterial expression system have hitherto been unsuccessful (26, 27), indicating the dependence of D1 folding on D2 including a glycan chain on Asn-175 (28). To date, soluble ICAM-1 has been produced in three different forms: full-length ectodomain D1-D5, D1D2, and D3-D5. Of these, D1D2 and D3-D5 have been crystallized both by themselves and in complex with other binding proteins (12, 14, 28–30). In contrast to D1 of ICAM-1, the first domain of ICAM-3 alone is stable independently of D2 (31) and was crystallized with the I domain of LFA-1 (32). Here we present a systematic approach based on the combined use of directed evolution and rational design for the design of mutations that would restore native folding of Ig superfamily domains in isolation. This study provides the first example of modular expression of a single protein domain that does not fold on its own. In light of the nature of the interaction of single Ig superfamily domains with different ligands, the ability to functionally express single Ig superfamily domains will contribute to structural studies of the interaction of these domains with their binding partners as well as elucidating their role in physiology.
EXPERIMENTAL PROCEDURES
Cells, Media, and Antibodies
All cells were propagated in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (Atlanta Biologicals) containing 2 mm l-glutamine, 0.5% Pen-strep (Invitrogen), and 50 μg/ml gentamycin (Invitrogen). The monoclonal antibodies (mAb) used in this study were RR1/1 (Bender MedSystems), LB2 (Santa Cruz), My13 (Santa Cruz), R6.5 (ATCC), CBR-IC1/11 (33), and CA-7 (34).
Expression of ICAM-1 D1, D1D2, and D1-D5 in Yeast Surface Display System
Yeast strain EBY100 was used to implement the yeast display system. ICAM-1 of varying lengths, residues Gln-1 to Trp-84 (D1), residues Gln-1 to Leu-187 (D1D2), and residues Gln-1 to Arg-451 (D1-D5) were subcloned into the display plasmid named CAga2, which was modified from pCTCON (35) to express Aga2 at the C-terminal of the protein of interest (36). Yeast cells were transformed with the plasmids using Frozen-EZ Yeast Transformation kit (Zymo Research). For protein induction, yeast cells were grown at 30 °C with shaking in selective dextrose media (20 g/liter dextrose, 6.7 g/liter Difco yeast nitrogen base, 5 g/liter Bacto casamino acids, 5.4 g/liter Na2HPO4, 8.56 g/liter NaH2PO4·H2O) for 24 h and then switched into selective galactose media (20 g/liter galactose, 6.7 g/liter Difco yeast nitrogen base, 5 g/liter Bacto casamino acids, 5.4 g/liter Na2HPO4, 8.56 g/liter NaH2PO4·H2O) for 24–48 h. The resulting fusion protein from the N to C terminus contains ICAM-1, Myc tag, and Aga2.
Error-prone and Focused Mutagenesis Libraries
ICAM-1 cDNA library was constructed from error-prone PCR amplification of ICAM-1 cDNA using GeneMorph II Random Mutagenesis kit (Stratagene). In focused mutagenesis, four primers spanning ICAM-1 cDNA were synthesized with mixed nucleotides into selected positions for the substitution of a subset of 20 amino acids (described in detail under “Results”). The mixture of 1 μg of PCR products and 0.5 μg of linearized CAga2 vector was transformed into yeast cells using an electroporation method, as described previously (37). The diversity of the yeast library was estimated to be ∼2 × 106.
Immunofluorescence Flow Cytometry of Yeast Cells
Induced yeast cells were washed with 100 μl of labeling buffer (PBS, 0.5% BSA, 10 mm MgCl2) and then incubated with the ligands at 10 μg/ml in 50 μl of the labeling buffer for 20 min with shaking at 30 °C. Ligands used in this study were anti-ICAM-1 mAb and LFA-1 I domain mutant containing a pair of cysteines (K287C/K294C) (38) to induce high affinity (HA) toward ICAM-1. The HA I domain was modified for the addition of biotin at the N terminus, mixed with streptavidin-fluorescein isothiocyanate (BIOSOURCE International), and purified by gel filtration column (Superdex S200) to isolate the multimeric complex (39). Yeast cells incubated with primary antibodies were washed in 100 μl of the labeling buffer and incubated with secondary antibodies at 5 μg/ml in 50 μl of the labeling buffer for 20 min at 4 °C. Finally, cells were washed once in 100 μl and suspended in 100 μl of the labeling buffer for flow cytometry (FACSCalibur, BD Biosciences).
Yeast Library Sorting
Sorting of the ICAM-1 yeast libraries was performed with a magnetic cell sorter (MACS LS column, Miltenyi Biotec). The ICAM-1 libraries induced in 1 ml of selective galactose media was spun down, washed in 1 ml of the labeling buffer, and incubated in 200 μl of the labeling buffer containing 5 μg/ml anti-ICAM-1 antibodies (My13, R6.5, and RR1/1) for 20 min with shaking at 30 °C. After incubation with primary ligands, cells were washed once with 1 ml of the labeling buffer and incubated in the mixture of 80 μl of the labeling buffer and 20 μl of mouse anti-human IgG microbeads (Miltenyi Biotec) for 20 min at 4 °C. Yeast cells were sorted as described previously (37). To isolate individual clones for further testing, mixed yeast clones were grown in selective dextrose medium with 2% (w/v) agar plate for 48–72 h at 30 °C.
Immunofluorescence Flow Cytometry of Mammalian Cells
Human embryonic kidney (HEK)-293 or COS-7 cells were harvested by trypsinization and washed once in PBS with 0.5% BSA. To detect ICAM-1 expression, cells were incubated for 30 min with 50 μl of PBS with 0.5% BSA containing 10 μg/ml of anti-ICAM-1 antibodies followed by incubation with 5 μg/ml secondary antibodies in 50 μl of PBS with 0.5% BSA for 30 min at 4 °C. Finally, cells were washed once in 100 μl and suspended in 200 μl of the labeling buffer for flow cytometry. To measure HRV binding, cells were incubated with either HRV14 or HRV16 for 1 h at 4 °C with gentle agitation, washed in 100 μl of the PBS with 0.5% BSA, and incubated with either mAb-17 (a kind gift from T. Smith (40)) for HRV14 or anti-HRV16 guinea pig serum (ATCC) for 1 h at 4 °C. Cells were then washed and incubated with phycoerythrin-labeled polyclonal goat anti-mouse antibody or anti-guinea pig antibody for detection.
Expression of Soluble I Domains and ICAM-1 D1
The HA I domain and ICAM-1 D1 variants were expressed in Escherichia coli BL21 DE3 cells (Invitrogen) using the pET20b vector (Novagen). For the isolation of inclusion bodies, cells were resuspended in washing buffer (20 mm Tris-buffered saline, pH 8.0, with 23% w/v sucrose, 1 mm EDTA, 0.5% w/v Triton X-100) and disrupted by sonication. The suspension was centrifuged to pellet the inclusion body. Then the supernatant was removed, and the pellet was resuspended in washing buffer and centrifuged, which was repeated for 3–6 cycles until the pellet was mostly of the inclusion body. The inclusion bodies were solubilized using a denaturing buffer containing 6 m guanidine hydrochloride, 50 mm Tris, pH 8.0, and 1 mm dithiothreitol. After this, the polypeptide was refolded by dilution at less than 25 μg/ml in 20 mm Tris, pH 8.0, in the presence of 10% glycerol, 1 mm MgCl2, 6 mm cysteamine hydrochloride, and 3 mm cystamine hydrochloride to promote the formation of correct disulfide bonds. After the refolding step, the refolding solution was filtered through a 0.22-μm filter, concentrated, and subjected to gel filtration chromatography using Superdex 75 column connected to AKTA Purifier (GE Healthcare).
Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were carried out using a Biacore 2000 instrument with CM-5 sensor chips using N-hydroxysuccinimide/N-ethyl-N′(dimethyl-aminopropyl) carbodiimide chemistry for the immobilization. To achieve functional coupling of the HA I domain, we introduced a cysteine residue to the N terminus that was subsequently incubated with maleimide-polyethylene oxide-biotin (Pierce) for biotinylation following the manufacturers protocol. The free biotin was removed from the HA I domain by gel filtration. The chip was activated with N-hydroxysuccinimide/N-ethyl-N′(dimethyl-aminopropyl) followed by the addition of 50 μg/ml streptavidin (Promega) in 10 mm sodium acetate buffer, pH 4.9. Ethanolamine hydrochloride was used to block the remaining activated groups. The biotinylated HA I domain or biotin as control was added to the streptavidin-immobilized chip at 50 μg/ml. The injection buffer and the reservoir buffer were the same throughout: 10 mm HEPES, pH 7.4, 150 mm NaCl, 0.005% Polysorbate-20 (GE Healthcare), 10 mm MgCl2. For analytes, 2-fold serial dilutions of ICAM D1 variants or D1-D5 (produced from a CHO stable cell line, as described previously (41)) were injected at 10 μl/min, 25 °C starting at 1 μm unless otherwise noted. The BIAevaluation software (GE Healthcare) was used for the calculation of kon, koff, and KD by curve-fitting the data with a Langmuir 1:1 binding model or with a steady state binding model if steady state was reached.
Purification of HRV
Two different HRV serotypes, HRV14 and HRV16, were used in this study. The rhinoviruses were purified according to a previous protocol (42, 43). Briefly, HeLa cells were grown to confluency and infected with HRV at a multiplicity of infection of 5 plaque-forming units/cell. Cells were harvested and subjected to three cycles of freeze-thaw followed by homogenization and centrifugation. Virus was precipitated overnight at 4 °C in 0.5 m NaCl and polyethylene glycol (PEG 8000). The resulting precipitate was then collected by centrifugation, resuspended in HEPES buffer, pH 7.5, containing 0.25 m NaCl, and treated with DNase, RNase, and trypsin. The virus was then spun at 45,000 rpm for 2 h in an SW41 rotor on 30% sucrose cushion followed by a spin at 36,000 rpm for 90 min in an SW41 rotor on 0–40% sucrose gradient. Virus bands were collected and quantified with a spectrophotometer using an extinction coefficient of 7.7 at 260 nm for 1 mg/ml solution. A final spin at 45,000 rpm for 2 h in an SW41 rotor was carried out to pellet the virus.
Generation of Stable Cell Lines
The cDNA encoding full-length ICAM-1 was cloned into the pcDNA-3.1 vector (Invitrogen) using the restriction sites NheI and BamHI. HEK-293 or COS-7 cells were transfected with the wild-type or mutant ICAM-1-pcDNA-3.1 by the calcium phosphate method. Briefly, 2.5 μg of plasmid DNA in 375 μl of 2× Hepes-buffered saline was mixed with an equal volume of 0.3 m CaCl2. This solution was mixed and added dropwise to the cells at 50–60% confluency in a 6-well plate. After 6 h, cells were changed into fresh media and left for 2 days. To select for stable cells expressing ICAM-1, hygromycin B (Invitrogen) was added to the culture media at 200 μg/ml. After 2 weeks, the concentration of hygromycin was reduced to 100 μg/ml. Hygromycin-resistant foci were picked and transferred to fresh plates for expansion. Cells were maintained thereafter in 25 μg/ml hygromycin.
Cytopathic Effect (CPE) Assay
Cells were seeded in each well of a 96-well plate to reach 100% confluency on the day of the assay. Culture medium was removed from the wells, and dilutions of virus were added to the wells. The virus was then added to culture wells in a total volume of 100 μl. Plates were incubated at 35 °C for the times specified. At the endpoints indicated, the remaining viable cells were stained with 0.25% crystal violet prepared in 20% methanol and washed extensively with water. 1% SDS was added to solubilize the dye, which was then quantified by measuring the absorbance at 570 nm. Results represent the average values of the absorbance at 570 nm, expressed as a percentage of the control well with no virus added.
RESULTS
Directed Evolution of ICAM-1 D1
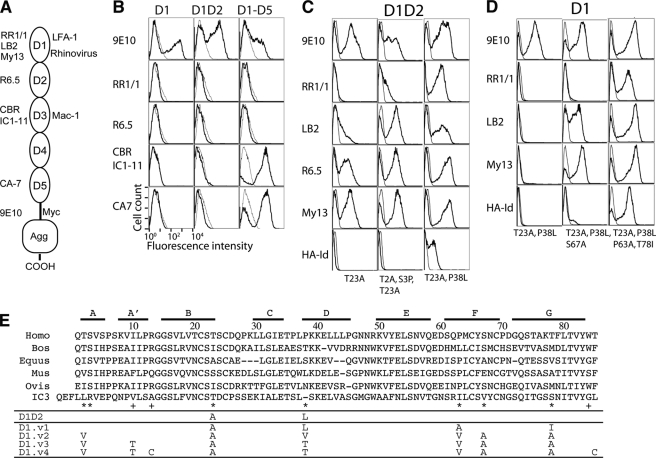
Directed evolution of ICAM-1 D1 was implemented in a yeast display system, where the protein of interest is displayed on the cell surface as a fusion to a yeast cell wall protein, agglutinin (44). Conformation-specific mAb against different domains of ICAM-1 (RR1/1, LB2, and My13 for probing D1, R6.5 for D2, CBR-IC1/11 for D3, and CA7 for D5 as depicted in Fig. 1A) as well as an anti-Myc monoclonal antibody (9E10) were used to assess functional expression of ICAM-1 on the yeast cell surface. Before generating a yeast library of ICAM-1 D1, we first tested for the expression of wild-type ICAM-1 D1, D1D2, and D1-D5. All three ICAM-1 fragments were displayed on the yeast surface as indicated by the binding of mAb 9E10 (Fig. 1B). To the clones expressing D1-D5, only the antibodies against D3-D5 showed binding, but unexpectedly there was no binding of any of the antibodies specific to D1 or D2 (Fig. 1B). ICAM-1 containing D1D2 has been expressed functionally as soluble forms in mammalian cells (26, 45) and in insect cells (28) as well as membrane-associated forms (46). The lack of D1D2 expression or native fold in yeast cells may indicate that D1D2 requires mammalian folding machinery to achieve native structure.
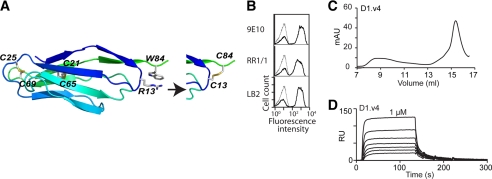
FIGURE 1.
Directed evolution approach to engineer a functional single Ig superfamily domain of ICAM-1. A, ICAM-1 contains five extracellular Ig superfamily domains, recognized by different antibodies. Domains 1 and 3 of ICAM-1 serve as ligands for integrins LFA-1 (αLβ2) and Mac-1 (αMβ2) respectively. mAb RR1/1, My13, and LB2 bind to D1; R6.5 binds to D2; CBR 1C1–11 binds to D4; CA-7 binds to D5. HA-Id, high affinity I domain. Agg, agglutinin. B, ICAM-1 expression in wild-type D1, D1D2, and D1-D5 as detected by immunofluorescence flow cytometry with the indicated antibodies is shown. mAb 9E10 is against the Myc tag that is fused to ICAM-1 at the C terminus. C and D, the isolation of functional mutants of D1 and D1D2 is shown. The binding of antibodies and high affinity I domain (tetrameric HA I domain (39)) to induced yeast cells was measured by immunofluorescence flow cytometry. Throughout the figures, ligand binding to induced yeast clones is shown as a histogram drawn in a thick line and to uninduced clones as a control in a thin line. The mutations found in the clones are indicated. E, shown is evolution of engineered ICAM-1 D1 variants, aligned with ICAM-1 D1 of different species and human ICAM-3 D1. Above the sequence alignment are shown the β strands, A–G. The asterisk and plus symbols identify the positions, respectively, where mutations were found from library screening and where the mutations were designed.
A D1D2 yeast library was constructed by error-prone PCR and screened for binding to mAb My13, R6.5, and RR1/1. After two rounds of sorting with the mixture of mAb My13 and R6.5, the library began to show binding to the mAb LB2, R6.5, and My13 and much less binding to RR1/1 (data not shown). From this library, six clones were individually tested for binding to antibodies and high affinity LFA-1 I domain. Sequencing analysis revealed that all six clones contained the mutation of T23A. Of the three D1-specific antibodies (My13, LB2, RR1/1), T23A alone induced high level binding to My13 and R6.5, with lower binding to LB2 and almost no binding to RR1/1 (Fig. 1C). Two of the six clones contained additional mutations of T2A and S3P, which led to an increase in the binding to mAb LB2 and RR1/1 (Fig. 1C). Recognizing that perhaps the binding of mAb RR1/1 was the most stringent indicator of native conformation of D1, we screened the initial D1D2 mutagenesis library with RR1/1. After three-rounds of sorting, ten clones were tested individually, of which the highest binders to RR1/1 and the LFA-1 HA I domain were found to contain the mutations of T23A and P38L (Fig. 1C).
With T23A and P38L introduced into the single domain D1 expressed in yeast, however, none of the D1-specific antibodies showed binding (Fig. 1D). This may indicate that the mutations resulting in native folding of D1D2 were not sufficient for D1 to fold on its own. The interface between D1 and D2 involves hydrophobic and electrostatic interactions (Fig. 2A) and additional stability in D1 may be required to compensate for interactions lost by removing D2. Using a D1 mutant containing T23A and P38L as a template, a mutagenesis library of D1 was constructed. After five rounds of sorting with mAb RR1/1, all the clones that showed high level binding to the antibodies and HA I domain were found to be identical, containing the mutations of T23A, P38L, P63A, and T78I (designated as D1.v1; Fig. 1E). Several clones that showed much lower binding to RR1/1 and HA I domain were found to contain the mutations of T23A, P38L, and S67A (Fig. 1D).
FIGURE 2.
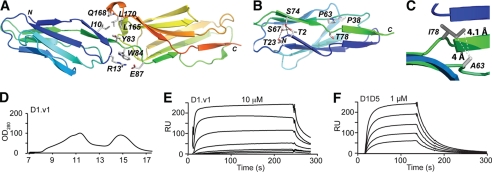
Isolation of a functional ICAM-1 D1 by directed evolution. A, ICAM-1 D1D2 (PDB code 1IAM (28)) is drawn in a schematic representation (PyMOL, DeLano Scientific LLC) with the residues at the domain interface shown in sticks. N and C termini are labeled. B, ICAM-1 D1 is shown with the wild-type residues drawn in sticks displayed for the positions where mutations were found. H-bond networks are shown with dotted lines. C, shown is a schematic diagram of ICAM-1 D1 with modeled van der Waals interaction after the mutations of P63A and T78I. D, ICAM-1 D1.v1 elution from Superdex-75 size exclusion column is shown. E–F, SPR measurement of the binding of the ICAM-1 D1.v1 (E) and D1-D5 (F) to the high affinity LFA-1 I domain immobilized on the chip is shown. RU, relative units. ICAM-1 was injected over the chip in a series of 2-fold dilutions, starting from the indicated concentrations.
Rationalization of the Effect of Mutations in Native Folding of ICAM-1 D1
The majority of mutations found over the course of engineering functional D1 were at the residues that form a hydrogen bond network, Thr-2, Thr-23, and Ser-67 (Fig. 2B). Although hydrogen bonding is considered to be one of the primary stabilizing interactions together with van der Waals and electrostatic interactions, hydrogen bonding may be less effective in a buried, solvent-excluded region than hydrophobic, van der Waals contacts. The mutation of these residues to Ala and Leu converts hydrogen bonds into hydrophobic contacts. Mutating Pro-63 and Thr-78 into Ala and Ile, respectively, creates a new van der Waals contact between them (Fig. 2C). It is interesting to note that for all the mutations that contributed to functional expression of D1D2 and D1, identical or similar types of substitutions are found in ICAM-1 of other species or other ICAM family members (Fig. 1E).
Soluble Expression of ICAM-1 D1 and Functional Assay by SPR
The newly generated D1.v1 was expressed from bacteria in inclusion bodies, refolded in cysteamine/cystamine redox buffer to promote correct disulfide bond formation, and purified by gel filtration chromatography (Fig. 2D). ICAM-1 D1 was found to elute at ∼15 ml on an Superdex-75 column, with the aggregate eluting at ∼11 ml. Using surface plasmon resonance (Biacore), the affinity (equilibrium dissociation constant, KD) for ICAM-1 D1.v1 was assessed for its binding to the LFA-1 HA I domain (Fig. 2E). The KD of D1.v1 to HA I domain was determined to be 6 μm, compared with 304 nm for ICAM-1 D1-D5 measured in this study (Fig. 2F) and a previously measured value of 277 nm for ICAM-1 D1-D5 (39). 20-Fold lower affinity of D1.v1 to the high affinity I-domain than D1-D5 may be attributable to the formation of aggregates during the course of the assay, leading to a decrease in the concentration of binding-competent, monomeric D1. Indeed, D1.v1 was found to aggregate within hours of storage at room temperature and lead to precipitation with an increase in concentration. This illustrates that the stability of a protein in solution cannot be predicted entirely by the stability of a protein displayed on a cell surface.
Focused Mutagenesis of D1 in Search of Stable Variants
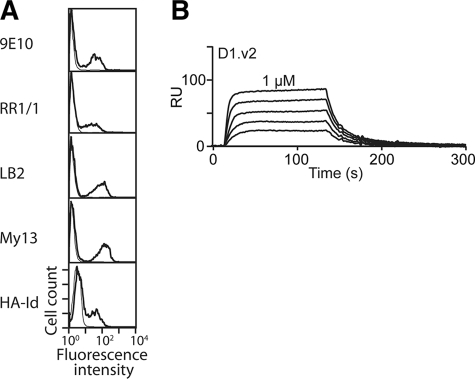
The selection pressure on ICAM-1 D1 is limited to attaining native conformation when it is fused to Aga2 and displayed on the surface of yeast cells. We reasoned that expanding the types of substitutions for the positions found by directed evolution approach to further enhance van der Waals contacts may promote the solution stability of D1. Therefore, the next version of the D1 library was constructed not by error-prone PCR but by using primers with degenerate nucleotides for the codons corresponding to the mutation sites found in functional D1D2 and D1.v1. Focusing the incorporation of random mutations into a limited position permits exhaustive searching of optimal substitutions, which may further enhance the stability of Ig superfamily fold. This method of “focused” mutagenesis (37) is an efficient means to overcome some of the limits imposed by the size of the random mutagenesis library and to compensate for mutation biases intrinsic to error-prone PCR. For example, to screen substitutions for Thr, the codon NCN (N = A, G, T, or C) would allow substitution of any of Ser, Pro, Thr, and Ala, whereas ANN would encode any of Ile, Met, Thr, Asn, Lys, Ser, and Arg. For Thr-2, Thr-23, Pro-38, Pro-63, and Thr-78, the codon of VYX (where V = A, G, or C; Y = C or T, and X = wild-type nucleotide) was used to code for any of Thr, Ile, Pro, Leu, Ala, and Val. For Ser-67 and Ser-74, the sequence NYX was used which encodes any of Ser, Thr, Ile, Pro, Leu, Ala, or Val. Four primers were synthesized spanning entire ICAM-1 cDNA sequences and combined together by PCR. Given the degenerate codons for seven positions, the size of the diversity becomes (3 × 2 × 1)5 × (4 × 2 × 1)2 = 497,664, which is smaller than the size of yeast library, ∼2 × 106. The focused mutagenesis library was again sorted with mAb RR1/1. After three rounds of sorting, the high level antibody binding clones contained T2V, T23A, P38V, P63V, S67A, and T78A and was designated as D1.v2 (Fig. 1E). The binding of mAb 9E10, RR1/1, My13, and LB2 and the HA I domain to D1.v2 expressed on yeast surface (Fig. 3A) was comparable with their binding to D1.v1. D1.v2 was expressed and purified in the same manner as D1.v1. When expressed in solution, D1.v2 was significantly more stable than D1.v1 at room temperature, and its affinity to the HA I domain was determined to be 221 ± 5.6 nm (Fig. 3B), comparable with that of the D1-D5 (Fig. 2E).
FIGURE 3.
Functional expression of ICAM-1 by focused mutagenesis. A, antibody and high affinity I domain (HA-Id) binding to D1.v2 is shown. B, SPR measurement of the binding of the ICAM-1 D1.v2 to the HA I domain is shown. The analytes were injected in a series of 2-fold serial dilutions starting from 1 μm. RU, relative units.
Structure-based Rational Design of ICAM-1 D1
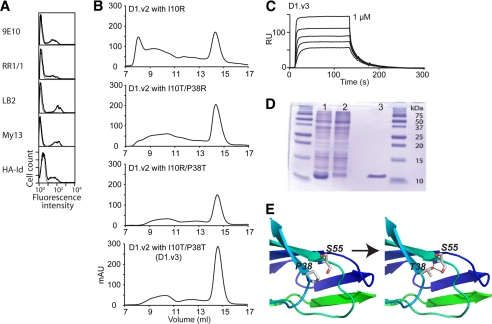
The interface of D1 and D2 consists of van der Waals contacts by a number of hydrophobic residues, Ile-10, Leu-165, and Leu-170, and the methylene groups of Tyr-83 and Gln-168 as well as electrostatic attraction between Arg-13 and Glu-87 (Fig. 2A). Without domain D2, Ile-10 is exposed to the solvent. We introduced a mutation of I10R or I10T to replace the hydrophobic residue Ile-10. To further reduce the number of hydrophobic residues exposed to the solvent, we also substituted Arg or Thr for Pro-38, where Val was selected from the library. Previously, P38R was found to increase the affinity of ICAM-1 for the LFA-1 I domain without adversely affecting ICAM-1 expression (39). A substitution of Thr for Pro-38 was selected to examine if the hydroxyl group of Thr in place of the methyl group of Val would enhance solution stability of D1. Additionally, P38T was predicted to create a hydrogen bond with the hydroxyl group of Ser-55 in an area that is solvent-exposed (Fig. 4E). When D1.v2 variants with Thr or Arg substitutions for Ile-10 and Pro-38 were expressed on yeast, their binding to anti-ICAM-1 antibodies and the HA I domain was comparable with D1.v2 (D1.v2 with I10R is shown in Fig. 4A). We then expressed D1.v2 with I10R, I10T/P38R, I10R/P38T, or I10T/P38T and analyzed solution stability by the efficiency of folding, the ratio of aggregate to monomer using gel filtration chromatography, and the binding affinity to LFA-1 I domain by SPR. During the refolding process, these D1 variants produced markedly less precipitant compared with the previous D1 variants; in gel filtration, they eluted at a distinct monomeric peak at 14.9 ml with a much lower amount of aggregate. The yield of the later D1 variants from the same culture volume increased significantly relative to D1.v1 (Fig. 4B). We designated D1.v2 containing I10T and P38T as D1.v3. After gel filtration, D1.v3 appeared as a distinct band at ∼10 kDa (Fig. 4D, lane 3). When the affinity of D1.v3 was tested for binding to the HA I domain, the KD was found to be 123 ± 12.7 nm (Fig. 4C). Refolded D1 variants purified by gel filtration were functional and homogenous in conformation, indicated by a complete shift of the D1 elution peak when mixed with the HA I domain in ion exchange chromatography (supplemental Fig. 1).
FIGURE 4.
Rational design of ICAM-1 D1 to convert surface-exposed hydrophobic residues into hydrophilic ones. A, shown are the histograms of the binding of anti-ICAM-1 mAb and the HA I domain to ICAM-1 D1.v2-containing I10R mutation. HA-Id, high affinity I domain. B, elution profiles are shown of D1.v2 containing I10R, I10T/P38R, I10R/P38T, and I10T/P38T from a Superdex-75 size exclusion column. mAU, milliabsorbance units. C, SPR measurement of the binding of the ICAM-1 D1.v3 to the HA I domain is shown. The analytes were injected in a series of 2-fold serial dilutions starting from 1 μm. RU, relative units. D, SDS-PAGE of induced (1) and uninduced (2) BL21 cell lysates and ICAM-1 D1.v3 after gel filtration column (3). E, schematic diagram of ICAM-1 D1 with modeled H-bond between Thr-38 and Ser-55 after P38T mutation.
To further tighten the C-terminal face of the Ig superfamily fold after the loss of D2, cysteines were substituted for Arg-13 and Trp-84. The distance between Cβ atoms of Arg-13 and Trp-84 is 4 Å, which is within the range for a disulfide bond (Fig. 5A). Before expressing D1.v3 with R13C and W84C (designated as D1.v4), we examined whether it could be expressed functionally in yeast. The binding of mAb 9E10, RR1/1, and LB2 to D1.v4 (Fig. 5B) was comparable with the previous clones, indicating that two cysteines formed a disulfide bond in yeast expression. Similarly, D1.v4 was expressed from bacteria and purified in a gel filtration column. It should be noted that the mutation of Trp-84 (the only one in D1) to cysteine decreases the extinction coefficient of D1 from 10,010 to 4,440 cm−1 m−1, resulting in a lower absorbance for the same molar concentration of D1. The KD of D1.v4 to the HA I domain was determined to be 362.5 ± 19.1 nm (Fig. 5D), which is comparable with those of D1.v2, D1.v3, and D1-D5. Overall, all the D1 variants exhibited faster kinetics of binding to the HA I domain with the on-rate (kon) ranges between 5 and 7 × 105 s−1 m−1, whereas D1-D5 exhibited a kon = 9 × 104 s−1 m−1, presumably in part due to the smaller size of D1.
FIGURE 5.
Rational design of ICAM-1 D1 to introduce extra disulfide-bond. A, a schematic diagram of ICAM-1 D1 is shown with two native disulfide bonds between β-strands, B and F, and Arg-13 and Trp-84, for which an extra disulfide-bond is designed by substitution of two cysteines. B, shown are the histograms of the binding of anti-Myc antibody, 9E10, and anti-ICAM-1 mAbs, RR1/1 and LB2. C, elution profiles of D1.v4 from a Superdex-75 size exclusion column are shown. mAU, milliabsorbance units. D, SPR measurement of the binding of the ICAM-1 D1.v4 to the HA I domain is shown. The analytes were injected in a series of 2-fold serial dilutions starting from 1 μm. RU, relative units.
Expression of Wild-type and Mutant ICAM-1 in HEK-293 and COS-7 Cells to Be Permissive for Rhinovirus Replication
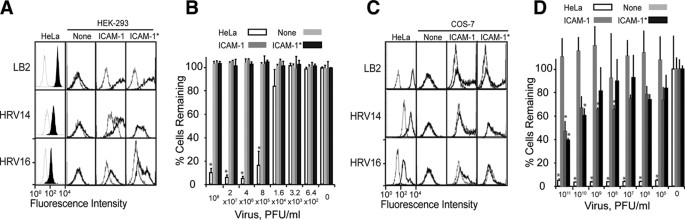
Throughout the engineering of ICAM-1 D1, the recovery of native function of D1 has been focused on its recognition of conformation-specific antibodies and LFA-1 I domain with flow cytometry and by SPR. Another important role of ICAM-1 D1 is the receptor for human rhinovirus (HRV). To examine if the mutations introduced into D1 preserve HRV binding and replication, the mutations in D1.v2 with P38R (Fig. 2A) were introduced into full-length ICAM-1 in pcDNA-3.1. In search of human host cells that do not express ICAM-1 and are non-permissive to rhinovirus replication, HEK-293 cells were found to satisfy both conditions with undetectable binding of ICAM-1-specific mAb LB2 (Fig. 6A). HEK-293 cells were transfected with the plasmids carrying full-length wild-type ICAM-1 or mutant ICAM-1 and cultured for 3–4 weeks in the presence of 25–200 μg/ml of hygromycin. The majority of cells expressed ICAM-1 stably with the level of ICAM-1 expression comparable between the clones expressing wild-type and mutant ICAM-1, confirmed by mAb LB2 binding (Fig. 6A). However, the binding of rhinovirus, particularly HRV14, was lower to the mutant ICAM-1 than to wild-type ICAM-1. To our surprise, however, the expression of the wild-type or mutant ICAM-1 in 293 cells failed to render them permissive to rhinovirus replication, in contrast to a clear cytopathic effect on HeLa cells after 48 h (Fig. 6B).
FIGURE 6.
Expression of full-length wild-type and mutant ICAM-1 in HEK-293 and COS-7 cells. A, shown is immunofluorescence staining of HeLa and HEK-293 cells that were transfected with the wild-type ICAM-1 and ICAM-1 with the mutations in D1.v2 with P38R (ICAM-1*) after incubation with mAb LB2, HRV14, and HRV16. Non-transfected HEK-293 cells were indicated as None. Histograms in thin open line are of antibody binding to cells without the primary antibody or viruses used as a control. B, shown is a CPE assay of HeLa and HEK-293 cells in a 96-well plate after incubation with HRV16 for 48 h, quantified by crystal violet staining of adherent cells. The data represent the average values of the absorbance at 570 nm, normalized to the control well with no virus added as 100%. The CPE assay was carried out in triplicate, and the error bars indicate S.D. (*, versus matching control, p < 0.05 by Student's t test). PFU, plaque-forming units. C, shown is immunofluorescence staining of HeLa and COS-7 cells that were transfected with the wild-type ICAM-1 and ICAM-1 with the mutations in D1.v2 with P38R (ICAM-1*) after incubation with mAb LB2, HRV14, and HRV16. Histograms in thin open lines are of antibody binding to cells without the primary antibody or viruses used as a control. D, shown is a CPE assay of HeLa and COS-7 cells in a 96-well plate after incubation with HRV16 for 96 h, quantified by crystal violet staining of adherent cells. The data represent the average values of the absorbance at 570 nm, normalized to the control well with no virus added as 100%. The CPE assay was carried out in triplicate, and the error bars indicate S.D. (*, versus matching control, p < 0.05 by Student's t test).
This demonstrates that ICAM-1 expression is a necessary condition for the host cells to be permissive to rhinovirus replication but is not sufficient, as other host components need to be orchestrated for the virus life cycle to be completed. Therefore, we tested a second cell type, African green monkey (COS-7) cells, which have previously been used for generation of stable cell lines with ICAM-1 and assay of viral protein synthesis (41). As before, the wild-type and mutant ICAM-1 in pcDNA-3.1 were introduced into COS-7 cells and maintained for 2–4 weeks in the presence of 200 μg/ml hygromycin. When tested for the expression of ICAM-1, the mAb LB2 binding was comparable in cells expressing the wild-type versus the mutant ICAM-1. Although the level of HRV16 binding to COS-7 cells was lower than that to HEK-293, COS-7 showed 30–60% cell death of both wild-type ICAM-1 and mutant in the range of 108–1011 plaque-forming units/ml (Fig. 6D).
DISCUSSION
ICAM-1 is a multifunctional cell surface molecule playing a role as a receptor for two members of the integrin family (LFA-1 and Mac-1) and human rhinovirus. Although all five Ig superfamily domains of ICAM-1 appear in high resolution structures as independently folded units, the first domain depends on the second domain to fold correctly. The ability to functionally express an individual domain can greatly contribute to understanding the biological role of a molecule as a whole through biochemical and structural studies with its binding partners. When a single domain of interest fails to express in its native conformation, a systematic method to engineer modular domains would be a highly useful tool in many biological studies. The goal of this study is to achieve a functional, single domain of ICAM-1 and to develop a systematic approach that can be applicable to other molecules containing the Ig superfamily fold, which is one of the most abundant protein folds found in cell surface molecules. We have successfully engineered stable and functional ICAM-1 D1 utilizing directed evolution to identify initial leads, structural rationalization of new interactions created by mutations, and fine-tuning of the types of substitutions in the rationally selected positions.
The mutations that led to the native conformation of D1 were isolated through successive rounds of mutagenesis; first, a D1D2 mutagenesis library was screened by conformation-specific antibodies, and the mutations therein were introduced into the template for the construction of a D1 library. Given the finding that all four mutations (T23A, P38L, P63A, T78I) were necessary for functional expression of D1 alone on the surface of yeast, it may have been difficult to isolate all of them directly of out of a D1 mutagenesis library without an intermediate step of D1D2 engineering. During the course of the library selection, we found that T23A is necessary for the binding of all D1-specific antibodies. With T23A alone, mAb My13 showed high level binding, indicating that its epitope does not depend on fully native conformation. The binding of mAb LB2 to this mutant was low, which increased greatly with the additional mutations of T2A and S3P. The epitope of mAb RR1/1 was found to be most dependent on the native conformation of D1, which directly related to that of the HA I domain. When the sequences of all the D1 variants were aligned, the mutations that led to creating or enhancing antibody binding were located at two clusters. One cluster, consisting of four hydrophilic residues (Thr-2, Thr-23, Ser-67, and Ser-74), forms a hydrogen bond network where the side-chain hydroxyl groups of Thr-2, Thr-23, and Ser-67 are buried from the solvent. The second cluster is formed by Pro-63 and Thr-78, where their side chains are within a range for van der Waals contacts (e.g. the distance between Pro-63-Cβ and Thr-78-Cγ is 4 Å), and the hydroxyl group of Thr-78 is buried and forms a hydrogen bond with the carbonyl group of Pro-6. The mutations of P63A and T78I may enhance the van der Waals interaction between them (Fig. 2C). Last, P38L was one of the two critical mutations that led to the functional expression of D1 in the context of D1D2. Previously, it was noted that Pro-38 has non-optimal ϕ-ψ angles that place it in the generously allowed region in a Ramachandran plot and its substitution led to higher expression (39). Although Pro-36 and Pro-38 are placed in the loop connecting β-strands C and D, the amino acids for these positions in ICAM-1 of other species are highly variable (Fig. 1E).
It is important to note that all the substitutions for the wild-type residues isolated in functional expression of human ICAM-1 D1 are found in the ICAM-1 of other species. For example, in the equine ICAM-1 D1, the cluster forming a hydrogen bond network at the N-terminal end in human ICAM-1 is composed of Ile, Ala, and Ala for the human equivalent positions of Thr-2, Thr-23, and Ser-67, respectively (Fig. 1E). Furthermore, in equine ICAM-1, Val is found for human Thr-78. Therefore, two clusters of van der Waals interactions created by the mutations in human ICAM-1 D1 are present in the wild-type sequence of the equine ICAM-1. The conversion of hydrogen bond networks into van der Waals contacts is also found in the first domain of human ICAM-3, whose peptide backbone is closely superimposable (<0.5 Å root mean square deviation) with that of ICAM-1 and was stably expressed without the second domain D2 from mammalian cells (32). Structural rationalization of the effect of the mutations and the fact that the forces strengthened by the mutations are found in other ICAM members and ICAM-1 of other species present strong evidence that the conversion of the hydrogen bond networks in a buried region into the van der Waals contact is effective in enhancing the stability of Ig superfamily domain fold. However, when expressed in solution, D1 with the mutations isolated by directed evolution was prone to aggregation, resulting in lower affinity than the wild-type D1-D5 to the HA I domain by SPR. We speculated that with the relatively small size of diversity in the yeast library, the searching for optimal substitutions to create effective van der Waals contact would be far from exhaustive. With the coding sequence of D1 synthesized from four overlapping primers with degenerate codons for the selected positions, the diversity of focused mutagenesis was approximately a half-million, far smaller than the size of our yeast library. Soluble expression of D1 identified from focused mutagenesis library was more stable than the D1 from random mutagenesis library, judging from much less precipitants of D1.v2 after 4 °C storage (data not shown). In addition, the affinity of D1.v2 to HA I domain was comparable to that of the wild-type D1-D5.
Rational design by structural inspection also contributed to the engineering of functional ICAM-1 D1. In an effort to mutate solvent-exposed hydrophobic residues to hydrophilic ones, we substituted Thr for Ile-10 and Pro-38, which was found to decrease the amount of aggregate during refolding. The choice of Thr was also based on the observation that β-branched amino acids are preferred in β-strands (47). Successive substitution of Thr for Ile-10 and Pro-38 led to enhanced stability of D1 with little aggregation over several weeks at 4 °C storage. Furthermore, to enhance the packing of the last β strand to the rest of the domain, we designed a pair of cysteines to substitute for Arg-13 and Trp-84 that are optimally placed for disulfide bond formation. When expressed in yeast, D1.v3 with two additional cysteines exhibited the levels of expression and binding to D1-specific antibodies comparable to those of D1.v3 (Fig. 5B), indicating disulfide bond formation between Cys-13 and Cys-84.
ICAM-1 D1 with the mutations isolated here was fully functional in its binding to LFA-1 I domain and several conformation-specific antibodies. Another important ligand for ICAM-1 is rhinovirus, where the virus binding to ICAM-1 is limited to the D1 with the N-terminal end of Ig superfamily fold inserting into the recessed surface in the viral capsid known as the canyon (48). To examine if the D1 alone is capable of mediating virus binding to ICAM-1, we attempted to express ICAM-1 D1.v2 with native transmembrane and cytoplasmic domains of ICAM-1. Despite functional expression of D1 in yeast surface and refolding of D1, when expressed in HEK-293, the expression of D1 alone was not detectable. This may reflect the difference in the machinery involved in folding of D1 in yeast and mammalian cells. Therefore, we examined whether the mutations introduced in D1 variants would affect virus binding in the context of full-length ICAM-1 expressed in HEK-293 cells and COS-7 cells. Interestingly, although virus binding was seen to wild-type or mutant ICAM-1-transfected HEK-293 cells, no cell death was observed in either case. This may likely be due to the absence of an ancillary component required for viral replication, as other studies have observed viral entry in BHK cells but an absence of viral protein production (49). In contrast to the HEK-293 cells, wild-type and mutant ICAM-1 stably transfected COS-7 cells were both rendered permissive to HRV infection, exhibiting 30–60% cell death. Incomplete cell death is attributable to the presence of cells with no or low level expression of ICAM-1 to allow HRV binding. It is also interesting to note that although the mutant ICAM-1 showed comparable binding to antibodies and LFA-1 I domain as the wild-type ICAM-1, virus binding, particularly of HRV14, to the mutant was significantly lower than to the wild type. This may be attributed to the loss of conformational flexibility at the N terminus of D1 due to the substitution of hydrophobic residues. It has been shown by cryoelectron microscopy image reconstruction that the N-terminal face of the ICAM-1 D1, formed by B-C, D-E, and F-G loops, inserts into the recessed surface of the virus capsid, creating extensive ionic networks (48, 50). Although none of the residues of ICAM-1 at the interface with HRV14 and HRV16 (50) were mutated in this study, mutations close to the N-terminal face may induce a slight change in the conformations of the three interface loops from native structure.
The development of selected Ig superfamily domains as “decoys” has wide-ranging implications for development of these molecules as therapeutic agents not only against cellular adhesion molecules such as LFA-1 but also for agents against viruses. Rational design and directed evolution may be used to engineer decoys to alter their interaction with one partner but not with the other. Although the aim of the current study was to engineer ICAM-1 D1 that retains native functions and Ig superfamily folds, it is anticipated that the same approach can be used to isolate mutations that would increase its affinity to LFA-1 I domain. A similar strategy was demonstrated in previous studies in which ICAM-1 D1-D5 (39) and LFA-1 I domain (37) were engineered for higher affinity to their binding partners. Many enveloped and non-enveloped viruses recognize Ig superfamily domains in transmembrane receptor for entry into the host cells, including human immunodeficiency virus binding to CD4 (25), poliovirus binding to poliovirus receptor (24), and Coxsackievirus A to ICAM-1 (11), where the N-terminal Ig superfamily is the main interaction site with the virus. Engineered D1 that is suitable for producing a stable complex with a particular serotype of rhinovirus may be useful for the production of high resolution crystals, which will contribute to the design of anti-viral compounds for inhibition of virus binding to ICAM-1. When the Ig superfamily domains interact with one or several serotypes, Ig superfamily domains engineered for highest affinity to the virus may be developed into antiviral compounds. An advantage of using receptor domains for developing antivirals is that viruses will not be able to mutate to avoid binding to the cellular receptor without losing the ability to enter the cell.
Supplementary Material
Acknowledgments
We thank Carol Bator and Dr. Michael Rossmann for providing the HRV16 strain and helpful insights on virus production and purification and Drs. Umesh Katpally and Thomas Smith for providing mAb-17.
This work was supported, in whole or in part, by National Institutes of Health Grants CA31798 and AI079532.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ICAM
- intercellular adhesion molecule
- LFA-1
- lymphocyte function-associated antigen-1
- mAb
- monoclonal antibody
- PBS
- phosphate-buffered saline
- HEK
- human embryonic kidney
- SPR
- surface plasmon resonance
- BSA
- bovine serum albumin
- CPE
- cytopathic effect
- HRV
- human rhinovirus
- HA
- high affinity.
REFERENCES
- 1.Dustin M. L., Springer T. A. (1988) J. Cell Biol. 107, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marlin S. D., Springer T. A. (1987) Cell 51, 813–819 [DOI] [PubMed] [Google Scholar]
- 3.Makgoba M. W., Sanders M. E., Ginther, Luce G. E., Dustin M. L., Springer T. A., Clark E. A., Mannoni P., Shaw S. (1988) Nature 331, 86–88 [DOI] [PubMed] [Google Scholar]
- 4.Smith C. W., Marlin S. D., Rothlein R., Toman C., Anderson D. C. (1989) J. Clin. Invest. 83, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diamond M. S., Staunton D. E., de Fougerolles A. R., Stacker S. A., Garcia-Aguilar J., Hibbs M. L., Springer T. A. (1990) J. Cell Biol. 111, 3129–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackford J., Reid H. W., Pappin D. J., Bowers F. S., Wilkinson J. M. (1996) Eur. J. Immunol. 26, 525–531 [DOI] [PubMed] [Google Scholar]
- 7.van de Stolpe A., van der Saag P. T. (1996) J. Mol. Med. 74, 13–33 [DOI] [PubMed] [Google Scholar]
- 8.Berendt A. R., Simmons D. L., Tansey J., Newbold C. I., Marsh K. (1989) Nature 341, 57–59 [DOI] [PubMed] [Google Scholar]
- 9.Tomassini J. E., Colonno R. J. (1986) J. Virol. 58, 290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staunton D. E., Merluzzi V. J., Rothlein R., Barton R., Marlin S. D., Springer T. A. (1989) Cell 56, 849–853 [DOI] [PubMed] [Google Scholar]
- 11.Shafren D. R., Dorahy D. J., Ingham R. A., Burns G. F., Barry R. D. (1997) J. Virol. 71, 4736–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casasnovas J. M., Stehle T., Liu J. H., Wang J. H., Springer T. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4134–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiménez D., Roda-Navarro P., Springer T. A., Casasnovas J. M. (2005) J. Biol. Chem. 280, 5854–5861 [DOI] [PubMed] [Google Scholar]
- 14.Yang Y., Jun C. D., Liu J. H., Zhang R., Joachimiak A., Springer T. A., Wang J. H. (2004) Mol. Cell 14, 269–276 [DOI] [PubMed] [Google Scholar]
- 15.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., et al. (2001) Nature 409, 860–92111237011 [Google Scholar]
- 16.Williams A. F., Barclay A. N. (1988) Annu. Rev. Immunol. 6, 381–405 [DOI] [PubMed] [Google Scholar]
- 17.Diamond M. S., Staunton D. E., Marlin S. D., Springer T. A. (1991) Cell 65, 961–971 [DOI] [PubMed] [Google Scholar]
- 18.Springer T. A., Dustin M. L., Kishimoto T. K., Marlin S. D. (1987) Annu. Rev. Immunol. 5, 223–252 [DOI] [PubMed] [Google Scholar]
- 19.Greve J. M., Davis G., Meyer A. M., Forte C. P., Yost S. C., Marlor C. W., Kamarck M. E., McClelland A. (1989) Cell 56, 839–847 [DOI] [PubMed] [Google Scholar]
- 20.Olson N. H., Kolatkar P. R., Oliveira M. A., Cheng R. H., Greve J. M., McClelland A., Baker T. S., Rossmann M. G. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolatkar P. R., Bella J., Olson N. H., Bator C. M., Baker T. S., Rossmann M. G. (1999) EMBO J. 18, 6249–6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao C., Bator-Kelly C. M., Rieder E., Chipman P. R., Craig A., Kuhn R. J., Wimmer E., Rossmann M. G. (2005) Structure 13, 1019–1033 [DOI] [PubMed] [Google Scholar]
- 23.Mendelsohn C. L., Wimmer E., Racaniello V. R. (1989) Cell 56, 855–865 [DOI] [PubMed] [Google Scholar]
- 24.Racaniello V. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11378–11381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryu S. E., Kwong P. D., Truneh A., Porter T. G., Arthos J., Rosenberg M., Dai X. P., Xuong N. H., Axel R., Sweet R. W. (1990) Nature 348, 419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greve J. M., Forte C. P., Marlor C. W., Meyer A. M., Hoover-Litty H., Wunderlich D., McClelland A. (1991) J. Virol. 65, 6015–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin S., Martin A., Staunton D. E., Springer T. A. (1993) Antimicrob. Agents Chemother. 37, 1278–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bella J., Kolatkar P. R., Marlor C. W., Greve J. M., Rossmann M. G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Kim T. D., Carman C. V., Mi L. Z., Song G., Springer T. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15358–15363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimaoka M., Xiao T., Liu J. H., Yang Y., Dong Y., Jun C. D., McCormack A., Zhang R., Joachimiak A., Takagi J., Wang J. H., Springer T. A. (2003) Cell 112, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klickstein L. B., York M. R., Fougerolles A. R., Springer T. A. (1996) J. Biol. Chem. 271, 23920–23927 [DOI] [PubMed] [Google Scholar]
- 32.Song G., Yang Y., Liu J. H., Casasnovas J. M., Shimaoka M., Springer T. A., Wang J. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Issekutz A. C., Rowter D., Springer T. A. (1999) J. Leukoc. Biol. 65, 117–126 [DOI] [PubMed] [Google Scholar]
- 34.Rothlein R., Mainolfi E. A., Czajkowski M., Marlin S. D. (1991) J. Immunol. 147, 3788–3793 [PubMed] [Google Scholar]
- 35.Boder E. T., Midelfort K. S., Wittrup K. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10701–10705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu X., Kang S., Chen X., Shoemaker C. B., Jin M. M. (2009) J. Biol. Chem. 284, 16369–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin M., Song G., Carman C. V., Kim Y. S., Astrof N. S., Shimaoka M., Wittrup D. K., Springer T. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5758–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimaoka M., Lu C., Palframan R. T., von Andrian U. H., McCormack A., Takagi J., Springer T. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6009–6014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song G., Lazar G. A., Kortemme T., Shimaoka M., Desjarlais J. R., Baker D., Springer T. A. (2006) J. Biol. Chem. 281, 5042–5049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith T. J., Chase E. S., Schmidt T. J., Olson N. H., Baker T. S. (1996) Nature 383, 350–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marlin S. D., Staunton D. E., Springer T. A., Stratowa C., Sommergruber W., Merluzzi V. J. (1990) Nature 344, 70–72 [DOI] [PubMed] [Google Scholar]
- 42.Oliveira M. A., Zhao R., Lee W. M., Kremer M. J., Minor I., Rueckert R. R., Diana G. D., Pevear D. C., Dutko F. J., McKinlay M. A. (1993) Structure 1, 51–68 [DOI] [PubMed] [Google Scholar]
- 43.Owens R. M., Wang C., You J. A., Jiambutr J., Xu A. S., Marala R. B., Jin M. M. (2009) J. Recept. Signal Transduct. Res. 29, 195–201 [DOI] [PubMed] [Google Scholar]
- 44.Boder E. T., Wittrup K. D. (1997) Nat. Biotechnol. 15, 553–557 [DOI] [PubMed] [Google Scholar]
- 45.Martin S., Casasnovas J. M., Staunton D. E., Springer T. A. (1993) J. Virol. 67, 3561–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staunton D. E., Gaur A., Chan P. Y., Springer T. A. (1992) J. Immunol. 148, 3271–3274 [PubMed] [Google Scholar]
- 47.Street A. G., Mayo S. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9074–9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossmann M. G. (1989) Viral Immunol. 2, 143–161 [DOI] [PubMed] [Google Scholar]
- 49.Grunert H. P., Wolf K. U., Langner K. D., Sawitzky D., Habermehl K. O., Zeichhardt H. (1997) Med. Microbiol. Immunol. 186, 1–9 [DOI] [PubMed] [Google Scholar]
- 50.Xiao C., Tuthill T. J., Bator Kelly C. M., Challinor L. J., Chipman P. R., Killington R. A., Rowlands D. J., Craig A., Rossmann M. G. (2004) J. Virol. 78, 10034–10044 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.