Abstract
Huntingtin-associated protein-1 (Hap1) is a neuronal protein that associates with huntingtin, the Huntington disease protein. Although Hap1 and huntingtin are known to be involved in intracellular trafficking, whether and how the impairment of Hap1-associated trafficking leads to neurological pathology and symptoms remain to be seen. As Hap1 is enriched in neuronal cells in the brain, addressing this issue is important in defining the role of defective intracellular trafficking in the selective neuropathology associated with Hap1 dysfunction. Here, we find that Hap1 is abundantly expressed in orexin (hypocretin)-producing neurons (orexin neurons), which are distinctly distributed in the hypothalamus and play an important role in the regulation of feeding and behavior. We created conditional Hap1 knock-out mice to selectively deplete Hap1 in orexin neurons via the Cre-loxP system. These mice show process fragmentation of orexin neurons and reductions in food intake, body weight, and locomotor activity. Sucrose density gradient fractionation reveals that loss of Hap1 in the mouse brain also reduces the distribution of trafficking protein complexes and cargo proteins in the fractions that are enriched in synaptosomes. These results suggest that Hap1 is critical for the transport of multiple proteins to the nerve terminals to maintain the integrity of neuronal processes and that selective disruption of the processes of orexin neurons can cause abnormal feeding and locomotor activity.
Keywords: Brain, Huntington Disease, Mouse, Neurodegeneration, Trafficking, Feeding, Hypothalamus, Neurites, Orexin
Introduction
Huntingtin-associated protein-1 (Hap1) was first identified as a cytoplasmic protein that interacts with huntingtin (htt)2 (1). Like htt, which is critical for embryonic development, Hap1 is also essential for animal survival, as deletion of the Hap1 gene in the mouse leads to a feeding defect, retarded growth, and early postnatal death (2, 3). However, unlike htt, which is ubiquitously expressed, Hap1 is enriched in the brain and is abundant in the hypothalamus, and loss of Hap1 leads to hypothalamic degeneration (3). Suppressing Hap1 in the hypothalamus of adult mice via stereotaxic injection of short hairpin RNA also reduces food intake and body weight (4).
Although the above findings suggest an important role for Hap1 in hypothalamic function, how Hap1 regulates this important function remains to be investigated. A growing body of evidence suggests that both htt and Hap1 participate in intracellular trafficking (5–7). Hap1 has been associated with the microtubule-dependent transporters dynactin p150 (8, 9) and kinesin light chain (10), as well as with the transport of various vesicular proteins or membrane receptors, including BDNF-containing vesicles (5), epidermal growth factor receptor (11), type 1 inositol (1,4,5)-triphosphate receptor (12), γ-aminobutyric acid, type A, receptor (13), and tropomyosin-related kinase A receptor tyrosine kinase (TrkA) (14). Importantly, mutant htt can affect Hap1-mediated BDNF trafficking in neuronal cells (5).
Despite all these interesting findings, whether impairment of Hap1-associated intracellular trafficking can cause neurological symptoms is unclear. Given the abundant expression of Hap1 in the hypothalamus, we wanted to investigate whether decreasing Hap1 expression in hypothalamic neurons could alter neuronal intracellular trafficking and animal behavior.
The hypothalamus consists of many cell types that play a critical role in regulating sleep, appetite, and energy homeostasis. In mammals, neurons in the lateral hypothalamic (LH) area are particularly important in these functions (15). The orexin (hypocretin)-producing neurons (orexin neurons) are a small group of neurons located specifically within the LH, and they project widely throughout the entire brain (16–18). Thus, orexin neurons can be readily identified via immunocytochemistry and provide us with an ideal system to examine whether Hap1 deficiency in these neurons can affect neuronal structure and function. To this end, we used the Cre-LoxP system to selectively deplete the expression of Hap1 in orexin neurons. We found that such depletion leads to decreased food intake and locomotor activity in mice. Importantly, we also discovered that a lack of Hap1 can affect neuronal process extension and reduce the distribution of trafficking protein complexes in sucrose density gradient fractions enriched in synaptosomes. These findings are strong in vivo evidence for the critical role Hap1-associated intracellular trafficking plays in maintaining neuronal process integrity and normal animal behavior.
EXPERIMENTAL PROCEDURES
Animals
In an earlier study we generated Hap1 null mice via the conventional knock-out approach (3). To generate conditional Hap1 knock-out mice, exon1 of the mouse Hap1 gene was flanked by two loxP sites to allow the Cre-mediated deletion of exon1. The phosphoglycerate kinase-neomycin resistance cassette was inserted together with the loxP sequence in the 3′-flanking intron of exon1. All loxP sequences were in the same orientation to allow Cre-mediated simultaneous excision of exon1 and the neomycin cassette. The targeting vector was linearized prior to electroporation into SV129-derived embryonic stem cells. Positive embryonic stem cells with correct locus targeting were identified by screening genomic DNA using PCR and confirmed with Southern blotting. These cells were then injected into C57BL/6J blastocysts to generate chimeric mice. Chimeric males were crossed with wild-type female mice (C57BL/6) to generate heterozygous floxed Hap1 (Hap1+/loxp) mice (C57BL/6/SV129), and homozygous mice (Hap1loxp/loxp) were produced by mating of the heterozygous mice. Those mice were crossed with mice (C57BL/6) expressing a Cre recombinase under the control of human prepro-orexin promoter (19) in the orexin neurons. The resulting heterozygous mice were used to generate homozygous conditional knock-out (Orexin-Cre+/Hap1loxp/loxp, referred to as Orexin-Hap1−/−) or heterozygous (Orexin-Cre+/Hap1+/loxp, referred to as Orexin-Hap1+/−) mice. Genotyping of these mice was performed via PCR to amplify the mouse Hap1 DNA fragment (from 3951 to 6957 nucleotides) using the forward (5′-TTTTGGAGGTCTGGTCTCGCTCTG-3′) and reverse (5′-AGGGGCCTGGATGCTGTAAAGG-3′) primers.
Expression analysis of Hap1 and behavioral tests of heterozygous Orexin-Hap1+/− and wild-type mice revealed no differences between these two groups. Because homozygous (Orexin-Hap1−/−) and heterozygous (Orexin-Hap1+/−) mice share the same mixed genetic background, mice of these two genotypes were used mainly to reveal differences related to Hap1 deficiency. All mice were bred and maintained in the animal facility at Emory University in accordance with institutional animal care and use guidelines.
Antibodies
Antibodies against Hap1, pHap1A, Ahi1, KCL, and dynactin p150 were generated in our laboratory and have been described previously (4, 14). Other antibodies used in this study were obtained from commercial sources. Antibodies against EEA1, GM130, and SNAP-25 were purchased from BD Transduction Laboratories (Franklin Lakes, NJ). Orexin-A antibody used in Western blotting and Htt 2166 antibody were from Chemicon International (Temecula, CA). Orexin-A antibody used in immunocytochemistry and BDNF antibody were from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against syntaxin, SVP38, and γ-tubulin were from Sigma. Beclin-1 antibody was from Cell Signaling Technology (Danvers, MA). PSD95 antibody was from Upstate Cell Signaling (Lake Placid, NY) and University of California, Davis/NINDS, National Institutes of Health/NIMH NeuroMab Facility (Davis, CA).
Feeding and Body Weight Measurements
Orexin-Hap1+/− and Orexin-Hap1−/− mice were weighed every 3 days after birth until 100 days old. Daily food and water consumptions were also recorded. More than 20 mice in each group were assayed.
Locomotor Activity
Locomotor activity was assessed using an automated system (San Diego Instruments, La Jolla, CA) with photobeams that recorded ambulations (consecutive beam breaks). Mice were placed individually in the chambers with free access to food and water in a 12-h light (7:00–19:00) and 12-h dark (19:00–7:00) cycle. Mice were allowed to acclimate for 3 h prior to recording. Activity was recorded once per h for 24 h. Fasting mice were given water only during the recording. A total of 18 male and 14 female mice were analyzed.
Immunofluorescence and Microscopy
Immunofluorescent staining of mouse brain sections was performed using the method described previously (20). After mice were anesthetized, brains were removed, cryoprotected in 30% sucrose at 4 °C, and sectioned at 10 μm with a cryostat at −24 °C and mounted onto gelatin-coated slides. The slides were stored at −80 °C until use. The tissues on slides were fixed with ice-cold 4% paraformaldehyde. After washes, the tissues were blocked with a buffer containing 3% bovine serum albumin and PBST (0.3% Triton X-100 in PBS) for 2 h at room temperature. Primary guinea pig antibody against Hap1 and goat antibody against orexin-A were incubated with the tissue at 4 °C overnight. Alexa 488- or rhodamine-conjugated secondary antibodies and nuclear Hoechst dye were then used to finalize the staining. For immunocytochemistry with 3,3′-diaminobenzidine staining, mice were intracardially perfused with 4% paraformaldehyde, and the isolated brains were cut into 40-μm sections with a cryostat (Leica 1920) at −20 °C. Sections were treated with 3% H2O2 for 10 min and then 1% sodium borohydride in PBS for 30 min at room temperature. After sufficient washes (four times for 15 min each time), the sections were blocked by PBST for 1 h and incubated with anti-orexin-A antibody overnight at room temperature. A biotin/avidin immunoassay (Vector Laboratories, Inc., Burlingame, CA) along with DAB staining was performed to visualize orexin neurons and neurites. We examined five wild-type (WT) mice and five homozygous orexin-Hap1 knock-out mutant mice. The hypothalamic sections were isolated from these mouse brains for immunostaining of orexin neurons. Light micrographs of series images were taken using a Zeiss (Oberkochen, Germany) microscope (Axiovert 200 MOT) and a ×20 or ×63 lens (LD-Achroplan ×20/0.4 or 63/0.75) with a digital camera (Orca-100; Hamamatsu Photonics, Bridgewater, NJ) and the Openlab software (Improvision, Lexington, MA). Brain sections of WT and Hap1 mutant mice containing approximately the same hypothalamic region were compared for orexin neuronal staining.
Sucrose Density Gradient Fractionation and Western Blotting
We performed sucrose density gradient fractionation using the method described previously (21, 22). Wild-type or Hap1 null mouse brains were homogenized in a buffer (100 mg/ml) containing 20 mm HEPES, 118 mm NaCl, 4.7 mm KCl, 1.2 mm MgSO4, 2.5 mm MgCl2, 1.53 mm KH2PO4, 212.7 mm glucose, 0.5 mm dithiothreitol, 100 μg/ml phenylmethylsulfonyl fluoride, 1× protease inhibitor mixture (Pierce). Post-nuclear supernatant was prepared by centrifugation of the homogenized brain lysates at 1300 × g for 5 min. A 5–45% (w/v) sucrose gradient was formed in a 12-ml tube containing 5 and 45% sucrose solutions with 10 mm HEPES, pH 7.4, 100 mm KCl, 5 mm MgCl2, 100 μg/ml phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture. One ml of the post-nuclear supernatant was loaded and centrifuged at 35,000 rpm in a Beckman rotor SW 41 for 120 min at 4 °C. A total of 12 fractions of 1 ml each was collected and subjected to SDS-gel electrophoresis and immunoblotting analysis. The samples were separated on a 4–12% Tris-glycine SDS-polyacrylamide gel (Invitrogen). Proteins were transferred onto a nitrocellulose membrane (0.45 μm). The membranes were blocked with 5% nonfat dry milk/PBS for 30 min at room temperature, and the blots were rinsed with 1× PBS for 5 min, with four changes of the 1× PBS, and incubated with primary antibodies in 3% bovine serum albumin/PBS overnight at 4 °C. Secondary horseradish peroxidase-conjugated anti-rabbit or -mouse IgGs were incubated with the blot in 5% milk/PBS for 1 h at room temperature. ECL-Plus (GE Healthcare) was then used to reveal immunoreactive bands on the blots.
Statistical Analysis
All data were expressed as means ± S.E. Statistical results were analyzed by GraphPad Prism (version 5) software, and statistical significance (p < 0.05) was assessed using Student's t test or one-way analysis of variance, followed when appropriate by a post hoc analysis using Dunnett's test.
RESULTS
Conditional Knock-out of Hap1 in Orexin-positive Neurons
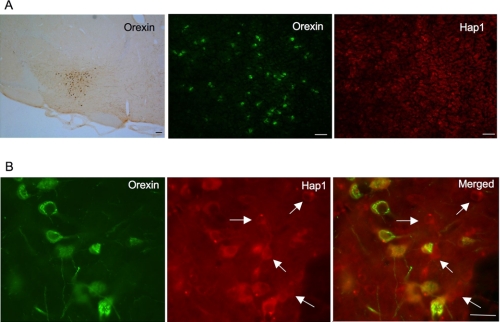
Orexin neurons are a small group of neurons located in the lateral hypothalamic region (Fig. 1A). Using immunofluorescent double labeling, we found that some Hap1-positive neurons also expressed orexin-A. High magnification graphs show that Hap1-positive neurons display cytoplasmic puncta as reported previously (23). Some of these cells show orexin-A staining, although others do not, indicating that a fraction of Hap1 neurons express orexin-A (Fig. 1B).
FIGURE 1.
Orexin-positive neurons express Hap1 in the hypothalamus. Orexin neurons are a small group of cells scattered in the lateral hypothalamic area, where Hap1 is also highly expressed. A, low magnification graphs showing DAB staining of orexin neurons (left panel), immunofluorescent staining of orexin-A (middle panel), and Hap1 (right panel). B, high magnification graphs showing that Hap1 is expressed in all orexin-positive neurons and a subset of non-orexin neurons (arrows). Scale bars, 20 μm in A and 10 μm in B. Male mice at the age of 7–8 months were used for the analysis.
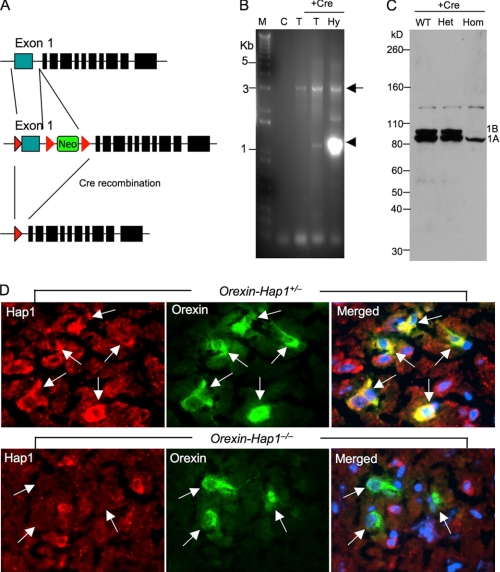
To selectively delete the Hap1 gene in hypothalamic neurons in mice, we first established gene-targeting mice in which exon1 of the Hap1 gene was flanked by loxP sites. The floxed Hap1 mice C57BL/6/SV129 carrying homozygous loxP-flanked Hap1 (Hap1Flox/Flox; Fig. 2A) were mated to Orexin-Cre C57BL/6 mice, which express Cre under the control of the human prepro-orexin promoter in orexin neurons (19). Genotyping of these mice from the tissue of tail or hypothalamus revealed the high efficiency and specificity of Cre recombination in the mouse hypothalamus (Fig. 2B). We then analyzed the expression of Hap1 via Western blotting and found that Hap1 in the hypothalamus of Orexin-Hap1−/− mice is apparently reduced compared with wild-type and Orexin-Hap1+/− mice (Fig. 2C). Because heterozygous Orexin-Hap1+/− mice show the same level of Hap1 as wild-type mice, but also carry the Cre and loxP transgenes, a genetic background similar to homozygous Orexin-Hap1−/− mice, we used Orexin-Hap1+/− mice as a control for comparison with Orexin-Hap1−/− mice to define the specific effects caused by Hap1 deficiency. To confirm the specific activity of the Cre recombination in orexin neurons, we labeled the mouse brain sections with antibodies against orexin-A and Hap1 (Fig. 2D). Hap1 protein was diminished in all orexin-positive neurons but remained in non-orexin cells in homozygous mice (Orexin-Hap1−/−), indicating an efficient and selective deletion of the Hap1 gene in these neurons.
FIGURE 2.
Selective depletion of Hap1 in orexin neurons. A, targeting vector showing exon1 of the mouse Hap1 gene that is flanked by loxP sites and a neomycin marker. After Cre recombination, exon1 of the Hap1 gene is deleted to inactivate it. B, genotyping of 4-month-old Orexin-Hap1+/− male mouse showing PCR products (3951–6957 nucleotides) from the wild-type allele (arrow) and Cre-loxp recombination allele (arrowhead). C, no DNA template; T, mouse tail; Hy, mouse hypothalamus. C, Hap1 Western blotting of the hypothalamic tissues from 6-month-old wild-type (WT), heterozygous (Orexin-Hap1+/−) (het), and homozygous (Orexin-Hap1−/−) (hom) knock-out male mice. Note that Hap1A (1A) is more abundant than Hap1B (1B) in WT and heterozygous knock-out mouse brains and that both Hap1A and Hap1B are reduced in homozygous knock-out mouse brain. D, double immunofluorescence staining showing the depletion of Hap1(red) in orexin neurons (green) (arrows) of the LH in Orexin-Hap1−/− mice. The merged images also show the nuclei (blue) of cells. Scale bar, 10 μm.
Orexin-Hap1−/− Mice Have Reduced Body Weight and Less Food Intake
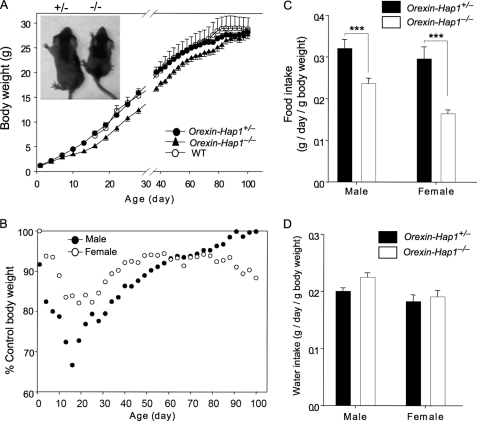
Suppression of Hap1 via adenoviral shRNA in various types of cells in the hypothalamic region can inhibit food intake and reduce body weight (4). Because orexin neurons also regulate food intake (24, 25), we wondered whether selective inhibition of Hap1 in orexin neurons would cause any abnormal feeding behavior. We monitored the body weight of Orexin-Hap1−/−mice along with control (wild-type or Orexin-Hap1+/−) mice from birth until the age of 100 days. At birth all the pups had similar weights, regardless of their genotype or gender (Fig. 3A). However, control mice gained more weight than Orexin-Hap1−/− mice, with the greatest differences seen at the ages of 2–4 weeks (Fig. 3B). For example, male Orexin-Hap1−/− mice had a body weight only 67% of the control mice at the age of 16 days; however, there was no significant difference seen between control and mutant males at the end of recording or at ages over 100 days, suggesting that Hap1 in orexin neurons may be more important for early animal development and growth.
FIGURE 3.
Reduced body weight and food intake in Orexin-Hap1−/− mice. A, Orexin-Hap1−/− mice grew more slowly than the control (WT or heterozygous Orexin-Hap1+/−) mice during the first 100 days. The WT, Orexin-Hap1+/−, and Orexin-Hap1−/− mice were all littermates. B, greatest body weight difference between Orexin-Hap1−/− and Orexin-Hap1+/− mice was seen at the age of 2–4 weeks. Orexin-Hap1−/− mice weighed only 67% of the control mice at day 16 for males and 82% at day 19 for females. C, Orexin-Hap1−/− mice ate less than the control Orexin-Hap1+/− mice (***, p < 0.001). D, two groups (Orexin-Hap1−/− and Orexin-Hap1+/−) of mice consumed similar amounts of water. Twenty mice per group for both males and females were examined.
We also recorded food and water consumption by male and female Orexin-Hap1−/− mice and their age- and sex-matched controls at the ages of 1–3 months. Because there was a body weight difference between the two groups examined, we normalized the daily diet consumption of each mouse by its body weight on the same day and found that there was still a significant reduction of food intake for Orexin-Hap1−/− mice (Fig. 3C). The male and female Orexin-Hap1−/− mice consumed 66 and 50%, respectively, of the food that control mice ate (Fig. 3C); however, the water consumption was similar for these two groups of mice (Fig. 3D). Thus, the elimination of Hap1 in orexin neurons appears to selectively inhibit food intake and lower the body weight of mice at ages <100 days.
Decreased Locomotor Activity in Orexin-Hap1−/− Mice
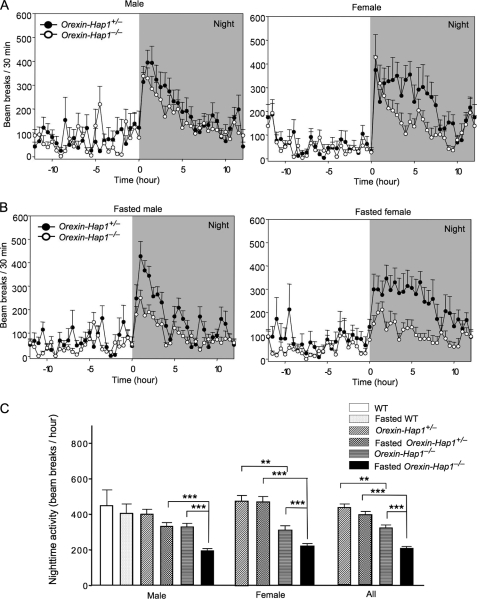
In addition to regulating feeding behavior, orexin neurons also modulate sleep/wakefulness or motor arousal (26). We therefore chose to compare the locomotor activity of Orexin-Hap1−/− mice with control mice. Both control and Orexin-Hap1−/− male mice showed little physical activity during the daytime and arose and became active during the night (Fig. 4A), which is typical rodent arousal behavior. Orexin-Hap1−/− mice, however, showed less activity than the control mice (Orexin-Hap1+/−) at night. This reduced activity was more prominent in female Orexin-Hap1−/− mice than in male Orexin-Hap1−/− mice. Because a fasting-induced reduction in locomotor activity was seen in other mutant mice with orexin neuronal defects (27), we also examined the locomotor activity of fasted mice. Fasting, which causes metabolic stress, led to a greater decrease in the locomotor activity of Orexin-Hap1−/− mice than the control mice, suggesting that Hap1 is also involved in the response of the body to metabolic stress. Again, female Orexin-Hap1−/− mice showed a more marked reduction in locomotor activity than female control mice (Fig. 4B). Statistical analysis of the nighttime activities of the mice also revealed a greater reduction in female Orexin-Hap1−/− mice than in male Orexin-Hap1−/− mice, regardless of their feeding status (Fig. 4C). Thus, orexin neurons with Hap1 deficiency were less able to stimulate locomotor arousal, and this defect was more prominent in female mice. Although the mechanism underlying this sex-dependent phenomenon remains to be investigated, our data suggest a role for Hap1 in orexin neuronal activities and associated animal behaviors.
FIGURE 4.
Decreased locomotor activity of Orexin-Hap1−/− mice. A and B, mouse locomotor activity was monitored and recorded in photobeam chambers for 24 h with normal food access (A) or food deprivation for 24 h (B) during recording. C, statistical analysis showed a significant reduction of nighttime activity for both male and female Orexin-Hap1−/− mice under normal and fasted conditions (**, p < 0.01; ***, p < 0.001). Eighteen male and 14 female mice at the age of 2–5 months were analyzed.
Impaired Orexin Neurite Extension in Orexin-Hap1−/− Mouse Brain
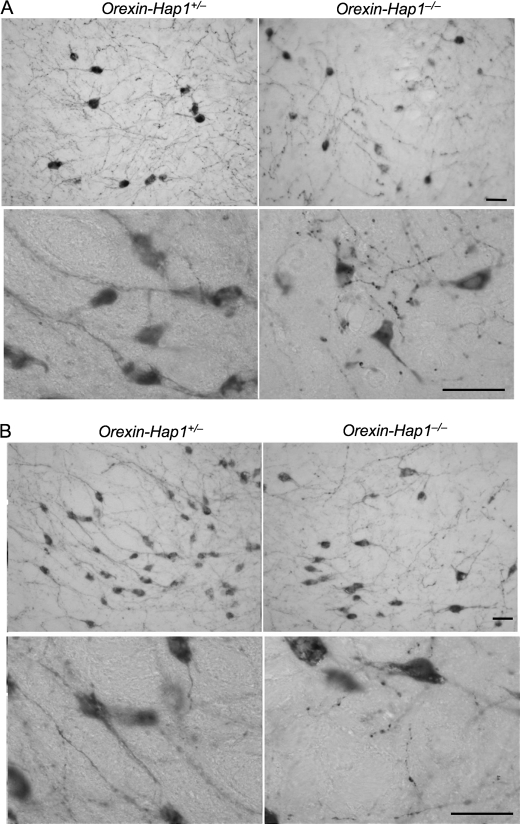
Early studies have shown that suppressing Hap1 expression in cultured PC12 cells can inhibit neurite extension (14); what remains unclear is whether Hap1 deficiency can also affect neuronal processes in the brain. Orexin neurons are a small group of neurons located in the lateral hypothalamic area, making it easy for us to observe the distribution of their neurites. In low power images of immunohistochemical staining of orexin-A, we noticed a higher density of processes of orexin neurons in control mice (Fig. 5). However, orexin neurons in the Orexin-Hap1−/− mouse brain displayed disrupted or fragmented neurites in the dorsomedial hypothalamic (Fig. 5A) and the lateral hypothalamic (Fig. 5B) nuclei, which is more prominent in the high power images. Although it is difficult to quantify neuritic density, we have confirmed this neuritic defect in Hap1-deficient orexin neurons by comparing 30 brain sections from five wild-type and five Hap1-deficient mice. Our finding provides strong in vivo evidence for the necessity of Hap1 in maintaining neuronal processes in the brain.
FIGURE 5.
Neuritic degeneration of orexin neurons in Orexin-Hap1−/− mice. Orexin-A immunohistochemical staining of the dorsomedial hypothalamic nucleus (A) and the lateral hypothalamic nucleus (B) of Orexin-Hap1+/− and Orexin-Hap1−/− mice. Low (×20, upper panels) and high (×65, lower panels) magnification images showing fragmented neurites of orexin neurons in Orexin-Hap1−/− mice. Scale bars, 10 μm.
Altered Distribution of Trafficking Proteins in Sucrose Density Gradient Fractions in Mouse Brain Lacking Hap1
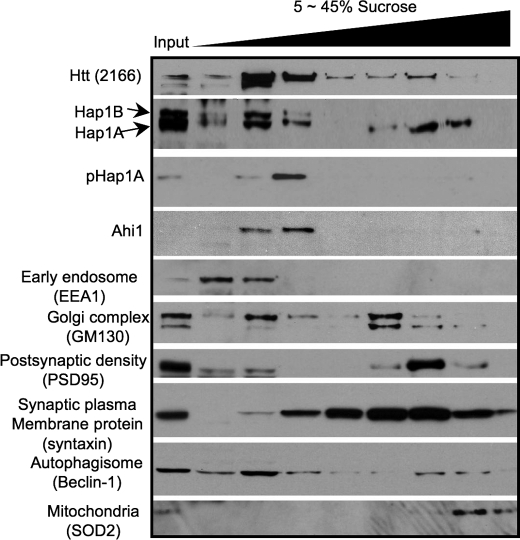
Intracellular trafficking is essential for supplying nutrients, molecules, and organelles to nerve terminals to maintain the normal function and morphology of neuronal processes. Hap1 is known to be involved in intracellular trafficking (6), and its deficiency may alter the trafficking of multiple proteins. Although our early studies have demonstrated the trafficking defect in live cultured primary neurons that lack Hap1 (10), the live cell imaging approach has limitations to reveal the changes of multiple trafficking proteins. For these reasons, we performed sucrose gradient fractionation, which can uncover altered distribution of various proteins in multiple compartments. Adult mouse brain homogenates were separated by centrifugation on sucrose density gradients (5–45%), and fractions were analyzed for the presence of Hap1 and organelle marker proteins. This assay revealed that, in wild-type mouse brains, Hap1 is widely distributed in various fractions but is more enriched in the fractions that contain endosome, Golgi complex, and synaptic vesicle marker proteins (Fig. 6). This distribution fits with the feature of a trafficking protein that associates with cargos for microtubule-dependent transport. Both Hap1 isoforms (Hap1A and Hap1B) are present in the low density fractions, but only Hap1A is present in the high density fractions (Fig. 6). Because synaptosome components syntaxin and PSD95 are distributed mainly in these high density fractions, the high density fractions generated by our procedure are indeed enriched in synaptosomes. The difference in the distribution of Hap1A and Hap1B in sucrose gradient fractions suggests that Hap1A is more likely to associate with synaptic vesicles or other organelles in nerve terminals. Furthermore, phosphorylated Hap1A (pHap1A) is seen in the low density fractions but not high density fractions. These differences are consistent with our early finding that Hap1A is more enriched in the neuronal processes and tips of PC12 cells and also support the hypothesis that dephosphorylated Hap1A is more likely to localize in nerve terminals (14).
FIGURE 6.
Hap1 is involved in different protein complexes. Hap1 isoforms (Hap1A and Hap1B) are distributed in the continuous sucrose density gradient fractions isolated from post-nuclear supernatant of adult wild-type mouse brain. Both Hap1 isoforms are seen in the low density fractions, but only Hap1A is present in the high density fractions. Other organelle marker proteins were also examined, as indicated. Input, post-nuclear supernatant loaded at half the volume of the fractions.
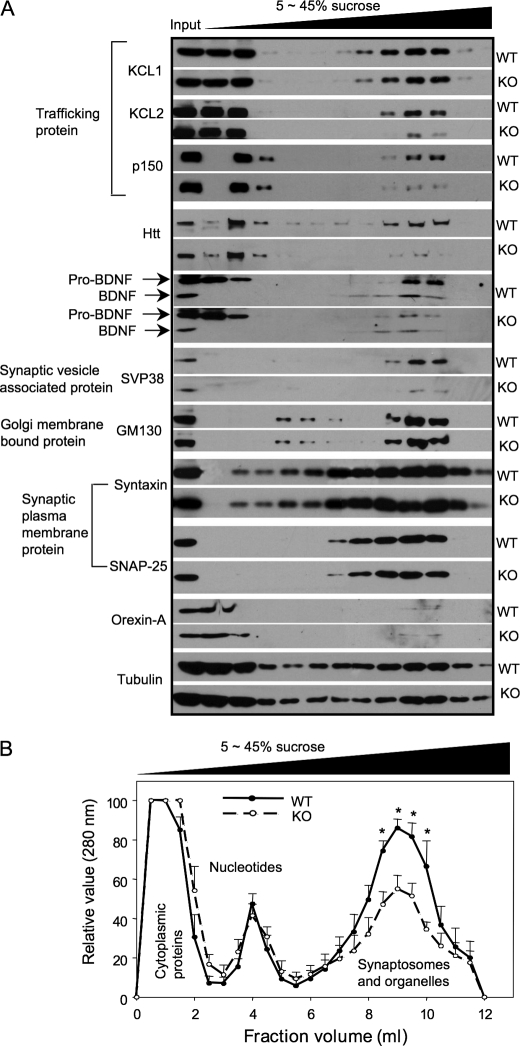
Next, we compared the distribution of a variety of proteins in the sucrose gradient fractions using the brain samples from wild-type and Hap1 null pups at postnatal days 1 and 2. This comparison demonstrates a decrease in the trafficking proteins, KLC and dynectin p150glued (p150), and in the fractions that are enriched in synaptic vesicle proteins (Fig. 7A). KLC1, KLC2, and P150 are microtubule-dependent transporters associated with motor molecules that are responsible for anterograde and retrograde transport, respectively. It seems that proteins that bind directly to Hap1 show a greater decrease in the high density fractions. There is less KLC2 than KLC1 in the high density sucrose fractions. This may be because KLC2 directly binds Hap1 (10) such that lack of Hap1 has more impact on its transport and distribution. Consistently, both htt and P150 are known as Hap1-interacting partners (1, 8, 9), and we also saw a decrease in their levels in the high density sucrose fractions. SVP38, a protein that associates with synaptic vesicles, is also decreased in fractions containing synaptosomes from Hap1 null mouse brain lysate. Consistent with the earlier finding that Hap1 is involved in the transport of BDNF vesicles (5), loss of Hap1 reduces the distribution of BDNF and its precursor, pro-BDNF, in high density fractions that contain synaptosomes in nerve terminals. However, the synaptic plasma membrane proteins, syntaxin and SNAP-25, were unchanged in the high density gradient fractions of Hap1 null mouse brain (Fig. 7A). In addition, orexin-A levels and distribution were not different between WT and Hap1 knock-out samples, perhaps because orexin is not transported via Hap1-associated transporters. These differences suggest that only trafficking proteins associated with Hap1 are reduced in the synaptosome fractions, perhaps because the lack of Hap1 could not facilitate their transport or stabilize them during their trafficking. Consistently, GM130, Golgi membrane-bound protein, and tubulin showed no change in their distribution in the fractions from Hap1 null mouse brain lysate.
FIGURE 7.
Loss of Hap1 alters the distribution of trafficking proteins and cargos. Brain samples from WT and Hap1 null (knock-out (KO)) mice at postnatal days 1 and 2 were analyzed. A, distribution of microtubule-dependent trafficking proteins (KCL, p150), htt, synaptic vesicle proteins, and tubulin in 5–45% sucrose gradient fractions. Note that in Hap1 null brain samples there is a decrease in the distribution of KLC2, p150, htt, BDNF, and SVP38 in the high density fractions, which are enriched in synaptic plasma membrane proteins. B, quantification analysis of the amounts of proteins in the fractions isolated via sucrose gradient density centrifugation. The data are presented as mean ± S.E. (n = 5). *, p < 0.05.
The above finding suggests that Hap1 may form protein complexes with the trafficking proteins KLC and dynactin p150 to facilitate their distribution in neuritic processes and nerve terminals. Because these trafficking proteins are involved in the transport of a variety of cargos or proteins, it is possible that a lack of Hap1 can reduce the amount of various proteins in the synaptosomal fractions. To test this idea, we measured the protein content of different sucrose gradient fractions from wild-type and Hap1 null mouse brains. Although protein contents in the cytoplasmic and nucleotide fractions showed no differences between WT and Hap1 null samples, there was a decreased amount of protein in the synaptosome and organelle fractions from Hap1 null mouse brain (Fig. 7B). Combined with the decreased trafficking proteins seen in the high density fractions enriched in synaptosomes when Hap1 is lost, it is likely that Hap1 deficiency reduces the intracellular trafficking of multiple proteins or cargos to nerve terminals, which might affect the neuronal process integrity and neuronal interactions.
DISCUSSION
Early studies using cellular models have demonstrated that Hap1 and htt are involved in intracellular trafficking (5); however, the in vivo role Hap1-associated trafficking plays in neurological phenotypes remains unknown. By selectively eliminating Hap1 expression in orexin neurons in the mouse brain, particularly in a subset of hypothalamic neurons, we identified a disruption of neuronal processes of orexin neurons, as well as decreased feeding and locomotor behavior in mice. Furthermore, we found that the loss of Hap1 can diminish the distribution of trafficking proteins in synaptosomal fractions. These findings demonstrate for the first time that impaired Hap1-associated intracellular trafficking, even in a subset of neurons, can lead to neurological symptoms, suggesting that intracellular trafficking dysfunction contributes to the neuropathological conditions related to Hap1 functional deficiency.
Although Hap1 knock-out mice have shown that Hap1 plays an essential role in early development and animal survival (2, 3), these mice die postnatally, precluding studies of Hap1 deficiency in adult mice. Nevertheless, these early studies indicate an important role of Hap1 in feeding behavior, as Hap1 knock-out mice show retarded growth and feeding defects. Other important evidence to support the critical role of Hap1 in feeding also comes from the facts that Hap1 is abundantly expressed in hypothalamic neurons and that loss of Hap1 can lead to hypothalamic neuronal degeneration (3). Furthermore, suppressing Hap1 via shRNA in adult mouse hypothalamus can inhibit feeding behavior (4). These early findings prompted us to focus on the function of Hap1 in hypothalamic neurons.
The hypothalamus is a central region that regulates feeding, arousal, and many other fundamental activities in vertebrates. Orexin (hypocretin)-producing neurons are specifically localized within and around the LH area and dorsomedial hypothalamic nucleus. These neurons were identified as regulators of feeding behavior and states of sleep/wakefulness (28–30). It is worth noting that every orexin neuron expresses Hap1, but only a subset of Hap1-expressing neurons produces orexin. Despite this, eliminating Hap1 in orexin neurons could alter the feeding behavior and locomotor activity of mice, suggesting that Hap1 is indeed important for the function of orexin-producing neurons, which can regulate animal behavior.
Orexin neurons release orexins, which serve as neurotransmitters and ligands for two orphan G protein-coupled receptors in other neurons associated with diverse cognitive and physiological functions (31). Initial studies suggest that orexins have a major role in the regulation of sleep/wake transitions. More recent studies indicate orexins also regulate food intake, addiction, and stress (24, 25). Although the sucrose gradient fractionation (Fig. 7) did not show altered orexin level in Hap1 knock-out mouse brain, it remains to be tested whether the released amount of orexin is reduced in Hap1-deficient orexin neurons. However, given neuritic fragmentation of orexin neurons and the importance of orexin neuronal connections with a wide range of neuronal cells, the degeneration of orexin neuronal processes is likely to contribute to the phenotypes seen in our conditional Hap1 knock-out mice. It has been found that the number of orexin neurons and expression of orexin in the mouse hypothalamus are increased from the postnatal period to maturation (32, 33). Also, feeding response to orexin A was seen in young, but not old, rats (34, 35). All these findings suggest that the development of orexin neurons is associated with energy balance and feeding in young animals. In support of this idea, Hap1 deficiency decreases the body weight of young mice before the age of 100 days.
Orexin neurons function through their connection with a variety of different types of neurons in the hypothalamus, which forms the circuitry to regulate body behavior, sleep/wakefulness, and feeding (30, 31). This circuitry is dependent on the long processes of orexin neurons, which interact with various types of cells. Thus, the integrity of these neuronal processes is critical for the normal function of orexin neurons. Intracellular trafficking is particularly important for maintaining the integrity of neuronal processes. This is because the nerve terminal function relies on the transport of molecules and cargos from the cell body to the axons and dendrites. The long range transport in axons is particularly vulnerable to many insults and injuries. Accordingly, disturbances to the transport of cargos through axons can lead to a number of neurological disorders, such as Alzheimer disease, amyotrophic lateral sclerosis, and Huntington disease (31, 41). Deletion of Hap1 could disrupt neuronal trafficking in the orexin neurons, which in turn may affect the formation of neurites during development or the neuritic integrity of mature neurons, leading to the behavioral phenotypes seen in our orexin neuronal Hap1 knock-out mice.
Although the selective elimination of Hap1 in orexin neurons allowed us to observe the association between the degeneration or dysfunction of these neurons and neurological symptoms, the depletion of Hap1 in this limited number of neurons makes it difficult to use biochemical assays to investigate the underlying mechanism. Hap1 null mouse brains offered us sufficient material to perform sucrose gradient fractionation to uncover any changes in the distribution of trafficking proteins. As Hap1 is involved in the trafficking of vesicles and different membrane receptors (5, 6), it is likely that Hap1 deficiency can cause a broader defect on the trafficking of various proteins in neurons. Our findings demonstrate that loss of Hap1 can reduce the distribution of its interacting proteins, p150, KLC, and htt, in the high density fractions that are enriched in synaptosomal proteins. As a result, the amount of proteins in the synaptosomal fractions is also reduced. Although this broader effect is perhaps unsurprising, as Hap1 binds to different microtubule transporters (KLC, p150 dynactin) and htt, which can also interact with other trafficking proteins (7), it does provide in vivo evidence that Hap1 is involved in the formation of various protein complexes for transporting different molecules and cargos. However, it remains to be investigated how Hap1 deficiency affects intracellular trafficking and how this trafficking defect can cause the degeneration of neuronal processes.
In this study, we focused on Hap1 function in orexin neurons and the resultant mouse behavior. However, our findings also have implications for other pathological conditions associated with Hap1 dysfunction. First, because mutant htt binds tightly to Hap1 (1), mutant htt can affect the function of Hap1 and its associated intracellular trafficking. Early studies also demonstrated that mutant htt affects intracellular trafficking (36, 37) and Hap1-dependent BDNF transport in neuronal cells (5). Because of the altered distribution of multiple trafficking proteins in the synaptosome-enriched fractions from Hap1 null mouse brains, it is likely that the trafficking of a number of important molecules may also be impaired in Huntington disease neuronal cells. Second, Hap1 is found to tightly associate with Ahi1, a protein encoded by the Abelson helper integration site 1 (AHI1) gene, and stabilizes the level of Ahi1 (38). Mutations of the Ahi1 gene can cause the autosomal recessive Joubert syndrome, a brain development disorder (39, 40). Recent studies suggest that Ahi1 may also be involved in intracellular trafficking (41). Thus, the defect in Hap1-mediated intracellular trafficking could also contribute to the abnormal early brain development and neurological symptoms of Joubert syndrome. Understanding how Hap1-associated intracellular trafficking causes neuropathology in these and possibly other diseases will help us find effective therapeutics and treatments.
Acknowledgments
We thank Drs. Takeshi Sakurai and Taizo Matsuki, University of Tsukuba, Japan, for providing the Orexin-Cre transgenic mice. We also thank Drs. David Weinshenker and Andrew Escayg and their colleagues for help with the animal behavioral studies and Dr. Stephen Warren and colleagues for assistance with the sucrose gradient fractionation study.
This work was supported, in whole or in part, by National Institutes of Health Grants NS036232 (to X.-J. L.) and NS045016 (to S. L.).
- htt
- huntingtin
- KLC
- kinesin light chain
- LH
- lateral hypothalamic nucleus
- PBS
- phosphate-buffered saline
- BDNF
- brain-derived neurotrophic factor
- WT
- wild type.
REFERENCES
- 1.Li X. J., Li S. H., Sharp A. H., Nucifora F. C., Jr., Schilling G., Lanahan A., Worley P., Snyder S. H., Ross C. A. (1995) Nature 378, 398–402 [DOI] [PubMed] [Google Scholar]
- 2.Chan E. Y., Nasir J., Gutekunst C. A., Coleman S., Maclean A., Maas A., Metzler M., Gertsenstein M., Ross C. A., Nagy A., Hayden M. R. (2002) Hum. Mol. Genet. 11, 945–959 [DOI] [PubMed] [Google Scholar]
- 3.Li S. H., Yu Z. X., Li C. L., Nguyen H. P., Zhou Y. X., Deng C., Li X. J. (2003) J. Neurosci. 23, 6956–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheng G., Chang G. Q., Lin J. Y., Yu Z. X., Fang Z. H., Rong J., Lipton S. A., Li S. H., Tong G., Leibowitz S. F., Li X. J. (2006) Nat. Med. 12, 526–533 [DOI] [PubMed] [Google Scholar]
- 5.Gauthier L. R., Charrin B. C., Borrell-Pagès M., Dompierre J. P., Rangone H., Cordelières F. P., De Mey J., MacDonald M. E., Lessmann V., Humbert S., Saudou F. (2004) Cell 118, 127–138 [DOI] [PubMed] [Google Scholar]
- 6.Li X. J., Li S. H. (2005) Trends Pharmacol. Sci. 26, 1–3 [DOI] [PubMed] [Google Scholar]
- 7.Caviston J. P., Holzbaur E. L. (2009) Trends Cell Biol. 19, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelender S., Sharp A. H., Colomer V., Tokito M. K., Lanahan A., Worley P., Holzbaur E. L., Ross C. A. (1997) Hum. Mol. Genet. 6, 2205–2212 [DOI] [PubMed] [Google Scholar]
- 9.Li S. H., Gutekunst C. A., Hersch S. M., Li X. J. (1998) J. Neurosci. 18, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGuire J. R., Rong J., Li S. H., Li X. J. (2006) J. Biol. Chem. 281, 3552–3559 [DOI] [PubMed] [Google Scholar]
- 11.Li Y., Chin L. S., Levey A. I., Li L. (2002) J. Biol. Chem. 277, 28212–28221 [DOI] [PubMed] [Google Scholar]
- 12.Tang T. S., Tu H., Chan E. Y., Maximov A., Wang Z., Wellington C. L., Hayden M. R., Bezprozvanny I. (2003) Neuron 39, 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kittler J. T., Thomas P., Tretter V., Bogdanov Y. D., Haucke V., Smart T. G., Moss S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rong J., McGuire J. R., Fang Z. H., Sheng G., Shin J. Y., Li S. H., Li X. J. (2006) J. Neurosci. 26, 6019–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellinger L. L., Bernardis L. L. (2002) Physiol. Behav. 76, 431–442 [DOI] [PubMed] [Google Scholar]
- 16.Peyron C., Tighe D. K., van den Pol A. N., de Lecea L., Heller H. C., Sutcliffe J. G., Kilduff T. S. (1998) J. Neurosci. 18, 9996–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nambu T., Sakurai T., Mizukami K., Hosoya Y., Yanagisawa M., Goto K. (1999) Brain Res. 827, 243–260 [DOI] [PubMed] [Google Scholar]
- 18.Date Y., Ueta Y., Yamashita H., Yamaguchi H., Matsukura S., Kangawa K., Sakurai T., Yanagisawa M., Nakazato M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuki T., Nomiyama M., Takahira H., Hirashima N., Kunita S., Takahashi S., Yagami K., Kilduff T. S., Bettler B., Yanagisawa M., Sakurai T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C. E., Tydlacka S., Orr A. L., Yang S. H., Graham R. K., Hayden M. R., Li S., Chan A. W., Li X. J. (2008) Hum. Mol. Genet. 17, 2738–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uldry M., Steiner P., Zurich M. G., Béguin P., Hirling H., Dolci W., Thorens B. (2004) EMBO J. 23, 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muddashetty R. S., Keliæ S., Gross C., Xu M., Bassell G. J. (2007) J. Neurosci. 27, 5338–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutekunst C. A., Li S. H., Yi H., Ferrante R. J., Li X. J., Hersch S. M. (1998) J. Neurosci. 18, 7674–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koob G. F. (2008) Neuron 59, 11–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter M. E., Borg J. S., de Lecea L. (2009) Curr. Opin. Pharmacol. 9, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willie J. T., Chemelli R. M., Sinton C. M., Yanagisawa M. (2001) Annu. Rev. Neurosci. 24, 429–458 [DOI] [PubMed] [Google Scholar]
- 27.Yoshida G., Li M. X., Horiuchi M., Nakagawa S., Sakata M., Kuchiiwa S., Kuchiiwa T., Jalil M. A., Begum L., Lu Y. B., Iijima M., Hanada T., Nakazato M., Huang Z. L., Eguchi N., Kobayashi K., Saheki T. (2006) Neurosci. Res. 55, 78–86 [DOI] [PubMed] [Google Scholar]
- 28.Chemelli R. M., Willie J. T., Sinton C. M., Elmquist J. K., Scammell T., Lee C., Richardson J. A., Williams S. C., Xiong Y., Kisanuki Y., Fitch T. E., Nakazato M., Hammer R. E., Saper C. B., Yanagisawa M. (1999) Cell 98, 437–451 [DOI] [PubMed] [Google Scholar]
- 29.Lin L., Faraco J., Li R., Kadotani H., Rogers W., Lin X., Qiu X., de Jong P. J., Nishino S., Mignot E. (1999) Cell 98, 365–376 [DOI] [PubMed] [Google Scholar]
- 30.Sakurai T. (2007) Nat. Rev. Neurosci. 8, 171–181 [DOI] [PubMed] [Google Scholar]
- 31.Kirouac G. J., Parsons M. P., Li S. (2005) Brain Res. 1059, 179–188 [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto Y., Ueta Y., Hara Y., Serino R., Nomura M., Shibuya I., Shirahata A., Yamashita H. (2000) Brain Res. Mol. Brain Res. 78, 108–119 [DOI] [PubMed] [Google Scholar]
- 33.Sawai N., Ueta Y., Nakazato M., Ozawa H. (2010) Neurosci Lett. 468, 51–55 [DOI] [PubMed] [Google Scholar]
- 34.Akimoto-Takano S., Sakurai C., Kanai S., Hosoya H., Ohta M., Miyasaka K. (2005) Neuroendocrinology 82, 256–263 [DOI] [PubMed] [Google Scholar]
- 35.Kotz C. M., Mullett M. A., Wang C. (2005) Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R359–R366 [DOI] [PubMed] [Google Scholar]
- 36.Gunawardena S., Her L. S., Brusch R. G., Laymon R. A., Niesman I. R., Gordesky-Gold B., Sintasath L., Bonini N. M., Goldstein L. S. (2003) Neuron 40, 25–40 [DOI] [PubMed] [Google Scholar]
- 37.Morfini G. A., You Y. M., Pollema S. L., Kaminska A., Liu K., Yoshioka K., Björkblom B., Coffey E. T., Bagnato C., Han D., Huang C. F., Banker G., Pigino G., Brady S. T. (2009) Nat. Neurosci. 12, 864–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheng G., Xu X., Lin Y. F., Wang C. E., Rong J., Cheng D., Peng J., Jiang X., Li S. H., Li X. J. (2008) J. Clin. Invest. 118, 2785–2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon-Salazar T., Silhavy J. L., Marsh S. E., Louie C. M., Scott L. C., Gururaj A., Al-Gazali L., Al-Tawari A. A., Kayserili H., Sztriha L., Gleeson J. G. (2004) Am. J. Hum. Genet. 75, 979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferland R. J., Eyaid W., Collura R. V., Tully L. D., Hill R. S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., Shugart Y. Y., Ruvolo M., Walsh C. A. (2004) Nat. Genet. 36, 1008–1013 [DOI] [PubMed] [Google Scholar]
- 41.Hsiao Y. C., Tong Z. J., Westfall J. E., Ault J. G., Page-McCaw P. S., Ferland R. J. (2009) Hum. Mol. Genet. 18, 3926–3941 [DOI] [PMC free article] [PubMed] [Google Scholar]