Abstract
The netrin-like (NTR) domain is a feature of several extracellular proteins, most notably the N-terminal domain of tissue inhibitors of metalloproteinases (TIMPs), where it functions as a strong inhibitor of matrix metalloproteinases and some other members of the metzincin superfamily. The presence of a C-terminal NTR domain in procollagen C-proteinase enhancers (PCPEs), proteins that stimulate the activity of astacin-like tolloid proteinases, raises the possibility that this might also have inhibitory activity. Here we show that both long and short forms of the PCPE-1 NTR domain, the latter beginning at the N-terminal cysteine known to be critical for TIMP activity, show no inhibition, at micromolar concentrations, of several members of the metzincin superfamily, including matrix metalloproteinase-2, bone morphogenetic protein-1 (a tolloid proteinase), and different ADAMTS (a disintegrin and a metalloproteinase with thrombospondin motifs) proteinases from the adamalysin family. In contrast, we report that the NTR domain within PCPE-1 leads to superstimulation of bone morphogenetic protein-1 activity in the presence of heparin and heparan sulfate. These observations point to a new mechanism whereby binding to cell surface-associated or extracellular heparin-like sulfated glycosaminoglycans might provide a means to accelerate procollagen processing in specific cellular and extracellular microenvironments.
Keywords: ADAM, ADAMTS, Collagen, Extracellular Matrix, Heparin, Matrix Metalloproteinase, Tolloid Proteinase
Introduction
It is now well established that different associations of modules in multidomain proteins can provide increased diversity and specificity of function in many areas of cell biology, including receptor signaling, enzyme (particularly proteinase) activity, and extracellular matrix function. The NTR6 domain (1), found solely in extracellular proteins, occurs in TIMPs (2), agrin (3, 4), netrins (5), complement proteins C3/C4/C5 (6), secreted frizzled-related proteins (7), growth and differentiation factor binding proteins (8), ADAMTSL-5 (9), and procollagen C-proteinase enhancers (PCPE-1 and -2) (10, 11).
In the TIMPs, the NTR module corresponds to the N-terminal domain, which forms a tight 1:1 complex with the active site of MMPs, thereby inhibiting their activity (12, 13). Within such complexes, the TIMP N-terminal cysteine residue plays a key role in expelling the catalytic water molecule bound to the zinc atom in the MMP active site (12, 13), such that addition of an alanine residue at the N terminus abolishes TIMP inhibition (14). The identification of NTR domains in proteins other than TIMPs prompted the suggestion that these proteins might also be involved in the regulation of metalloproteinase activity (1). In agrin (4), the NTR domain (also called NtA) also occurs at the N terminus of the molecule, and its three-dimensional structure (an OB-fold) is strikingly similar to that of TIMP-1 (3). In contrast, however, the first residue in agrin, just before the TIMP-like cysteine, is an asparagine, and there have been no reports of TIMP-like inhibitory activity of the agrin NTR domain.
In all of the other NTR domain-containing proteins, this domain occurs in the C-terminal region, suggesting that proteolytic release of the NTR domain would be a prerequisite for possible TIMP-like activity. One such protein is PCPE-1, an ECM protein that is devoid of intrinsic proteolytic activity but that stimulates the action of tolloid proteinases in a substrate-specific manner (15, 16). PCPE-1 and PCPE-2 consist of two CUB (module found in complement subcomponents Clr/Cls, Uegf, and BMP-1) domains separated from a C-terminal NTR domain by a protease-sensitive linker. Some years ago, NTR domain-containing fragments from PCPE-1 with apparent TIMP-like activity were identified in the culture medium of brain tumor cell lines (17). Unlike TIMP-2, however, which inhibited MMP-2 cleavage of a quenched fluorescent peptide with an IC50 value of 1.6 nm, these fragments appeared to be rather weak MMP-2 inhibitors (IC50 = ∼500 nm) (17). Since then, the three-dimensional structure of the PCPE-1 NTR domain was solved (10), but no evidence of inhibitory activity was obtained, against a range of metalloproteinases including MMPs-1, -2, -3, and -9, and tumor necrosis factor-α converting enzyme (TACE/ADAM-17), as well as the serine proteinases trypsin, plasmin, kallikrein, and thrombin. It should be noted, however, that both the fragments studied by Mott et al. (17) and the constructions of Liepinsh et al. (10) included several residues N-terminal to the TIMP-like terminal cysteine, which might be expected to interfere with inhibitory activity. Thus the role of the NTR domain in PCPE-1 remains unclear.
Information on the roles of the NTR domains in the other NTR domain-containing proteins is limited. There have been no reports of proteinase inhibitory activity in the NTR domains of netrins, complement proteins C3/C4/C5, or the growth and differentiation factor binding proteins WFIKKN1 and WFIKKN2. In contrast, this domain is frequently implicated in binding to heparin or related macromolecules. TIMP-3, for example, binds to sulfated glycosaminoglycans of the ECM (18) or to other sulfated oligosaccharides (19), interactions that enhance its inhibitory activity against MMP-2 (20), ADAMTS-2 (21), ADAMTS-4 (19, 22), and ADAMTS-5 (19). Similarly, PCPE-1 binds to heparin (23) via its NTR domain, as does netrin-1 to sulfated glycosaminoglycans of the ECM (24).
Here we show that the NTR domain of PCPE-1, in both long and short forms, is not an inhibitor of representative members of the metzincin superfamily, including MMPs, ADAMTSs, and the tolloid proteinase BMP-1. On the other hand, this domain is involved in relatively weak binding to MMP-2 and to BMP-1. Strikingly, in the presence of both PCPE-1 and heparin or heparan sulfate, we found that the activity of BMP-1 is further stimulated, an effect requiring the presence of the NTR domain, with implications for cell surface and ECM control of procollagen processing.
EXPERIMENTAL PROCEDURES
Reagents
Unless otherwise stated, all of the reagents were obtained from Sigma-Aldrich, including the two forms of heparin (H-3149 and H-3393), both with a molecular mass of 17–19 kDa (according to the manufacturer), from porcine intestinal mucosa. In addition, heparin and heparan sulfate were obtained from Iduron (Manchester, UK), also from porcine mucosa. Acid-soluble rat tail type I collagen was from BD Biosciences and Brij 35 (oxidant-free) from Uptima.
Recombinant Proteins
Recombinant human BMP-1, BMP-1-FLAG, Myc-tagged mini-procollagen III, PCPE-1, PCPE-1His, and its CUB2-NTR fragment were expressed in 293-EBNA cells and purified as described (16, 25, 26). For some experiments, recombinant PCPE-1 and the C-propeptide trimer of procollagen III were produced in insect cells (baculovirus system), as also described (23, 27). In addition to BMP-1, other recombinant human proteinases were produced as follows: MMP-2 in NSO cells (28) and ADAMTS-2, ADAMTS-4, and ADAMTS-5 in 293-EBNA cells (29–31). In all cases, the protein concentrations were determined by the Bradford assay (Pierce).
Production of NTRs and NTRt
By homology with the TIMPs, the NTR domain of human PCPE-1 begins with the cysteine residue at position 318 in the full-length protein (including the signal sequence; see Fig. 1) (1). To produce this fragment (called NTRshort or NTRs), a construct coding for the human PCPE-1 signal sequence followed immediately by the NTR domain was produced by PCR, using the QuikChange XL site-directed mutagenesis kit (Stratagene), followed by ligation with T4 DNA ligase (rapid DNA ligation kit; Fermentas), from a pCEP4 vector containing the full-length coding region (plus an 8-histidine tag) (25), using the following primers: 5′-GCCCTGGGCAAAAGGCAGCAG-3′ (antisense, 3′ extremity of signal peptide) and 5′-TGCCCAAAGCAGTGCCGCCGG-3′ (sense, 5′ extremity of NTR domain). After sequence verification (Genome Express, Grenoble, France), HEK 293-EBNA cells were transfected as described (16), and conditioned medium was collected every 2 days for up to 3 weeks. NTRs was then purified from the medium by nickel affinity chromatography (nickel-nitrilotriacetic acid; Qiagen), as described (25), followed by heparin affinity chromatography (Heparin Sepharose 6 Fast Flow; GE Healthcare). Briefly, fractions from the nickel-nitrilotriacetic acid column were diluted 3× in 20 mm HEPES, pH 7.4, then loaded onto the heparin column, and eluted with a 0.15 to 0.75 m NaCl gradient. Pooled fractions were then concentrated using Amicon Ultra centrifugal filter devices (molecular mass cut-off, 10 kDa). For the preparation of the other form of the NTR domain used in this study, called NTRtrypsin or NTRt, containing the entire NTR domain plus part of the CUB2-NTR linker region, full-length PCPE-1His or CUB2-NTR was subjected to limited proteolysis with immobilized trypsin, and the released NTRt was purified as described (26). It should be noted that trypsin treatment removes the His tag at the C terminus of both PCPE-1His and CUB2-NTR.
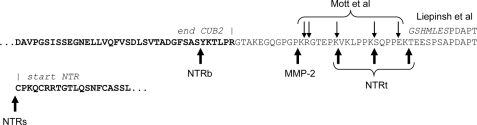
FIGURE 1.
Amino acid sequence of human PCPE-1 in the region of the CUB2-NTR linker, showing the start positions of the NTR containing fragments previously identified by Mott et al. (17) and the N-terminal sequence of the construct used by Liepinsh et al. (10), as well as the MMP-2 cleavage site determined here and the start positions of the different forms of NTR used in this study.
Circular Dichroism Spectroscopy
To evaluate the folding of NTRs and NTRt, far UV (180–260 nm) circular dichroism measurements were carried out using thermostatted 0.2-mm-path length quartz cells in an Applied Photophysics Chirascan instrument, calibrated with aqueous d-10-camphorsulfonic acid. Proteins (∼ 0.4 mg/ml) were analyzed at 25 or 65 °C in 20 mm Tris-H2SO4, 150 mm NaF, pH 7.4. The spectra were measured with a wavelength increment of 0.5 nm, an integration time of 8 s, and a bandpass of 1 nm.
Cleavage of PCPE-1 by MMP-2
To determine the MMP-2 cleavage sites in PCPE-1, recombinant MMP-2 was first autoactivated by preincubation for 15 min at 37 °C and then incubated (at a final concentration of 150 nm) for 8 h at 37 °C with PCPE-1His (1.45 μm) in 25 mm Tris-HCl, pH 7.5, 0.15 m NaCl, 5 mm CaCl2 in a final volume of 50 μl. The reaction products were separated by 12% SDS-PAGE (under reducing conditions) and then transferred to a polyvinylidene difluoride membrane. After Coomassie Blue detection, N-terminal sequencing was carried out at the common sequencing center of IFR128 Biosciences (Lyon, France).
Effect of PCPE-1 NTR on MMP Activity
Potential inhibitory activity of the insect cell-derived NTR fragment was assayed with purified MMP-1 (collagen substrate), MMP-2 (gelatin substrate), and MMP-3 and MMP-7 (both casein substrates), as described (32). Potential inhibition of MMP-2 activity by NTRs and NTRt fragments was also followed using the quenched fluorescent substrate Mca-PLGL-Dpa-AR-NH2 (Calbiochem), as described (33). The reactions were carried out in microtiter plates in a total volume of 100 μl. Briefly, MMP-2 (final concentration, 17 nm) was first activated by preincubation for 10 min at 37 °C in buffer A (100 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm CaCl2, 0.02% octylglucoside). NTRs or NTRt was then added to a final concentration of 1 μm (as well as 0.25 mm Pefabloc in the case of NTRt, to inhibit potential residual trypsin activity), and the reactions were initiated by the addition of substrate to a final concentration of 1.14 μm. Enzyme activity was measured, using a TECAN spectrofluorimeter, from the increase in fluorescence over the first 5 min of the reaction. Excitation and emission wavelengths were set at 328 and 390 nm, respectively.
The effects of NTRs and NTRt on MMP-2 cleavage of denatured collagen were also tested using acid-soluble rat tail tendon collagen I, after denaturation for 30 min at 60 °C. The reactions were carried out in buffer A for 1 h at 37 °C using 0.37 μm collagen, 17 nm MMP-2, and up to 2 μm NTRs or NTRt and then analyzed by SDS-PAGE (12% acrylamide in the separating gel) in the presence of 20 mm dithiothreitol. The gels were stained with Coomassie Blue.
Effect of PCPE-1 NTR on BMP-1 Activity
To determine the effects of the different forms of PCPE-1 NTR on the activity of BMP-1, fluorescently labeled mini-procollagen III substrate (26) (150 nm) was incubated with 12 nm BMP-1-FLAG and 1 μm NTR (with 0.25 mm Pefabloc in the case of NTRt) in 50 mm HEPES, pH 7.4, 0.15 m NaCl, 5 mm CaCl2, 0.02% Brij-35 for 2 h at 37 °C (final volume, 20 μl). The reactions were stopped by the addition of 10 μl of 3× Laemmli sample buffer (without bromphenol blue), followed by heating at 100 °C for 5 min and analysis by discontinuous SDS-PAGE (7.5% acrylamide in the separating gel). Quantitation was done on a Molecular Dynamics Storm 860 imager using the red laser (635 nm).
Activity was also analyzed using the quenched fluorescent substrate Mca-YVADAPK(Dnp)-OH (Bachem) in microtiter plates at a concentration of 20 μm with 0–2 μm NTR (and 0.25 mm Pefabloc in the case of NTRt) in 50 mm Tris-HCl, pH 7.4, 0.15 m NaCl, 5 mm CaCl2, 0.02% octylglucoside. BMP-1-FLAG was then added to a final concentration of 36 nm, in a total volume of 100 μl, and subsequent changes in fluorescence were monitored at 405 nm (excitation at 330 nm) in a TECAN spectrofluorimeter for 30 min at 37 °C.
Effects of PCPE-1 NTR on ADAMTS-2 Activity
ADAMTS-2 activity was assayed by release of the N-propeptides from a 14C-labeled pN-collagen I substrate, as described (34). Briefly, substrate (240 nm) was incubated with recombinant ADAMTS-2 (2 nm) for 18 h at 26 °C in 50 mm Tris, pH 7.5, 500 mm NaCl, 2 mm CaCl2, 2.5 mm N-ethylmaleimide, 0.5 mm phenylmethylsulfonyl fluoride, 0.02% Brij 35 (final volume, 125 μl) in the presence of increasing concentrations of NTR (0–2.5 μm). The amount of N-propeptide released was measured by scintillation counting of the supernatant after ethanol precipitation of collagen product and uncleaved substrate. The measured activities were corrected for nonspecific activity by the subtracting values for control assays carried out in the presence of 50 mm EDTA.
Effects of PCPE-1 NTR on the Activities of ADAMTS-4 and ADAMTS-5
Activities of ADAMTS-4 (form 2, without the spacer domain (30)) and ADAMTS-5 (form 4; without the cysteine-rich, spacer and second thrombospondin domains (31)) were assayed using the quenched fluorescent substrate FAM-AELQGRPISIAK-TAMRA, as described (22). Briefly, protease (ADAMTS-4 or ADAMTS-5, 1 nm) was preincubated for 2 h at 37 °C with different concentrations of NTR fragment in 50 mm Tris-HCl, pH 7.5, 0.15 m NaCl, 10 mm CaCl2, 0.02% NaN3, 0.05% Brij-35. Fluorescent substrate was then added to a concentration of 0.5 μm (final volume, 200 μl), and activity was measured by the increase in fluorescence at 535 nm after excitation at 485 nm using a Gemini SpectraMax (Molecular Devices) spectrofluorimeter. Peptide hydrolysis was measured for 2 h at 37 °C.
Surface Plasmon Resonance
Protein interactions were studied by surface plasmon resonance experiments using a Biacore T100 instrument (GE Healthcare) at the protein production and analysis facility of IFR128 Biosciences. BMP-1 was immobilized in 10 mm sodium acetate buffer, pH 4.5, on a CM5 sensor chip (series S, noncertified) using amine coupling chemistry. The control channel was prepared with the same activation/deactivation procedure except that the BMP-1 solution was replaced by buffer alone. Prior to injection, the analytes were dialyzed against HBS (10 mm HEPES, pH 7.4, 0.15 m NaCl) containing 5 mm CaCl2, which was also used as running buffer after the addition of 0.05% P-20. Sensorgrams were recorded at 25 °C with flow rates of 30 or 50 μl/min. Regeneration of active and control channels was performed using 50% ethylene glycol followed by 2 m NaCl. MMP-2 was immobilized in the same way, but the surface was regenerated using 2 m guanidium chloride.
For the analysis of kinetic experiments, double referencing (subtraction of the control sensorgram and then subtraction of a sensorgram obtained with buffer alone) were used. Because it was not possible to fit the association and dissociation curves shown in Fig. 4A using the usual binding models, responses during the association phase were plotted against 1/t, and a linear fit was used to calculate steady state values at infinite time (36). These values were then plotted against PCPE-1 concentrations, and the curve was fitted using the equation RUeq = (Rmax1 × [PCPE-1]/(KD1 + [PCPE-1])) + (Rmax2 × [PCPE-1]/(KD2 + [PCPE-1])) with Kaleidagraph software.
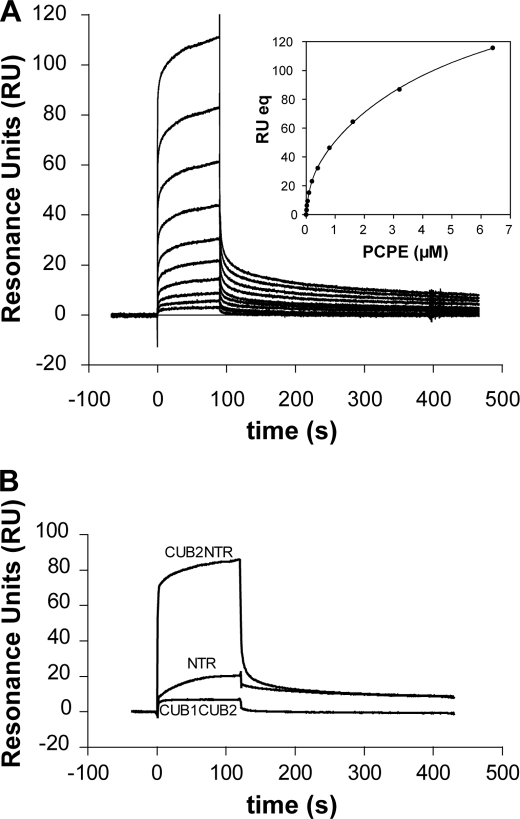
FIGURE 4.
Binding of PCPE-1 to BMP-1 studied by surface plasmon resonance (Biacore). A, concentration dependence of binding of full-length PCPE-1 (12.5, 25, 50, 100, 200, 400, 800, 1600, 3200, and 6400 nm) to immobilized BMP-1 (500 RU). Inset, fit of the response at infinite time (RU eq) as a function of PCPE-1 concentration using the equation given under “Experimental Procedures.” B, interactions of PCPE-1 fragments CUB1CUB2, NTRt, and CUB2-NTR (all at 500 nm) with immobilized BMP-1 (1463 RU).
For experiments with heparin, streptavidin (5000–6000 RU) was first immobilized as described (35) on both active and control channels. Biotinylated heparin was then injected on the active channel to reach a level of immobilization of 40–50 RU, and the surface was conditioned with two pulses of 1 m NaCl. Protein solutions for injections were prepared in 10 mm HEPES, pH 7.4, 0.15 m NaCl, and 0.05% P20, which was also used as running buffer. A flow rate of 30 μl/min was used, and regeneration was achieved with injections of 1 or 2 m NaCl.
Co-immunoprecipitation of PCPE-1 with BMP-1
To study the possible interaction of BMP-1 with PCPE-1, 200 nm BMP-1-FLAG was preincubated with an equimolar quantity of PCPE-1His in 50 mm Tris, pH 7.5, 0.15 m NaCl, 5 mm CaCl2 (total volume, 200 μl) for 1 h at 4 °C. Meanwhile 50 μl of anti-FLAG M2 affinity gel slurry (Sigma) was equilibrated in the same reaction buffer. The protein mixture was then incubated with the anti-FLAG-agarose for 3 h at 4 °C, rotating continuously. The suspension was then loaded into 500-μl spin cups (Pierce) and washed five times with 50 μl of 50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100 by pulse centrifugation, and then proteins were eluted with 100 mm glycine, pH 2.5. The eluates were analyzed by 10% SDS-PAGE under nonreducing conditions. The gels were then blotted onto polyvinylidene difluoride membranes, and the proteins were detected using a polyclonal rabbit PCPE-1 antibody (36) followed by an horseradish peroxidase-labeled anti-rabbit secondary antibody. The blots were developed using a Chemi-Doc luminescence visualization scanner (Bio-Rad).
Effect of Heparin on Stimulation of BMP-1 Activity by PCPE-1
To investigate the effects of heparin and heparan sulfate on BMP-1 activity in the presence and absence of PCPE-1 or its fragments, BMP-1-FLAG (5 or 10 nm) was incubated (in LoBind Eppendorf tubes) at 37 °C for 10–30 min (or longer in the absence of PCPE-1) with mini-procollagen III (200 or 400 nm, added last), with or without PCPE-1His, CUB1CUB2, NTRt, or a mixture of CUB1CUB2 and NTRt (each added at equimolar concentrations with respect to mini-procollagen III) and up to 800 μg/ml heparin (or 200 μg/ml heparan sulfate) in 50 mm HEPES, pH 7.4, 0.15 m NaCl, 5 mm CaCl2, 0.02% Brij 35 (total volume, 25 or 50 μl). In the case of CUB1CUB2 or NTRt, 0.25 mm Pefabloc was also present. The reactions were stopped on ice with the addition of 250 mm EDTA, pH 7.4, to a final concentration of 25 mm and then either processed directly for SDS-PAGE by the addition of 5× Laemmli sample buffer or first trichloroacetic acid/acetone precipitated and pretreated with 8 m urea for 5 min at 100 °C (26) prior to the addition of sample buffer and then heated for 3 min at 100 °C. The reaction products were then separated on Ready Gel precast acrylamide gels (Bio-Rad; 7.5% or 4–20% acrylamide in the separating gel; nonreducing conditions) followed by staining with Coomassie Blue. Alternatively, for quantitation, the gels were stained with SYPRO Ruby (Sigma) and then scanned on a Molecular Dynamics STORM 860 imager (blue laser; wavelength, 450 nm). The extent of cleavage of the mini-procollagen III substrate was determined using ImageQuant 5.2 software and expressed as the total intensity in the C-propeptide trimer and N-terminal fragment peaks divided by the total intensity of substrate and products.
RESULTS
Effects of PCPE-1 NTR on MMPs
Following the report of Mott et al. (17) identifying NTR domain-containing fragments of PCPE-1 as apparently weak inhibitors of MMP-2, we first sought to explore these observations using different forms of NTR domain prepared either by proteolysis or by directed mutagenesis. Initially, using a baculovirus system for expression of recombinant human PCPE-1 in insect cells (23), we purified a form of the NTR domain from the medium that arose by spontaneous proteolysis of the full-length protein. By N-terminal sequencing, we determined that this form (called NTRbaculovirus or NTRb) also included the entire linker between the CUB2 and NTR domains as well as the six C-terminal residues from the CUB2 domain (Fig. 1). We found no detectable inhibition by NTRb of purified MMP-1 (collagen substrate), MMP-2 (gelatin substrate), and MMP-3 and MMP-7 (both casein substrates) at concentrations up to 5 μm (data not shown).
Because the baculovirus form of PCPE-1 NTR included the entire linker between the CUB2 and NTR domains, in contrast to the forms identified by Mott et al. (17) where cleavage occurred within the linker region, we also tested the effects of a form of the PCPE-1 NTR domain (NTRt) prepared by limited proteolysis of full-length PCPE-1 with immobilized trypsin. N-terminal sequencing showed that NTRt consisted of three forms starting in the CUB2-NTR linker region at Val294, Ser300, or Thr306 (numbering for full-length PCPE-1 including the signal peptide; Fig. 1). Furthermore, because structural studies on TIMP·MMP complexes have shown the importance of the N-terminal cysteine in the mechanism of inhibition (2, 12–14), we also produced, by targeted mutagenesis, an additional form of PCPE-1 NTR, called NTRs, starting precisely at the first cysteine residue (Cys318; Fig. 1), as confirmed by N-terminal sequencing.
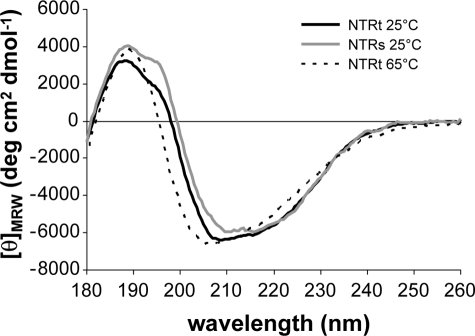
By circular dichroism spectroscopy, NTRs and NTRt were found to have similar spectra in the 180–260-nm range (Fig. 2) and similar to that of recombinant PCPE-1 NTR previously used for structural studies (1, 10). In addition, this domain was found to be remarkably stable to temperature, with a CD spectrum that remained essentially unchanged in the temperature range 25–65 °C (Fig. 2). We conclude that both NTRs and NTRt were correctly folded.
FIGURE 2.
Circular dichroism spectra of the PCPE-1 NTR domain. The spectra for NTRs and NTRt were measured at 25 °C, and that for NTRt was also measured at 65 °C.
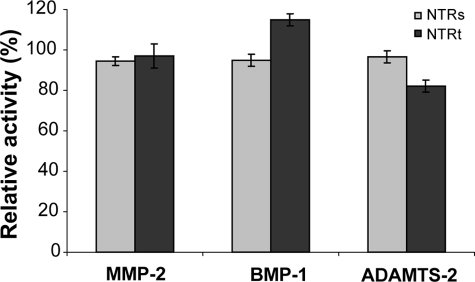
We then tested the effects of NTRs and NTRt on the activity of purified MMP-2, using a quenched fluorescent peptide substrate. In both cases, we found less than 5% inhibition of MMP-2 activity at a NTR concentration of 1 μm (Fig. 3). We also tested the effects of both forms of NTR on MMP-2 cleavage of denatured collagen I but found no evidence of inhibition at a NTR concentration of 2 μm (supplemental Fig. S1).
FIGURE 3.
Effects of NTRs and NTRt on the activities of MMP-2, BMP-1, and ADAMTS-2. MMP-2 activity was assayed on a quenched fluorogenic peptide (NTR concentration, 1 μm), BMP-1 activity was assayed on fluorescently labeled mini-procollagen III (NTR concentration, 1 μm), and ADAMTS-2 activity was assayed on radioactively labeled pN-collagen I (NTR concentration, 2.5 μm). The activities are expressed relative to activity in the absence of NTR, for each enzyme/substrate combination. The error bars show standard deviations.
We conclude that the NTR domain of PCPE-1 is not an inhibitor of MMP-2. In agreement with this lack of inhibition of MMP-2, NTR binding to immobilized MMP-2 (1832 RU) was assayed using Biacore technology, and both forms of NTR were found to bind only weakly to MMP-2, with dissociation constants at least 1 μm (data not shown).
In addition, as previously reported (37), we found that full-length PCPE-1 was itself a substrate for MMP-2, although cleavage was relatively inefficient, requiring long incubations at high enzyme concentrations. By N-terminal sequencing, we found the cleavage site to be localized at the Pro286–Lys287 bond in the CUB2-NTR linker region (Fig. 1), i.e. N-terminal to the serine protease sites identified by Mott et al. (17).
Effects of PCPE-1 NTR on Other Metzincins
We also tested the effects of NTRs and NTRt on other members of the metzincin superfamily, not previously assayed elsewhere (10, 17), including BMP-1, and ADAMTS-2, -4, and -5. Using fluorescently labeled mini-procollagen III as a substrate for BMP-1, we found no evidence of inhibition by either form of NTR at a concentration of 1 μm (Fig. 3). This lack of inhibitory activity was confirmed using a quenched fluorescent peptide substrate (supplemental Fig. S2). Using a pN-collagen substrate (i.e. procollagen without the C-propeptide region), we also tested the effect of NTRs and NTRt on N-terminal procollagen processing by ADAMTS-2 (Fig. 3). Again, no significant inhibition was seen by either form of NTR up to concentrations of 2.5 μm. Similar results were obtained for ADAMTS-4 and -5, using a quenched fluorescent peptide substrate (supplemental Fig. S2). We conclude that the NTR domain of PCPE-1 is not an inhibitor of BMP-1 or ADAMTS-2, -4, or -5.
Role of NTR within PCPE-1
It has previously been reported (38) that PCPE-1 binds to mTLL-1, which, like BMP-1, is also a member of the tolloid family but which consists of an additional EGF-CUB-CUB motif at the C terminus. To determine whether PCPE-1 also binds to BMP-1, we studied the interaction by surface plasmon resonance (Biacore Technology). As shown in Fig. 4A, full-length PCPE-1 was found to bind to immobilized BMP-1, in a concentration-dependent manner. Although a weak signal could already be detected when a 12.5 nm solution of PCPE-1 was injected, saturation was not reached even at 6.4 μm PCPE, showing that several binding processes were occurring. Because the experimental curves (fast association and dissociation phases followed by slower phases) could not be fitted with the usual binding models, we used steady state analysis with extrapolation to infinite time to obtain responses at equilibrium (36). As expected, better fits to the data were obtained assuming two binding sites (χ2 = 3 RU2) than one binding site (χ2 = 253 RU2). The calculated KD values for the two binding sites model were 129 ± 19 nm (accounting for 16% of the signal) and 5.2 ± 0.5 μm (accounting for 84% of the signal). This interaction of PCPE-1 with BMP-1 was confirmed by co-immunoprecipitation (data not shown) using BMP-1-FLAG and PCPE-1His at concentrations of 200 nm.
To study which domains of PCPE-1 were involved in the interaction with BMP-1, we measured interactions with PCPE-1 fragments. As shown in Fig. 4B, binding of the CUB1CUB2 fragment was essentially nonexistent, whereas binding of NTRt to immobilized BMP-1 was clearly apparent. (Similar observations were made by dot blot analysis, data not shown.) It should be borne in mind that the Biacore signal is proportional to the mass of analyte bound. Therefore, because the mass of NTRt (∼16 kDa) is approximately one-half that of CUB1CUB2 (30 kDa), binding of the NTR domain would appear greater after correcting for differences in molecular mass. However, the signal with the CUB2-NTR fragment (33 kDa) was even higher and comparable with binding of the full-length protein. Because CUB2 itself does not appear to bind BMP-1, from the lack of binding of the CUB1CUB2 fragment, this suggests that binding of PCPE-1 to BMP-1 occurs via the NTR domain, with a further contribution from the CUB2-NTR linker region. Interestingly, as also suggested elsewhere (39), this linker region is probably intrinsically disordered, as predicted by the program RONN (40) (supplemental Fig. S3), and could therefore become ordered on binding to BMP-1.
Heparin Superstimulation of PCPE-1
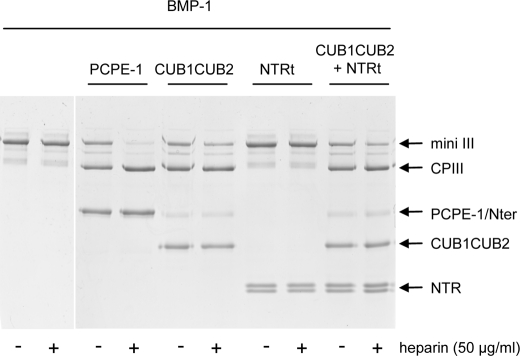
The NTR domain of PCPE-1 is known to bind to heparin (23). In addition, heparin has been reported to increase the inhibition of MMP-2 (20) and ADAMTS-2 (21) by TIMP-3. In view of these observations, we investigated the effect of heparin on the stimulation of BMP-1 activity by PCPE-1. Strikingly, using the standard BMP-1 assay with mini-procollagen III as substrate (16), we found that stimulation of BMP-1 activity by PCPE-1 was even further increased by the presence of 50 μg/ml heparin (Fig. 5). The same result was obtained using three different types of heparin (see “Experimental Procedures”) whether BMP-1, PCPE-1, and heparin were preincubated together or not, prior to the addition of mini-procollagen III. Also, control experiments showed that the effect was not due to prevention by heparin of nonspecific absorption of PCPE-1 to the walls of the reaction tube (data not shown). In contrast, using the same molar concentration of CUB1CUB2 fragment instead of full-length PCPE-1, BMP-1 activity was enhanced, as expected, but there was little additional effect of heparin. The same observation was obtained using an equimolar mixture of CUB1CUB2 and NTRt. Also, there was no stimulation by heparin of BMP-1 activity in the absence of PCPE-1 or CUB1CUB2. We conclude that the NTR domain contiguously linked to the rest of the PCPE-1 molecule is required for “superstimulation” of BMP-1 activity by heparin in the presence of PCPE-1.
FIGURE 5.
Superstimulation of PCPE-1 enhancing activity by heparin. Effect of heparin (50 μg/ml; Sigma H-3393) on the activity of BMP-1 (10 nm) on mini-procollagen III (mini III, 400 nm) in the presence and absence of PCPE-1His, its CUB1CUB2 or NTRt fragments, or a mixture of CUB1CUB2 and NTRt (all at 400 nm). Incubations were carried out at 37 °C for 15 min. The migration positions of the released C-propeptide trimer (CPIII) and the N-terminal fragment (Nter) are indicated. Discontinuous SDS-PAGE (4–20% acrylamide in the separating gel) was carried out in nonreducing conditions, followed by staining with Coomassie Blue.
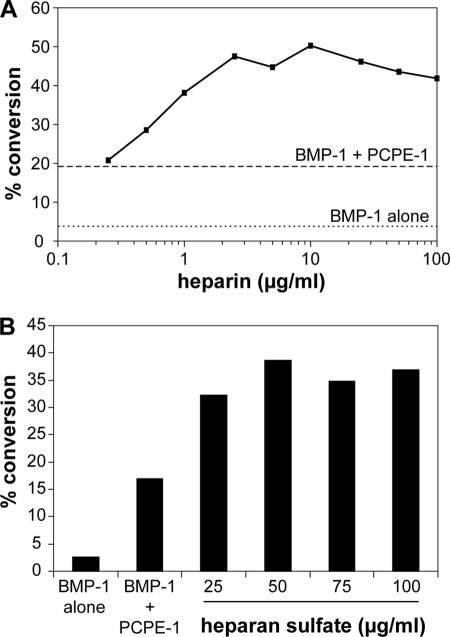
To attempt to understand the mechanism of heparin stimulation, we determined the concentration dependence using a fixed amount of BMP-1 (typically 5 nm), equimolar amounts (200 nm) of miniprocollagen III and PCPE-1 (for maximum enhancement (23)), and increasing concentrations of heparin. As shown in Fig. 6A, whereas PCPE-1 led to a ∼6-fold increase in BMP-1 activity, this was further increased by at least 2-fold in the presence of heparin. The extent of superstimulation increased with heparin concentration, reaching a plateau at ∼2.5 μg/ml and remaining constant up to 800 μg/ml (data shown up to 100 μg/ml). We also studied the effect of heparan sulfate, which again gave rise to superstimulation (Fig. 6B) of BMP-1 activity in the presence of PCPE-1.
FIGURE 6.
A, concentration dependence of the effect of heparin on PCPE-1 enhancing activity, determined by SDS-PAGE followed by SYPRO Ruby staining, showing the percentage of conversion of the mini-procollagen III substrate (200 nm) in the presence of BMP-1 alone (5 nm), BMP-1 + PCPE-1His (200 nm), and BMP-1 + PCPE-1His + increasing concentrations of heparin. The incubations were carried out at 37 °C for 10 min. B, concentration dependence of the effect of heparan sulfate on PCPE-1 enhancing activity. The experimental conditions were as in A.
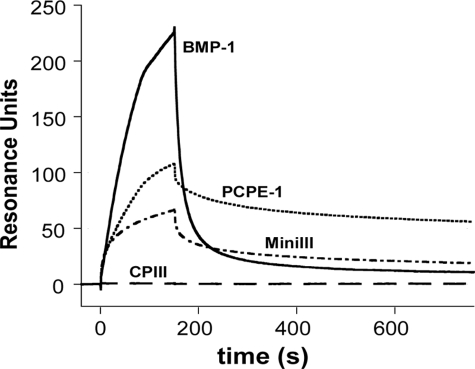
To further investigate the mechanism of superstimulation, we studied the binding of heparin to each of the molecules involved: PCPE-1, mini-procollagen III, and BMP-1. By surface plasmon resonance using biotinylated heparin stably immobilized to a streptavidin surface, we observed binding of both BMP-1 and mini-procollagen III when injected at a concentration of 50 nm (Fig. 7). This was also the case for PCPE-1, which has previously been reported to bind to heparin, via the NTR domain, with nanomolar affinity (23, 41). Interestingly, whereas mini-procollagen III bound heparin in a rather stable manner with a slow dissociation phase, BMP-1 displayed fast association and dissociation phases and was washed off from the surface very rapidly. Dissociation constants were found to be in the range of 10–150 nm for PCPE-1 and mini-procollagen III and 0.5–2 μm for BMP-1. In contrast, the C-propeptide trimer of procollagen III did not bind at all to the immobilized heparin, suggesting that the binding site for heparin is located in the triple-helical or telopeptide regions of mini-procollagen III.
FIGURE 7.
Interaction of PCPE-1, mini-procollagen III (miniIII), the procollagen III C-propeptide trimer (CPIII), and BMP-1 with heparin. Biotinylated heparin was first bound to immobilized streptavidin to reach 44 RU and then solutions containing 50 nm of each protein were injected in 10 mm HEPES, 0.15 m NaCl, 0.05% P20 for 150 s at 30 μl/min.
DISCUSSION
PCPE-1 NTR Is Not an Inhibitor of MMP-2, BMP-1, or ADAMTS-2, -4, or -5
The identification of NTR containing fragments of PCPE-1 that appeared to act as weak inhibitors of MMP-2 (17), together with the observed structural similarities with TIMPs (1, 10, 39), prompted the hunt for possible high affinity targets of this domain. The presence in these fragments of residues N-terminal to the cysteine at the start of the NTR domain, known to abrogate the inhibitory activity of TIMPs against MMPs (14), suggested that if it is indeed a proteinase inhibitor, then its mechanism of action must be somewhat unusual. Using a form of PCPE-1 NTR with 12 residues N-terminal to the first cysteine, Liepinsh et al. (10) were unable to detect inhibitory activity against a range of MMPs, ADAM-17, and also serine proteinases. ADAM-17 is of particular interest, because it has been shown to be inhibited by TIMP-3 (42), and in this case addition of an alanine residue N-terminal to the first cysteine still results in inhibitory activity (43), although the mechanism of inhibition is unclear (44). TIMP-3 is also known to inhibit ADAMTS-2, -4, and -5 (21, 45). In addition, early observations (46) suggested that PCPE-1 without the NTR domain was more active in enhancing procollagen C-terminal processing than the full-length protein, suggesting possible inhibition by NTR of tolloid proteinases. For these reasons, we decided to investigate possible inhibitory activity, on a range of metalloproteinases, of two forms of PCPE-1 NTR, one partially overlapping the fragments identified by Mott et al. (17) and the other beginning precisely at the first cysteine. With both forms, however, we were unable to detect inhibitory activity (at micromolar concentrations) against members of the three main families within the metzincin superfamily (47): the matrixins, the astacins, and the adamalysins, namely MMP-2, BMP-1, and ADAMTS-2, -4, and -5.
Previous studies (10, 39) on the PCPE-1 NTR domain have revealed its structural similarities with TIMP-1 and TIMP-2 but also its differences, notably in relation to the structural core of PCPE-1 NTR, which is more dynamically active (39). Moreover, structure-based alignment shows the disulfide bonding pattern to be somewhat different between PCPE-1 and TIMP-2 (10). Also, residue 2 in TIMPs, normally Ser or Thr that enters into the S1′ specificity pocket in MMPs, is replaced by Pro or Arg in PCPE-1 NTR, depending on the alignment (1, 10). With regard to ECM binding, PCPE-1 NTR is more closely related to the N-terminal domain of TIMP-3, both having relatively high predicted isoelectric points (9.4 and 9.3, respectively), but the results presented here show that this is not sufficient to inhibit adamalysins as in the case of TIMP-3.
Superstimulation of PCPE-1 Enhancing Activity by Heparin
Here we show for the first time that BMP-1 binds to PCPE-1. This observation is consistent with a previous report (38) on the binding of PCPE-1 to mTLL-1, a longer form of tolloid proteinase, found by co-immunoprecipitation, and also with data using BMP-1 deletion constructs (48). Here we show that PCPE-1 binding to BMP-1 is relatively weak, with micromolar affinity, in comparison with the nanomolar affinity of the PCPE-1/mini-procollagen III interaction (26). In addition, unlike the binding to mini-procollagen III, which involves the CUB1CUB2 region, binding of PCPE-1 to BMP-1 mainly involves the NTR domain. In view of its relatively weak affinity, however, it is most unlikely that the BMP-1/PCPE-1 interaction alone (i.e. in the absence of additional cooperative interactions) will play a significant role at the concentrations used in typical in vitro assays.
A further new finding presented here is the observation that heparin (or heparan sulfate) leads to the superstimulation of BMP-1 activity on mini-procollagen III in the presence of PCPE-1. In addition, we found essentially no superstimulation by heparin of BMP-1 activity in the presence of the CUB1CUB2 fragment of PCPE-1 nor in the presence of an equimolar mixture of CUB1CUB2 and NTRt. These observations point to a role for the NTR domain of PCPE-1, when present in the intact full-length molecule, in the mechanism of BMP-1 superstimulation.
Previous observations (49), in the absence of PCPE-1, showed that tolloid proteinase activity on procollagen I (450 kDa) can be enhanced in the presence of high molecular mass (500 kDa) dextran sulfate. This effect was attributed to the formation of a complex between the procollagen and the highly sulfated polysaccharide, probably involving several molecules of procollagen, which could be isolated by centrifugation for 5 min at 15,000 × g, thereby increasing the local concentration of the substrate (49). The effect required both a high degree of sulfation (as also found in heparin) and also a high molecular mass, because it was not observed with 500-kDa nonsulfated dextran nor with 17–19-kDa heparin. Our results confirm the lack of effect of heparin alone on BMP-1 activity in the absence of PCPE-1.
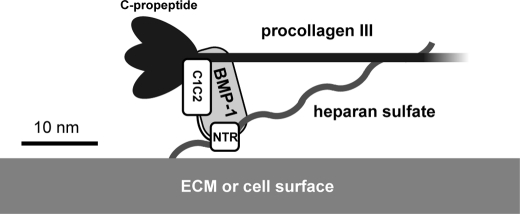
PCPE-1 is known to bind to heparin, via its NTR domain (23, 41), with nanomolar affinity (41). Here we confirm this observation by surface plasmon resonance (Fig. 7) and show that although binding of heparin to mini-procollagen III is also relatively tight, binding of BMP-1 to heparin is much weaker. In contrast, the isolated C-propeptide trimer of procollagen III did not bind to heparin, indicating that the heparin-binding site is in the triple-helical or telopeptide region. By analogy with the known heparin binding sequence GIKGHR found near the C terminus of the collagen α1(I) chain triple-helical region (50), we propose that the site of interaction of heparin with procollagen III is an identical sequence at the same position (residues 1104–1109, numbered from the start of the signal sequence), which also occurs in mini-procollagen III. In addition, PCPE-1 has previously been shown to bind to mini-procollagen III, involving the CUB1CUB2 and C-propeptide regions, with nanomolar affinity (26), forming a 1:1 complex (23). Finally, here we show that PCPE-1 binds to BMP-1 mainly via its NTR domain, with micromolar affinity. Taken together, these data point to the possible formation of a complex involving all of the reactants, as depicted in Fig. 8.
FIGURE 8.
Possible mechanism by which heparin-like glycosaminoglycans might superstimulate the enhancing activity of PCPE-1 in the C-terminal processing of procollagen III. PCPE-1 binds to procollagen III in the region of the C-propeptides with nanomolar affinity. This complex then binds to cell surface or extracellular matrix associated heparin-like glycosaminoglycans via interactions with PCPE-1 NTR and with procollagen III. In addition, BMP-1 interacts with PCPE-1 NTR, the CUB2-NTR linker, and heparin. These interactions might increase the local concentrations of the reactants and/or create a new recognition site that further facilitates the action of BMP-1, resulting in superstimulation. Approximate molecular dimensions are based on known low resolution structures (27, 64, 65). C1C2, CUB1CUB2.
In this model, in the absence of heparin, PCPE-1 would bind tightly to mini-procollagen III via its CUB1CUB2 region, thereby facilitating cleavage by BMP-1, as described elsewhere (26, 36). In the presence of heparin (Fig. 8), cooperative effects involving heparin binding to PCPE-1 NTR and mini-procollagen III (also weak binding to BMP-1), as well as interactions of BMP-1 with NTR and the CUB2-NTR linker, would increase the local concentration of the reactants and/or change the way in which the enzyme is presented to the substrate, resulting in superstimulation. In contrast, in the absence of the NTR domain (and the CUB2-NTR linker), interaction of the CUB1CUB2·mini-procollagen III complex with heparin would not be sufficient to lead to the above effects. Even if the NTR were present but no longer covalently attached to the rest of the PCPE-1 molecule, there would be no reason for it to bind to the same heparin molecule as the CUB1CUB2·mini-procollagen III complex. The model is therefore consistent with the results shown in Fig. 5.
In favor of such a model, we note that superstimulation reached a plateau at a heparin concentration of 2.5 μg/ml, equivalent to ∼140 nm, which is comparable with the concentrations of mini-procollagen III and PCPE-1 used in the assays (200 nm) and therefore suggestive of a 1:1:1 heparin·PCPE-1·mini-procollagen III complex. In this regard, it is of interest to note that there was little indication of reduced superstimulation at high heparin concentrations (Fig. 6A), in contrast to the bell shaped curves seen by Butler et al. (20) and Wang et al. (21) in studying heparin potentiation of the interaction of TIMP-3 with MMP-2 or ADAMTS-2, respectively. These data on TIMP-3 were interpreted in terms of an optimum heparin concentration that allows simultaneous binding of different reactants to the same heparin molecule, unlike the binding to separate heparin molecules that would occur at high heparin concentrations. The fact that a bell-shaped curve was not observed in the experiments reported here is consistent with the idea that a single heparin molecule binds to a pre-existing mini-procollagen III·PCPE-1 complex. Such a complex would be relatively small (<200 kDa), compared with the large dextran sulfate·procollagen I complex (>1000 kDa) studied by Hojima et al. (49), which would explain our inability to pellet the complex by centrifugation (data not shown) in the conditions used by Hojima et al. (49).
These observations point to a possible role for the NTR domain in localizing the site of procollagen processing to specific extracellular matrix or cell surface microenvironments rich in heparin-like glycosaminoglycans, as previously suggested by Kessler and co-workers (41, 51). Sulfated glycosaminoglycans have been shown in many cases to influence proteinase activity, often in close proximity to the cell surface. The most well documented examples are heparin stimulation of the inhibition of thrombin by antithrombin (52, 53) or by protein C inhibitor (54). Stimulation of activity has also been observed, as in the case of plasmin activation by plasminogen activators (55–57). Here we report the superstimulation of a proteinase enhancer by heparin (and heparan sulfate). This further adds to the rich arsenal of macromolecules known to affect the activities of tolloid proteinases, including PCPEs, twisted gastrulation (58), secreted frizzled related proteins (59–61), olfactomedin (62), and, most recently, fibronectin (63).
Supplementary Material
Acknowledgments
We thank Dominique Mazzocut for help with the N-terminal sequencing, Eve Pecheur for use of the TECAN spectrofluorimeter, and Sylvie Ricard-Blum for help with preliminary studies on the BMP-1-PCPE-1 interaction.
This work was supported, in whole or in part, by National Institutes of Health Grant AR40994. This work was also supported by La Ligue Contre le Cancer, the Agence Nationale de la Recherche, the Région Rhône-Alpes, the Centre National de la Recherche Scientifique, and the Université Claude Bernard Lyon 1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- NTR
- netrin-like
- BMP-1
- bone morphogenetic protein-1
- ECM
- extracellular matrix
- MMP
- matrix metalloproteinase
- PCPE
- procollagen C-proteinase enhancer
- RU
- resonance unit(s)
- TIMP
- tissue inhibitor of metalloproteinases.
REFERENCES
- 1.Bányai L., Patthy L. (1999) Protein Sci. 8, 1636–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagase H., Visse R., Murphy G. (2006) Cardiovasc. Res. 69, 562–573 [DOI] [PubMed] [Google Scholar]
- 3.Stetefeld J., Jenny M., Schulthess T., Landwehr R., Schumacher B., Frank S., Rüegg M. A., Engel J., Kammerer R. A. (2001) Nat. Struct. Biol. 8, 705–709 [DOI] [PubMed] [Google Scholar]
- 4.Bezakova G., Ruegg M. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 295–308 [DOI] [PubMed] [Google Scholar]
- 5.Cirulli V., Yebra M. (2007) Nat. Rev. Mol. Cell Biol. 8, 296–306 [DOI] [PubMed] [Google Scholar]
- 6.Bramham J., Thai C. T., Soares D. C., Uhrín D., Ogata R. T., Barlow P. N. (2005) J. Biol. Chem. 280, 10636–10645 [DOI] [PubMed] [Google Scholar]
- 7.Bovolenta P., Esteve P., Ruiz J. M., Cisneros E., Lopez-Rios J. (2008) J. Cell Sci. 121, 737–746 [DOI] [PubMed] [Google Scholar]
- 8.Kondás K., Szláma G., Trexler M., Patthy L. (2008) J. Biol. Chem. 283, 23677–23684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerhard D. S., Wagner L., Feingold E. A., Shenmen C. M., Grouse L. H., Schuler G., Klein S. L., Old S., Rasooly R., Good P., Guyer M., Peck A. M., Derge J. G., Lipman D., Collins F. S., Jang W., Sherry S., Feolo M., Misquitta L., Lee E., Rotmistrovsky K., Greenhut S. F., Schaefer C. F., Buetow K., Bonner T. I., Haussler D., Kent J., Kiekhaus M., Furey T., Brent M., Prange C., Schreiber K., Shapiro N., Bhat N. K., Hopkins R. F., Hsie F., Driscoll T., Soares M. B., Casavant T. L., Scheetz T. E., Brown-stein M. J., Usdin T. B., Toshiyuki S., Carninci P., Piao Y., Dudekula D. B., Ko M. S., Kawakami K., Suzuki Y., Sugano S., Gruber C. E., Smith M. R., Simmons B., Moore T., Waterman R., Johnson S. L., Ruan Y., Wei C. L., Mathavan S., Gunaratne P. H., Wu J., Garcia A. M., Hulyk S. W., Fuh E., Yuan Y., Sneed A., Kowis C., Hodgson A., Muzny D. M., McPherson J., Gibbs R. A., Fahey J., Helton E., Ketteman M., Madan A., Rodrigues S., Sanchez A., Whiting M., Madari A., Young A. C., Wetherby K. D., Granite S. J., Kwong P. N., Brinkley C. P., Pearson R. L., Bouffard G. G., Blakesly R. W., Green E. D., Dickson M. C., Rodriguez A. C., Grimwood J., Schmutz J., Myers R. M., Butterfield Y. S., Griffith M., Griffith O. L., Krzywinski M. I., Liao N., Morin R., Palmquist D., Petrescu A. S., Skalska U., Smailus D. E., Stott J. M., Schnerch A., Schein J. E., Jones S. J., Holt R. A., Baross A., Marra M. A., Clifton S., Makowski K. A., Bosak S., Malek J. (2004) Genome Res. 14, 2121–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liepinsh E., Banyai L., Pintacuda G., Trexler M., Patthy L., Otting G. (2003) J. Biol. Chem. 278, 25982–25989 [DOI] [PubMed] [Google Scholar]
- 11.Hopkins D. R., Keles S., Greenspan D. S. (2007) Matrix Biol. 26, 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomis-Rüth F. X., Maskos K., Betz M., Bergner A., Huber R., Suzuki K., Yoshida N., Nagase H., Brew K., Bourenkov G. P., Bartunik H., Bode W. (1997) Nature 389, 77–81 [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Catalan C., Bode W., Huber R., Turk D., Calvete J. J., Lichte A., Tschesche H., Maskos K. (1998) EMBO J. 17, 5238–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wingfield P. T., Sax J. K., Stahl S. J., Kaufman J., Palmer I., Chung V., Corcoran M. L., Kleiner D. E., Stetler-Stevenson W. G. (1999) J. Biol. Chem. 274, 21362–21368 [DOI] [PubMed] [Google Scholar]
- 15.Adar R., Kessler E., Goldberg B. (1986) Collagen Rel. Res. 6, 267–277 [DOI] [PubMed] [Google Scholar]
- 16.Moali C., Font B., Ruggiero F., Eichenberger D., Rousselle P., François V., Oldberg A., Bruckner-Tuderman L., Hulmes D. J. (2005) J. Biol. Chem. 280, 24188–24194 [DOI] [PubMed] [Google Scholar]
- 17.Mott J. D., Thomas C. L., Rosenbach M. T., Takahara K., Greenspan D. S., Banda M. J. (2000) J. Biol. Chem. 275, 1384–1390 [DOI] [PubMed] [Google Scholar]
- 18.Yu W. H., Yu S., Meng Q., Brew K., Woessner J. F., Jr. (2000) J. Biol. Chem. 275, 31226–31232 [DOI] [PubMed] [Google Scholar]
- 19.Troeberg L., Fushimi K., Khokha R., Emonard H., Ghosh P., Nagase H. (2008) FASEB J. 22, 3515–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler G. S., Apte S. S., Willenbrock F., Murphy G. (1999) J. Biol. Chem. 274, 10846–10851 [DOI] [PubMed] [Google Scholar]
- 21.Wang W. M., Ge G., Lim N. H., Nagase H., Greenspan D. S. (2006) Biochem. J. 398, 515–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wayne G. J., Deng S. J., Amour A., Borman S., Matico R., Carter H. L., Murphy G. (2007) J. Biol. Chem. 282, 20991–20998 [DOI] [PubMed] [Google Scholar]
- 23.Moschcovich L., Bernocco S., Font B., Rivkin H., Eichenberger D., Chejanovsky N., Hulmes D. J., Kessler E. (2001) Eur. J. Biochem. 268, 2991–2996 [DOI] [PubMed] [Google Scholar]
- 24.Kappler J., Franken S., Junghans U., Hoffmann R., Linke T., Müller H. W., Koch K. W. (2000) Biochem. Biophys. Res. Commun. 271, 287–291 [DOI] [PubMed] [Google Scholar]
- 25.Blanc G., Font B., Eichenberger D., Moreau C., Ricard-Blum S., Hulmes D. J., Moali C. (2007) J. Biol. Chem. 282, 16924–16933 [DOI] [PubMed] [Google Scholar]
- 26.Kronenberg D., Vadon-Le Goff S., Bourhis J. M., Font B., Eichenberger D., Hulmes D. J., Moali C. (2009) J. Biol. Chem. 284, 33437–33446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernocco S., Finet S., Ebel C., Eichenberger D., Mazzorana M., Farjanel J., Hulmes D. J. (2001) J. Biol. Chem. 276, 48930–48936 [DOI] [PubMed] [Google Scholar]
- 28.Murphy G., Willenbrock F., Ward R. V., Cockett M. I., Eaton D., Docherty A. J. (1992) Biochem. J. 283, 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colige A., Ruggiero F., Vandenberghe I., Dubail J., Kesteloot F., Van Beeumen J., Beschin A., Brys L., Lapière C. M., Nusgens B. (2005) J. Biol. Chem. 280, 34397–34408 [DOI] [PubMed] [Google Scholar]
- 30.Kashiwagi M., Enghild J. J., Gendron C., Hughes C., Caterson B., Itoh Y., Nagase H. (2004) J. Biol. Chem. 279, 10109–10119 [DOI] [PubMed] [Google Scholar]
- 31.Gendron C., Kashiwagi M., Lim N. H., Enghild J. J., Thøgersen I. B., Hughes C., Caterson B., Nagase H. (2007) J. Biol. Chem. 282, 18294–18306 [DOI] [PubMed] [Google Scholar]
- 32.Galloway W. A., Murphy G., Sandy J. D., Gavrilovic J., Cawston T. E., Reynolds J. J. (1983) Biochem. J. 209, 741–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knight C. G., Willenbrock F., Murphy G. (1992) FEBS Lett. 296, 263–266 [DOI] [PubMed] [Google Scholar]
- 34.Colige A., Beschin A., Samyn B., Goebels Y., Van Beeumen J., Nusgens B. V., Lapière C. M. (1995) J. Biol. Chem. 270, 16724–16730 [DOI] [PubMed] [Google Scholar]
- 35.Ricard-Blum S., Féraud O., Lortat-Jacob H., Rencurosi A., Fukai N., Dkhissi F., Vittet D., Imberty A., Olsen B. R., van der Rest M. (2004) J. Biol. Chem. 279, 2927–2936 [DOI] [PubMed] [Google Scholar]
- 36.Ricard-Blum S., Bernocco S., Font B., Moali C., Eichenberger D., Farjanel J., Burchardt E. R., van der Rest M., Kessler E., Hulmes D. J. (2002) J. Biol. Chem. 277, 33864–33869 [DOI] [PubMed] [Google Scholar]
- 37.Dean R. A., Overall C. M. (2007) Mol. Cell Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 38.Ge G., Zhang Y., Steiglitz B. M., Greenspan D. S. (2006) J. Biol. Chem. 281, 10786–10798 [DOI] [PubMed] [Google Scholar]
- 39.Williamson R. A., Panagiotidou P., Mott J. D., Howard M. J. (2008) Mol. Biosyst. 4, 417–425 [DOI] [PubMed] [Google Scholar]
- 40.Yang Z. R., Thomson R., McNeil P., Esnouf R. M. (2005) Bioinformatics 21, 3369–3376 [DOI] [PubMed] [Google Scholar]
- 41.Ricard-Blum S., Moschcovich L., Weiss T., Wineman E., Kessler E. (2006) Federation of the European Connective Tissue Societies, XXth Meeting, Oulu, Finland, July 1–5, 2006, Federation of the European Connective Tissue Societies [Google Scholar]
- 42.Amour A., Slocombe P. M., Webster A., Butler M., Knight C. G., Smith B. J., Stephens P. E., Shelley C., Hutton M., Knäuper V., Docherty A. J., Murphy G. (1998) FEBS Lett. 435, 39–44 [DOI] [PubMed] [Google Scholar]
- 43.Wei S., Kashiwagi M., Kota S., Xie Z., Nagase H., Brew K. (2005) J. Biol. Chem. 280, 32877–32882 [DOI] [PubMed] [Google Scholar]
- 44.Wisniewska M., Goettig P., Maskos K., Belouski E., Winters D., Hecht R., Black R., Bode W. (2008) J. Mol. Biol. 381, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 45.Kashiwagi M., Tortorella M., Nagase H., Brew K. (2001) J. Biol. Chem. 276, 12501–12504 [DOI] [PubMed] [Google Scholar]
- 46.Kessler E., Adar R. (1989) Eur. J. Biochem. 186, 115–121 [DOI] [PubMed] [Google Scholar]
- 47.Stöcker W., Grams F., Baumann U., Reinemer P., Gomis-Rüth F. X., McKay D. B., Bode W. (1995) Protein Sci. 4, 823–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petropoulou V., Garrigue-Antar L., Kadler K. E. (2005) J. Biol. Chem. 280, 22616–22623 [DOI] [PubMed] [Google Scholar]
- 49.Hojima Y., Behta B., Romanic A. M., Prockop D. J. (1994) Anal. Biochem. 223, 173–180 [DOI] [PubMed] [Google Scholar]
- 50.Di Lullo G. A., Sweeney S. M., Korkko J., Ala-Kokko L., San Antonio J. D. (2002) J. Biol. Chem. 277, 4223–4231 [DOI] [PubMed] [Google Scholar]
- 51.Weiss T., Wineman E., Moschcovich L., Mesilaty S., Moali C., Hulmes D. J., Rousselle P., Kessler E. (2008) Federation of the European Connective Tissue Societies, XXIst Meeting, Marseille, France, July 9–13, 2008, Federation of the European Connective Tissue Societies [Google Scholar]
- 52.Li W., Johnson D. J., Esmon C. T., Huntington J. A. (2004) Nat. Struct. Mol. Biol. 11, 857–862 [DOI] [PubMed] [Google Scholar]
- 53.Dementiev A., Petitou M., Herbert J. M., Gettins P. G. (2004) Nat. Struct. Mol. Biol. 11, 863–867 [DOI] [PubMed] [Google Scholar]
- 54.Li W., Adams T. E., Nangalia J., Esmon C. T., Huntington J. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4661–4666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edelberg J. M., Conrad H. E., Pizzo S. V. (1991) Biochemistry 30, 10999–11003 [DOI] [PubMed] [Google Scholar]
- 56.Takada Y., Urano T., Takada A. (1994) Thromb. Res. 73, 301–311 [DOI] [PubMed] [Google Scholar]
- 57.Brunner G., Reimbold K., Meissauer A., Schirrmacher V., Erkell L. J. (1998) Exp. Cell Res. 239, 301–310 [DOI] [PubMed] [Google Scholar]
- 58.Scott I. C., Blitz I. L., Pappano W. N., Maas S. A., Cho K. W., Greenspan D. S. (2001) Nature 410, 475–478 [DOI] [PubMed] [Google Scholar]
- 59.Lee H. X., Ambrosio A. L., Reversade B., De Robertis E. M. (2006) Cell 124, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muraoka O., Shimizu T., Yabe T., Nojima H., Bae Y. K., Hashimoto H., Hibi M. (2006) Nat. Cell Biol. 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 61.Kobayashi K., Luo M., Zhang Y., Wilkes D. C., Ge G., Grieskamp T., Yamada C., Liu T. C., Huang G., Basson C. T., Kispert A., Greenspan D. S., Sato T. N. (2009) Nat. Cell Biol. 11, 46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inomata H., Haraguchi T., Sasai Y. (2008) Cell 134, 854–865 [DOI] [PubMed] [Google Scholar]
- 63.Huang G., Zhang Y., Kim B., Ge G., Annis D. S., Mosher D. F., Greenspan D. S. (2009) J. Biol. Chem. 284, 25879–25888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernocco S., Steiglitz B. M., Svergun D. I., Petoukhov M. V., Ruggiero F., Ricard-Blum S., Ebel C., Geourjon C., Deleage G., Font B., Eichenberger D., Greenspan D. S., Hulmes D. J. (2003) J. Biol. Chem. 278, 7199–7205 [DOI] [PubMed] [Google Scholar]
- 65.Berry R., Jowitt T. A., Ferrand J., Roessle M., Grossmann J. G., Canty-Laird E. G., Kammerer R. A., Kadler K. E., Baldock C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8561–8566 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.