Abstract
The acGFPL is the first-identified member of a novel, colorless and non-fluorescent group of green fluorescent protein (GFP)-like proteins. Its mutant aceGFP, with Gly replacing the invariant catalytic Glu-222, demonstrates a relatively fast maturation rate and bright green fluorescence (λex = 480 nm, λem = 505 nm). The reverse G222E single mutation in aceGFP results in the immature, colorless variant aceGFP-G222E, which undergoes irreversible photoconversion to a green fluorescent state under UV light exposure. Here we present a high resolution crystallographic study of aceGFP and aceGFP-G222E in the immature and UV-photoconverted states. A unique and striking feature of the colorless aceGFP-G222E structure is the chromophore in the trapped intermediate state, where cyclization of the protein backbone has occurred, but Tyr-66 still stays in the native, non-oxidized form, with Cα and Cβ atoms in the sp3 hybridization. This experimentally observed immature aceGFP-G222E structure, characterized by the non-coplanar arrangement of the imidazolone and phenolic rings, has been attributed to one of the intermediate states in the GFP chromophore biosynthesis. The UV irradiation (λ = 250–300 nm) of aceGFP-G222E drives the chromophore maturation further to a green fluorescent state, characterized by the conventional coplanar bicyclic structure with the oxidized double Tyr-66 Cα=Cβ bond and the conjugated system of π-electrons. Structure-based site-directed mutagenesis has revealed a critical role of the proximal Tyr-220 in the observed effects. In particular, an alternative reaction pathway via Tyr-220 rather than conventional wild type Glu-222 has been proposed for aceGFP maturation.
Keywords: Crystal Structure, Crystallography, Fluorescence, Post-translational Modification, Protein Structure, X-ray Crystallography, Chromophore Biosynthesis, Green Fluorescent Protein, X-ray Structure
Introduction
Green fluorescent proteins (GFP)2 and the GFP-like proteins (FPs) have become in recent years very useful tools in many areas of cell biology, biotechnology, and medicine. These proteins exhibit a wide spectral range of fluorescence, from blue to far-red. It became possible to effectively use FPs as single or coupled biomarkers for multicolor labeling of proteins, subcellular compartments, and specific tissue regions. Utilization of FPs enabled monitoring of a variety of characteristics, such as cellular pH and ion concentration, tracking of expression, intracellular localization, and trafficking of proteins of interest in the cell or whole organism and following their interactions with other cellular components (1–5).
Chromoproteins are another large group of non-fluorescent counterparts of FPs that share with them the principal fold but not spectral properties (6, 7). However, a number of artificially created, genetically engineered variants of chromoproteins do exhibit fluorescence (6, 8). Members of both fluorescent and non-fluorescent families possess visible coloration corresponding to an absorption range of 450–610 nm. Extensive diversity of their photophysical characteristics arises mostly from variations in the chemical structure of the internal chromophore group and in the stereochemistry of its adjacent environment.
The first identified member of a novel colorless non-fluorescent group of GFP-like proteins was acGFPL from jellyfish Aequorea coerulescens (9). Its discovery suggested that many other species might express similar colorless FPs. It was found that a glycine replacement of Glu-222, a conserved residue adjacent to the chromophore, plays a key role in creating a fluorescent form of acGFPL. A variant of acGFPL, named aceGFP (commercial name AcGFP1 from Clontech), obtained by mutation of five residues that included mutating residue Glu-222 to a glycine, exhibited bright green fluorescence. The reverse single mutation G222E in aceGFP resulted in the colorless variant aceGFP-G222E that demonstrated no visible coloration and fluorescence, similarly to the wild type acGFPL. Detailed characterization of the purified aceGFP-G222E showed that only a minor fraction (about 3%) of this protein contains mature chromophore in the protonated state (absorption maximum at 390 nm), whereas the major fraction exhibits no absorption/fluorescence in the visible region, indicating the presence of an immature chromophore. Importantly, this immature protein can be converted into a green fluorescent state (similar to aceGFP, with excitation and emission maxima at 480 and 505 nm, respectively) by UV light illumination (λ = 250–300 nm). We have focused on the stereochemical features responsible for these unique properties. We present here the results of a high resolution x-ray study of the mutant variants of the colorless non-fluorescent progenitor acGFPL. The proteins that were investigated include the green fluorescent aceGFP (resolution 1.5 Å), the colorless aceGFP-G222E (resolution 1.14 Å), and UV-photoconverted aceGFP-G222E-(UV) (resolution 1.75 Å). The structural aspects of this study have been supported by site-directed mutagenesis and by spectroscopic characterization of three new mutant variants, namely aceGFP-Y220L, aceGFP-Y220F, and aceGFP-G222E-Y220L.
EXPERIMENTAL PROCEDURES
Expression, Purification, and Crystallization
The pQE30 plasmids with the corresponding vectors of aceGFP, aceGFP-Y220L/F, aceGFP-G222E, and aceGFP-G222E-Y220L were transformed into Escherichia coli JM109 (DE3) cells. The proteins were expressed in 3 liters of LB medium by overnight incubation at 37 °C. Cells were resuspended in 20 mm Tris, pH 7.8, 200 mm NaCl and disrupted by sonication. Purification of each protein was performed on metal-affinity (TALON resin, Clontech) and size-exclusion (Superdex-200 (16/60), Amersham Biosciences) columns. All major purification steps were performed in 20 mm Tris, pH 7.8, 200 mm NaCl buffer.
Prior to crystallization, aceGFP and aceGFP-G222E were transferred to 10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 2.5 mm EDTA buffer and concentrated to 10–15 mg/ml in 10-kDa molecular mass cutoff concentration units (Vivascience). Crystals were obtained by the hanging drop vapor diffusion method at 20 °C. Each crystallization drop consisted of 1 μl of protein solution mixed with an equal volume of reservoir solution and was incubated against 500 μl of reservoir solution. The hexagonal bipyramidal crystals of aceGFP, with visible green coloration, were obtained from 0.2 m potassium/sodium tartrate, 0.1 m sodium citrate, 2 m ammonium sulfate, pH 5.6, and the prismatic crystals of aceGFP-G222E were obtained from 0.2 m ammonium acetate, 0.1 m Bis-Tris, pH 6.5, 25% polyethylene glycol 3350. Crystal growth time varied from 1 week to 2 months. A crystal of the colorless aceGFP-G222E, surrounded by the mother liquor in its crystallization drop, was photoconverted to the green fluorescent state by 2 h of UV irradiation (254 nm) at 20 °C.
Crystal Structure Determination and Refinement
X-ray diffraction data for aceGFP, aceGFP-G222E, and the UV-photoconverted aceGFP-G222E-(UV) were collected from single crystals flash-cooled in a 100 K nitrogen stream. Prior to cooling, the crystals were transferred to a cryo-protecting solution containing 20% glycerol and 80% reservoir solution. Data were collected with a MAR300 CCD detector at the SER-CAT beamline 22ID (Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL) and were processed with HKL2000 (10).
The crystal structure of aceGFP was solved by the molecular replacement method with MOLREP (11, 12), using the coordinates of a GFP monomer (PDB ID: 1GFL (13)) without its chromophore. The refined coordinates of aceGFP were used to solve the aceGFP-G222E structure. Structure refinement was performed with PHENIX (14), alternating with manual correction of the model using COOT (15). Water molecules were located with ARP/wARP (16), and structure validation was performed with PROCHECK (17). Crystallographic data and refinement statistics are presented in the Table 1. The coordinates and structure factors of the aceGFP, aceGFP-G222E, and aceGFP-G222E-(UV) (after UV irradiation) were deposited in the Protein Data Bank under accession codes 3LVA, 3LVC, and 3LVD, respectively.
TABLE 1.
Crystallographic data and refinement statistics
| Protein |
|||
|---|---|---|---|
| aceGFP (PDB ID: 3LVA) | aceGFP-G222E (PDB ID: 3LVC) | aceGFP-G222E (UV-photoconverted) (PDB ID: 3LVD) | |
| Crystallographic data | |||
| Space group | P6122 | P212121 | P212121 |
| Cell dimensions (Å) | a, b = 74.5, c = 325.9 | a = 59.9, b = 80.4, c = 95.8 | a = 59.5, b = 80.2, c = 95.4 |
| Z/(Z′) | 24 (2) | 8 (2) | 8 (2) |
| Estimated solvent content (%) | 50 | 43 | 42 |
| Temperature (K) | 100 | 100 | 100 |
| Wavelength (Å) | 1.00 | 1.00 | 1.00 |
| Resolution range (Å) | 29.1–1.50 (1.57–1.50)a | 29.7–1.14 (1.17–1.14)a | 28.4–1.75 (1.81–1.75)a |
| Total reflections measured | 805,382 | 757,611 | 223,690 |
| Unique reflections observed (F > 0) | 83,894 | 168,358 | 46,602 |
| Multiplicity | 9.6 (8.8)a | 4.5 (3.9)a | 4.8 (4.8)a |
| I/σ (I) | 30.0 (2.4)a | 16.8 (3.1)a | 17.7 (2.7)a |
| Rmerge | 0.068 (0.718)a | 0.074 (0.481)a | 0.084 (0.583)a |
| Completeness (%) | 96.5 (96.0)a | 100.0 (100.0)a | 99.9 (100.0)a |
| Refinement statistics | |||
| Non-H atoms in model | |||
| Protein | 3,756 (2 × (2–238) residues) | 3,636 (2 × (1–229) residues) | 3,636 (2 × (1–229) residues) |
| Water | 419 | 486 | 317 |
| SO4 | 5 | ||
| Glycerol | 24 | 30 | 24 |
| Rwork | 0.144 (97.9%)b | 0.139 (99.0%)b | 0.160 (96.9%)b |
| Rfree | 0.201 (2.1%)b | 0.157 (1.0%)b | 0.193 (3.1%)b |
| Mean B factor/(r.m.s.d.)c (Å2) | |||
| Protein atoms | |||
| Main chain | 23.3 (2.0) | 12.1 (0.9) | 18.8 (1.9) |
| Side chain | 27.5 (3.5) | 17.2 (2.9) | 23.9 (3.9) |
| Chromophore | 19.3 (1.5) | 8.6 (0.7) | 15.5 (1.6) |
| r.m.s.d.c bond length (Å) | 0.006 | 0.009 | 0.009 |
| r.m.s.d.c bond angles (°) | 0.83 | 1.38 | 0.99 |
| Ramachandran statistics (%) | |||
| (for non-Gly/Pro residues) most favorable/additional allowed | 92.8/7.2 | 92.3/7.7 | 92.1/7.9 |
a Values for the data in the highest resolution shell.
b Percentage of the data reserved for working and free sets.
c r.m.s.d., root mean square deviation.
Mutagenesis and Spectroscopic Characterization
Preparation of the structure-based variants aceGFP-Y220F/L and aceGFP-222E-Y220L by site-directed mutagenesis was performed by PCR using the overlap extension method with primers containing the appropriate target substitutions (18). Absorption and excitation-emission spectra of the purified proteins were recorded with a Varian Cary 100 UV-vis spectrophotometer and Varian Cary Eclipse fluorescence spectrophotometer, respectively.
RESULTS
Overall Structure
A monomer (molecular mass ∼27 kDa) of aceGFP and aceGFP-G222E adopts a fold typical for all GFP-like proteins that consists of an 11-stranded β-barrel, with loop caps from both sides and a chromophore embedded in the middle of an internal α-helix going along the β-barrel axis. In the crystalline state, both proteins form dimers composed of two such monomers arranged in an antiparallel fashion, with an angle of ∼40° between the β-barrel axes.
Chromophore Structure of the Green aceGFP
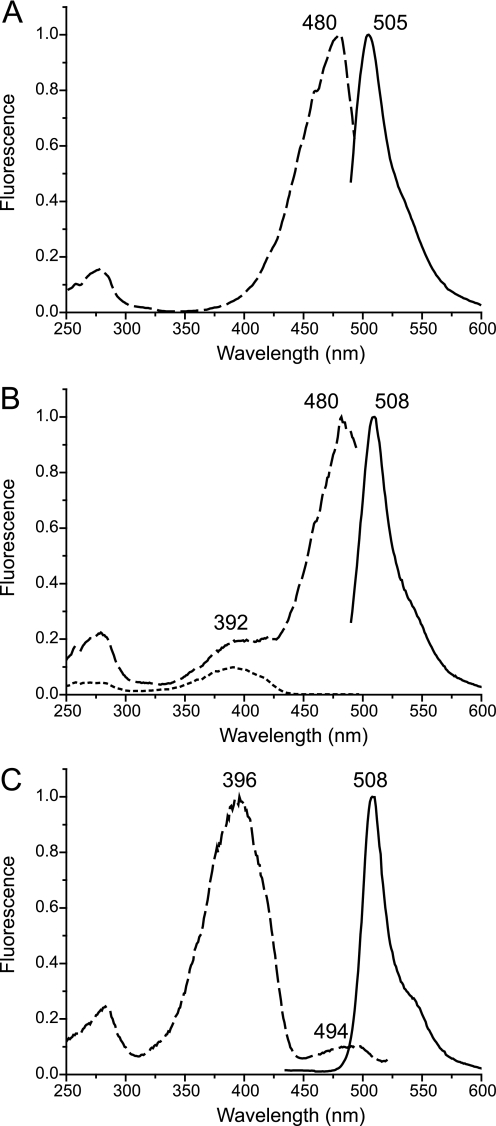
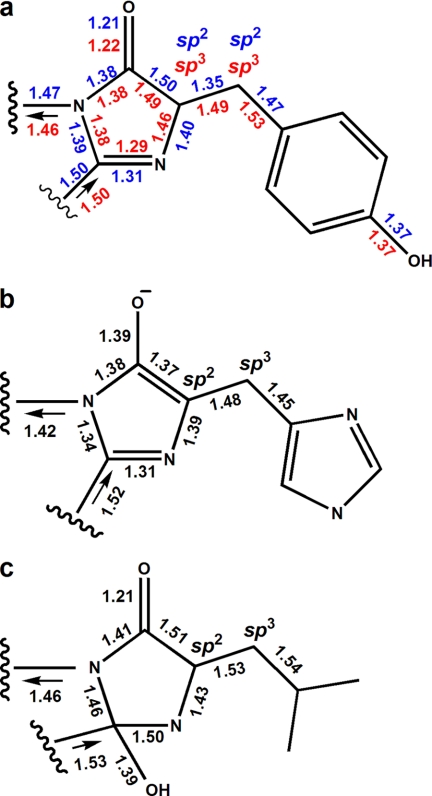
The aceGFP variant exhibits bright emission in the green spectral range (λex = 480 nm, λem = 505 nm; Fig. 1A). In contrast to wild type green fluorescent proteins, the important position 222, proximal to the chromophore, is occupied in aceGFP by an unusual Gly residue instead of the highly conserved catalytic Glu. Nevertheless the chromophore structure, matured from the Ser-65–Tyr-66–Gly-67 triad (Fig. 2), is identical to that present in wild type avGFP from Aequorea victoria (PDB ID: 1W7S) and cgGFP from Clytia gregaria (2HPW). The chromophore adopts a two-ring coplanar structure typical for GFP, consisting of a five-member imidazolone heterocycle having a p-hydroxybenzylidene substituent with the phenolic ring of Tyr-66 in cis-orientation to the Cα–N(66) bond. Tyr-66 exhibits trigonal planar geometry of bonds attached to the Cα atom and a double character of the Cα=Cβ bond, indicating the sp2 hybridization state of both atoms. Ser-65 is characterized by the sp3 hybridization of Cα, the single character of the Cα–N bond, and the trans-configuration of the preceding peptide bond.
FIGURE 1.
Normalized excitation (dashed lines) and emission (solid lines) spectra for aceGFP (A), aceGFP-G222E after UV-induced photoconversion (dotted line is excitation before UV irradiation) (B), and aceGFP-G222E-Y220L (C).
FIGURE 2.
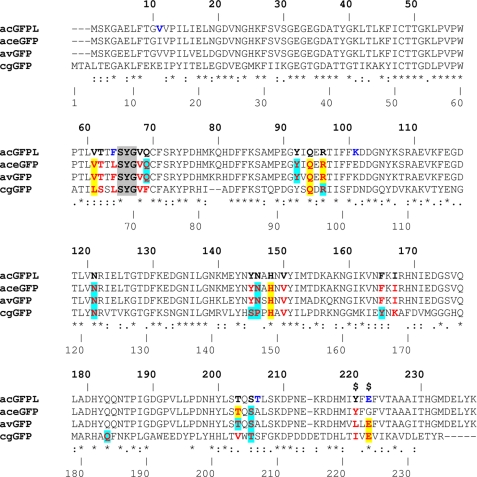
Sequence alignment of the wild type colorless progenitor acGFPL (A. coerulescens) with its green fluorescent variant aceGFP (five sites of mutation are shown in dark blue), WT avGFP (A. victoria), and WT cgGFP (C. gregaria). The upper (bold) and lower numbers correspond to those in the crystal structures of acGFPL/aceGFP/avGFP and cgGFP, respectively. The residues in the immediate chromophore environment are shown in red. The residues forming direct bonds and the residues mediated by water H-bonds (d ≤ 3.3Å) with chromophore are highlighted in yellow and light blue, respectively, and those making van der Waals contacts (d ≤ 3.9Å) are not highlighted. Two critical sites responsible for coloration are marked by a dollar sign.
Chromophore Structure of the Colorless aceGFP-G222E
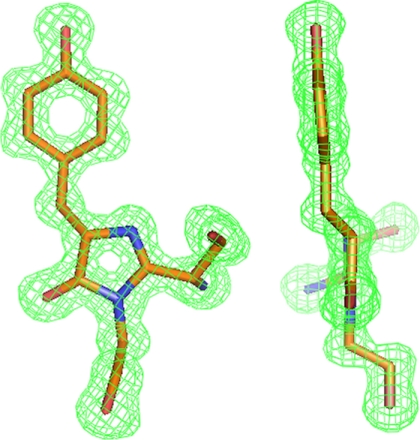
The replacement of the unusual Gly-222 with the generally conserved Glu residue in aceGFP traps the protein in an immature colorless state (9). Indeed, the crystal structure of aceGFP-G222E showed that the chromophore moiety is not fully matured. Here the posttranslational modification has terminated at the backbone cyclized state characterized by the non-coplanar, non-conjugated two-ring structure, with Tyr-66 in the native (non-oxidized) form. The single character of the Tyr-66 Cα–Cβ and Cβ–Cγ bonds, the tetrahedral geometry of bonds attached to the Cα atom, and the characteristic value of the torsion angle χ1 = −55° around the Cα–Cβ all unequivocally define the sp3 hybridization state of both the Cα and the Cβ atoms (Figs. 3 and 4a).
FIGURE 3.
Front and side views of the aceGFP-G222E chromophores in Fo − Fc electron density (cutoff ρ = 3.0 σ), exhibiting tetrahedral bond arrangement around the sp3 hybridized state of the Tyr-66 Cα and Cβ atom. This figure was produced with PyMOL (34).
FIGURE 4.
Chromophore geometry in the mutant variants. a, Ser-65–Tyr-66–Gly-67 in aceGFP (numbers/symbols shown in blue; oxidized Tyr-66 with sp2 hybridized Cα/Cβ) and aceGFP-G222E (numbers/symbols shown in red; native Tyr-66 with sp3 hybridized Cα/Cβ) (current study). b, Thr-65–His-66–Gly-67 in the reduced BFPsol (PDB ID: 2FWQ (32). c, Thr-65–Leu-66–Gly-67 in the EGFP variant (PDB ID: 1S6Z (31)).
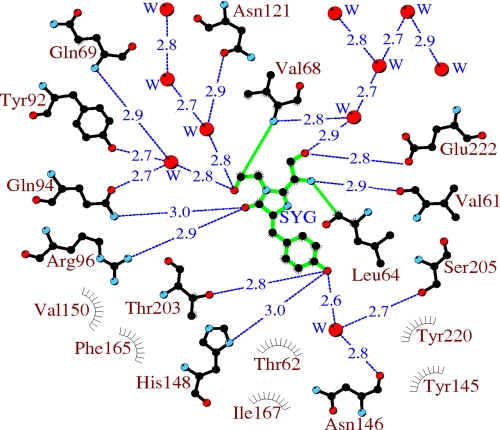
The environment of the chromophore in aceGFP-G222E can be defined as 19 side chains arranged in a manner practically identical to those in aceGFP, except for the extra Glu-222 side chain (Fig. 5). Glu-222 and another highly conserved residue, Arg-96, play an active catalytic role in the GFP-like proteins, facilitating oxidative chemistry in the posttranslational modification process (19–22). Most of the proximal residues are involved in an extensive H-bond network interacting with the chromophore. The proximal waters (presumed reaction products of the maturation process) also participate in the network, mediating interactions between the residues. The network is apparently functionally important as a potential proton/electron wire in the maturation process.
FIGURE 5.
A schematic diagram illustrating the environment of aceGFP-G222E. Hydrogen bonds (≤3.3 Å) are shown as blue dashed lines, waters (W) are shown as red spheres, and van der Waals contacts (≤3.9 Å) as gray “eyelashes.” This figure was produced with LIGPLOT/HBPLUS (35, 36).
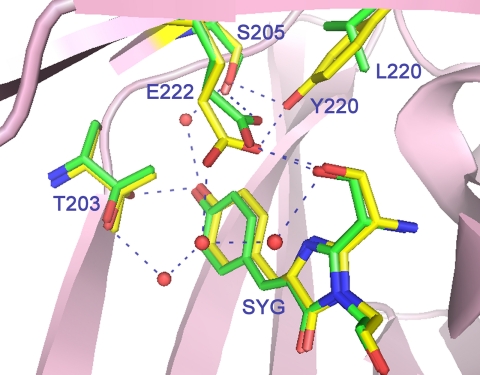
Interestingly, the carboxyl group of Glu-222 in the immature aceGFP-G222E exhibits ∼2 Å displacement relative to the corresponding position in the wild type green avGFP and cgGFP. This shift is stabilized by an H-bond with the Tyr-220 side chain and can be reproduced with ∼20, ∼30, and ∼35° torsion rotations around the respective Cα–Cβ, Cβ–Cγ, and Cγ–Cδ bonds of the side chain of Glu-222. In WT avGFP and cgGFP, the carboxyl Oϵ1 and Oϵ2 atoms of Glu-222 form hydrogen bonds with the side chains of Ser-205 and the chromophore Ser-65, respectively. The corresponding water-mediated H-bond pathway OH(Tyr-66)–OHO(Wat)–OH(Ser-205)–Oϵ1(Glu-222)Oϵ2–OH(Ser-65) was proposed to be important for promoting rapid proton transfer from a neutral chromophore to Glu-222 (23). In the colorless aceGFP-G222E, this pathway is replaced by a longer chain OH(Tyr-66)–OHO(Wat)–OH(Ser-205)–OH(Tyr-220)–Oϵ1(Glu-222)Oϵ1–OH(Ser-65), which also incorporates Tyr-220. Here the corresponding Oϵ1 atom of Glu-222 makes two H-bonds with the hydroxyl of Tyr-220 and the chromophore Ser-65, and Oϵ2 interacts with the proximal water, mediating interactions in the H-bond network (Fig. 6).
FIGURE 6.
The vicinity of the chromophore in aceGFP-G222E (yellow) and avGFP (green). The side chain of Glu-222 in aceGFP-G222E is shifted by as much as 2 Å relative to its conventional position. This figure was produced with PyMOL (34).
Chromophore Structure of the UV-photoconverted aceGFP-G222E
Irradiation of the colorless aceGFP-G222E variant with UV light (λ = 250–300 nm) causes its irreversible photoconversion to a green fluorescent state that is similar to the one found in aceGFP (9). In contrast to a non-irradiated precursor, the chromophore structure in the UV-converted variant is fully matured and has spectral properties practically identical to that in aceGFP (Fig. 1, A and B), exhibiting coplanar arrangement of the imidazolone and phenolic rings with a conjugated system of π-electrons. Except for the chromophore itself, photoconverted and non-photoconverted aceGFP-G222E, as well as aceGFP, have practically identical three-dimensional structural arrangement of the residues nearest to the chromophore. No decarboxylation of Glu-222, characteristic for UV-induced photoconversion of WT avGFP (24) and photoactivatable GFP (25, 26), was observed in aceGFP-G222E.
Mutagenesis of Tyr-220
A comparison of the three-dimensional structures of aceGFP/aceGFP-G222E with WT avGFP/cgGFP revealed an important amino acid difference at position 220. This position, located in the immediate environment of the chromophore, is occupied by Tyr and Leu/Ile, respectively (Fig. 2). In aceGFP-G222E, the Tyr-220 phenolic ring shifts the Glu-222 carboxylate by 2 Å from the standard position observed in avGFP/cgGFP. The replacement Y220L in the colorless aceGFP-G222E resulted in a green fluorescent product aceGFP-G222E-Y220L with excitation-emission spectra similar to those of the wild type avGFP (λex = 396 and 494 nm, λem = 508 nm; Fig. 1C). In the green fluorescent aceGFP, lacking Glu-222, substitutions Y220L and especially Y220F resulted in very slowly maturing mutants that required 1–2 weeks of incubation to develop fluorescence. At the same time, matured aceGFP-Y220L and aceGFP-Y220F proteins have excitation and emission spectra indistinguishable from those of parental aceGFP (Fig. 1A). Moreover, molar extinction coefficients and fluorescence quantum yields for these mutants were found to be very close to that of aceGFP (50,000 m−1cm−1 and 0.55, respectively (9). Thus, Tyr-220 appears to be important to ensure fast chromophore maturation of aceGFP, but its presence has a negligible effect on the spectral properties of the matured protein.
DISCUSSION
The immature chromophore of aceGFP-G222E exhibits non-coplanar arrangement of the imidazolone and phenolic rings, indicating the lack of their conjugation (Figs. 3 and 4a). This state is characterized by the non-oxidized single Tyr-66 Cα–Cβ bond, with both atoms in sp3 hybridization. A distinct feature of the colorless aceGFP-G222E, as well as of aceGFP, is the presence of Tyr-220, as opposed to Leu and Ile in WT avGFP and cgGFP, respectively. In aceGFP-G222E, the side chain of Tyr-220 pushes the negatively charged carboxyl group of Glu-222 by ∼2 Å from its conventional position. As a result, the H-bonded wire between Glu-222 and the chromophore Tyr-66 is extended by insertion of the interactions with Tyr-220. Such a shift apparently causes local distortion of the electrostatic field, which terminates the chromophore maturation process at an intermediate stage in which the cyclization of the protein backbone has occurred, but Tyr-66 still remains in the native, non-oxidized form. To check the proposed charge effect, we replaced the “pushing” Tyr-220 residue with a smaller Leu, aiming to restore the location of the Glu-222 side chain to that observed in avGFP and cgGFP. Indeed, this replacement resulted in a fully matured product aceGFP-G222E-Y220L, with spectral properties similar to those of the wild type avGFP (Fig. 1C).
UV irradiation of the immature aceGFP-G222E drives the post-translational modification process further and results in a fully matured chromophore having the GFP conventional planar bicyclic structure and the conjugated system of π-electrons. The conserved Glu-222, Arg-96, and possibly, Tyr-220, are expected to participate in catalysis of this process. The UV-converted variant exhibits bright green fluorescence similar to that observed in green aceGFP. Surprisingly, despite the critical Glu-222/Gly-222 difference, both proteins have almost identical spectral characteristics (λex = 480 nm, λem = 505 nm; Fig. 1, A and B). The absence of Glu-222 or its displacement from the position standard in GFPs correlates with the excitation peak at ∼480 nm, which is presumably due to the deprotonated state of the chromophore (27). Indeed, replacement of Glu-222 with a Gly in WT avGFP shifts the excitation maximum in spectra from 398 to 481 nm, leaving the emission peak at ∼507 nm (28). On the other hand, the proposed restoration of the conventional position of Glu-222 side chain in aceGFP-G222E-Y220L shifts the excitation peak from 480 to 390 nm, characteristic for WT avGFP (28) (Fig. 1C). The latter property has been proposed to relate with the protonated chromophore state, stabilized by electrostatic repulsion between Glu-222 carboxylate and the chromophore (27, 29, 30).
A distinctive feature of aceGFP is the presence of an unusual Gly residue substituting for the catalytic Glu-222, invariant in all wild type GFP-like proteins. Glu-222 was found to be one of the key residues in biosynthesis of the GFP chromophore. It has been suggested that this residue plays a role of the general base, facilitating proton abstraction from the chromophore moiety (19–22). The absence of Glu-222 turned out not to be critical for complete maturation of the aceGFP chromophore to a conventional planar π-conjugated bicyclic system (Fig. 4a), which might indicate the possibility of an alternative path for the chemical reaction. In this particular variant, the hydroxyl of Tyr-220 resides within ∼1 Å distance from the position occupied by one of the oxygens of the carboxylate of Glu-222 in WT avGFP and apparently acts in a similar way as a proton acceptor. To check a possible role of this residue in aceGFP, we substituted Tyr-220 by Leu, characteristic for avGFP, and by Phe. The resultant variants, aceGFP-Y220L and aceGFP-Y220F, have the spectra and the intrinsic brightness similar to aceGFP, but in contrast to the latter protein, show a very slow maturation rate. This experiment supports our suggestion of an alternative catalytic pathway of chromophore maturation via Tyr-220, rather than via conventional Glu-222.
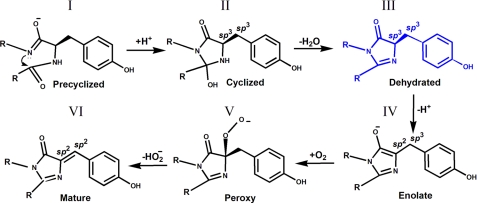
The details of the mechanism of chromophore formation in GFPs have been a subject of considerable controversy. Several crystal structures trapping various intermediate states have been reported, leading to a number of proposed reaction schemes of chromophore biosynthesis (21, 31, 32). One of the plausible pathways of chromophore maturation (Fig. 7) has been proposed by Barondeau et al. (32). Currently, three intermediates, I, III, IV, and the mature state VI of the chromophore from the proposed pathway, have been experimentally detected by x-ray crystallography. The precyclized state I was trapped in the anaerobically prepared GFP variants with modified chromophore-forming sequences Gly-65–Gly-66–Gly-67 and Ala-65–Ser-66–Ala-67 (19, 33). The dehydrated intermediate III has now been observed by us in the non-modified chromophore Ser-65–Tyr-66–Gly-67 of the colorless aceGFP-G222E. The distinguishing feature of this state is the intact geometry of Tyr-66, characterized by sp3 hybridization of both the Cα and the Cβ atoms and by the tetrahedral arrangements of their covalent bonds (Fig. 4a). The structure corresponding to the following partially oxidized enolate intermediate IV with sp2 and sp3 hybridization of the respective Cα and Cβ atoms (Fig. 4b) has been detected for the blue fluorescent BFPsol matured variant after reducing the previously mutated chromophore Thr-65–His-66–Gly-67 with dithionite (32).
FIGURE 7.
Proposed reaction scheme for GFP chromophore biosynthesis according to Barondeau et al. (32).
Pouwels et al. (21) proposed a different pathway of chromophore formation in which the dehydration reaction of the hydroxylated imidazolone is the last chemical step leading to a mature chromophore. The state preceding the matured form has been attributed to the non-dehydrated chromophore structure (Fig. 4c) trapped in the crystal of the colorless EGFP variant. This variant has the chromophore-forming sequence Thr-65–Leu-66–Gly-67 with an unusual non-aromatic Y66L substitution (31). The observed form reveals the hydroxylated imidazolone and partially oxidized Leu-66 Cα–Cβ bond with sp2 and sp3 hybridization of the respective atoms. The authors' assignment of the Cα(Leu-66) atom hybridization to sp2 is consistent with the observed trigonal planar geometry of three attached bonds but somewhat disagrees with their single character. This non-dehydrated state with oxidized Cα(66)_sp2 atom cannot succeed or precede our dehydrated form III with non-oxidized Cα(66)_sp3 (Fig. 7). This might indicate the complexity of chromophore biosynthesis, suggesting more than one possible reaction pathway defined by the concrete chromophore-forming sequence and the immediate stereochemical environment. The unusual aliphatic nature of the chromophore Leu-66 in the EGFP variant could also make a dramatic difference in the maturation process when compared with that in GFP-like proteins that contain an aromatic residue at position 66.
Supplementary Material
Acknowledgments
We acknowledge the use of beamline 22-ID of the Southeast Regional Collaborative Access Team (SER-CAT), located at the APS, Argonne National Laboratory. Use of the APS was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract Number W-31-109-Eng-38.
This work was supported in part by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research, and with federal funds from the National Institutes of Health, NCI, under Contract HHSN261200800001E. This project also was supported in part by the Russian Foundation for Basic Research (Grants 09-04-00212 and 08-04-01702-a), Molecular and Cell Biology Program Russian Academy of Sciences, and by Grant MD-2780.2009.4 from the President of the Russian Federation.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3LVA, 3LVC, and 3LVD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GFP
- green fluorescent protein
- FP
- GFP-like protein
- EGFP
- enhanced GFP
- WT
- wild type
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Verkhusha V. V., Lukyanov K. A. (2004) Nat. Biotechnol. 22, 289–296 [DOI] [PubMed] [Google Scholar]
- 2.Chudakov D. M., Lukyanov S., Lukyanov K. A. (2005) Trends Biotechnol. 23, 605–613 [DOI] [PubMed] [Google Scholar]
- 3.Zubova N. N., Savitsky A. P. (2005) Uspekhi Biol. Khim. 45, 1–66 [Google Scholar]
- 4.Remington S. J. (2006) Curr. Opin. Struct. Biol. 16, 714–721 [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Shyy J. Y., Chien S. (2008) Annu. Rev. Biomed. Eng. 10, 1–38 [DOI] [PubMed] [Google Scholar]
- 6.Gurskaya N. G., Fradkov A. F., Terskikh A., Matz M. V., Labas Y. A., Martynov V. I., Yanushevich Y. G., Lukyanov K. A., Lukyanov S. A. (2001) FEBS Lett. 507, 16–20 [DOI] [PubMed] [Google Scholar]
- 7.Prescott M., Ling M., Beddoe T., Oakley A. J., Dove S., Hoegh-Guldberg O., Devenish R. J., Rossjohn J. (2003) Structure 11, 275–284 [DOI] [PubMed] [Google Scholar]
- 8.Bulina M. E., Chudakov D. M., Mudrik N. N., Lukyanov K. A. (2002) BMC. Biochem. 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurskaya N. G., Fradkov A. F., Pounkova N. I., Staroverov D. B., Bulina M. E., Yanushevich Y. G., Labas Y. A., Lukyanov S., Lukyanov K. A. (2003) Biochem. J. 373, 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 11.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 12.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 13.Yang F., Moss L. G., Phillips G. N., Jr. (1996) Nat. Biotechnol. 14, 1246–1251 [DOI] [PubMed] [Google Scholar]
- 14.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta Crystallogr. D. Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 16.Perrakis A., Sixma T. K., Wilson K. S., Lamzin V. S. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 448–455 [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 18.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 19.Barondeau D. P., Putnam C. D., Kassmann C. J., Tainer J. A., Getzoff E. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12111–12116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barondeau D. P., Kassmann C. J., Tainer J. A., Getzoff E. D. (2006) J. Am. Chem. Soc. 128, 4685–4693 [DOI] [PubMed] [Google Scholar]
- 21.Pouwels L. J., Zhang L., Chan N. H., Dorrestein P. C., Wachter R. M. (2008) Biochemistry 47, 10111–10122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sniegowski J. A., Lappe J. W., Patel H. N., Huffman H. A., Wachter R. M. (2005) J. Biol. Chem. 280, 26248–26255 [DOI] [PubMed] [Google Scholar]
- 23.Shu X., Leiderman P., Gepshtein R., Smith N. R., Kallio K., Huppert D., Remington S. J. (2007) Protein Sci. 16, 2703–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Thor J. J., Gensch T., Hellingwerf K. J., Johnson L. N. (2002) Nat. Struct. Biol. 9, 37–41 [DOI] [PubMed] [Google Scholar]
- 25.Patterson G. H., Lippincott-Schwartz J. (2002) Science 297, 1873–1877 [DOI] [PubMed] [Google Scholar]
- 26.Henderson J. N., Gepshtein R., Heenan J. R., Kallio K., Huppert D., Remington S. J. (2009) J. Am. Chem. Soc. 131, 4176–4177 [DOI] [PubMed] [Google Scholar]
- 27.Heim R., Prasher D. C., Tsien R. Y. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12501–12504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrig T., O'Kane D. J., Prendergast F. G. (1995) FEBS Lett. 367, 163–166 [DOI] [PubMed] [Google Scholar]
- 29.Brejc K., Sixma T. K., Kitts P. A., Kain S. R., Tsien R. Y., Ormö M., Remington S. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2306–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palm G. J., Zdanov A., Gaitanaris G. A., Stauber R., Pavlakis G. N., Wlodawer A. (1997) Nat. Struct. Biol. 4, 361–365 [DOI] [PubMed] [Google Scholar]
- 31.Rosenow M. A., Huffman H. A., Phail M. E., Wachter R. M. (2004) Biochemistry 43, 4464–4472 [DOI] [PubMed] [Google Scholar]
- 32.Barondeau D. P., Tainer J. A., Getzoff E. D. (2006) J. Am. Chem. Soc. 128, 3166–3168 [DOI] [PubMed] [Google Scholar]
- 33.Barondeau D. P., Kassmann C. J., Tainer J. A., Getzoff E. D. (2005) Biochemistry 44, 1960–1970 [DOI] [PubMed] [Google Scholar]
- 34.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 35.Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 36.McDonald I. K., Thornton J. M. (1994) J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.