Abstract
We report the presence of oligosaccharide structures on a glutamine residue present in the VL domain sequence of a recombinant human IgG2 molecule. Residue Gln-106, present in the QGT sequence following the rule of an asparagine-linked consensus motif, was modified with biantennary fucosylated oligosaccharide structures. In addition to the glycosylated glutamine, analysis of a lectin-enriched antibody population showed that 4 asparagine residues: heavy chain Asn-162, Asn-360, and light chain Asn-164, both of which are present in the IgG1 and IgG2 constant domain sequences, and Asn-35, which was present in CDRL1, were also modified with oligosaccharide structures at low levels. The primary sequences around these modified residues do not adhere to the N-linked consensus sequon, NX(S/T). Modeling of these residues from known antibody crystal structures and sequence homology comparison indicates that non-consensus glycosylation occurs on Asn residues in the context of a reverse consensus motif (S/T)XN located on highly flexile turns within 3 residues of a conformational change. Taken together our results indicate that protein glycosylation is governed by more diversified requirements than previously appreciated.
Keywords: Antibodies, Glycoprotein Structure, Glycosylation, Post-translational Modification, Protein Motifs, N-Glycosylation, Q-Glycosylation, Glutamine, Non-consensus motif
Introduction
The modification of proteins with oligosaccharide structures linked through the side chain of Asn residues is classically associated with the consensus sequence motif, NX(S/T), where X is not proline (1, 2). The architecture of the OST enzyme complex and the dolichol pyrophosphate-GlcNAc2Man9Glc3 donor oligosaccharide dictate the properties of the C-terminal amino acids in the N-glycosylation consensus sequence. Mechanistic studies involving mutation of the +2 amino acid following the Asn residue have shown that a hydrogen acceptor is necessary to render the Asn residue sufficiently nucleophilic to displace the GlcNAc2Man9Glc3 oligosaccharide from the dolichol donor (3, 4). Further insight into the mechanism of N-glycosylation was obtained by replacing the +2 amino acid in the consensus sequon with threonine analogues (5, 6). The results from these studies indicated that the OST enzyme complex did not tolerate changes in the position of the threonine methyl group nor the introduction of charge at the +2 residue. The presence of a Thr in the +2 position is associated with a higher fraction of occupancy at a particular N-glycosylation sequon (7) and a greater likelihood of occupancy in general (8). In addition to the necessary requirement for the presence of a Ser or Thr to occupy the +2 position of a sequon, the absence of Pro in the +1 position has been found to be absolutely necessary for N-glycan occupancy (9). This has been attributed to the rigidity that is imparted to the peptide backbone resulting from the cyclic structure of Pro (3).
N-Glycosylation has been mechanistically understood to occur as the side chains of amino acids in the sequon are reoriented in the OST active site such that the side chain of the +1 residue is positioned away from the target Asn side chain amide and the Ser/Thr side chain hydroxyl in the +2 position. The role of conformational flexibility was highlighted in studies where Cys residues that constrained the conformational degrees of freedom were incorporated N- and C-terminal to consensus sequences in model peptides (10). In the preceding case, the rigidity resulting from the formation of disulfides proximal to the consensus Asn was negatively correlated with N-glycan occupancy in acceptor peptides. The Asx-turn motif was found to be associated with N-glycan occupancy based on studies of the solution conformational properties of a series of tripeptides as well as their competency as an acceptor substrate for the OST complex (11, 12). These findings were subsequently validated by assessing the substrate specificity of a constrained synthetic peptide, which adopted an Asx-turn or β-turn motif (13, 14).
Petrescu et al. (8) surveyed the neighboring amino acids and structural features found on glycosylated Asn residues on proteins deposited in the Protein Data Bank (15). There is a greater likelihood of finding aromatic, hydrophobic amino acids immediately before the glycosylated Asn residue as well as small hydrophobic and larger hydrophobic amino acids in the +1 and +3 positions, respectively. There was also a preference for finding Pro in the vicinity of the occupied residue except for the complete absence in the +1 position and reduced frequency in the +3 position. From a structural standpoint, it was found that there was some preference for finding occupied Asn residues on turns and bends but that there was a marked preference for finding occupied Asn residues in structural transitions where the transition occurred at the Asn residue itself or in the +2 or −2 position with respect to the Asn (8). In subsequent work, it was found that the probability of Asn occupancy was highly dependent on the distance of the Asn side chain amide to the Ser/Thr side chain hydroxyl in the +2 position. The greatest frequency of N-glycosylation occurred when this distance was ∼7.3 Å (16).
Although an understanding of the types of secondary structures associated with N-glycosylation is important for assessing the probability of glycosylation at a given consensus Asn, it is important to note that proteins are typically unstructured at the time of modification. The OST enzyme complex is membrane bound and forms a ternary complex with the 60 S ribosomal subunit and the Sec61 protein translocation channel in the rough endoplasmic reticulum lumen (17, 18). N-Glycans are attached to the nascent polypeptide chain on the lumenal side of the endoplasmic reticulum (ER)2 as it is secreted from the ribosomal peptidyl transferase site (P site), which is located on the cytoplasmic side of the ER (19). The minimal length of an extended polypeptide chain necessary to traverse the distance through the Sec61 protein translocation channel between the P site and the OST complex is 65 and 75 residues. This relatively short distance has lead to the concept of protein N-glycosylation as a co-translational or, perhaps more accurately, a co-translocational event (20–22). The coincidental occurrence of translation and N-glycosylation implies that protein folding does not influence the occurrence of oligosaccharides at a particular site. Indeed, the point has been made in previous studies that examination of the structural context of N-glycosylation is important for providing an understanding of evolutionarily conserved glycosylation motifs (8), however, structural aspects do not necessarily drive the modification event.
We recently documented the presence of N-glycosylation on asparagine residues not adhering to the canonical motif NX(S/T), where X is not proline (23). This unexpected modification was located on asparagine 162 in the CH1 domain of human antibodies. Building on this previous finding we asked the question of whether this was an isolated phenomena or something that occurred widely on other non-consensus asparagine residues in IgG. In our follow up studies, we enriched non-consensus N-glycan structures on a recombinant human antibody. By exploiting the differential activity of endoglycosidases to consensus and non-consensus N-glycans and applying classic lectin affinity enrichment techniques, we have been able to more fully probe the tolerance of the OST enzyme complex to non-canonical motifs and acceptor residues. Our approach has led to the discovery of a glutamine residue modified with oligosaccharide structures, a finding that stands in contradiction to our current understanding of the limitations that protein sequence imposes on the enzymatic activity of cellular glycosylation machinery. Of no less importance are the implications that arise out of the discovery of 3 additional non-consensus Asn-linked glycosylation sites on a recombinant human IgG2 antibody, one of which was also observed on antibodies obtained from human serum. From our data set, we have delineated the secondary structural motifs that are correlated with non-consensus glycosylation (NCG) based on known crystal structures of antibody constant domains and homology modeling of the occupied Gln and Asn residues. We propose the non-consensus sequence motif (S/T)XN, where N is glycosylated, X may be any amino acid, is necessary but not sufficient for N-glycosylation when S/T is not present in the +2 position. Taken together our current results enable further inquiry into this highly unusual modification in a targeted manner by providing parameters for in silico prediction of NCG based on sequence and secondary structural motifs.
MATERIALS AND METHODS
Recombinant Antibodies
The IgG2 antibodies used in this study were human recombinant molecules stably expressed in Chinese hamster ovary cells and purified using conventional techniques (24). Purified antibodies were formulated in sodium acetate buffer at pH 5.0.
Endo- and Exoglycosidase Digestion
The CH2 domain consensus N-glycans at Asn-296 (equivalent to 314 in Kabat numbering (25)) were removed from ∼300 mg of human recombinant IgG2 antibody or the IgG component of pooled normal human serum (Sigma). The samples were diluted in 30 ml of 50 mm Tris-HCl and deglycosylated with 300,000 units of PNGase F (New England Biolabs, Ipswich, MA) for 8 h at 37 °C with orbital agitation at 60 rpm. Terminal N-acetylneuraminic acid on antibody oligosaccharide structures that have been observed on non-consensus N-glycans were removed by addition of 2 units of sialidase A (Glyko, Novato, CA) and further incubation as described above for 2 h. After treatment with endo- and exoglycosiases, the volume of the antibody pool was increased to 100 ml with the addition of phosphate-buffered saline (PBS) and bound to a 5-ml HiTrap MabSelect SuRe protein A column (GE Healthcare) at a flow rate of 2.0 ml/min. The bound antibody was washed with 5 column volumes of PBS to deplete the treated pool of released oligosaccharides prior to lectin chromatography. Bound antibody was eluted with 50 mm sodium citrate at pH 3.5 and the pH of the eluate was increased to 7.5 by addition of 1.0 m Tris-HCl at pH 8.0. The protein A eluate was vacuum filtered with a Steriflip cartridge (Millipore, Bilerica, MA) and the volume of the eluted pool was brought to 100 ml with PBS.
Lectin Affinity Chromatography
The deglycosylated, protein A purified antibody was passed over a 2-ml affinity column of immobilized Erythrina cristagalli (Vector Labs, Burlingame, CA), which is specific for terminal galactose, at 0.1 ml/min. The lectin-bound antibody was washed with 5 column volumes of PBS at 0.5 ml/min and eluted with 0.2 m lactose-PBS at 0.5 ml/min. Lectin eluates containing antibody were concentrated 10-fold in Centricon/Centriprep spin filters (Millipore) with a 30-kDa molecular mass cut-off and buffer exchanged into 20 mm sodium acetate, pH 5.0. The final protein concentration was typically 2 mg/ml.
Liquid Chromatography-Mass Spectroscopy (LC-MS) of Reduced Heavy and Light Chains
Reversed-phase separation of antibody heavy and light chains and subsequent mass measurement was carried out as described previously (23).
Peptide Map Analysis
Human antibody was reduced and alkylated prior to peptide map analysis according to previously established methods (23). When removal of non-consensus N-glycans was required, 1500 units of PNGase F was added to 100 μg of reduced and alkylated antibody and subsequently incubated at 37 °C for 3 h. Urea was then added to samples at a final concentration of 2.0 m as well as recombinant trypsin (Roche Diagnostics) at a ratio of 1:10 (w/w) and incubated at 37 °C for 4 h. Peptides were separated using a Varian Polaris ether C18 column (1.0 × 250 mm) at 50 °C on a Waters Aquity HPLC (Waters, Milford, MA) at a flow rate of 70 μl/min. The mobile phases used in the separation were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The peptides were bound to the column in 0.5% B and the buffer composition was maintained for 10 min, at which time a linear gradient to 50% B in 90 min was initiated to elute the peptides. The column was brought to 90% B over 12 min and maintained at 90% B for 5 min. Following the column wash, the mobile phase composition was brought to the initial conditions in 3 min and equilibrated for 40 min prior to the next injection. The identification of peptides was determined using a Thermo LTQ XL mass spectrometer (Thermo Scientific, Waltham, MA) set to perform collision-induced dissociation (CID) MS2 and MS3 in a data-dependent manner.
Site Identification of Non-consensus N-Glycans
Prior to reduction and alkylation of antibody, 0.3 units of endoglycosidase F2 enzyme was added (Glyko) and samples were incubated at 37 °C for 16 h. Subsequent sample preparation and peptide map separation was carried out as described above. The eluate from the HPLC column was split using an Advion Nanomate fraction collection robot (Advion Biosciences, Ithaca, NY). Briefly, the flow rate of 70 μl/min was split and 150 nl was analyzed on-line with a Thermo LTQ XL mass spectrometer with electron transfer dissociation (ETD) capability (Thermo Scientific), whereas the remainder was collected in a 96-well plate for off-line analysis. Endo F2-digested glycoconjugates containing a HexNAc-Fuc disaccharide were analyzed by MS using the Nanomate in static-nanospray infusion mode. The candidate glycopeptide oligosaccharide linkage was established using a combined approach involving CID-MS2 at 16–35 volts followed by ETD-MS3 or CID-MS3 of the putative glycopeptides. The dominant fragments observed from CID-MS2 analysis of the endo F2-treated glycopeptides were product ions corresponding to the facile loss of the core-linked fucose residue. The dominant CID-MS2 product was further fragmented by ETD-MS3 or CID-MS3 when ETD did not yield informative fragments ions and the modified amino acid site was determined by a careful comparison of the fragment ions observed in the putative glycopeptides and their non-glycosylated counterparts.
IgG2 Homology Model
Structural homology models of the human IgG2 were generated in-house using the Molecular Operating Environment (Chemical Computing Group, Montreal, Canada) and PyMOL for both constant and variable regions. Constant domains were also modeled with Swiss-Model (26). The scaffolds for creating the homology model were selected based on sequence similarity and multiple structure factors across the chains from a single Fv structure for the non-CDR regions, and based on CDR length, sequence similarity, and structure diversity for the CDR regions, utilizing known antibody structures from both in-house efforts and from the RCSB Protein Data Bank. The Fc domain structure is modeled from antibody 1HZH in the RCSB Protein Data Bank (27). Percent solvent accessibility was calculated using ASAview (28) and the reported values are expressed as relative solvent accessibility.
RESULTS
Lectin Enrichment and Reduced Mass Analysis of Lectin-enriched NCG Sites
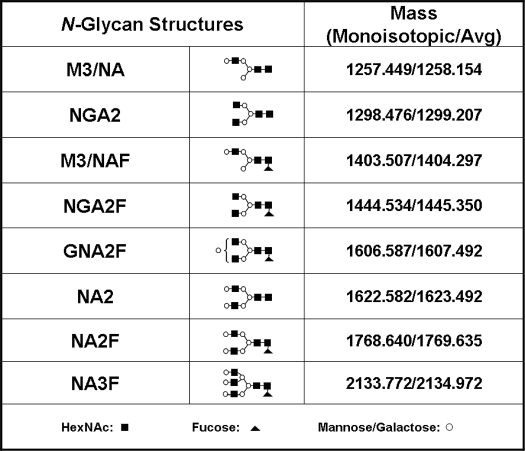
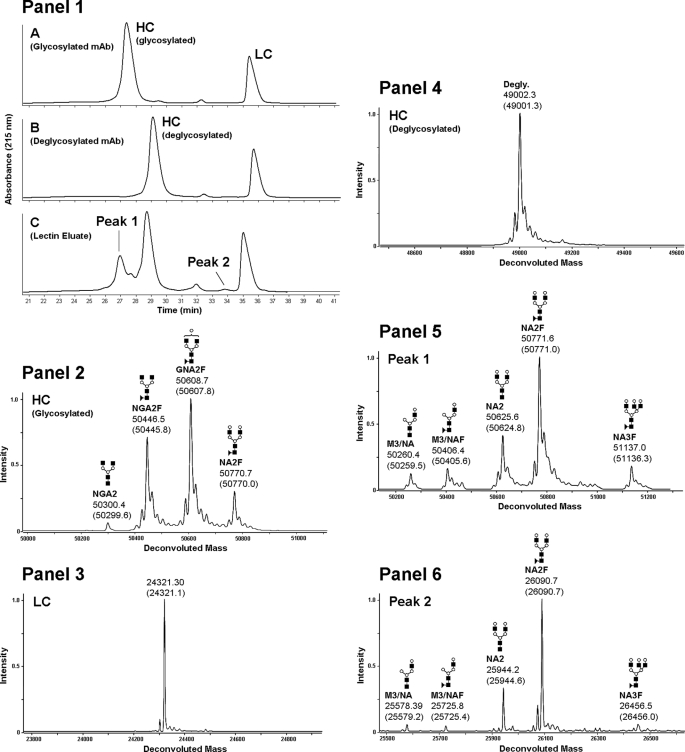
Consensus glycosylation sites were removed with endoglycosidases and non-consensus sites were enriched with lectin affinity chromatography as described under “Materials and Methods.” The presence of NCG on both the heavy and light chain of a recombinant IgG2 antibody was detected by LC-MS after lectin enrichment. The experimental masses of the glycosylated, deglycosylated, and lectin-enriched antibody samples were compared with the theoretical values of the antibody HC and LC fragments. We found that the experimental masses of the lectin-enriched material could be interpreted as HC and LC modified with various oligosaccharide structure shown in Table 1. The structures themselves have not been elucidated, rather, they are inferred on the basis of mass and agreement with structures typically found in recombinant antibodies expressed in Chinese hamster ovary cells (29). The reduced glycosylated heavy chain (Fig. 1, Panel 1, A) eluted before the deglycosylated heavy chain (Fig. 1, Panel 1, B) by reversed-phase LC-MS. The antibody eluted from the lectin column contained an early eluting heavy chain population with a retention time that was consistent with glycosylated heavy chain (Fig. 1, Panel 1, C). The mass spectrum of the glycosylated heavy chain peak (Fig. 1, Panel 2) consisted primarily of species with NGA2F, GNA2F, and NA2F oligosaccharides (Table 1). The mass spectrum of the reduced antibody light chain was found to be consistent with the expected, theoretical mass (Fig. 1, Panel 3). After PNGase F and sialidase A treatment, the reduced heavy chain mass was consistent with the expected mass for the deglycosylated species (Fig. 1, Panel 4), demonstrating that enzymatic treatment efficiently removed the N-linked glycans present on the CH2 consensus site Asn-296. The masses of the early eluting HC peak (Fig. 1, Panel 1, C, with heavy chain modified with M3/NA, M3/NAF, NA2, NA2F, and NA3F oligosaccharide structures (Fig. 1, Panel 5) and the mass of the later eluting heavy chain peak was consistent with deglycosylated heavy chain (data not shown). A low level peak was observed eluting before the light chain in the lectin eluate (Fig. 1, Panel 1, C, peak 2). The masses of this species were consistent with the expected mass of the light chain modified with M3/NA, M3/NAF, NA2, NA2F, and NA3F oligosaccharide structures (Fig. 1, Panel 6). These results suggested that the enriched samples likely contained oligosaccharides on antibody domains other that CH2 that were not affected by PNGase F treatment. The different glycan profiles observed on the original antibody and the lectin eluate samples were reminiscent of substantial differences often observed between glycan structures present on consensus sites in the variable region and those observed on the CH2 domain consensus site (30).
TABLE 1.
Oligosaccharide structures observed on non-consensus Asn and Gln residues
Monoisotopic and average mass additions observed on peptides and reduced antibody fragments are shown and the structures themselves are inferred on the basis of mass alone.
FIGURE 1.
Reduced mass analysis of glycosylated, deglycosylated, and lectin-enriched recombinant human IgG2 antibody. The UV chromatogram of the reversed phase separations of reduced glycosylated, deglycosylated, and lectin-enriched antibody samples are shown in Panel 1, A–C, respectively. The glycosylated HC mass spectrum is shown in Panel 2, the light chain mass spectrum is shown in Panel 3 and the deglycosylated antibody HC spectrum is shown in Panel 4. The lectin-enriched antibody HC mass spectrum from reversed phase peak 1 (Panel 1, C) is shown in Panel 5 and the lectin-enriched antibody LC mass spectrum from reversed phase peak 2 (Panel 1, C) is shown in Panel 6. The theoretical masses for each species are listed in parenthesis below the observed masses.
Peptide Map Analysis of Lectin Eluate
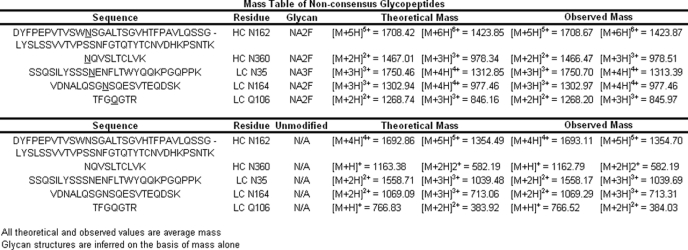
Tryptic peptide maps were undertaken to determine N-glycosylation sites that were enriched by lectin affinity purification. The presence of the deglycosylated tryptic peptide in the CH2 domain of IgG2 antibody containing Asn-296 and the complete absence of the glycosylated form of the same peptide indicated that binding of deglycosylated antibody to the ricin-agarose lectin column was due to oligosaccharide structures present elsewhere on the molecule. In agreement with our prior results (23), the major glycosylated species observed was the CH1 tryptic peptide corresponding to IgG2 amino acids 151–213 (25), which contained the putative non-consensus N-glycosylation site at Asn-162 (equivalent to residue 162, Kabat numbering; data not shown). A comparison of peptide data from the lectin-enriched antibody and the non-enriched starting material revealed 4 additional peptides with masses consistent with oligosaccharide structures on Asn or Gln residues that were not part of the canonical N-glycosylation sequence motif. The candidate glycopeptide sequences and mass data are summarized in Table 2 and included Asn-linked glycopeptide in CH3 and CL antibody domains as well as an Asn-linked glycopeptides in CDRL1 and an apparent Gln-linked glycopeptide in the VL antibody domain. The CH1 and CH3 domain tryptic glycopeptides described above were also observed in the lectin-enriched antibody sample derived from pooled normal human serum (data not shown), indicating that NCG also occurs in vivo. The lower apparent level of NCG observed in human serum may be due to the abundant consensus CDR glycosylation, which is typically present on 30% of circulating antibodies (29). It is possible that CDR glycans were not completely removed by PNGase F digestion under native conditions and these populations were then isolated during lectin capture along with antibody populations modified with non-consensus glycans thus reducing the efficiency of the enrichment.
TABLE 2.
Multiply charged masses for non-consensus glycopeptides and the corresponding unmodified peptides
The glycosylated residue is underlined in the peptide sequence and enumerated in the residue column. The theoretical multiply charged masses were calculated for each observed mass for comparison.
Identification of Glycosylated Residues
ETD-MS fragmentation has previously been used to identify O-glycosylation sites (31–33), however, the significantly larger size of N-glycans relative to O-glycans makes it much more difficult to determine the glycan-amino acid site of attachment on the intact glycopeptide. To simplify site identification of non-consensus N- and Q-linked glycans, samples were digested with endoglycosidase F2, which cleaves specifically after the 1st GlcNAc residue in the core structure of N-glycosylated oligosaccharides resulting in a fucosylated N-acetylglucosamine (HexNAc-Fuc) disaccharide at the amino acid site of attachment or a peptide with a single HexNac residue if the glycan lacks a core fucose.
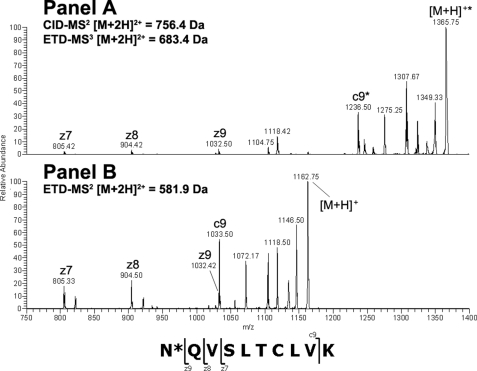
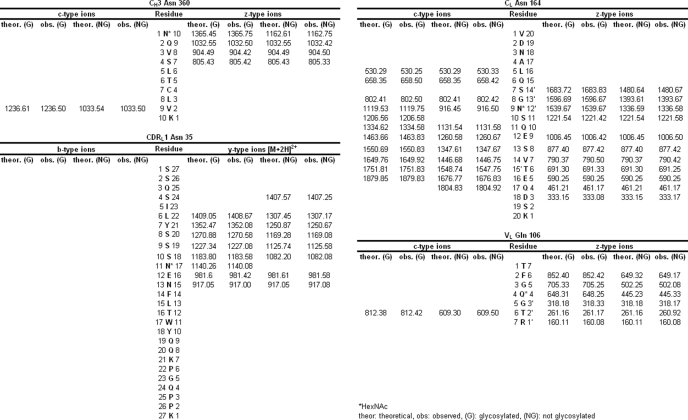
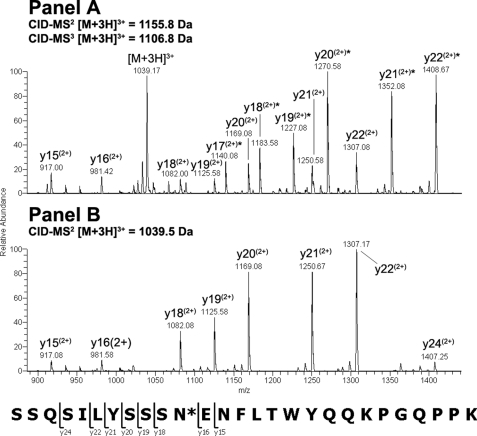
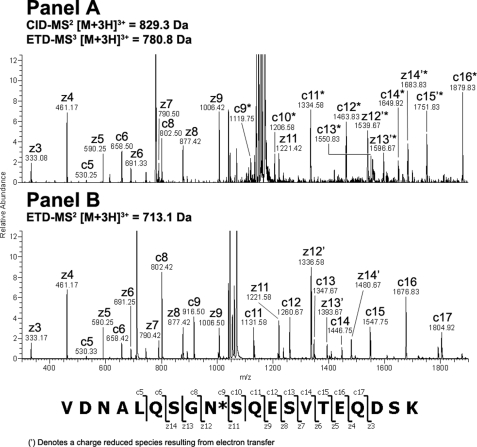
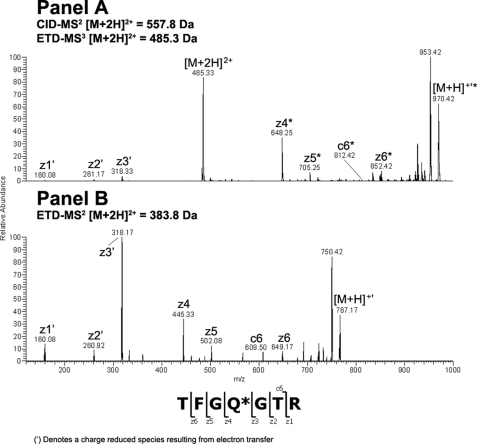
Endo-F2 digestion of CH3 domain tryptic peptide residues 360–369 modified with an A2F oligosaccharide resulted in an [M + 2H]2+ ion at m/z = 756.40 Da. Application of the CID-MS2/ETD-MS3 analysis described above on the CH3 glycopeptide and the corresponding unmodified peptide (Fig. 2, Panels A and B, respectively) resulted in a clear z-ion series for both species. A comparison of the theoretical and observed masses for the c- and z-type ions resulting from ETD fragmentation of the glycosylated and non-glycosylated peptides indicated that the glycan was attached to the peptide N-terminal Asn at position 360 (384, Kabat numbering) in the IgG2 CH3 domain based on the observed mass addition of 203 Da on the glycopeptide (Table 3). Endo-F2 digestion of CDRL1 domain tryptic peptide residues 25–51 modified with an A3F oligosaccharide resulted in an [M + 3H]3+ ion at m/z = 1155.80 Da, which was consistent with mass of the CDRL1 peptide modified with a HexNAc-Fuc disaccharide. Adequate sequence information could not be obtained on the modified species using ETD fragmentation so the −fucose product from the CID-MS2 scan event was further fragmented by CID-MS3 and compared with the CID-MS2 spectra of the unmodified peptide (Fig. 3, Panels A and B, respectively). A comparison of the modified and unmodified peptide clearly indicated that Asn-35 (29, kabat numbering) was glycosylated based on the observed addition in mass of ∼203 Da evident in the y-ion series (Table 3). There was no evidence for the modification of the Asn residue in position 37 based on the CID-MS3 spectrum of the glycopeptide. Endo-F2 digestion of CL domain tryptic peptide residues 156–175 modified with an A2F oligosaccharide resulted in an [M + 3H]3+ ion at m/z = 829.30 Da. A fragment ion comparison of this species and the unmodified peptide using the methodology described above (Fig. 4, Panels A and B, respectively) indicated that the glycan was attached at the Asn residue in position 164 (158, Kabat numbering) based on the observed mass addition of ∼203 Da on the glycopeptide relative to the unmodified peptide (Table 3). It was also evident that the other Asn residue on the glycopeptides, in position 158, was not occupied as there was no evidence of product ions showing an addition in mass consistent with a HexNAc modification in c-type ions from c5 to c8 in the ETD-MS3 spectrum of the glycopeptide. Endo-F2 treatment of the VL domain tryptic peptide resulted in an [M + 2H]2+ ion at m/z = 557.80 Da. Application of the CID-MS2/ETD-MS3 analysis used previously on the glycopeptide and the corresponding unmodified peptide (Fig. 5, Panels A and B, respectively) resulted in a clear z-ion series for both species. A comparison of the modified and unmodified peptide clearly indicated that the Gln residue at position 106 (100, Kabat numbering) was glycosylated based on the observed addition in mass of ∼203 Da evident in the z-ion series beginning at z4 (Table 3). The assignment of the modified residue was unambiguous due to the z-ion series that covered the entire sequence of the peptide, and the absence of any Asn residues in the sequence that could be modified with an oligosaccharide.
FIGURE 2.
ETD-MS2/MS3 comparison of glycosylated and non-glycosylated CH3 peptide amino acids 360–369 after treatment with endo-F2. The modified [M + 2H]2+ peptide ion at m/z = 756.4 Da was first analyzed by CID-MS2 to remove the core fucose. The dominant [M + 2H]2+ product ion a at m/z = 683.4 Da was analyzed by ETD-MS3 and the resulting spectrum was compared with the ETD-MS2 spectrum from the unmodified [M + 2H]2+ peptide ion at m/z = 581.9 Da.
TABLE 3.
Table of ETD and CID fragment ions
The observed masses of the MS3 fragment ions for each Endo-F2-digested glycopeptide and the MS2 fragments from the corresponding unmodified peptides were compared to the theoretical masses. The mass added to the modified residue corresponds to a HexNAc monosaccharide, which is maintained at the site of modification after the loss of the core-linked fucose in the MS2 scan event.
FIGURE 3.
CID-MS2/MS3 comparison of glycosylated and non-glycosylated CDRL1 peptide amino acids 25–51 after treatment with endo-F2. The modified [M + 3H]3+ peptide ion at m/z = 1155.8 Da was first analyzed by CID-MS2 to remove the core fucose. The dominant [M + 3H]3+ product ion a at m/z = 1106.8 Da was further analyzed by CID-MS3 and the resulting spectrum was compared with the CID-MS2 spectrum from the unmodified [M + 3H]3+ peptide ion at m/z = 1039.5 Da.
FIGURE 4.
ETD-MS2/MS3 comparison of glycosylated and non-glycosylated CL peptide amino acids 156–175 after treatment with endo-F2. CID-MS2 of the [M + 3H]3+ peptide ion at m/z = 829.3 Da resulted in the removal of the core fucose from the CL glycopeptide. The dominant [M + 3H]3+ product ion a at m/z = 780.8 Da was further analyzed by ETD-MS3 and the resulting spectrum was compared with the ETD-MS2 spectrum from the unmodified [M + 2H]2+ peptide ion at m/z = 713.1 Da.
FIGURE 5.
ETD-MS2/MS3 comparison of glycosylated and non-glycosylated VL peptide amino acids 103–109 after treatment with endo-F2. The modified [M + 2H]2+ peptide ion at m/z = 557.8 Da was first analyzed by CID-MS2 to remove the core fucose. The dominant [M + 2H]2+ product ion a at m/z = 485.3 Da was subsequently analyzed by ETD-MS3 and the resulting spectrum was compared with the ETD-MS2 spectrum from the unmodified [M + 2H]2+ peptide ion at m/z = 383.8 Da.
Enzymatic Release of Non-consensus N-Glycans
As discussed above we found that NCG present in the CH1 domain of human antibodies at Asn-162 had an apparent resistance to digestion by PNGase F under native, non-denaturing conditions, whereas the consensus site on the CH2 domain Asn at position 296 was easily deglycosylated under the same conditions. Glycan cleavage from non-consensus sites required the samples to be first denatured at an elevated temperature in the presence of 4 m guanidine HCl, and subsequently reduced and alkylated. The apparent enrichment of several non-consensus oligosaccharide structures provided us with an opportunity to test this observation in a more thorough manner. Previous work by Fan and Lee (34) on the substrate specificity of PNGase F to chemically synthesized N-glycosylated peptides has shown that the glycolytic activity of the enzyme is dramatically reduced when the +2 amino acid is not Ser or Thr. The pre-treatment levels of non-consensus glycopeptides in the lectin-enriched eluate were quantitated by extracted ion current comparison of the modified and unmodified peptides using the observed masses shown in Table 2. The levels of NCG in the starting material (pre-lectin enrichment) were inferred based on the fold-enrichment of CH1 NCG as a consequence of the lectin enrichment. The CH1 NCG structures were enriched ∼25-fold following lectin affinity chromatography (Table 4) and this factor was used to estimate the starting levels of all other NCG as they were not detectable in the pre-lectin enrichment starting material. We investigated the substrate specificity of PNGase F to endogenous, non-consensus glycans present on the recombinant antibody used in this study by treating the lectin-enriched samples with PNGase F prior to denaturing reduction and alkylation or after denaturing reduction and alkylation and monitored the reactions by extracted ion current quantitation of the glycopeptides in the tryptic peptide maps. Addition of PNGase F prior to denaturing reduction and alkylation was generally not effective at releasing non-consensus glycans as the reduction in the levels of the various glycopeptides decreased less than 15% for 4 out of 5 glycopeptides compared with the pre-treatment levels (Table 4). However, when PNGase F was added after the sample was denatured in 4.0 m guanidine and subsequently reduced and alkylated, the levels of 4 out of 5 of the non-consensus glycopeptides dropped to less than 2% of their pre-treatment levels (Table 4).
TABLE 4.
Oligosaccharide occupancy levels observed on non-consensus Asn and Gln residues before and after lectin enrichment
The pre-treatment level of NCG is inferred from the fold-enrichment of the glycosylated CH1 peptide due to lectin affinity chromatography. Reduction of occupancy levels was observed after treatment of the sample with PNGase F either prior to or after denaturation at elevated temperatures in the presence of 4 m guanidine HCl followed by reduction and alkylation as shown in the 4th and 5th columns, respectively.
| Glycan site | % Gly |
|||
|---|---|---|---|---|
| Unenriched | Enriched | Enriched de-Gly-R/Aa | Enriched R/A-de-Gly | |
| HC Asn-162 | 1.07 | 24.50 | 24.35 | 2.94 |
| HC Asn-360 | 0.12b | 2.73 | 2.58 | 0.03 |
| LC Asn-35 | 0.01b | 0.23 | 0.14 | NDc |
| LC Asn-164 | 0.02b | 0.44 | 0.38 | ND |
| LC Gln-106 | 0.02b | 0.59 | 0.47 | ND |
a R/A, reduced and alkylated.
b Value extrapolated from HC Asn-162 fold-enrichment.
c ND, not deteted.
Structural Motifs and Solvent Accessibility of Glycosylated Non-consensus Residues
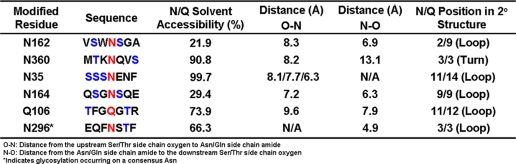
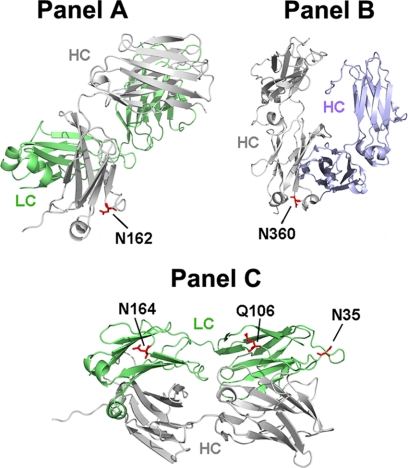
A homology model of the recombinant human IgG2 antibody that was the subject of this study was generated based on known crystal structures of IgG1 and IgG2 antibodies with high sequence homology found in the RCSB Protein Data Bank. The solvent accessibility of each residue was determined by modeling the exposed surface of each amino acid to a water molecule probe (35). Each of the NCG sites reported here was found to be solvent accessible with values ranging from 22 to 99% (Table 5). Asn-162, in the CH1 domain is the least solvent accessible non-consensus site with a calculated value of 22% (Fig. 6, Panel A). This residue was found on the second position of an 8-residue loop (Table 3). Asn-360, which is located in the CH3 domain, was found to have a calculated solvent exposure of 91% (Fig. 6, panel B) and was located in the 3rd position on a 3-residue solvent accessible turn (Table 5). The calculated solvent accessibility of Asn-35 on CDRL1 was 99.7% (Fig. 6, Panel C) and this residue was in the 11th position of a 14-residue loop (Table 5). The CL domain Asn in position 164 has limited solvent accessibility, ∼29% (Fig. 6, panel C) and is located in the 9th position of a 9-residue loop (Table 5). The solvent accessibility of Gln-106 located in the VL domain was found to 74% (Fig. 6, panel C) and placement of this residue was in the second to last position of a 12-residue loop (Table 5). These results are in agreement with a recent report that surveyed structural features of consensus glycosylation observed in the PDB and found that glycosylation occurred on residues with surprisingly little solvent accessibility and in regions near changes in secondary structure (8). The position of the Asn/Gln amide with respect to the hydroxyl group of Ser/Thr residues located N- or C-terminal to Asn was also determined using the homology model based on the IgG2 crystal structure and these distances are summarized in Table 5. A Ser or Thr residue was found in the −2 position with respect to the non-consensus glycosylated Asn residue in all occurrences of this modification. It should be noted that all Asn residues in the recombinant IgG2 sequence occurring in loops or turns with a Ser or Thr in the −2 position were glycosylated to some degree.
TABLE 5.
Summary of the structural context and amino acid sequence surrounding each NCG site and the consensus glycosylation site in IgG Fc
The solvent exposure of each occupied Asn residue was calculated by modeling the exposed surface of each amino acid to a water molecule probe. The distance from the upstream or downstream Ser/Thr side chain oxygen atom to the occupied Asn/Gln side chain amide was calculated from an IgG2 homology model. The Ser/Thr amino acids from which distance measurements were taken are shown in blue and the occupied Asn/Gln residues are shown in red. The relative position of the occupied Asn/Gln residue within the secondary structural element is shown in the 6th column.
FIGURE 6.
Crystal structure of IgG2 Fab and Fc domains showing the location of non-consensus oligosaccharide structures. In Panel A, the IgG2 Fab crystal structure is rotated so that the HC Fd (gray) is in the foreground and the LC (green) is in the background and the non-consensus site at Asn-162 is shown in red. Panel B shows IgG2 Fc homodimer crystal structure with one chain colored gray and the other modified chain colored blue and the NCG site at Asn-360 shown in red. In Panel C, the IgG2 Fab crystal structure is oriented to show the occupied Asn and Gln residues (red) at positions 164, 106, and 35, respectively, from left, occurring on the LC (green).
DISCUSSION
Using a combination of differential deglycosylation, lectin enrichment, and sensitive mass spectrometric analyses, we found evidence for glycosylation events occurring outside the well established consensus motif. Although validation of this enrichment strategy on other protein types is necessary to ultimately assess the general utility of the above techniques, clearly, they have been successful for analyzing non-consensus structures on antibodies. Without question, the most surprising result to come out of our current study has been the discovery of oligosaccharide structures on a Gln residue. Such a finding has never been described in nature nor resulted from in vitro studies using model peptides and purified intact OST enzyme complexes. Interestingly, with exception of the Gln residue, which is occupied, the modification follows the consensus sequence motif for N-glycosylation, NX(S/T). Although Gln shares chemical properties with Asn, it was thought that the addition of an extra methyl group, which adds ∼1.5 Å to the side chain length relative to Asn, would make OST binding and thus even fractional occupancy of a Gln residue highly unlikely. However, it now seems that the factors that govern fractional oligosaccharide occupancy are more fluid than previously thought. It is then perhaps reasonable to expect that replacement of an Asn on a constitutively modified sequon with a Gln residue might have some very low level of occupancy that would not be observable without the enrichment and detection strategies that we have employed in the current work.
In this study, we have also sought to define the structural and conformational contexts that are associated with NCG. Our results indicate that Ser/Thr amino acids that are located in the −2 position relative to the occupied Asn are mechanistically important for non-consensus N-glycosylation. The complete lack of a C-terminal Ser/Thr residue following CDRL1 Asn-35 and our prior results in which the mutation of the non-consensus CH1 sequence from VSWN162SGA to VSWN162AGA resulted in a 2-fold increase in glycosylation at Asn-162 (23) indicate that the occurrence of NCG does not require a C-terminal Ser/Thr. Additional evidence highlighting the lack of importance of a Ser/Thr located C-terminal to a non-consensus Asn is drawn from a measurement of the distance between the CH3 Asn-360 side chain amide and the Ser side chain hydroxyl in the +3 position that is 13.1 Å, well outside of typical values observed for the amide-hydroxyl distance in consensus sequons (16). Although it has been determined that residues in the +3 position can inhibit glycosylation in consensus sequons to some degree (36), it is highly unlikely that they would participate mechanistically, over a relatively great distance, in a positive manner. All NCG sites that are known contain a Ser or Thr residue in the −2 position and it now seems apparent that these residues may perhaps function as a hydrogen acceptor when there is no Ser or Thr residue present in the +2 position. Petrescu et al. (16) surveyed the Structural Assessment of Glycosylation Sites data base to determine the distance between the nitrogen where N-glycosylation takes place and the side chain oxygen of the sequon serine/threonine located in the +2 position. The N-O distance was found distributed in the 4–10-Å range, with a mean of 7.3 Å. The distance from the N-terminal Thr side chain oxygen in the −2 position to the Asn-360 amide was 8.2 Å, which is in line with the 7.3 Å average distance between these atoms in the consensus sequence (16). The distance from the Ser/Thr side chain hydroxyl located N-terminal to the non-consensus Asn was determined for all of the non-consensus sites (Table 5) and all distances are within 1 Å of the average value cited by Petrescu et al. (16) for consensus sequons. We believe that our results offer convincing evidence for the existence of a non-consensus N-glycosylation sequence motif (S/T)XN, where N is glycosylated, X may be any amino acid. This motif seems to be necessary but not sufficient for N-glycosylation when S/T is not present in the +2 position.
Our results indicate that the non-consensus N-glycosylation motif is merely a backwards consensus N-glycosylation motif. Certain amino acids flanking the consensus N-glycosylation site are correlated with site occupancy (8) and we believe that this correlation can be translated to apply to NCG in a limited manner. It was established by Petrescu et al. (8) that the presence of a large aromatic residue, particularly Trp and to a lesser extent Tyr in the −1 position was correlated with a greater probability of occupancy on a consensus Asn. Sequence analysis of all non-consensus sites indicates that the non-consensus sequon, Asn-162, which is the most abundant non-consensus site, is indeed preceded by a Trp in the −1 position (Table 5), whereas the lower level non-consensus glycopeptides do not contain a −1 Trp. At first glance, this result argues against the idea that the non-consensus sequon is merely a backwards consensus sequon. We have shown in our previous study (23), however, that mutation of the −1 Trp to Ala in the CH1 domain sequence VSWN162SGA resulted in a 4-fold increase in NCG observed at Asn-162. In the case of our hypothetical non-consensus sequon, the +1 residue with respect to the glycosylated Asn corresponds to the −1 residue in the consensus sequence. We surmise that the presence of Trp in the −1 position of the CH1 non-consensus sequence SWN negatively affects glycan occupancy on Asn-162 by interfering with the interaction between the −2 Ser and the glycosylated Asn. This result on the non-consensus sequon would be consistent with the work of Kasturi et al. (37), which demonstrated that the presence of Trp in the +1 position had an inhibitory effect on N-glycan occupancy of the consensus sequon NXS in an in vitro system.
Through the study of known antibody constant domain secondary structures, we have determined that NCG occurred on Asn residues that were present on highly flexible loops and turns. The amino acid length of loop/turn structures on which an Asn residue was present varied from 3 to 14 residues in length. The wide distribution of lengths prompted an investigation into the centrality of the occupied Asn/Gln residues within the loop/turn structure. We found that NCG occurred exclusively on domains that were within 3 residues of a transition in the secondary domain structure that is consistent with structural contexts typically associated with consensus glycosylation (8). An association of consensus and NCG events with protein secondary structural features is highly relevant for in silico prediction of glycosylation. Recent results have clarified the complementary roles of the two subunits of the OST complex, STT3A and STT3B, in co-translational as well as post-translational glycosylation events (38). It is widely understood that the co-translational glycosylation occurs in the absence of the protein secondary structure, whereas post-translational glycosylation is concurrent with folding events, as is the case for human coagulation factor VII, which has been shown to be glycosylated well after translation, while it is being folded in the lumenal space of the ER (39). It is not known whether or not NCG is mediated by the post-translational machinery of the OST enzyme complex. We can speculate, however, that if NCG is mediated by the STT3B subunit, then the structural features associated with this modification may have a direct impact on non-consensus occupancy. The relatively long time frame associated with the folding of various antibody domains, particularly CH1, which contains the most abundant non-consensus site (40), and the long residence time in the ER-lumen that is implied by this process, could favor NCG events. The implication is that proteins that fold on a very fast time scale may not reside in the ER-lumen for a sufficient period of time for NCG to occur. However, multidomain proteins that undergo extensive post-translational folding may be more likely to be glycosylated at non-consensus residues merely because there is a longer period of time in which these proteins sample the lumenal space and thus a greater likelihood that they will transiently interact with the post-translational glycosylation machinery.
In the present study, we have extended the understanding of the phenomena of NCG and, with the discovery of a glycosylated glutamine residue, added to the repertoire of residues that may be modified with oligosaccharides. Our discovery of 4 NCG sites has allowed us to survey the distance between amino acid side chains thought to be involved mechanistically, propose a non-consensus N-glycosylation sequence motif, and specify secondary structural characteristics associated with this unusual modification. The cataloging of further NCG sites is ongoing and will continue to contribute to the evolving view of the fidelity of this ubiquitous protein modification.
Acknowledgments
We are grateful to Jennifer Kerr and Maria A. Vanushkina for assistance with lectin affinity chromatography and Julia Bach for help with protein A purification. We thank Dean Pettit for critical review of this manuscript and continuing support of this work.
Footnotes
- ER
- endoplasmic reticulum
- NCG
- non-consensus glycosylation
- PBS
- phosphate-buffered saline
- PNGase F
- peptide:N-glycosidase
- HPLC
- high pressure liquid chromatography
- CID
- collision-induced dissociation
- ETD
- electron transfer dissociation
- LC
- liquid chromatography
- MS
- mass spectrometry
- HC
- heavy chain
- LC
- light chain.
REFERENCES
- 1.Bause E., Hettkamp H. (1979) FEBS Lett. 108, 341–344 [DOI] [PubMed] [Google Scholar]
- 2.Marshall R. D. (1974) Biochem. Soc. Symp. 40, 17–26 [PubMed] [Google Scholar]
- 3.Bause E., Legler G. (1981) Biochem. J. 195, 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silberstein S., Gilmore R. (1996) FASEB J. 10, 849–858 [PubMed] [Google Scholar]
- 5.Bause E., Breuer W., Peters S. (1995) Biochem. J. 312, 979–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breuer W., Klein R. A., Hardt B., Bartoschek A., Bause E. (2001) FEBS Lett. 501, 106–110 [DOI] [PubMed] [Google Scholar]
- 7.Kasturi L., Eshleman J. R., Wunner W. H., Shakin-Eshleman S. H. (1995) J. Biol. Chem. 270, 14756–14761 [DOI] [PubMed] [Google Scholar]
- 8.Petrescu A. J., Milac A. L., Petrescu S. M., Dwek R. A., Wormald M. R. (2004) Glycobiology 14, 103–114 [DOI] [PubMed] [Google Scholar]
- 9.Bause E. (1983) Biochem. J. 209, 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bause E., Hettkamp H., Legler G. (1982) Biochem. J. 203, 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker E. N., Hubbard R. E. (1984) Prog. Biophys. Mol. Biol. 44, 97–179 [DOI] [PubMed] [Google Scholar]
- 12.Imperiali B., Shannon K. L. (1991) Biochemistry 30, 4374–4380 [DOI] [PubMed] [Google Scholar]
- 13.Imperiali B., Shannon K. L., Rickert K. W. (1992) J. Am. Chem. Soc. 114, 7942–7944 [Google Scholar]
- 14.Imperiali B., Shannon K. L., Unno M., Rickert K. W. (1992) J. Am. Chem. Soc. 114, 7944–7945 [Google Scholar]
- 15.Berman H., Henrick K., Nakamura H. (2003) Nat. Struct. Biol. 10, 980. [DOI] [PubMed] [Google Scholar]
- 16.Petrescu A. J., Wormald M. R., Dwek R. A. (2006) Curr. Opin. Struct. Biol. 16, 600–607 [DOI] [PubMed] [Google Scholar]
- 17.Chavan M., Yan A., Lennarz W. J. (2005) J. Biol. Chem. 280, 22917–22924 [DOI] [PubMed] [Google Scholar]
- 18.Harada Y., Li H., Li H., Lennarz W. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6945–6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitley P., Nilsson I. M., von Heijne G. (1996) J. Biol. Chem. 271, 6241–6244 [DOI] [PubMed] [Google Scholar]
- 20.Glabe C. G., Hanover J. A., Lennarz W. J. (1980) J. Biol. Chem. 255, 9236–9242 [PubMed] [Google Scholar]
- 21.Kelleher D. J., Kreibich G., Gilmore R. (1992) Cell 69, 55–65 [DOI] [PubMed] [Google Scholar]
- 22.Nilsson I., von Heijne G. (1993) J. Biol. Chem. 268, 5798–5801 [PubMed] [Google Scholar]
- 23.Valliere-Douglass J. F., Kodama P., Mujacic M., Brady L. J., Wang W., Wallace A., Yan B., Reddy P., Treuheit M. J., Balland A. (2009) J. Biol. Chem. 284, 32493–32506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla A. A., Hubbard B., Tressel T., Guhan S., Low D. (2007) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 848, 28–39 [DOI] [PubMed] [Google Scholar]
- 25.Kabat E. A., Wu T. T., Perry H. M., Gottesman K. S., Foeller C. (1991) Sequences of Proteins of Immunological Interest, 5th Ed., National Institutes of Health, United States Department of Health and Human Services, Bethesda, MD [Google Scholar]
- 26.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 27.Saphire E. O., Parren P. W., Pantophlet R., Zwick M. B., Morris G. M., Rudd P. M., Dwek R. A., Stanfield R. L., Burton D. R., Wilson I. A. (2001) Science 293, 1155–1159 [DOI] [PubMed] [Google Scholar]
- 28.Ahmad S., Gromiha M., Fawareh H., Sarai A. (2004) BMC Bioinformatics 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jefferis R. (2009) Nat. Rev. Drug Discov. 8, 226–234 [DOI] [PubMed] [Google Scholar]
- 30.Mimura Y., Ashton P. R., Takahashi N., Harvey D. J., Jefferis R. (2007) J. Immunol. Methods 326, 116–126 [DOI] [PubMed] [Google Scholar]
- 31.Chalkley R. J., Thalhammer A., Schoepfer R., Burlingame A. L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8894–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikesh L. M., Ueberheide B., Chi A., Coon J. J., Syka J. E., Shabanowitz J., Hunt D. F. (2006) Biochim. Biophys. Acta 1764, 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valliere-Douglass J. F., Brady L. J., Farnsworth C., Pace D., Balland A., Wallace A., Wang W., Treuheit M. J., Yan B. (2009) Glycobiology 19, 144–152 [DOI] [PubMed] [Google Scholar]
- 34.Fan J. Q., Lee Y. C. (1997) J. Biol. Chem. 272, 27058–27064 [DOI] [PubMed] [Google Scholar]
- 35.Lee B., Richards F. M. (1971) J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 36.Mellquist J. L., Kasturi L., Spitalnik S. L., Shakin-Eshleman S. H. (1998) Biochemistry 37, 6833–6837 [DOI] [PubMed] [Google Scholar]
- 37.Kasturi L., Chen H., Shakin-Eshleman S. H. (1997) Biochem. J. 323, 415–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Canada C., Kelleher D. J., Gilmore R. (2009) Cell 136, 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolt G., Kristensen C., Steenstrup T. D. (2005) Glycobiology 15, 541–547 [DOI] [PubMed] [Google Scholar]
- 40.Feige M. J., Groscurth S., Marcinowski M., Shimizu Y., Kessler H., Hendershot L. M., Buchner J. (2009) Mol. Cell 34, 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]