Abstract
Restitution of intestinal epithelial barrier damage involves the coordinated remodeling of focal adhesions in actively migrating enterocytes. Defining the extracellular mediators and the intracellular signaling pathways regulating those dynamic processes is a key step in developing restitution-targeted therapies. Previously we have determined that activation of the chemokine receptor CXCR4 by the cognate ligand CXCL12 enhances intestinal epithelial restitution through reorganization of the actin cytoskeleton. The aim of these studies was to investigate the role of calcium effectors in CXCL12-mediated restitution. CXCL12 stimulated release of intracellular calcium in a dose-dependent manner. Inhibition of intracellular calcium flux impaired CXCL12-mediated migration of IEC-6 and CaCo2 cells. Pharmacological blockade and specific shRNA depletion of the phospholipase-C (PLCβ3) isoform attenuated CXCL12-enhanced migration, linking receptor activation with intracellular calcium flux. Immunoblot analyses demonstrated CXCL12 activated the calcium-regulated focal adhesion protein proline-rich tyrosine kinase-2 (Pyk2) and the effector proteins paxillin and p130Cas. Interruption of Pyk2 signaling potently blocked CXCL12-induced wound closure. CXCL12-stimulated epithelial cell migration was enhanced on laminin and abrogated by intracellular calcium chelation. These results suggest CXCL12 regulates restitution through calcium-activated Pyk2 localized to active focal adhesions. Calcium signaling pathways may therefore provide a novel avenue for enhancing barrier repair.
Keywords: Calcium/Cellular Regulation, Cell/Epithelial, Cytokines/Chemokines, Extracellular Matrix/Basal Lamina, Receptors/Chemokine, Signal Transduction/Protein Kinases/Tyrosine, Tissue/Organ Systems/Epithelium, Phospholipase C, Pyk2
Introduction
A battery of molecules stimulate epithelial restitution in vitro and in vivo, including cytokines, luminal peptides, probiotics, and as we have reported, chemokines (1–11). Restitution in vivo is dependent on several factors absent from cell-culture model systems, including mucin-producing goblet cells, extracellular matrix-producing fibroblasts, immune cells, and the luminal microbiota. Within that complex environment, deletion of genes specifically within the intestinal epithelium has proven useful in deciphering roles for transforming growth factor-β1 (TGF-β1)3 receptor, epidermal growth factor receptor, cadherin, laminin, and Vav in integrity and repair of the gut mucosa (12–16). More recent reports have begun to link mucosal fibroblasts and T cells with key roles in injury repair (17–22). Despite these findings, the mechanisms by which those molecules elicit their functions, in either reductionist cell-culture models, or complex in vivo systems, remain incompletely characterized.
Chemokines are abundantly and ubiquitously produced host defense molecules that participate in activation and directional trafficking of leukocytes. The chemokine receptors CXCR4, CCR5, CCR6, and CX3CR1 are expressed by the cells of the human intestinal epithelium (23–26). Chemokines produced by intestinal epithelial cells play an important role in orchestrating physiological and pathological inflammation, consistent with a role in amplifying intestinal inflammation. Genetic deletion of the murine CXCL8 orthologue increases susceptibility to colitis, a finding recapitulated in mice genetically deficient in the chemokine receptors CCR5, CCR6, or CXCR3 (27–30). The chemokine stromal cell-derived factor-1, known as CXCL12, is up-regulated in hypoxic tumors and regulates dermal injury repair (31, 32). Our findings add to the current model and show that chemokines alter epithelial permeability and secretory functions in intestinal epithelial cell culture model systems (3–5, 23, 33, 34). CXCR4 and CXCL12 deletion results in embryonic lethality in knock-out mice, indicating that reductionist epithelial model systems are needed to decipher the roles for those molecules in mucosal injury repair (35, 36). Together these data suggest broader roles for chemokines, and the cells they regulate, in mucosal injury and host defense.
Chemokine binding to G-protein-coupled chemokine receptors mobilizes intracellular calcium and regulates cell mobility (37). Although calcium is an established regulator of the actin cytoskeleton, its roles in intestinal epithelial restitution remain poorly characterized (38, 39). Our laboratory has shown that the chemokines CXCL12 and CCL20 activate their cognate receptors, CXCR4 and CCR6, respectively, to strengthen intestinal epithelial wound healing. Recently, we determined that the inducibly regulated inflammatory chemokine CCL20, and the antimicrobial peptide human β-defensin-2, regulates epithelial restitution in part through mobilization of intracellular calcium (3–5). Although calcium regulates a variety of cellular effectors important in enterocyte migration, (17, 40, 41), the host defense factors regulating these signaling pathways remain incompletely understood (39). In this report, we document the signaling events whereby the G-protein-linked chemokine receptor CXCR4 regulates epithelial cell migration through phospholipase-C (PLC)-β3-mediated calcium mobilization and activation of focal adhesion-localized proline-rich tyrosine kinase-2 (Pyk2).
EXPERIMENTAL PROCEDURES
Materials
Recombinant human CXCL12 was produced and purified from Escherichia coli, as previously defined (42). TGF-β1 was purchased from R&D Systems (Minneapolis, MN). Antibodies to specifically detect phosphorylated p130Cas at tyrosine residue 410, paxillin tyrosine residue 118, or Pyk2 tyrosine residue 402 were from Cell Signaling Technologies (Danvers, MA). Polyclonal antibody to detect phosphorylation of Pyk2 on tyrosine 580 was from Invitrogen. For immunofluorescence detection of Pyk2, tyrosine 402 phospho-specific antibody from Santa Cruz Biotechnology (Santa Cruz, CA) was used. Antibodies specific for total paxillin or p130Cas were obtained from BD Transduction Laboratories (Bedford, MA). Vinculin antibody was purchased from Abcam (Cambridge, MA). Phospholipase-C-β3 (PLCβ3) specific antibody was from Santa Cruz Biotechnology. The specific pan-PLC inhibitor U-73122 was from EMD Biosciences (San Diego, CA). Rhodamine-conjugated goat anti-mouse or fluorescein isothiocyanate-labeled donkey anti-rabbit antibodies were obtained from Jackson ImmunoResearch (West Grove, PA). AlexaFluor 488 goat anti-rabbit antibody was from Invitrogen. BAPTA-AM, lysophosphatidic acid, and Polybrene were purchased from Sigma.
Cell Culture
The normal, non-transformed rat small intestinal IEC-6 cell line (CRL-1592) was purchased from the American Type Culture Collection (ATCC, Rockville, MD) and cultured using Dulbecco's modified Eagle's medium (4 g/liter glucose) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Omega Scientific, Tarzana, CA), 2 mm l-glutamine, 1.5g/liter NaHCO3, and 0.1 unit/ml bovine insulin (Invitrogen). The human intestinal carcinoma cell line CaCo2 was cultured in Dulbecco's modified Eagle's medium (4 g/liter glucose) supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2 mm l-glutamine, and 10 mm HEPES (Invitrogen).
Epithelial Wounding Models
Confluent IEC-6 cell monolayers cultured in 60-mm dishes were incubated 48 h in serum-free medium. Monolayers were wounded with a sterile razor blade and incubated in serum-free medium alone or the indicated stimuli for 18 h at 37 °C in 5% CO2, as described previously (3, 6). Photomicrographs at 4–5 locations per wound were taken using 100× magnification. The number of migrated cells was determined by counting nucleated cells that crossed the wound edge.
Polarized CaCo2 cells were grown to confluence in 6-well Transwell inserts (pore size, 0.4 μm, Corning, Danvers, MA). Cells were serum-starved 24 h and wounded with a 0.1- to 10-μl plastic pipette tip (USA Scientific, Ocala, FL) connected to a bench-top vacuum aspirator as we defined previously (4). Polarized monolayers that had been wounded were stimulated on a daily basis with serum-free medium containing CXCL12 or TGF-β1. Photomicrographs of the circular injury were taken using the 4× objective. The area of each wound was defined using MetaMorph software (Molecular Devices, Sunnyvale, CA). Wound closure was calculated by subtracting the remaining wound area on each day from the initial wound size. In separate assays, cell monolayers were pre-treated 30 min at 37 °C with the intracellular calcium chelator BAPTA-AM (10 μm) or treated at 37 °C with titrated doses of U-73122 and wounded, and epithelial migration was measured.
shRNA Construction and Transduction
PLCβ3-targeted shRNA was generated using the Lentilox3.7 (LL3.7) system (Addgene, Cambridge, MA). Rat specific PLCβ3 shRNA oligonucleotide sequences were: 5′-tGCTTCTGACTACATCCCAGATttcaagagaATCTGGGATGTAGTCAGAAGCtttttggaaac-3′ and 5′-tcgagtttccaaaaaGCTTCTGACTACATCCCAGATtctcttgaaATCTGGGATGTAGTCAGAAGCa-3′. Annealed oligonucleotides were separately cloned into HpaI/XhoI-digested pLL3.7-green fluorescent protein (LL3.7-GFP) or pLL3.7-puromycin (LL3.7-puro) expression vectors. Plasmid orientation was verified by PCR and DNA sequencing. Empty, scramble, or PLCβ3 LL3.7 vectors were transfected, along with the accessory plasmids, pVSVG, pREV, and pRRE, into HEK293T cells using TransIT-293 transfection reagent (Mirus, Madison, WI). Transfection medium was replaced, and viral particle-containing conditioned medium was collected and filtered through a 0.45-μm filter 48 h later. IEC-6 cells were transduced with lentiviral particles in the presence of 8 μg/ml Polybrene for 24 h. GFP expression was monitored by fluorescence microscopy 24 and 48 h later. Alternatively, cells were cultured in growth medium supplemented with 2.5 μg/ml puromycin to select those expressing the specific shRNA sequences. Lentiviral-targeted knockdown of PLCβ3 was confirmed by immunoblot analysis.
Calcium Mobilization
Intracellular calcium mobilization was measured using the Fluo-4NW Assay from Invitrogen according to the manufacturer's directions and as defined previously (43, 44). Briefly, IEC-6 or CaCo-2 cells were plated in 96-well white-walled plates (BD Biosciences) and grown to 90% confluence. Cells were serum-starved overnight and loaded with cell-permeant Fluo-4-AM diluted in calcium-free Hanks' balanced salt solution supplemented with 20 mm HEPES buffer provided by the manufacturer. CXCL12 was diluted in calcium-free Hanks' balanced salt solution/HEPES buffer and added to specified wells in the indicated concentrations. Intracellular calcium flux was measured by fluorescence spectroscopy every 5 s for a total of 220 s (Victor2 Wallac, PerkinElmer Life Sciences). Background fluorescence was measured 30 s before addition of ligand and the average background subtracted from each value.
Immunoblot Analysis
Epithelial cells were grown to 80% confluence, serum-starved for 24 h, and stimulated as indicated. Cells were solubilized in modified radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.25% (v/v) sodium deoxycholate, 1.0% (v/v) Nonidet P-40, 0.1% (v/v) SDS, and 1 mm EDTA) supplemented with Protease Inhibitor Mixture Set III (EMD Biosciences) and 10 mm sodium orthovanadate, 40 mm β-glycerolphosphate, 20 mm sodium fluoride, and 20 mm p-nitrophenyl phosphate phosphatase inhibitors. Lysates were passed through a 25-gauge needle and centrifuged at 550 rpm for 5 min at 4 °C to pellet nuclei. Protein concentrations were determined using the Bradford protein assay (BCA kit, Pierce). Proteins were size-separated using reducing SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride (Immobilon-P, Millipore) for immunoblot analysis using specific antibodies.
Immunofluorescence Microscopy
IEC-6 cells were grown to 100% confluence on glass eight-well chamber slides (LabTek) or glass coverslips and serum-starved 24 h. Cells were wounded with a sterile razor blade, rinsed with PBS, and stimulated with 20 ng/ml CXCL12 for 5 or 15 min. Cells were fixed with 2% (w/v) paraformaldehyde and blocked with 1% (v/v) donkey and 1% (v/v) goat serum in PBS with 0.3% (v/v) Triton X-100 for 1 h. Cells were incubated with primary antibodies overnight at 4 °C, washed, incubated 2 h at room temperature with the appropriate secondary antibody, and co-stained 5-min with 4′,6-diamidino-2-phenylindole (Molecular Probes, Invitrogen) to visualize cell nuclei. Images were obtained using a fluorescence microscope at 600× magnification.
Dominant-interfering Pyk2 Signaling
CaCo2 cells were grown in 6-well dishes to 50% confluence and infected with equal levels of adenovirus encoding kinase domain-deficient human Pyk2 (AdCRNK) or as control, GFP (AdGFP, a kind gift from Dr. Andrey Sorokin, Medical College of Wisconsin). Medium was replenished after 24 h, and GFP expression was monitored by fluorescence microscopy. Cells were then serum-starved 24 h and stimulated as indicated. Total protein lysates were collected for immunoblot analysis. Polarized CaCo2 monolayers were separately infected with either AdCRNK or AdGFP and wounded, and wound closure was assessed as described above.
Extracellular Matrix
Tissue culture dishes were coated with 10 μg/ml laminin (Sigma) or 10 μg/ml collagen-IV (R&D Systems). Laminin was derived from Englebreth-Holm cells known to produce a matrix rich in laminin-1 (45). Extracellular matrix proteins diluted in PBS were added to tissue culture dishes incubated 2 h at 37 °C, washed with PBS, and blocked with 1% (w/v) BSA.
Statistical Analysis
Differences between stimulated and control cells were analyzed using an unpaired Student's t test (SigmaStat, Jandel Scientific Software, San Rafael, CA). Multiple comparisons between groups were analyzed using a two-way analysis of variance with a Bonferroni post-hoc analysis used to identify pairwise differences (GraphPad Prism 4, La Jolla, CA). Statistical significance was set at p ≤ 0.05.
RESULTS
Calcium Regulates CXCL12-induced Migration of Model Intestinal Epithelial Cells
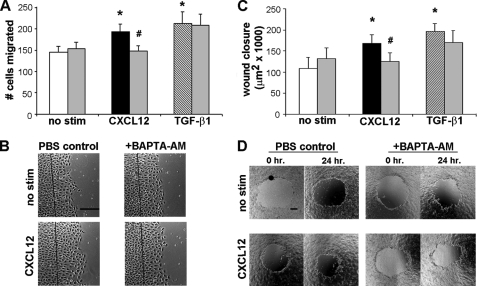
Our data indicate that the chemokine receptor CCR6 regulates restitution in part through the dose-dependent mobilization of intracellular calcium (5). This response was not observed in TGF-β1-stimulated migration, suggesting calcium activation is a potent effector of chemokine-directed restitution. To test that notion, we first sought to determine if the homeostatic chemokine CXCL12 similarly regulates epithelial restitution via mobilization of intracellular calcium. IEC-6 cells (Fig. 1A) cultured on plastic, or CaCo2 monolayers grown as a polarized epithelium (Fig. 1C), were pretreated with the calcium chelator BAPTA-AM to inhibit elevation of intracellular calcium. Monolayers were then wounded and stimulated with 20 ng/ml CXCL12; a concentration we have previously shown induces enterocyte migration. Chelation of intracellular calcium abrogated CXCL12-stimulated IEC-6 cell migration (Fig. 1, A and B). Further, CXCL12-stimulated wound closure in polarized CaCo2 monolayers was blocked by pretreatment with 10 μm BAPTA-AM (Fig. 1, C and D). In contrast, TGF-β1-stimulated IEC-6 migration, or CaCo2 wound closure, was minimally altered by calcium chelation. These data indicate that calcium is a key regulator of CXCL12-directed enterocyte migration in non-transformed IEC-6 cells, as well as polarized human CaCo2 model epithelia.
FIGURE 1.
CXCL12-stimulated migration is dependent on intracellular calcium. IEC-6 (A and B) or polarized CaCo2 (C and D) monolayers were loaded with the intracellular calcium chelator BAPTA-AM (10 μm, gray bars) or, as a control, PBS (white, black, and striped bars) for 30 min. Cell monolayers were wounded and either left untreated (no stim) or were stimulated with 20 ng/ml CXCL12 or 5 ng/ml TGF-β1 as a positive control. A, IEC-6 cell migration was enumerated after 18 h. C, CaCo2 monolayers were wounded, and the area of closure (μm2) was calculated after 24 h. CXCL12 and TGF-β1 increased cell migration and wound closure above the unstimulated control. Migration in response to CXCL12 was abrogated in BAPTA-AM-treated IEC-6 or CaCo2 cells. TGF-β1 induced migration was minimally, if at all, altered by intracellular calcium chelation. Representative wounds are shown in B and D. Values in A and C are mean ± S.E. of three experiments. The asterisk denotes statistically significant difference from untreated cells (p ≤ 0.05). #, statistical significance between CXCL12-stimulated PBS and BAPTA-AM treated monolayers (p ≤ 0.05). Scale bar, 200 μm.
CXCL12-stimulated Migration Is PLCβ-dependent
The Gβγ subunits of ligand activated Gαi-protein-coupled receptors increase intracellular calcium levels through activation of PLCβ isoforms (46). In agreement with that model, migration of CXCL12-stimulated IEC-6 cells was dose-dependently blocked using the pan-PLC inhibitor U-73122 (Table 1).
TABLE 1.
Pharmacological blockade of PLC dose-dependently inhibits migration of CXCL12-stimulated IEC-6 monolayers
Values are mean ± S.E., n = 4–8 individual experiments. Wounded cell monolayers were stimulated with 20 ng/ml CXCL12, and cell migration was enumerated 18 h later. Unstimulated cells were a treatment control.
| U-73122 (μg/ml) |
|||||
|---|---|---|---|---|---|
| 0 | 0.5 | 2.5 | 5 | 7.5 | |
| No. of cells migrated | |||||
| Unstimulated cells | 132.7 ± 7.0 | 136.9 ± 8.6 | 127.3 ± 29.8 | 128.1 ± 7.3 | 120.8 ± 7.8 |
| CXCL12 | 163.8 ± 10.4a | 169.6 ± 5.0a | 139.0 ± 12.2 | 139.2 ± 8.5 | 101.9 ± 9.1b |
a Statistically significant difference (p ≤ 0.05) between control and CXCL12-treated monolayers.
b Statistically significant difference (p ≤ 0.05) between CXCL12-stimulated monolayers pretreated with U-73122 compared to cells without the inhibitor.
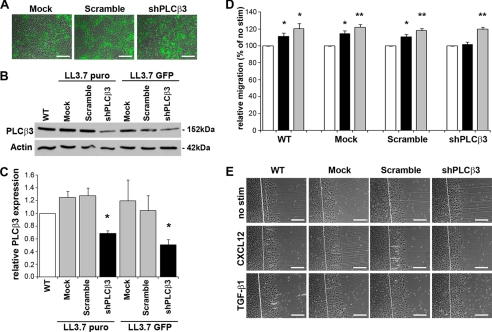
RNA interference approaches were used to decipher roles for specific G-protein-activated PLC isoforms expressed by intestinal epithelial cells. PLCβ3 has been shown to be abundantly expressed in rat intestinal epithelium (47) and is known to be activated by G-protein-coupled receptors (46). IEC-6 cells were separately transduced with equivalent levels of lentiviral particles expressing shPLCβ3, a scramble sequence, or empty vector (Fig. 2A). Immunoblot analysis demonstrated that depletion of PLCβ3 in IEC-6 cells using two separate lentiviral vectors, LL3.7-puro or LL3.7-GFP, significantly decreased endogenous levels of PLCβ3 protein compared with empty or scramble sequence-expressing vectors (Fig. 2, B and C). Consistent with the U-73122 inhibitor, epithelial migration stimulated by CXCL12 was abolished in PLCβ3-depleted cells (Fig. 2, D and E). In parallel with our data on chelation of intracellular calcium, knockdown of PLCβ3 had little, if any, impact on TGF-β1-stimulated restitution.
FIGURE 2.
PLCβ3 depletion blocks CXCL12-stimulated IEC-6 cell migration. A, fluorescence microscopy indicated equivalent transduction of lentiviral particles encoding GFP (green) and either rat-specific shRNA against PLCβ3, a Scramble sequence, or empty control vector (Mock) in IEC-6 cells. Data are representative GFP-fluorescence and phase-contrast overlay images. B, immunoblot analysis revealed that endogenous levels of protein were significantly depleted within shPLCβ3-targeted cells relative to Scramble and Mock transduced cells. C, densitometric analysis of PLCβ3 protein levels. Wild-type (WT) IEC-6 cells (white), control Scramble and Mock cells (gray), and shPLCβ3 cells (black) from LL3.7-puro or LL3.7-GFP lentiviral vectors. Data are the ratio of PLCβ3 protein levels relative to actin loading control. *, the significant knockdown of PLCβ3 compared with WT cells (p ≤ 0.05). Values in C are mean ± S.E. of three experiments. D, IEC-6 cell migration in PLCβ3 knockdown cells. IEC-6 cell migration was assessed 18 h after wounding and stimulation with 20 ng/ml CXCL12 (black bars) or 5 ng/ml TGF-β1 (gray bars). CXCL12-stimulated migration was decreased in shPLCβ3 knockdown cells and not significantly different from untreated control cells (white bars). Data are expressed as percentage of basal migration in unstimulated (no stim) control cells. *, significant difference (p ≤ 0.05); **, significant difference (p ≤ 0.01). Values are mean ± S.E. from four experiments. E, representative wound images from lentiviral transduced cells. Scale bar, 200 μm.
CXCL12 Stimulates Release of Intracellular Calcium
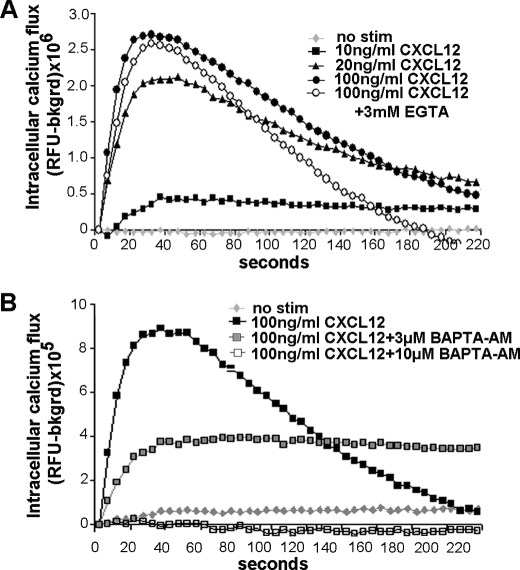
We next assessed whether intestinal cell migration stimulated by CXCL12 reflected ligand-specific mobilization of calcium. CXCL12 dose-dependently increased intracellular calcium in adherent IEC-6 cells (Fig. 3A). Calcium chelators, BAPTA-AM and EGTA, were used to assess if the rise in intracellular calcium was a result of release from internal stores or reflected extracellular entry through channels. Treatment with 3 mm EGTA had limited effect on the initial, rapid calcium flux, but markedly reduced the slower, prolonged elevation in calcium (Fig. 3A). In contrast, pretreatment with 10 μm BAPTA-AM completely abolished intracellular calcium mobilization (Fig. 3B). As expected, an intermediate 3 μm concentration of BAPTA-AM decreased CXCL12-induced calcium flux by ∼50%. Pretreatment with the Gαi-specific inhibitor pertussis toxin decreased the chemokine-stimulated response (data not shown). These data are consistent with CXCL12 evoking the rapid, receptor-mediated release of calcium from intracellular stores, with prolonged flux reflective of inflow through channels found on the plasma membrane. Moreover, depletion of PLCβ3 using shRNA, or pharmacological inhibition with U-73122, blocked chemokine-induced mobilization of intracellular calcium (data not shown). Chemokine-induced calcium mobilization was not solely a function of rat intestinal epithelial cells as CXCL12 stimulated a rapid, maximal increase in calcium within 40 s of ligand stimulation (data not shown) in CaCo2 cells.
FIGURE 3.
CXCL12 stimulates mobilization of intracellular calcium. A, CXCL12 in calcium-free buffer stimulated a dose-dependent increase in intracellular calcium. Treatment with the extracellular calcium chelator EGTA (3 mm) decreased persistent calcium mobilization in cells stimulated with 100 ng/ml CXCL12. B, CXCL12-stimulated calcium release is blocked by the cell-permeant chelator BAPTA-AM. Chemokine-induced calcium mobilization was completely blocked to control (no stim) levels by pretreatment with 10 μm BAPTA-AM. Pretreatment with 3 μm BAPTA-AM partially abrogated calcium mobilization. Values are relative Fluo-4 fluorescence units minus background signal from a representative of three separate experiments.
Intracellular Calcium Regulates CXCL12-induced Activation of Pyk2
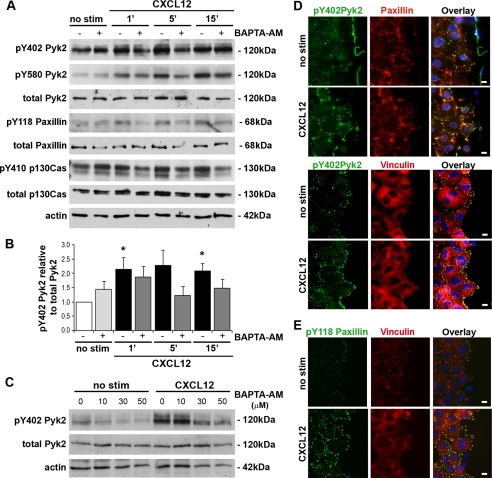
Pyk2, also known as RAFTK, CADTK, or CAK-β, is a calcium-regulated member of the focal adhesion non-receptor tyrosine kinase family (48, 49) previously shown to be phosphorylated after CXCL12 stimulation (50). Although Pyk2 has been shown to be activated in rat intestinal epithelial cell lines (51, 52) its role as a downstream effector of calcium-mediated restitution has yet to be determined. Immunoblot analyses indicated that CXCL12 consistently activated Pyk2 in IEC-6 cells stimulated with the motogenic 20 ng/ml concentration of the ligand (Fig. 4). Consistent with CXCL12 having a broader role in Pyk2 activity, chemokine stimulation resulted in increased phosphorylation at tyrosine 402 (Fig. 4, A and B) known to reflect overall activation of the kinase (49). Importantly, pretreatment with the 10 μm BAPTA-AM concentration shown to inhibit calcium mobilization and cell migration, consistently reduced Pyk2 phosphorylation at tyrosine 402 (Fig. 4, A and B). Increased concentrations of BAPTA-AM, which we have previously shown inhibit calcium mobilization and migration (5), completely blocked CXCL12-stimulated phosphorylation of Pyk2 (Fig. 4C). As shown in Fig. 4A, CXCL12 stimulated the rapid phosphorylation of the Pyk2 kinase domain at tyrosine 580 previously shown to depend upon intracellular calcium levels (52). Similarly, increasing concentrations of BAPTA-AM dose-dependently inhibited phosphorylation of Pyk2 at tyrosine 580 (not shown). Pyk2 interacts with the focal adhesion regulatory proteins paxillin and p130Cas (53). Immunoblot analysis indicated that CXCL12 stimulation resulted in a reproducible calcium-regulated increase in paxillin and p130Cas phosphorylation (Fig. 4A).
FIGURE 4.
CXCL12 stimulates Pyk2 phosphorylation and localization at focal adhesions. A, immunoblot analysis of IEC-6 cells indicated that CXCL12 (20 ng/ml) stimulated phosphorylation of Pyk2 tyrosine residues 402 (pY402) and 580 (pY580). Pretreatment with 10 μm BAPTA-AM (+) or PBS (−) revealed that chemokine-stimulated Pyk2 phosphorylation was variably decreased by the calcium chelator. CXCL12-stimulated phosphorylation of paxillin tyrosine 118 and p130Cas tyrosine 410 was also blocked by pretreatment with BAPTA-AM. Total Pyk2, paxillin, p130Cas, and actin were detected as loading controls. Data are representative of three separate analyses. B, densitometric analysis of Pyk2 phosphorylation. CXCL12-stimulated Pyk2 phosphorylation (black bars) was reduced by pretreatment with 10 μm BAPTA-AM (dark gray bars). *, a statistically significant difference (p ≤ 0.05) between CXCL12-stimulated Pyk2 phosphorylation relative to control (no stim) cells (white bar). Data are relative densitometric units normalized to total protein. Values are mean ± S.E. of three experiments. C, immunoblot analysis of IEC-6 cells pretreated with increasing concentrations of BAPTA-AM (0, 10, 30, or 50 μm) showed the dose-dependent inhibition of 5-min CXCL12-stimulated phosphorylation of Pyk2. Data representative of three separate analyses. D, localization of active Pyk2 in CXCL12-stimulated monolayers. Antibodies specific for Pyk2 (pY402), paxillin, or vinculin were used to define protein localization in wounded IEC-6 control cells (no stim) or cell monolayers 5 min after CXCL12 stimulation. Overlay images indicate co-localization (yellow) of active Pyk2 (green) with paxillin (top, red)- or vinculin (bottom, red)-defined focal adhesions. E, levels of active paxillin were increased after 15-min CXC12 stimulation. Antibodies specific for paxillin (pY118) or vinculin were used to define protein localization in wounded IEC-6 control (no stim) or CXCL12-stimulated cells. Overlay images indicate co-localization (yellow) of active paxillin (green) with vinculin (red)-delimited focal adhesions. Immunofluorescence images are representative of three or four independent experiments. Scale bar, 10 μm.
Immunofluorescence microscopy was used to define the localization of activated Pyk2 within wounded IEC-6 cells. Phosphorylated Pyk2 was markedly increased compared with unstimulated control cells 5 min after CXCL12 stimulation. Moreover, active Pyk2 co-localized with paxillin (Fig. 4D, top) and vinculin (Fig. 4D, bottom). In addition, active phosphorylated paxillin was increased 15 min after CXCL12 stimulation (Fig. 4E). In agreement with active Pyk2 localization, phosphorylated paxillin levels were increased at vinculin-delimited focal adhesions (Fig. 4E).
Dominant-interfering Pyk2 Decreases CXCL12-induced Cell Migration
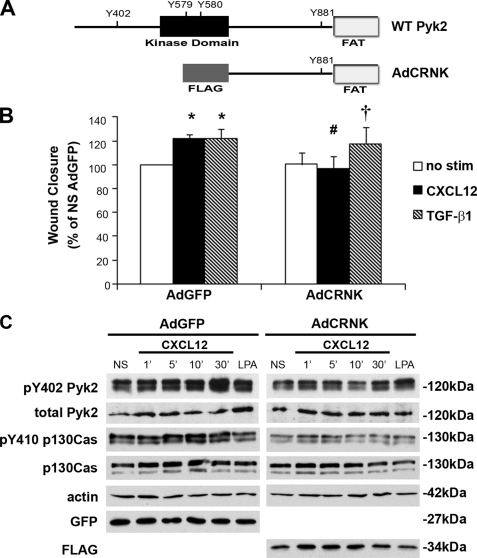
Next, we postulated that Pyk2 plays a key role in restitutive migration. For these studies, a previously described adenoviral vector (54, 55) encoding a truncated form of human Pyk2 (AdCRNK) was utilized (Fig. 5A). Wound closure examined in AdCRNK-infected CaCo2 cells, cultured as a polarized epithelium, deciphered roles for Pyk2 in restitution. AdCRNK significantly diminished wound closure to basal levels in CaCo2 cells treated with CXCL12 (Fig. 5B). These data are in contrast to CaCo2 cells infected with the AdGFP control virus in which wound closure was significantly increased in cells stimulated with CXCL12 or the TGF-β1 positive control. Further, wound closure in TGF-β1-treated cells was minimally altered in cells expressing the truncated Pyk2 protein.
FIGURE 5.
CXCL12-stimulated migration is blocked by dominant-interfering Pyk2. A, schematic representation of wild-type (WT) Pyk2 protein and an adenovirus-encoded dominant-interfering construct (AdCRNK) lacking the kinase domain. Key tyrosine residues (tyrosines 579, 580, 402, and 881) are identified. B, CXCL12, and the TGF-β1-positive control, stimulated significant wound closure in AdGFP-infected CaCo2 cells. Expression of the AdCRNK dominant-interfering construct blocked CXCL12-, but not TGF-β1-, induced migration. Data are percent wound closure relative to the unstimulated (no stim) AdGFP control cells. Values are mean ± S.E. of three experiments. *, statistically significant difference (p ≤ 0.05) in migration between unstimulated and CXCL12 or TGF-β1-treated cells. #, indicates a statistically significant difference between CXCL12-stimulated AdGFP and AdCRNK cells (p ≤ 0.05). †, indicates a statistically significant difference between unstimulated and TGF-β1-stimulated AdCRNK cells (p ≤ 0.05). C, immunoblot analysis for Pyk2 phosphorylated tyrosine 402 (pY402) from CaCo2 cells stimulated with 20 ng/ml CXCL12. CXCL12 increased phosphorylation of Pyk2 in control, AdGFP, infected cells. Expression of the dominant-interfering CRNK construct markedly reduced basal and CXCL12-stimuled Pyk2 phosphorylation. Control and CXCL12-stimulated p130Cas tyrosine 410 (pY410) phosphorylation was decreased in AdCRNK-infected cells. Actin and total, non-active Pyk2 and p130Cas protein levels were assessed as loading controls. Equal expression levels of the GFP control or the dominant-interfering protein were confirmed using immunoblot analysis of the virally encoded GFP or FLAG proteins. Data shown are representative of three experiments.
In agreement with our findings in IEC-6 cells, CXCL12 increased tyrosine phosphorylation of Pyk2 and p130Cas in CaCo2 model epithelia infected with AdGFP control virus (Fig. 5C). Consistent with reduced migration, AdCRNK significantly decreased basal and CXCL12-stimulated phosphorylation of Pyk2 (Fig. 5C and supplemental Table 1). Infection with AdCRNK significantly blocked p130Cas activation in response to CXCL12 treatment (Fig. 5C), as noted in prior reports for other G-protein-coupled receptor ligands (56). Minimal compensatory FAK activation was observed in AdCRNK-infected cells (data not shown).
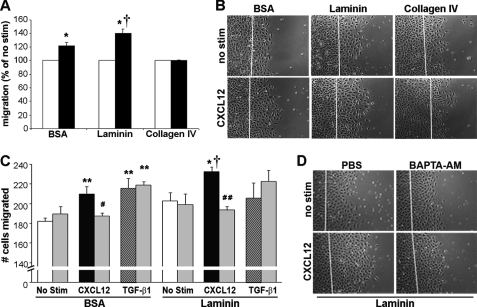
Laminin Enhances CXCL12-stimulated Migration
Focal adhesion proteins regulate cellular interactions with the extracellular matrix. We next asked if CXCL12-induced migration was modulated by extracellular matrix components known to play roles in restitution (57–59). Epithelial cell migration of cells cultured on the non-integrin substrate BSA was significantly increased by CXCL12 stimulation (Fig. 6), consistent with our previous findings on non-integrin substrates (3, 4). CXCL12-induced migration was significantly increased with the addition of laminin, relative to chemokine-stimulated migration on BSA-coated plates (Fig. 6, A and C). In contrast, CXCL12 was unable to stimulate further changes in migration of cells cultured on collagen-IV. These latter data suggest chemokine-stimulated migration on laminin does not reflect a global response to matrix proteins.
FIGURE 6.
CXCL12 increases epithelial migration on laminin. A, CXCL12-stimulated increased migration on laminin. IEC-6 cell migration was assessed on plates coated with the integrin substrates, laminin or collagen-IV, or as a non-integrin control, BSA. CXCL12 (black bar) increased migration on BSA- and laminin-coated dishes compared with the unstimulated control cells (no stim). CXCL12 was unable to enhance restitution on collagen-IV-coated dishes. B, representative images of wounded IEC-6 monolayers migrating on different extracellular matrix proteins. C, IEC-6 monolayers were grown on laminin- or BSA-coated dishes and wounded, and the number of cells that migrated over the wound were calculated after 18 h. Monolayers treated with 20 ng/ml CXCL12 (black bar) or 5 ng/ml TGF-β1 (hatched bar) increased migration relative to unstimulated monolayers (white bar). Pretreatment with the cell-permeant calcium chelator BAPTA-AM (10 μm, gray bars) blocked CXCL12-, but not TGF-β1-stimulated wound closure. Values in A and C are mean ± S.E. of three to six experiments. Asterisks denote statistically significant differences (*, p ≤ 0.05; **, p ≤ 0.01) from unstimulated cells. †, indicates the statistically significant difference (p ≤ 0.05) between CXCL12-stimulated migration on laminin- compared with BSA-coated plates. Pound symbols denote statistically significant differences (#, p ≤ 0.05; ##, p ≤ 0.01) in CXCL12-stimulated cell migration on laminin-coated dishes between PBS and BAPTA-AM pretreated cells. D, representative images of wounded IEC-6 monolayers migrating on laminin after BAPTA-AM or PBS pretreatment. Photomicrographs in B and D are representative of three to six separate experiments. Scale bar, 200 μm.
To delineate roles for calcium in CXCL12-evoked migration on laminin, cells were pretreated with 10 μm BAPTA-AM and treated with CXCL12 or TGF-β1. Chelation of intracellular calcium impaired CXCL12-directed, and not TGF-β1-induced, migration on laminin (Fig. 6C). Further, the role for calcium in increasing CXCL12-stimulated restitution on laminin was not restricted to IEC-6 cells. Indeed, chelation of calcium with 10 μm BAPTA-AM blocked wound closure in CXCL12-stimulated polarized CaCo2 monolayers (data not shown).
DISCUSSION
Calcium mobilization is an established regulator of actin rearrangement, cell migration, and integrin activation (39, 49, 60–62). With respect to cell migration, elevation in intracellular calcium regulates polarization of migrating cells and stabilization of the leading edge (39). Despite these studies, and others, underscoring the importance of calcium mobilization in regulating epithelial cell migration (63, 64), the mechanism of calcium-regulated sheet migration is not well defined. Taken together, our data indicate that CXCL12 regulates epithelial wound closure in part through the calcium-dependent focal adhesion kinase Pyk2.
Previously we showed that CCR6-mediated wound healing reflects ligand induction of calcium mobilization (5). With this report we have expanded upon those data defining calcium-dependent restitution stimulated by the inflammatory and inducible CCR6 ligands and focused on the homeostatic ligand for the chemokine receptor CXCR4 (3, 4). Chelation of intracellular calcium with BAPTA-AM potently blocked epithelial migration. Our results indicate that CXCL12 induced a dose-dependent calcium response in adherent epithelial cells. Calcium mobilization was potently inhibited by chelation of intracellular calcium. Extracellular chelation only minimally altered the chemokine-induced calcium response, consistent with release from internal stores. Moreover, calcium was shown to regulate CXCL12-stimulated migration in both IEC-6 and CaCo2 epithelial model systems, suggesting comparable signaling pathways regulate scrape- and aspiration-induced injury repair. Together, these data indicate that CXCR4 and CCR6 chemokine receptors utilize intracellular calcium to enhance enterocyte migration.
Pharmacological inhibition of PLC activity dose-dependently blocked CXCL12-stimulated migration, implicating a role for G-protein-coupled receptors in calcium signaling. Subsequent investigations employed an RNA interference genetic approach to knockdown levels of PLCβ3, a PLC isoform prominently expressed within the colonic epithelium (47), and shown to be activated by G-protein-coupled receptors (46). Depletion of PLCβ3 blocked restitution in CXCL12-treated cells. Basal and TGF-β1-induced migration responses remained intact in PLCβ knockdown cells, consistent with redundant signaling pathways regulating restitution. In agreement with its well characterized role in calcium signaling, pharmacological inhibition, or genetic depletion of PLC, decreased intracellular calcium mobilization elicited by CXCL12 treatment. PLCγ is an additional PLC isoform expressed in colonic epithelium that may also play a role in basal, chemokine, or growth factor-evoked enterocyte migration.
Pyk2 was subsequently shown to be a molecular target activated by calcium mobilization and as such may provide a biochemical link between G-protein-coupled receptors and the focal adhesion complex (48). Congruent with our data on calcium mobility, we determined that CXCL12 activates Pyk2. As implicated in a prior report (52), we found that CXCL12 fully activated the Pyk2 kinase domain in a calcium-dependent fashion. Activation of Pyk2 was inhibited dose-dependently using BAPTA-AM. In agreement with our previous studies showing that BAPTA-AM in excess of 10 μm potently disrupts enterocyte morphology and migration, we determined that 30 μm and 50 μm concentrations abolished Pyk2 phosphorylation. Pyk2 activation was observed using overlapping epithelial model systems and suggests the protein plays a role in restitution in both IEC-6 and CaCo2 monolayers. The signaling intermediates linking G-protein-coupled receptors with calcium and Pyk2 activation have yet to be fully defined. Separate reports implicate calcium/calmodulin and the myosin light chain kinase as key Pyk2 activators (65, 66). In addition to Pyk2, focal adhesion kinase (FAK) and paxillin have previously been shown to play roles in epithelial migration (15, 67). Despite those reports, it remains uncertain if FAK, paxillin, p130Cas, or Pyk2 focal adhesion proteins are primarily activated via external, “outside-in,” or internal, “inside-out,” signaling in adherent or actively migrating cells. Our data agree with a prior report using the dominant-interfering Pyk2 construct and suggest that outside-in and inside-out signaling have complementary roles in activation of those proteins (56). Thus, infection with AdCRNK markedly decreased basal and induced phosphorylation of Pyk2 and p130Cas but was unable to abolish the signal, suggesting the outside-in contribution to their activation remained intact. Alternatively, cell type-specific differences in integrin expression may account for the differing levels of CXCL12-activated focal adhesion proteins observed. Prior work indicates p130Cas and paxillin interact with active Pyk2 at focal adhesions (49, 68). Immunofluorescence microscopy indicated that CXCL12 stimulation led to the preferential localization of active Pyk2 at vinculin- and paxillin-delimited focal adhesions in migrating epithelial sheets. Together, our data indicate that CXCL12 has a role in reorganization of focal adhesions.
Enterocyte migration reflects dynamic and transient adhesion with underlying matrix, facilitated primarily by focal adhesion complexes that anchor the F-actin cytoskeleton to the basement membrane (15, 39, 49, 67, 69). Based upon the rapid localization of active Pyk2 to focal adhesions, combined with ablation of CXCL12-stimulated migration with a dominant-interfering construct, we reasoned the chemokine would regulate enterocyte migration on varying extracellular matrix components. In support of that notion, cellular migration on laminin was significantly increased in CXCL12-treated cells compared with control cells. Evaluation of the baseline number of IEC-6 cells migrating on collagen-IV indicated a slight increase in migration. Although not statistically significant, the elevated migration of IEC-6 cells cultured on collagen-IV is consistent with a prior report (57) using CaCo2 cells. The inability of CXCL12 to stimulate further migration on collagen-IV suggests increased migration on laminin does not simply reflect the addition of extracellular matrix proteins. Alternatively, the lack of CXCL12-stimulated migration on collagen-IV may result from outside-in activation of those integrin receptors. Intriguingly, laminin alone has been shown to increase wound healing in vivo (70). Our data demonstrate a similar trend in which basal restitution on laminin-coated dishes was markedly increased relative to cells on BSA. Inhibition of chemokine-stimulated migration on laminin suggests release of intracellular calcium activates laminin-specific receptors. Although it remains unclear as to how CXCL12 activates laminin-specific receptors, our data support the model that CXCL12 strengthens epithelial restitution in part through inside-out activation of focal adhesion complexes.
In agreement with our prior report on CCR6-mediated restitution (5), we found that restitution stimulated by TGF-β1 was refractory to interruption of calcium-signaling pathways. Thus, chelation of intracellular calcium with cell-permeant BAPTA-AM, pharmacological, or genetic inhibition of PLCβ signaling or dominant-interfering Pyk2 constructs failed to stop TGF-β1-induced restitution. Moreover, this response was observed in non-transformed IEC-6 cells and human CaCo2 cells cultured as a polarized model epithelium. Together, these data provide strong evidence that there is not only a redundancy in extracellular mediators regulating epithelial restitution, but also point to overlapping and distinct signaling pathways in that process. Given the importance of maintaining a strong epithelial barrier, it is not surprising that multiple extracellular mediators and intracellular pathways regulate epithelial migration. With that respect, we postulate that CXCR4 and CCR6 ligands provide, with TGF-β1, key non-redundant avenues for activating and enhancing epithelial injury repair.
Although our data suggest chemokines activate calcium signaling pathways independent of TGFβ receptors, additional growth factor-activated receptor tyrosine kinases have been shown to mobilize intracellular calcium (71). Further, our findings that CXCL12 significantly enhances migration on laminin support a previous report linking epidermal growth factor stimulation with restitution on that matrix protein (57). Chemokine receptors have been linked with transactivation of the epidermal growth factor receptor (72, 73) in varying cell types. Although not examined here, it is tempting, based on those reports, to speculate the calcium response observed in CXCL12-treated cells reflect in part transactivation of members of the epidermal growth factor receptor tyrosine kinase family. Taken together, our data significantly expand upon our previous model (3–5, 74) of CXCR4- and CCR6-mediated wound healing to include calcium mobilization and Pyk2 as key regulators of chemokine stimulated barrier repair.
Supplementary Material
Acknowledgments
We are grateful for the expert technical assistance of Sheena Faherty. The assistance of Cayla Lillge (University of Wisconsin-Platteville) and Marie-Elizabeth Barabas in the phospholipase C studies is greatly appreciated. We thank Dr. Noah P. Zimmerman and Luke J. Drury for critically reading the manuscript. We appreciate the assistance of Dr. Christopher Veldkamp and Dr. Brian Volkman, Dept. of Biochemistry, Medical College of Wisconsin, in generating recombinant CXCL12 for use in these studies.
This work was supported, in whole or in part, by National Institutes of Health Grant DK062066. This work was also supported by the American Recovery and Reinvestment Act of 2009.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- TGF-β1
- transforming growth factor-β1
- BSA
- bovine serum albumin
- PLC
- phospholipase-C
- Pyk2
- proline-rich tyrosine kinase-2
- BAPTA-AM
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis (acetoxymethyl ester)
- Ad
- adenovirus
- shRNA
- short hairpin RNA
- GFP
- green fluorescent protein
- PBS
- phosphate-buffered saline
- AdCRNK
- adenovirus encoding kinase domain-deficient human Pyk2.
REFERENCES
- 1.Svanes K., Ito S., Takeuchi K., Silen W. (1982) Gastroenterology 82, 1409–1426 [PubMed] [Google Scholar]
- 2.Podolsky D. K. (1999) Am. J. Physiol. 277, G495–G499 [DOI] [PubMed] [Google Scholar]
- 3.Smith J. M., Johanesen P. A., Wendt M. K., Binion D. G., Dwinell M. B. (2005) Am. J. Physiol. Gastrointest. Liver Physiol. 288, G316–G326 [DOI] [PubMed] [Google Scholar]
- 4.Moyer R. A., Wendt M. K., Johanesen P. A., Turner J. R., Dwinell M. B. (2007) Lab. Invest. 87, 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vongsa R. A., Zimmerman N. P., Dwinell M. B. (2009) J. Biol. Chem. 284, 10034–10045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dignass A., Lynch-Devaney K., Kindon H., Thim L., Podolsky D. K. (1994) J. Clin. Invest. 94, 376–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polk D. B., Tong W. (1999) Am. J. Physiol. 277, C1149–C1159 [DOI] [PubMed] [Google Scholar]
- 8.Beck P. L., Rosenberg I. M., Xavier R. J., Koh T., Wong J. F., Podolsky D. K. (2003) Am. J. Pathol. 162, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basson M. D. (2002) Am. J. Pathol. 161, 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madsen K., Cornish A., Soper P., McKaigney C., Jijon H., Yachimec C., Doyle J., Jewell L., De Simone C. (2001) Gastroenterology 121, 580–591 [DOI] [PubMed] [Google Scholar]
- 11.Resta-Lenert S., Barrett K. E. (2003) Gut 52, 988–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stappenbeck T. S., Gordon J. I. (2000) Development 127, 2629–2642 [DOI] [PubMed] [Google Scholar]
- 13.Wong M. H., Rubinfeld B., Gordon J. I. (1998) J. Cell Biol. 141, 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahoney Z. X., Stappenbeck T. S., Miner J. H. (2008) J. Cell Sci. 121, 2493–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen C. R., Yuan L., Basson M. D. (2008) Lab. Invest. 88, 1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu J. Y., Seno H., Miletic A. V., Mills J. C., Swat W., Stappenbeck T. S. (2009) J. Cell Sci. 122, 324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayburgh D. R., Barrett T. A., Tang Y., Meddings J. B., Van Eldik L. J., Watterson D. M., Clarke L. L., Mrsny R. J., Turner J. R. (2005) J. Clin. Invest. 115, 2702–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charrier L., Yan Y., Nguyen H. T., Dalmasso G., Laboisse C. L., Gewirtz A. T., Sitaraman S. V., Merlin D. (2007) J. Biol. Chem. 282, 16948–16958 [DOI] [PubMed] [Google Scholar]
- 19.Kagimoto Y., Yamada H., Ishikawa T., Maeda N., Goshima F., Nishiyama Y., Furue M., Yoshikai Y. (2008) J. Leukoc. Biol. 83, 165–172 [DOI] [PubMed] [Google Scholar]
- 20.Schwarz B. T., Wang F., Shen L., Clayburgh D. R., Su L., Wang Y., Fu Y. X., Turner J. R. (2007) Gastroenterology 132, 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F., Schwarz B. T., Graham W. V., Wang Y., Su L., Clayburgh D. R., Abraham C., Turner J. R. (2006) Gastroenterology 131, 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keely S., Glover L. E., MacManus C. F., Campbell E. L., Scully M. M., Furuta G. T., Colgan S. P. (2009) FASEB J. 23, 1338–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dwinell M. B., Eckmann L., Leopard J. D., Varki N. M., Kagnoff M. F. (1999) Gastroenterology 117, 359–367 [DOI] [PubMed] [Google Scholar]
- 24.Jordan N. J., Kolios G., Abbot S. E., Sinai M. A., Thompson D. A., Petraki K., Westwick J. (1999) J. Clin. Invest. 104, 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izadpanah A., Dwinell M. B., Eckmann L., Varki N. M., Kagnoff M. F. (2001) Am. J. Physiol. Gastrointest. Liver Physiol 280, G710–G719 [DOI] [PubMed] [Google Scholar]
- 26.Brand S., Sakaguchi T., Gu X., Colgan S. P., Reinecker H. C. (2002) Gastroenterology 122, 166–177 [DOI] [PubMed] [Google Scholar]
- 27.Sturm A., Baumgart D. C., d'Heureuse J. H., Hotz A., Wiedenmann B., Dignass A. U. (2005) Cytokine 29, 42–48 [DOI] [PubMed] [Google Scholar]
- 28.Andres P. G., Beck P. L., Mizoguchi E., Mizoguchi A., Bhan A. K., Dawson T., Kuziel W. A., Maeda N., MacDermott R. P., Podolsky D. K., Reinecker H. C. (2000) J. Immunol. 164, 6303–6312 [DOI] [PubMed] [Google Scholar]
- 29.Tokuyama H., Ueha S., Kurachi M., Matsushima K., Moriyasu F., Blumberg R. S., Kakimi K. (2005) Int. Immunol. 17, 1023–1034 [DOI] [PubMed] [Google Scholar]
- 30.Hyun J. G., Lee G., Brown J. B., Grimm G. R., Tang Y., Mittal N., Dirisina R., Zhang Z., Fryer J. P., Weinstock J. V., Luster A. D., Barrett T. A. (2005) Inflamm. Bowel Dis. 11, 799–805 [DOI] [PubMed] [Google Scholar]
- 31.Ceradini D. J., Kulkarni A. R., Callaghan M. J., Tepper O. M., Bastidas N., Kleinman M. E., Capla J. M., Galiano R. D., Levine J. P., Gurtner G. C. (2004) Nat. Med. 10, 858–864 [DOI] [PubMed] [Google Scholar]
- 32.Loh S. A., Chang E. I., Galvez M. G., Thangarajah H., El-ftesi S., Vial I. N., Lin D. A., Gurtner G. C. (2009) Plast. Reconstr. Surg. 123, 65S–75S [DOI] [PubMed] [Google Scholar]
- 33.Dwinell M. B., Ogawa H., Barrett K. E., Kagnoff M. F. (2004) Am. J. Physiol. Gastrointest. Liver Physiol. 286, G844–G850 [DOI] [PubMed] [Google Scholar]
- 34.Yang C. C., Ogawa H., Dwinell M. B., McCole D. F., Eckmann L., Kagnoff M. F. (2005) Am. J. Physiol. Cell Physiol. 288, C321–C328 [DOI] [PubMed] [Google Scholar]
- 35.Nagasawa T., Tachibana K., Kishimoto T. (1998) Semin. Immunol. 10, 179–185 [DOI] [PubMed] [Google Scholar]
- 36.Tachibana K., Hirota S., Iizasa H., Yoshida H., Kawabata K., Kataoka Y., Kitamura Y., Matsushima K., Yoshida N., Nishikawa S., Kishimoto T., Nagasawa T. (1998) Nature 393, 591–594 [DOI] [PubMed] [Google Scholar]
- 37.Rossi D., Zlotnik A. (2000) Annu. Rev. Immunol. 18, 217–242 [DOI] [PubMed] [Google Scholar]
- 38.Parekh A. B., Penner R. (1997) Physiol. Rev. 77, 901–930 [DOI] [PubMed] [Google Scholar]
- 39.Evans J. H., Falke J. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16176–1618117911247 [Google Scholar]
- 40.Turner J. R., Angle J. M., Black E. D., Joyal J. L., Sacks D. B., Madara J. L. (1999) Am. J. Physiol. 277, C554–C562 [DOI] [PubMed] [Google Scholar]
- 41.Russo J. M., Florian P., Shen L., Graham W. V., Tretiakova M. S., Gitter A. H., Mrsny R. J., Turner J. R. (2005) Gastroenterology 128, 987–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veldkamp C. T., Peterson F. C., Hayes P. L., Mattmiller J. E., Haugner J. C., 3rd, de la Cruz N., Volkman B. F. (2007) Protein Expr. Purif. 52, 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha C., Nigam S. K., Denker B. M. (1998) J. Biol. Chem. 273, 21629–21633 [DOI] [PubMed] [Google Scholar]
- 44.Wendt M. K., Drury L. J., Vongsa R. A., Dwinell M. B. (2008) Gastroenterology 135, 508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marinkovich M. P. (2007) Nat. Rev. Cancer 7, 370–380 [DOI] [PubMed] [Google Scholar]
- 46.Kiselyov K., Shin D. M., Muallem S. (2003) Cell Signal. 15, 243–253 [DOI] [PubMed] [Google Scholar]
- 47.Yassin R. R., Abrams J. T. (1998) Peptides 19, 47–55 [DOI] [PubMed] [Google Scholar]
- 48.Lev S., Moreno H., Martinez R., Canoll P., Peles E., Musacchio J. M., Plowman G. D., Rudy B., Schlessinger J. (1995) Nature 376, 737–745 [DOI] [PubMed] [Google Scholar]
- 49.Litvak V., Tian D., Shaul Y. D., Lev S. (2000) J. Biol. Chem. 275, 32736–32746 [DOI] [PubMed] [Google Scholar]
- 50.Ganju R. K., Brubaker S. A., Meyer J., Dutt P., Yang Y., Qin S., Newman W., Groopman J. E. (1998) J. Biol. Chem. 273, 23169–23175 [DOI] [PubMed] [Google Scholar]
- 51.Wu S. S., Chiu T., Rozengurt E. (2002) Am. J. Physiol. Cell Physiol. 282, C1432–C1444 [DOI] [PubMed] [Google Scholar]
- 52.Wu S. S., Jácamo R. O., Vong S. K., Rozengurt E. (2006) Cell Signal. 18, 1932–1940 [DOI] [PubMed] [Google Scholar]
- 53.Keogh R. J., Houliston R. A., Wheeler-Jones C. P. (2002) Biochem. Biophys. Res. Commun. 290, 1470–1477 [DOI] [PubMed] [Google Scholar]
- 54.Schaller M. D., Sasaki T. (1997) J. Biol. Chem. 272, 25319–25325 [DOI] [PubMed] [Google Scholar]
- 55.Sorokin A., Kozlowski P., Graves L., Philip A. (2001) J. Biol. Chem. 276, 21521–21528 [DOI] [PubMed] [Google Scholar]
- 56.Rufanova V. A., Alexanian A., Wakatsuki T., Lerner A., Sorokin A. (2009) J. Cell Physiol. 219, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basson M. D., Modlin I. M., Madri J. A. (1992) J. Clin. Invest. 90, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lotz M. M., Nusrat A., Madara J. L., Ezzell R., Wewer U. M., Mercurio A. M. (1997) Am. J. Pathol. 150, 747–760 [PMC free article] [PubMed] [Google Scholar]
- 59.Mammen J. M., Matthews J. B. (2003) Crit. Care Med. 31, S532–S537 [DOI] [PubMed] [Google Scholar]
- 60.Glogauer M., Arora P., Yao G., Sokholov I., Ferrier J., McCulloch C. A. (1997) J. Cell Sci. 110, 11–21 [DOI] [PubMed] [Google Scholar]
- 61.Clapham D. E. (1995) Cell 80, 259–268 [DOI] [PubMed] [Google Scholar]
- 62.Boucek M. M., Snyderman R. (1976) Science 193, 905–907 [DOI] [PubMed] [Google Scholar]
- 63.Rao J. N., Platoshyn O., Golovina V. A., Liu L., Zou T., Marasa B. S., Turner D. J., Yuan J. X., Wang J. Y. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 290, G782–G792 [DOI] [PubMed] [Google Scholar]
- 64.Rao J. N., Li L., Golovina V. A., Platoshyn O., Strauch E. D., Yuan J. X., Wang J. Y. (2001) Am. J. Physiol. Cell Physiol. 280, C993–C1007 [DOI] [PubMed] [Google Scholar]
- 65.Kohno T., Matsuda E., Sasaki H., Sasaki T. (2008) Biochem. J. 410, 513–523 [DOI] [PubMed] [Google Scholar]
- 66.Xu J., Gao X. P., Ramchandran R., Zhao Y. Y., Vogel S. M., Malik A. B. (2008) Nat. Immunol. 9, 880–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y. W., Sanders M. A., Basson M. D. (1998) J. Surg. Res. 77, 112–118 [DOI] [PubMed] [Google Scholar]
- 68.Astier A., Avraham H., Manie S. N., Groopman J., Canty T., Avraham S., Freedman A. S. (1997) J. Biol. Chem. 272, 228–232 [DOI] [PubMed] [Google Scholar]
- 69.Lotz M. M., Rabinovitz I., Mercurio A. M. (2000) Am. J. Pathol. 156, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malinda K. M., Wysocki A. B., Koblinski J. E., Kleinman H. K., Ponce M. L. (2008) Int. J. Biochem. Cell Biol. 40, 2771–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keely S. J., Uribe J. M., Barrett K. E. (1998) J. Biol. Chem. 273, 27111–27117 [DOI] [PubMed] [Google Scholar]
- 72.Venkatakrishnan G., Salgia R., Groopman J. E. (2000) J. Biol. Chem. 275, 6868–6875 [DOI] [PubMed] [Google Scholar]
- 73.Cabioglu N., Summy J., Miller C., Parikh N. U., Sahin A. A., Tuzlali S., Pumiglia K., Gallick G. E., Price J. E. (2005) Cancer Res. 65, 6493–6497 [DOI] [PubMed] [Google Scholar]
- 74.Zimmerman N. P., Vongsa R. A., Wendt M. K., Dwinell M. B. (2008) Inflamm. Bowel Dis. 14, 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.