Abstract
Ubiquitously expressed membrane type-1 matrix metalloproteinase (MT1-MMP), an archetype member of the MMP family, binds tissue inhibitor of metalloproteinases-2 (TIMP-2), activates matrix metalloproteinase-2 (MMP-2), and stimulates cell migration in various cell types. In contrast with MT1-MMP, the structurally similar MT6-MMP associates with the lipid raft compartment of the plasma membrane using a GPI anchor. As a result, MT6-MMP is functionally distinct from MT1-MMP. MT6-MMP is insufficiently characterized as yet. In addition, a number of its biochemical features are both conflicting and controversial. To reassess the biochemical features of MT6-MMP, we have expressed the MT6-MMP construct tagged with a FLAG tag in breast carcinoma MCF-7 and fibrosarcoma HT1080 cells. We then used phosphatidylinositol-specific phospholipase C to release MT6-MMP from the cell surface and characterized the solubilized MT6-MMP fractions. We now are confident that cellular MT6-MMP partially exists in its complex with TIMP-2. Both TIMP-1 and TIMP-2 are capable of inhibiting the proteolytic activity of MT6-MMP. MT6-MMP does not stimulate cell migration. MT6-MMP, however, generates a significant level of gelatinolysis of the fluorescein isothiocyanate-labeled gelatin and exhibits an intrinsic, albeit low, ability to activate MMP-2. As a result, it is exceedingly difficult to record the activation of MMP-2 by cellular MT6-MMP. Because of its lipid raft localization, cellular MT6-MMP is inefficiently internalized. MT6-MMP is predominantly localized in the cell-to-cell junctions. Because MT6-MMP has been suggested to play a role in disease, including cancer and autoimmune multiple sclerosis, the identity of its physiologically relevant cleavage targets remains to be determined.
Keywords: Lipid Raft, Membrane Enzymes, Metalloprotease, Protease, Tissue Inhibitors of Metalloprotease (TIMPs)
Introduction
The members of the matrix metalloproteinase (MMP)2 family degrade a wide spectrum of extracellular matrix proteins, growth factors, and cell receptors and play important roles in multiple diseases (1–3). In malignancy, MMPs, especially membrane-type MMPs (MT-MMPs), have been proposed to play key roles in tumor invasion and metastasis (4). As a result, it is now accepted that in depth mechanistic understanding of the MT-MMP functionality will ultimately lead to novel and effective therapies against invasive, metastatic malignancies (5).
The MT-MMP subfamily includes six members, named MT1-MMP (MMP-14), MT2-MMP (MMP-15), MT3-MMP (MMP-16), MT4-MMP (MMP-17), MT5-MMP (MMP-24), and MT6-MMP (MMP-25). MT1-MMP, MT2-MMP, MT3-MMP, and MT5-MMP are anchored to the plasma membrane via a transmembrane domain (1, 4). In contrast, the attachment of MT4-MMP and MT6-MMP to the plasma membrane takes place via a glycosylphosphatidylinositol (GPI) moiety that directs these proteinases to the caveola-enriched lipid raft compartment (6). Although there is a significant level of knowledge of the structural-functional relationships and regulation of MT1-MMP, an archetype member of the subfamily, exceedingly little is known about the biochemical and cellular properties of GPI-linked MT4-MMP and especially MT6-MMP. Originally, MT6-MMP was cloned from a fetal liver cDNA library and from leukocytes (7–9). Because of its abundance in leukocytes, the protease has also been called leukolysin. MT6-MMP was also found to be abnormally expressed by colon, prostate, urothelial, and brain tumors, suggesting its implication in the diversified malignancies (10). MT6-MMP is a membrane proteinase with an extracellular prodomain followed by the catalytic domain, the hinge region, the hemopexin domain, the stalk region, and a GPI anchor attached to the carboxyl end of the stalk. Similar to MT1-MMP, MT6-MMP is synthesized as an inactive precursor that is transformed into the functionally active proteinase by the cleavage action of furin-like proprotein convertases (1, 6).
The regulation mechanisms, the functional role, and the repertoire of physiologically relevant cleavage targets of MT6-MMP remain largely unknown. In addition to a limited number of extracellular matrix components, including fibronectin, gelatin, type IV collagen, and chondroitin and dermatan sulfate proteoglycan, the catalytic domain of MT6-MMP has been reported to cleave galectin-3, urokinase plasminogen activator receptor and myelin basic protein (6, 11, 12). The relevance of MT6-MMP proteolysis of these target proteins to the enzyme functionality in both normal development and disease progression, however, is not entirely clear as of now.
It is widely accepted that tissue inhibitors of MMPs (TIMPs) play an important role in the regulation of the net proteolytic activity of MMPs (13). Four individual TIMPs, including TIMP-1 and TIMP-2, are known in humans (14). There are two domains in both TIMP-1 and TIMP-2. The inhibitory N-terminal domain directly interacts, albeit with different affinity, with the active site of the MMP catalytic domain. The C-terminal domain of TIMP-1 forms a stoichiometric complex with the hemopexin domain of the MMP-9 proenzyme. TIMP-2 forms a similar complex with the MMP-2 proenzyme. TIMP-2 performs not only as an inhibitor but also as an essential component of the MT1-MMP-dependent activation pathway, leading to the MMP-2 mature enzyme (4, 15–18).
The existing data suggest that both TIMP-1 and TIMP-2 inhibit the proteolytic activity of the individual catalytic domain of MT6-MMP (6, 10, 19). There are conflicting results that point to the potential role of MT6-MMP in the mechanisms of MMP-2 activation (6, 20). Cellular MT6-MMP, however, was not observed in a complex with either TIMP-1 or TIMP-2, thus raising a question of how the activity of MT6-MMP is regulated (6, 21). To date, all of the biochemical studies have employed the individual catalytic domain rather than the full-length MT6-MMP enzyme or its short C-terminal truncations (6, 9, 10, 12, 19). As a result, the question of whether other domains of MT6-MMP do or do not affect the functionality of the proteinase remains to be answered.
To understand better the main parameters of the MT6-MMP functionality, we biochemically characterized the MT6-MMP samples directly isolated from the cells and reexamined the ability of cellular MT6-MMP to associate with TIMP-2, to activate MMP-2, and to function as an invasion-promoting, matrix-degrading proteinase. Because the current knowledge of MT6-MMP is exceedingly limited, we believe that the mechanistic observations we generated shed additional light on the function of the lipid raft-associated, GPI-linked MT6-MMP in cancer.
MATERIALS AND METHODS
General Reagents and Antibodies
All reagents were purchased from Sigma unless indicated otherwise. EZ-link sulfo-NHS-SS-biotin was from Pierce. (7-Methoxycoumarin-4-yl)-acetyl-Pro-Leu-Gly-Leu-(3-[2,4-dinitrophenyl]-l-2,3-diaminopropionyl)-Ala-Arg-NH2 (Mca-PLGL-Dpa-AR-NH2) was obtained from R&D Systems. GM6001 (a potent, wide range hydroxamate inhibitor of MMPs, including MT1-MMP) and the rabbit MT1-MMP (anti-hinge) antibody were purchased from Chemicon. A murine monoclonal E-cadherin antibody (catalog no. 610181) was purchased from BD Biosciences. Purified TIMP-1 and a rabbit MT6-MMP antibody were a kind gift from Dr. Rafael Fridman (Wayne State University, Detroit, MI). Recombinant TIMP-2 and the TIMP-2-free MMP-2 proenzyme were isolated from the conditioned medium of CHO cells and p2AHT2A72 cells, respectively. p2AHT2A72 cells were derived from the fibrosarcoma HT1080 cell line sequentially transfected with E1A and MMP-2 cDNAs, respectively (22, 23). The sheep TIMP-2 antibody was from Abcam. The secondary antibodies were purchased from Jackson ImmunoResearch. The secondary species-specific antibodies conjugated with Alexa Fluor 594 (red) and green Alexa Fluor 488 (green) were obtained at Molecular Probes. Human α1-antitrypsin (AAT) and a protease inhibitor mixture set III were from Calbiochem.
Recombinant MMPs
The individual catalytic domain of MT6-MMP (MT6CAT) and MT1-MMP (MT1CAT) were expressed in Escherichia coli, purified from the inclusion bodies, and refolded to restore its catalytic activity (11). The recombinant pro-forms of the catalytic domain of MMP-2 and MMP-9 were purified from the serum-free medium conditioned by stably transfected HEK293 cells and then activated using 4-aminophenylmercuric acetate (24). The MMP activity was determined using the fluorescent Mca-PLGL-Dpa-AR-NH2 peptide substrate. The concentrations of the catalytically active MT6CAT, MT1CAT, MMP-2, and MMP-9 were measured by titration against a standard GM6001 solution of a known concentration (11, 12, 25).
Cloning of FLAG-tagged MT6-MMP (MT6F)
The full-length human MT6-MMP cDNA gene (GenBankTM NM_022468) was a gift from Dr. Rafael Fridman (Wayne State University). The sequence coding for a FLAG tag was inserted between Gly-514 and Pro-515 in the stalk region of MT6-MMP to generate the MT6F construct. The authenticity of the recombinant constructs was confirmed by DNA sequencing. MT6F was recloned into the pcDNA3.1D/V5-His-TOPO-neo plasmid.
Cells
Human breast carcinoma MCF-7 cells (MCF7 cells) and human fibrosarcoma HT1080 cells (HT cells) were obtained from the ATCC (Manassas, VA). HT and MCF7 cells stably transfected with MT6F in the pcDNA3.1D/V5-His-TOPO-neo plasmid (MCF7-MT6F and HT-MT6F cells) and control cells transfected with the original pcDNA3.1D/V5-His-neo plasmid (MCF7-mock and HT-mock cells) were obtained as described earlier (26). To avoid clonal effects, 5–6 neomycin-resistant MT6F cell clones were combined and used in our further studies. HT and MCF7 cells stably transfected with MT1-MMP (HT-MT1 and MCF7-MT1 cells, respectively) were isolated and characterized earlier (27). Cells were routinely cultured in high glucose DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Fractionation of Cellular MT6-MMP
Confluent cells (1 × 108) were detached using an enzyme-free cell dissociation solution (Chemicon). Cells were suspended in serum-free DMEM and incubated for 2 h at 37 °C with 0.25 units/ml phosphatidylinositol-specific phospholipase C (PLC). Cells were collected by centrifugation at 300 × g for 5 min. The supernatant (PLC fraction) was clarified by centrifugation at 20,000 × g, 20 min. The following steps were performed at 4 °C. The collected cells were lysed in 10 ml of 20 mm Tris-HCl, pH 7.9, 150 mm NaCl, 5 mm MgCl2, 10% glycerol, a protease inhibitor mixture set III, 1 mm phenylmethylsulfonyl fluoride, and 0.5% Nonidet P-40 (Nonidet P-40 buffer). The extract was centrifuged (40 min; 20,000 × g). The supernatant (Nonidet P-40 fraction; 20–40 mg of total protein) and the PLC fraction were incubated for 4 h with a 50% FLAG M2 antibody-bead slurry (40 μl). The beads were collected by centrifugation and washed six times in the above buffer but with 0.1% Nonidet P-40 instead of 0.5% Nonidet P-40. The beads were next incubated for 1 h with 20 μl of FLAG peptide (0.2 mg/ml) to elute the FLAG-containing constructs. The beads were removed by centrifugation. The supernatant samples were analyzed further using SDS-PAGE followed by silver staining and LC/MS/MS; by Western blotting with the FLAG M2 (dilution 1:4,000), MT6-MMP (dilution 1:1,500), and TIMP-2 (dilution 1:1,000) antibodies; and by the enzymatic activity assays. The species-specific peroxidase-conjugated secondary antibodies (dilution 1:3,000) and a SuperSignal West Dura extended duration substrate Kit (Pierce) were used for the detection of the immunopositive protein bands.
Mass Spectrometry
Following SDS-PAGE, the MT6F-PLC and MT6F-Nonidet P-40 bands were excised and subjected to an in-gel tryptic digest. The digest peptides were extracted and then identified by LC/MS/MS using an LTQ XL Linear Ion Trap mass spectrometer (Thermo Scientific). MS/MS spectra were searched against the Swiss-Prot database using the SEQUEST Sorcerer software. The search led to the multiple peptides the sequence of which permitted an unambiguous identification of MT6-MMP.
Cell Surface Biotinylation and 2-Mercaptoethane Sulfonic Acid (MESNA) Treatment
Cells were surface-biotinylated using membrane-impermeable EZ-Link sulfo-NHS-SS-biotin (0.3 mg/ml), lysed in 0.5% Nonidet P-40 or RIPA buffer (20 mm Tris-HCl, pH 7.4, containing 150 mm NaCl, 0.1% SDS, 1% Triton X-100 and 1% sodium deoxycholate). The biotin-labeled plasma membrane proteins were pulled down using streptavidin-beads (27). The precipitates were dissolved in SDS sample buffer (0.125 m Tris-HCl, pH 6.8, 20% glycerol, 2% SDS, 0.005% bromphenol blue, and 5% 2-mercaptoethanol) and analyzed by Western blotting with the MT1-MMP and MT6-MMP antibody, followed by species-specific secondary antibodies conjugated with horseradish peroxidase and a TMB/M substrate.
For the uptake experiments, immediately following the completion of the biotinylation procedure, the biotinylated cells were incubated for 30–60 min at 37 °C in serum-free DMEM supplemented with 1% insulin/transferrin/selenium to allow the internalization of biotin-labeled MT1-MMP (27, 28). To remove the residual cell surface biotin, cells were incubated for 25 min on ice in Sorenson phosphate buffer (14.7 mm KH2PO4, 2 mm Na2HPO4, 120 mm sorbitol, pH 7.8) containing membrane-impermeable MESNA (150 mm). Cells were next extensively washed and lysed, and the lysates were precipitated using streptavidin-beads and analyzed as above.
Gelatin Zymography
Cells (2 × 105) were seeded for 24 h in DMEM plus 10% fetal bovine serum in wells of a 12-well plate. The medium was then replaced with serum-free DMEM (0.35 ml/well) supplemented with pro-MMP-2 (0.5 nm). When indicated, TIMP-1 or TIMP-2 (0.5–100 nm each) or GM6001 (50 μm) was added. In 18 h, the medium aliquots (25 μl) were analyzed by gelatin zymography in 0.1% gelatin, 10% acrylamide gels (18). 15% gelatin gels were used to detect the gelatinolytic activity of the purified 25-kDa MT6CAT construct.
In the in vitro assays, pro-MMP-2 (15 nm) was co-incubated for 2 h at 37 °C with the purified MT1CAT and MT6CAT constructs (0.15–15 nm) in 50 mm HEPES, pH 7.5, containing 10 mm CaCl2 and 50 μm ZnCl2. Where indicated, GM6001 (5 μm) was added to the reactions. The digest aliquots were then analyzed by gelatin zymography.
Cleavage of AAT
The cleavage reactions (22 μl each) were performed in 50 mm HEPES, pH 7.5, containing 10 mm CaCl2 and 50 μm ZnCl2. AAT (1.6 μm) was co-incubated for 3 h at 37 °C with MT1CAT and MT6CAT (16 nm each; 1:100 enzyme/substrate molar ratio), and cellular MT6F was isolated from the PLC fraction (16–80 nm; 1:20–1:100 enzyme/substrate molar ratio). The cleavage reactions were stopped using a 5× SDS sample buffer and analyzed by SDS-PAGE followed by Coomassie staining. Where indicated, GM6001 (5 μm) was added to the reactions to inhibit MMP activity.
Enzymatic Assay
MMP activity was measured in wells of a 96-well plate in 0.2 ml of 50 mm HEPES, pH 7.5, containing 10 mm CaCl2 and 50 μm ZnCl2. Mca-PLGL-Dpa-AR-NH2 (5 μm) was used as a fluorescent substrate. The concentration of MMPs in the reactions was 20–600 fmol. The steady-state rate of substrate hydrolysis was monitored continuously (λex = 320 nm and λem = 400 nm) at 37 °C for 3–75 min using a fluorescence spectrophotometer. Where indicated, TIMP-1 (2.5–50 nm), TIMP-2 (2.5–25 nm), and GM6001 (1 μm) were co-incubated for 30 min at ambient temperature with the MMP samples prior to the addition of the substrate. The samples were measured in triplicate. The results were highly reproducible without any significant day to day variations.
A Library of the Potential MMP Inhibitors
Our prototype CFL-1 (chelator fragment library-1) was described earlier (29). Our extended library now includes ∼500 potential MMP inhibitors.
Determination of the IC50 Values of Inhibitors
MT1CAT, MT6CAT, MMP-2, and MMP-9 (10 nm each) were preincubated for 30 min at ambient temperature with increasing concentrations of the individual compounds from the MMP inhibitor library. The residual activity of MMPs was then measured using Mca-PLGL-Dpa-AR-NH2. IC50 values of the inhibitors were calculated using GraphPad Prism as a fitting software.
Synthesis of BI-92G11 and BI-102C8
A solution of the corresponding acid (0.200 g, 0.933 mmol) and dimethylformamide (0.053 g, 0.933 mmol) in CH2Cl2 (5 ml) was cooled to 0 °C. Oxalyl chloride (2.05 mmol) was then added slowly. Vigorous gas evolution was observed. After stirring for 2 h at 0 °C, this solution was added to a solution of hydroxylamine hydrochloride (0.259 g, 3.73 mmol) and triethylamine (0.566 g, 5.60 mmol) in tetrahydrofuran (5 ml)/H2O (1 ml). After stirring for an additional 4 h at ambient temperature, the mixture was poured into 2 n HCl and extracted with CH2Cl2. The organic phase was dried over Na2SO4 and evaporated in vacuo. The residue was recrystallized from aqueous ethanol or purified via flash chromatography (30). BI-92G11: 1H NMR (500 MHz, CDCl3) δ10.56 (s, 1H), 8.89 (brs, 1H), 7.89 (d, 2H, J = 7.0 Hz), 7.75 (t, 1H, J = 7.0 Hz), 7.66 (t, 2H, J = 7.0 Hz), 3.49 (m, 2H), 3.03 (m, 2H). HRMS e/z (electrospray ionization time-of-flight). Found 184.0423 [MH]+, C8H9NO2S requires 184.0423. BI-102C8: 1H NMR (600 MHz, (CD3)2SO) δ10.72 (s, 1H), 8.95 (s, 1H), 7.48 (t, 1H, J = 7.5), 7.37–7.33 (m, 5H), 6.95–6.93 (d, 2H, J = 8.5), 4.07–4.04 (q, 2H, J = 6.9), 1.36–1.34 (t, 3H, J = 6.9).
Cell Migration
Cell migration experiments were performed in wells of a 24-well, 8-μm pore size, Transwell plate (Corning Costar). A 6.5-mm insert membrane was coated with 100 μl of type I collagen (100 μg/ml in DMEM) and then dried for 16 h. The collagen coating was rehydrated in 0.5 ml of DMEM for 30 min before the experiments. The inner chamber contained DMEM supplemented with 10% fetal bovine serum as chemoattractant. Cells (1 × 104) were seeded in the outer chamber in serum-free DMEM. GM6001 (25 μm) or DMSO alone (0.01%) were added to both inner and outer chambers 15 min before plating the cells. Cells were allowed to migrate for 3.5 h and then stained with 20% methanol, 0.2% Crystal Violet. Cells from the upper membrane surface were removed with a cotton swab. Incorporated dye from the migrated cells was extracted using 0.25 ml of 1% SDS. The A570 value of the extract was measured. Data are means ± S.E. from several individual experiments performed in triplicate. Statistical analysis was performed by the two-tailed unpaired t test.
In Situ Gelatin Zymography Using FITC-gelatin
To prepare FITC-gelatin (31), gelatin (2 mg/ml) was conjugated for 2 h at ambient temperature to 100 μg/ml FITC in 0.1 m carbonate-bicarbonate buffer, pH 9.1. Unreacted dye was removed by gel filtration through a G-25 Sephadex M column (GE Healthcare) equilibrated in PBS. The A280 and A494 values of the fractions were determined. The fractions with an FITC/protein molar ratio of ≥3 were used further (32). Sterilized 15-mm glass coverslips were coated for 2 h at 37 °C with 100 μl of FITC-gelatin (100 μg/ml). The excess of gelatin was removed by washing with prewarmed DMEM. Cells (2 × 104) were seeded onto the gelatin-coated coverslips and incubated for 16 h at 37 °C in DMEM with or without GM6001 (50 μm). The cells were then fixed with 4% paraformaldehyde for 10 min and mounted in the Vectashield medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole (DAPI) for the nuclear staining. The slides were then analyzed using an Olympus BX51 fluorescent microscope equipped with a MagnaFire digital camera.
Immunostaining of Cells
Cells were fixed for 10 min in 4% paraformaldehyde, permeabilized using 0.1% Triton X-100, washed with PBS containing 0.1% Tween 20, and blocked for 1 h in 1% casein. Cells were then stained for 1 h at ambient temperature using the rabbit antibody to MT6-MMP (dilution 1:250) and the monoclonal murine antibody to E-cadherin (dilution 1:1,000), followed by incubation for 1 h with the secondary antibodies (dilution 1:200) conjugated with Alexa Fluor 594 or Alexa Fluor 488, respectively. The slides were mounted in the Vectashield medium with DAPI. Images were acquired at an original magnification of ×600 using an Olympus BX51 fluorescence microscope equipped with a MagnaFire digital camera.
RESULTS
Cloning, Expression, and Analysis of the MT6-MMP Construct Tagged with a FLAG Tag
To facilitate the follow-on analysis of MT6-MMP and to generate the FLAG-tagged MT6-MMP construct (MT6F), the Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-Gly sequence coding for a FLAG tag (underlined) was inserted between Gly-514 and Pro-515 of the stalk region of MT6-MMP. The comparison of the MT6F construct with MT1-MMP, an archetype member of the MT-MMP subfamily, is schematically shown in Fig. 1A.
FIGURE 1.
Constructs and expression of MT6-MMP. A, left, the MT1-MMP and MT6F constructs. A FLAG tag was inserted between Gly-514 and Pro-515 in the stalk region of MT6-MMP. An additional Gly residue was inserted at the C-end of FLAG. S, signal peptide; PRO, prodomain; CAT, catalytic domain; H, hinge region; PEX, hemopexin domain; ST, stalk region (the start of the stalk region is shown by an arrow); TM, transmembrane domain; CT, cytoplasmic tail. Right, MCF7-mock and MCF7-MT6F cells were lysed in 1% SDS. The lysates (3 μg of total protein) were analyzed by Western blotting (WB) with the MT6-MMP antibody. B, MT6F and MCF7-mock (mock) cells were stained with the MT6-MMP antibody, followed by Alexa Fluor 594-conjugated anti-rabbit antibody. The nuclear DNA was stained with DAPI.
The MT6F construct was transfected into breast carcinoma MCF-7 cells, and after selection procedures, the stably transfected MCF7-MT6F cells were obtained. We specifically selected MCF-7 cells for our biochemical experiments because they normally do not express this proteinase. A total cell lysate was prepared using 1% SDS. The solubilized material was analyzed by Western blotting with an MT6-MMP antibody. The results clearly demonstrated that MCF7-MT6F cells expressed significant levels of MT6F. In contrast, MT6-MMP immunoreactivity was not detected in MCF7-mock cells transfected with the original plasmid without the MT6F insert (Fig. 1A). Two species of MT6F, a 60-kDa minor and a 55-kDa major species, were observed in MCF7-MT6F cells. The molecular mass of these species correlated well with the expected size of the proenzyme and the mature enzyme of MT6F, suggesting that the cellular furin-like proprotein convertases processed the de novo synthesized MT6F construct during its trafficking to the cell surface. MT6-MMP immunoreactivity was predominantly localized in the cell-cell contact regions (Fig. 1B).
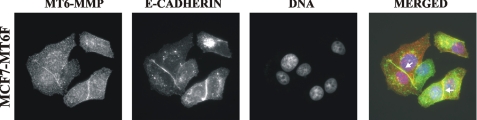
To support the predominant presence of MT6-MMP at the cell-cell contact regions, we then performed immunostaining of MCF7-MT6F cells using the antibodies to MT6-MMP and E-cadherin (the classic homophilic adhesion molecule that is normally present in cell-cell junctions) (33, 34). There was a clear co-localization of MT6F with E-cadherin in cell-cell junction regions in MCF7-MT6F cells (Fig. 2). The functional significance of the predominant association of MT6-MMP with the cell-cell junctions remains to be identified.
FIGURE 2.
Co-localization of MT6-MMP with E-cadherin in MCF7-MT6F cells. Cells were stained with the MT6-MMP and E-cadherin antibodies (red and green, respectively). The nuclei were stained with DAPI. The arrows point to the cell-cell junction regions, in which MT6-MMP and E-cadherin are co-localized. The slides were observed using a fluorescent microscope (magnification, ×600).
Fractionation of Cellular MT6-MMP
To analyze cell compartmentation of MT6F in more detail, we used several pull-down and detection procedures. Thus, cells were surface-biotinylated and then extracted using the Nonidet P-40 buffer and, alternatively, the RIPA buffer. The concentration of 0.5% Nonidet P-40 in the Nonidet P-40 buffer was insufficient to solubilize the lipid rafts and to release the GPI-anchored cellular MT6F. In turn, the RIPA buffer solubilized well the lipid raft-associated MT6F. The extracted Nonidet P-40 and RIPA samples were precipitated using the FLAG M2 antibody and streptavidin-beads, respectively.
MCF7-MT6F cells were also co-incubated with PLC to destroy the GPI linker and to liberate cell surface-associated MT6F. The solubilized fraction and the residual cells were separated by centrifugation. The cell samples were then extracted with the Nonidet P-40 buffer. Both the Nonidet P-40 extract and the PLC-solubilized samples were precipitated using the FLAG M2 antibody-beads. The samples were then analyzed using Western blotting with FLAG M2 and MT6-MMP antibodies. MCF-7 cells that were transfected with MT1-MMP (MCF7-MT1 cells) were used as an additional control.
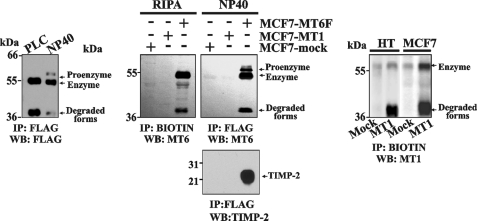
Because the mature MT6-MMP enzyme was associated with the lipid rafts, we expected that the Nonidet P-40 extraction procedure would predominantly liberate the intracellular pool of MT6-MMP. The analysis of both the PLC and the Nonidet P-40 samples supported this suggestion. Thus, the Western blotting analysis with the FLAG M2 antibody detected the presence of the 55-kDa mature enzyme and 40-kDa degraded forms in the PLC-extracted samples. In turn, the 60-kDa proenzyme was detected in the Nonidet P-40 samples in addition to the 55-kDa enzyme and the minor amounts of the 40-kDa proteolyzed forms of MT6-MMP. The level of degraded MT6-MMP was minor relative to that of MT1-MMP in both HT1080 and MCF-7 cells (Fig. 3).
FIGURE 3.
Analysis of cellular MT6-MMP. Left, MCF7-MT6F cells were treated with PLC followed by Nonidet P-40 extraction of the cells. The PLC and Nonidet P-40 fractions were precipitated (IP) using the FLAG M2 antibody-beads. The precipitates were analyzed by Western blot (WB) with the FLAG M2 antibody. Middle left panel, MCF7-mock, MCF7-MT6F, and MCF7-MT1 cells were surface-biotinylated. Cells were lysed in RIPA buffer. The biotin-labeled proteins were pulled down using streptavidin-beads. The samples were analyzed using Western blot with the MT6-MMP antibody. Middle right panels, MCF7-mock, MCF7-MT6F, and MCF7-MT1 cells were extracted with Nonidet P-40. The extracts were precipitated using the FLAG M2 antibody-beads. The precipitates were analyzed by WB with the MT6-MMP (MT6; top) and TIMP-2 (bottom) antibodies. Right panel, HT-mock, HT-MT1, MCF7-mock, and MCF7-MT1 cells were surface-biotinylated. Cells were lysed in RIPA buffer. The biotin-labeled proteins were pulled down using streptavidin-beads. The samples were analyzed using Western blot with the MT1-MMP antibody.
Overall, our results imply that, as MT1-MMP, MT6-MMP is activated by the furin-like proprotein convertases during its trafficking to the cell surface. As a result, MT6-MMP is predominantly represented at the cell surface by the lipid raft-associated mature enzyme and its proteolyzed forms, whereas the Nonidet P-40 extracted material included the residual amounts of the intracellular pool of the MT6-MMP proenzyme.
TIMP-2 Is Associated with Cellular MT6-MMP
To determine also if TIMP-2 was present in the cellular MT6F samples, MCF7-MT6F, MCF7-MT1, and MCF7-mock cells were extracted using the Nonidet P-40 buffer. The extracts were immunoprecipitated using the FLAG M2 antibody-beads. The precipitated MT6F samples clearly displayed the presence of TIMP-2, suggesting that cellular MT6F can form a complex with this inhibitor. In contrast, neither FLAG nor TIMP-2 immunoreactivity was observed if the MCF7-mock and MCF7-MT1 samples were analyzed (Fig. 3).
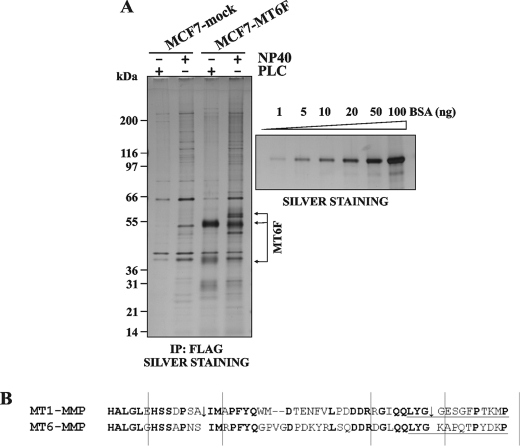
Mass Spectrometry Analysis of the Isolated MT6-MMP Samples
Both the Nonidet P-40 extract and the PLC samples were immunoprecipitated using the FLAG M2 antibody-beads. The precipitates were then analyzed by SDS-gel electrophoresis followed by silver staining. To unambiguously determine the identity of the bands, the stained proteins were excised and subjected to the in-gel trypsin digestion, followed by LC/MS/MS. Only a few of the background, nonspecific protein bands were detected in the control MCF7-mock cells. The MT6-MMP bands were readily identified in the MCF7-MTF6 cell samples. The PLC samples exhibited largely the 55-kDa MT6-MMP mature enzyme and a minor level of the degraded, 40-kDa, MT6-MMP forms. In turn, the Nonidet P-40 samples, which predominantly included the intracellular pool of MT6-MMP, were represented by the 60-kDa proenzyme of MT6-MMP in addition to the 55-kDa enzyme. The degradation products were not detected in the Nonidet P-40 samples from MCF7-MT6F cells (Fig. 4A). The difference in the sequence of the catalytic domain-hinge region could be a possible explanation of the low degradation levels of MT6-MMP relative to MT1-MMP (Fig. 4B).
FIGURE 4.
Isolation of the PLC and Nonidet P-40 fraction of MT6F. A, MCF7-mock and MCF7-MT6F cells were treated with PLC, followed by Nonidet P-40 extraction of the cells. The PLC and Nonidet P-40 fractions were precipitated using the FLAG M2 antibody-beads. The precipitates were separated by SDS-PAGE and silver-stained. The MT6F bands were subjected to the LC/MS/MS analysis. The calibration curve of bovine serum albumin (BSA; 1–100 ng) is on the right. According to this curve, the levels of MT6F (isolated from 1 × 107 cells) were ∼60 and ∼30 ng in the PLC and the Nonidet P-40 fractions, respectively. B, the partial peptide sequence alignment of the catalytic domain-hinge region of MT1-MMP and MT6-MMP. Identical residues are in boldface type. The arrows indicate the self-proteolyzed bonds in MT1-MMP (44–46, 52). The hinge sequence is underlined. IP, immunoprecipitation.
Using the bovine serum albumin calibration curve, we estimated the amounts of MT6-MMP we isolated from the MCF7-MT6F cells. These amounts equaled ∼60 and ∼30 ng of MT6-MMP in the PLC and Nonidet P-40 samples, respectively, which originated from 1 × 107 MCF7-MT6F cells and which, if combined, corresponded to a total number of ∼120,000 MT6F molecules/cell. These isolated amounts were sufficient to measure the catalytic activity of the protease.
MT6-MMP Is Catalytically Active
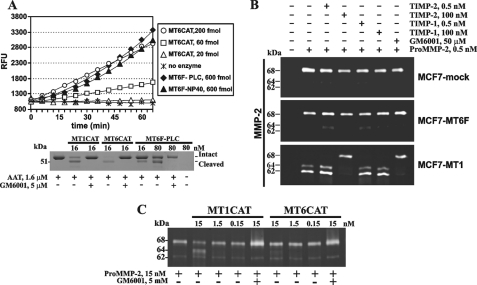
For the measurement of the catalytic activity of the MT6F fractions we used the Mca-PLGL-Dpa-AR-NH2 fluorescent peptide substrate. The catalytic activity of both the PLC and Nonidet P-40 fractions (30–35 ng, 600 fmol each) was directly compared with that of MT6CAT (20–200 fmol) (Fig. 5A). The concentration of the catalytically active MT6CAT in its purified samples was quantified by active site titration against the known concentrations of GM6001 (12). According to our analysis, the activity of 600 fmol of MT6F in both PLC and Nonidet P-40 fractions was similar and, in addition, equal to the activity of 200 fmol of MT6CAT. These data suggest that the PLC and Nonidet P-40 fractions represented the active enzyme of MT6-MMP and that ∼30% of the isolated material was fully catalytically potent.
FIGURE 5.
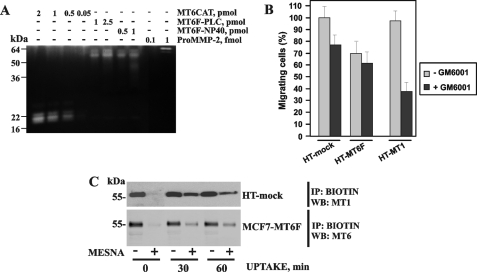
Proteolytic activity of MT6-MMP. A, top, the proteolytic activity of MT6CAT (20–200 fmol), MT6F-PLC, and MT6F-Nonidet P-40 (600 fmol each) was measured using Mca-PLGL-Dpa-AR-NH2 as a substrate (5 μm). Bottom, MT6F-PLC is catalytically active and cleaves AAT. AAT (1.6 μm) was co-incubated with the indicated concentration of MT1CAT, MT6CAT, and MT6F-PLC. Where indicated, GM6001 (5 μm) was added to the reactions. The reactions were analyzed by SDS-PAGE, followed by Coomassie staining. RFU, relative fluorescence unit. B, gelatin zymography of the condition medium aliquots from MCF7-mock, MCF7-MT1, and MCF7-MT6F cells. Where indicated, pro-MMP-2 (0.5 nm), TIMP-1 (0.5–100 nm), TIMP-2 (0.5–100 nm), and GM6001 (50 μm) were added to the cells. C, the individual catalytic domain of MT6-MMP exhibits a limited ability to activate pro-MMP-2 in the cell-free system. The indicated concentrations of MT1CAT and MT6CAT were co-incubated with pro-MMP-2 (15 nm). The samples were then analyzed by gelatin zymography. Where indicated, GM6001 (5 μm) was added to the reactions to block MMP activity.
To strengthen our results further, we tested if MT6F-PLC was capable of cleaving AAT, a common and convenient substrate for testing the functional activity of the individual MMPs in vitro (35). Several individual MMPs, including MMP-1, MMP-3, MMP-7, MMP-9, MMP-26, and MT1-MMP, have been reported to cleave AAT and to destroy its serpin activity (35–40). The individual MMPs cleave 55-kDa AAT near the C terminus and generate the 51-kDa N-terminal fragment as well as a C-terminal fragment of ∼4 kDa (38). In agreement with these data, MT1CAT, MT6CAT, and MT6F-PLC cleaved the 55-kDa AAT and generated, as a result, the 51-kDa cleavage product (Fig. 5). MT6CAT was superior in these tests relative to MT1CAT and especially MT6F-PLC. The latter, however, clearly demonstrated its ability to specifically cleave AAT. MT6F-PLC activity against AAT was severalfold lower compared with that of MT6CAT, thus confirming the data we obtained with the fluorescent peptide substrate.
Cellular MT6-MMP Does Not Activate MMP-2 Efficiently
Cells were incubated with 30 ng/ml (0.5 nm) purified pro-MMP-2, and then gelatin zymography of the medium aliquots was used to identify the status of pro-MMP-2. As expected, MCF7-MT1 cells readily activated the 68-kDa MMP-2 proenzyme and generated, as a result, the 64-kDa intermediate (the minor band) and the 62-kDa mature enzyme (the major band). GM6001 and TIMP-2 fully blocked the activation of pro-MMP-2 by MCF7-MT1 cells, whereas TIMP-1 did not demonstrate any significant effect. In contrast with MCF7-MT1 cells, both MCF7-mock and MCF7-MT6F cells did not activate pro-MMP-2 (Fig. 5B).
To determine if MT6-MMP exhibits an intrinsic capacity to process and activate the MMP-2 proenzyme, we co-incubated pro-MMP-2 with increasing concentrations of MT1CAT and MT6CAT. As expected, MT1CAT readily generated the 64-kDa activation intermediate of MMP-2 in the cleavage reactions. These results are consistent with the well established ability of MT1-MMP to proteolytically cleave the prodomain region of the MMP-2 proenzyme in both the cell system and the cell-free system (41, 42). In turn, the ability of MT6CAT to process pro-MMP-2 was low, and as a result, insignificant levels of the processed, 64-kDa, MMP-2 species were observed in the samples. GM6001 inhibited the processing of pro-MMP-2 by MT1CAT and MT6CAT (Fig. 5C). Based on these data, we conclude that MT6-MMP has a low ability, especially when compared with MT1-MMP, to accomplish the activation of the MMP-2 proenzyme. Because of this low intrinsic capability of the individual catalytic domain of MT6-MMP to activate MMP-2, it is exceedingly difficult to observe any meaningful levels of pro-MMP-2 activation using the cells that express MT6-MMP.
Gelatinolytic Activity of Purified MT6-MMP
To determine if cellular MT6F isolated from the PLC and Nonidet P-40 fractions was catalytically active, we compared their gelatinolytic activities with those of MT6CAT and MMP-2. The results showed that both the PLC and Nonidet P-40 factions of MT6F were capable of gelatin hydrolysis. The specific gelatinolytic activity of MT6F in these fractions was comparable with that of MT6CAT. It became clear, however, that both cellular MT6F fractions and MT6CAT were at least 1,000-fold less active in gelatin zymography tests compared with MMP-2 (Fig. 6A).
FIGURE 6.
Gelatinolytically active MT6-MMP does not support cell migration. A, gelatin zymography of MT6F-PLC and MT6F-Nonidet P-40 (0.5–2.5 pmol), MT6CAT (0.05–2 pmol), and pro-MMP-2 (0.1–1 fmol). B, migration assay. HT-mock, HT-MT1, and HT-MT6F cells (1 × 104) were allowed to migrate through type I collagen-coated Transwell inserts. Where indicated, GM6001 (50 μm) was added to the cells. The migration efficiency was calculated relative to HT-mock cells (100%). C, the uptake of MT6-MMP by cells. To prevent proteolysis of cellular MT1-MMP and MT6-MMP, HT and MCF7-MT6F cells (the top and bottom panels, respectively) were co-incubated with GM6001 (50 μm) for 16 h. The cells were then surface-biotinylated using membrane-impermeable, cleavable EZ-Link NHS-SS biotin and incubated for 30–60 min at 37 °C to stimulate the uptake of biotin-labeled plasma membrane proteins by the cells. Biotin-labeled protein was captured on streptavidin-beads, and the captured material was analyzed by Western blotting with the MT1-MMP and MT6-MMP antibodies. Where indicated, MESNA was used to release a biotin moiety from the cell surface-associated proteins. One representative experiment is shown. Multiple additional experiments generated similar results. IP, immunoprecipitation; WB, Western blot.
Cellular MT6-MMP Does Not Stimulate Cell Migration
Because MCF-7 cells do not efficiently migrate, we used highly migratory HT1080 cells to assess a potential effect of MT6-MMP on cell locomotion. For this purpose, HT1080 cells were stably transfected with the MT6F construct (HT-MT6F cells). The migration efficiency of HT-MT6F cells was compared in the presence and absence of GM6001 with that of HT1080-mock cells (HT-mock cells) transfected with the original plasmid and HT1080 cells transfected with MT1-MMP (HT-MT1 cells). There was an ∼30% reduction of the migration efficiency of HT-MT6F cells compared with HT-mock cells. In contrast with HT1080-mock and HT1080-MT1, GM6001 had no effect on migration of HT-MT6F cells (Fig. 6B). Based on these tests, we concluded that MT6-MMP does not stimulate cell migration. From the migration perspectives, lipid raft-associated MT6-MMP performs similarly to the tailless MT1-MMP lacking the cytoplasmic tail domain. In contrast to the invasion-promoting wild-type MT1-MMP, the tailless MT1-MMP construct is primarily associated with the lipid raft compartment and does not stimulate cell migration (43).
The Uptake Rate of Cellular MT1-MMP and MT6-MMP
To determine if the uptake rate of cellular MT6-MMP affects its ability to support cell migration, we compared the internalization rate of cellular MT1-MMP and MT6-MMP. For this purpose, we used HT-mock and MCF7-MT6F cells. The cells were surface-biotinylated with membrane-impermeable, cleavable, EZ-Link NHS-SS-biotin. Biotinylation was followed by incubation of the cells at 37 °C to initiate protein uptake. Cells were next transferred on ice to arrest protein trafficking and then treated with MESNA to release the biotin moiety from the residual cell surface-associated MT1-MMP and MT6-MMP molecules. The biotin-labeled internalized MMPs were protected from MESNA. The labeled MT1-MMP and MT6-MMP pools were then captured on streptavidin-beads, and the captured material was analyzed by Western blotting. These tests demonstrated that a major portion of cell surface-associated MT1-MMP was already internalized following a 30-min incubation. After 60 min, the levels of the biotin-labeled MT1-MMP were lower in HT-mock cells. In contrast, only a small fraction of MT6-MMP was protected from MESNA at 30–60 min, thus suggesting that MT6-MMP was inefficiently internalized, especially when compared with MT1-MMP (Fig. 6C). Because of its association with the lipid rafts, the bulk of cell surface MT6-MMP was still present on the cell surface following a 30–60-min incubation. As a result, we conclude that the observed low internalization rate cannot contribute to the inability of MT6-MMP to support cell migration.
TIMP-1 and TIMP-2 Efficiently Inhibit MT6-MMP
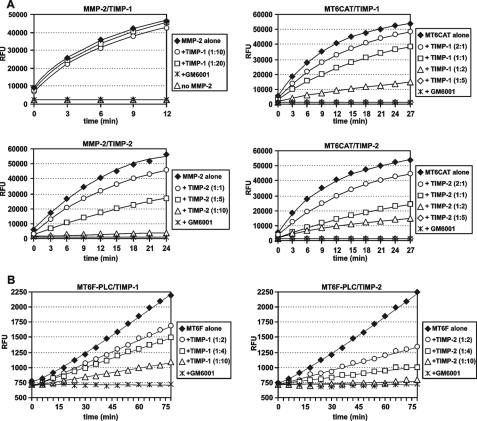
We next compared the inhibitory efficiency of TIMP-1 and TIMP-2 against MT6CAT relative to that of MMP-2. For these purposes, MMP-2 and MT6CAT were co-incubated for 30 min with the indicated amounts of the inhibitors. The residual activity of MMP-2 and MT6CAT was then measured using Mca-PLGL-Dpa-AR-NH2 as a substrate. Under our experimental conditions, we recorded a nearly complete and complete inhibition of MT6CAT at the enzyme/TIMP-1 molar ratio of 1:2 and 1:5, respectively, whereas no significant inhibition of MMP-2 was observed at the MMP-2/TIMP-1 molar ratio of 1:20 (Fig. 7A). Our data correlate well with the earlier observations by others whose reports indicated that TIMP-1 was a more potent inhibitor of MT6-MMP compared with MMP-2 (10). In turn, TIMP-2 was equally efficient in inhibiting MT6CAT and MMP-2. Indeed, a complete inhibition of both enzymes was observed at a 5–10-molar excess of TIMP-2 (Fig. 7A).
FIGURE 7.
TIMP-1 and TIMP-2 inhibit MT6CAT and MT6F. A, MMP-2 (20 ng) and MT6CAT (40 ng) were co-incubated with TIMP-1 and TIMP-2 at the indicated enzyme/inhibitor molar ratio. The residual activity was measured using Mca-PLGL-Dpa-AR-NH2 as a substrate. B, MT6F-PLC was co-incubated with TIMP-1 and TIMP-2 at the indicated enzyme/inhibitor molar ratio. The residual activity was measured using Mca-PLGL-Dpa-AR-NH2 as a substrate. RFU, relative fluorescence units.
Similar results were obtained when the efficiency of TIMP-1 and TIMP-2 were measured using the PLC fraction of cellular MT6F. Thus, a nearly complete inhibition of the purified MT6F construct was observed at a 5–10-fold molar excess of both TIMP-1 and TIMP-2 (Fig. 7B). Overall, we conclude that MT6-MMP is similarly sensitive to the inhibition by both TIMP-1 and TIMP-2. These parameters discriminate MT6-MMP from MT1-MMP, which is highly sensitive to TIMP-2 inhibition but insensitive to TIMP-1 (42).
Selective Inhibitors of MT6-MMP
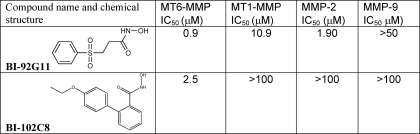
To get a clearer idea of the inhibitor profile of MT6-MMP compared with other MMPs, we screened the our inhibitor library of the potential MMP inhibitors using MT1CAT, MT6CAT, MMP-2, and MMP-9. Mca-PLGL-Dpa-AR-NH2 was used as a substrate. The inhibitory kinetic parameters, including the IC50 values, of the identified hits were then determined. The secondary screen employing the cleavage of AAT was used to confirm the inhibitory efficiency of the hit compounds (not shown). We readily identified a number of hits, from which BI-102C8 and BI-92G11 were the most selective against MT6-MMP when compared with MT1-MMP, MMP-2, and MMP-9 (Table 1). These data emphasize the structural differences existing between the catalytic domain of MT6-MMP and other MMPs. Our findings also suggest that the selective MT6-MMP inhibitors we identified and especially BI-92G11, a submicromolar range inhibitor of MT6-MMP, can be used both as valuable molecular tools in the MMP studies and as a valid starting point for further iterative optimization leading to the pharmacological inhibitors of MT6-MMP in disease, including cancer and multiple sclerosis (6, 11, 12).
TABLE 1.
Chemical structure and IC50 values of the MT6-MMP inhibitors
The inhibitory potency of the individual compounds from the MMP inhibitor library (∼500 compounds) was determined using the individual MMPs (MT1-MMP, MT6-MMP, MMP-2, and MMP-9) and Mca-PLGL-Dpa-AR-NH2 as a cleavage substrate.
Cellular MT6-MMP Degrades Gelatin
To test if cellular MT6-MMP degrades gelatin, we used in situ zymography performed with FITC-conjugated gelatin. For this purpose, we plated MCF7-mock, MCF7-MT6F, and MCF7-MT1 cells on the FITC-labeled gelatin. MMP activity caused digestion of the FITC-gelatin, which was visualized as dark zones without fluorescence. MCF7-mock did not cause any noticeable hydrolysis of the FITC-gelatin. In contrast, MCF7-MT6F and MCF7-MT1 cells were comparably active in the cleavage of the FITC-gelatin (Fig. 8). GM6001 completely abolished gelatinolytic activity of MCF7-MT6F and MCF7-MT1 cells, thus confirming that the MMP activity was directly involved in this gelatin cleavage.
FIGURE 8.
In situ FITC-gelatin zymography. MCF7-mock, MCF7-MT1, and MCF7-MT6F cells were seeded in serum-free DMEM on FITC-gelatin-coated coverslips. In 16 h, cells were fixed, and the nuclei were stained with DAPI. The slides were observed using a fluorescent microscope (magnification, ×400).
DISCUSSION
GPI-linked MT6-MMP is one of the least studied members of the MMP family. Because of the GPI anchor, MT6-MMP is directly associated with the lipid rafts in the plasma membranes. The association with this specific compartment affects the functionality of cell surface-associated MT6-MMP, thus making it different from that of conventional MT-MMPs, including MT1-MMP, the most well studied member of the MMP family.
There was conflicting evidence about the ability of MT6-MMP to be regulated by TIMPs, to play a role in the activation of MMP-2 (a target of MT1-MMP activation in multiple cell and tissue types), and to support cell migration. It was not clear in the earlier works whether cellular MT6-MMP was or was not functionally active, and as a result, it was exceedingly difficult to conclude whether MT6-MMP was or was not capable of MMP-2 activation and how distinct the MT6-MMP functionality was from that MT1-MMP. Earlier studies by others suggested that MT6-MMP could play a role in cellular migration and invasion of the extracellular matrix and basement membranes and that its activity may be tightly regulated by the members of the TIMP family. On the other hand, there was no direct evidence that a direct complex of TIMPs, including TIMP-1 or TIMP-2, could exist with cellular MT6-MMP.
Because of these conflicting results, our goal was to analyze the biochemical characteristics of cellular MT6-MMP. To facilitate the isolation and analysis, the FLAG-tagged MT6-MMP chimera was expressed in the cells that do not exhibit any detectable expression of this proteinase. Based on our multiple and diversified pull-down and extraction approaches supplemented by LC/MS/MS, we are now confident that the proteolytically active, mature MT6-MMP enzyme is presented on the cell surface, whereas minor amounts of the residual proenzyme are predominantly present inside the cells. MT6-MMP was not significantly proteolyzed, especially if compared with MT1-MMP (27). Self-proteolysis of cellular MT1-MMP takes place at the DPSA↓I256 and QLYG↓G285 sites of the C-terminal portion of the catalytic domain and in the hinge region, respectively. As a result, the inactive, 40–45-kDa, membrane-attached MT1-MMP form (44–46) is generated. These putative self-proteolytic sites are modified in MT6-MMP, and these parameters explain why cellular MT6-MMP is not significantly proteolyzed (Fig. 4B).
Cellular MT6-MMP exists in its partially saturated complex with TIMP-2. Both TIMP-1 and TIMP-2 are capable of inhibiting the proteolytic activity of MT6-MMP. The ability of MT6-MMP to hydrolyze collagen is low. However, this ability is still sufficient to induce a significant level of gelatinolysis of the FITC-labeled gelatin. Because the rate of internalization of the lipid raft compartment is low compared with the clathrin-coated pits (47), the lipid raft-associated cellular MT6-MMP is inefficiently internalized, especially if compared with MT1-MMP. Similarly, a low internalization rate was previously recorded for the tailless MT1-MMP mutant missing the C-terminal cytoplasmic tail. As a result of this truncation, the tailless MT1-MMP relocates to the lipid raft compartment and loses its ability to support cell migration in the conventional cell motility tests (28, 43).
According to our multiple co-localization and pull-down experiments (not shown), MT6-MMP does not efficiently interact with the known targets of MT1-MMP, including tissue transglutaminase and CD44 (48–50). According to our immunostaining studies, MT6-MMP is predominantly localized in cell-cell junction regions. The functional significance of the association of MT6-MMP with the specific cell membrane regions, however, is not yet understood.
We also did not observe any interaction of MT6-MMP with cell adhesion signaling receptors, including epidermal growth factor receptor. Regardless of the presence of the consensus 14-3-3-binding motif (underlined, T121WRVRSFPQSSQL133) in MT6-MMP (however, in the extracellular portion of the proteinase), our pull-down experiments have demonstrated that the interactions of cellular MT6-MMP with the 14-3-3 protein do not exist.
Despite the presence of its active, mature enzyme species on the cell surface and its ability to complex TIMP-2, cellular MT6-MMP was not capable of activating MMP-2 under our experimental conditions. The cell-free, in-solution, tests that employed the purified components demonstrated that the individual catalytic domain of MT6-MMP, however, was capable of cleaving the prodomain sequence of pro-MMP-2 in a way similar to that of MT1-MMP, albeit significantly less efficiently. As a result of this low intrinsic MMP-2-activating capacity of the MT6-MMP catalytic domain, it is exceedingly difficult, but not entirely impossible, to record the activation of MMP-2 by using the MT6-MMP-overexpressing cells (6, 10, 20, 21).
As a purified enzyme, MT6-MMP, however, is a potent proteinase that is capable of efficiently cleaving a diversified set of the peptide substrates3 and proteins, including myelin basic protein (12). Our data suggest that the cleavage of myelin basic protein and its splice variant (golli myelin basic protein) by MT6-MMP plays a role in both inflammation and the onset of multiple sclerosis and, potentially, other neuroimmune diseases (11).
Because the transmembrane domain of MT1-MMP could be functionally substituted by the GPI anchor of MT6-MMP and because the GPI-anchored MT1-MMP activated MMP-2 on the cell surface and promoted cell growth in a three-dimensional type I collagen matrix (51), it may be suggested that the effects of the lipid raft compartment on the MMP proteolysis are limited. Overall, based on our results and the data of others, it is reasonable to suggest that both specific membrane tethering and proteolytic activity encoded by MT1-MMP are required for its ability to promote cell locomotion (51) and that the lipid raft localization alone is insufficient to explain multiple functional differences between MT6-MMP and MT1-MMP. It is likely that the unique structural and biochemical properties also lead to an unconventional performance of cellular MT6-MMP. Because MT6-MMP has been suggested to play a role in cancer and multiple sclerosis (6, 11), it remains to be determined how these unique biochemical and structural properties of MT6-MMP regulate its function at the cell surface.
This work was supported, in whole or in part, by National Institutes of Health Grants CA83017 and CA77470 (to A. Y. S.).
B. I. Ratnikov, P. Cieplak, A. M. Eroshkin, M. D. Kazanov, J. Pierce, Q. Sun, B. Stec, A. L. Osterman, A. Y. Strongin, and J. W. Smith, manuscript in preparation.
- MMP
- matrix metalloproteinase
- AAT
- α1-antitrypsin
- GPI
- glycosylphosphatidylinositol
- DAPI
- 4′,6-diamidino-2-phenylindole
- EZ-link sulfo-NHS-SS-biotin
- sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate
- HT cells
- human fibrosarcoma HT1080 cells
- Mca-PLGL-Dpa-AR-NH2
- methoxycoumarin-4-yl)-acetyl-Pro-Leu-Gly-Leu-(3-[2,4-dinitrophenyl]-l-2,3-diaminopropionyl)-Ala-Arg-NH2
- MCF7 cells
- human breast carcinoma MCF-7 cells
- MESNA
- 2-mercaptoethane sulfonic acid
- MT-MMP
- membrane-type MMP
- MT1-MMP and MT6-MMP
- membrane type-1 and membrane type-6 MMP, respectively
- MT1CAT and MT6CAT
- the individual catalytic domain of MT1-MMP and MT6-MMP, respectively
- MT6F
- MT6-MMP tagged with a FLAG tag
- PLC
- phosphatidylinositol-specific phospholipase C
- TIMP
- tissue inhibitor of metalloproteinases
- DMEM
- Dulbecco's modified Eagle's medium
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- FITC
- fluorescein isothiocyanate
- RIPA
- radioimmune precipitation assay.
REFERENCES
- 1.Egeblad M., Werb Z. (2002) Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 2.López-Otín C., Bond J. S. (2008) J. Biol. Chem. 283, 30433–30437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf K., Wu Y. I., Liu Y., Geiger J., Tam E., Overall C., Stack M. S., Friedl P. (2007) Nat. Cell Biol. 9, 893–904 [DOI] [PubMed] [Google Scholar]
- 4.Nagase H., Woessner J. F., Jr. (1999) J. Biol. Chem. 274, 21491–21494 [DOI] [PubMed] [Google Scholar]
- 5.Seiki M., Yana I. (2003) Cancer Sci. 94, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohail A., Sun Q., Zhao H., Bernardo M. M., Cho J. A., Fridman R. (2008) Cancer Metastasis Rev. 27, 289–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pei D. (1999) Cell Res. 9, 291–303 [DOI] [PubMed] [Google Scholar]
- 8.Kojima S., Itoh Y., Matsumoto S., Masuho Y., Seiki M. (2000) FEBS Lett. 480, 142–146 [DOI] [PubMed] [Google Scholar]
- 9.Velasco G., Cal S., Merlos-Suárez A., Ferrando A. A., Alvarez S., Nakano A., Arribas J., López-Otín C. (2000) Cancer Res. 60, 877–882 [PubMed] [Google Scholar]
- 10.Sun Q., Weber C. R., Sohail A., Bernardo M. M., Toth M., Zhao H., Turner J. R., Fridman R. (2007) J. Biol. Chem. 282, 21998–22010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiryaev S. A., Remacle A. G., Savinov A. Y., Chernov A. V., Cieplak P., Radichev I. A., Williams R., Shiryaeva T. N., Gawlik K., Postnova T. I., Ratnikov B. I., Eroshkin A. M., Motamedchaboki K., Smith J. W., Strongin A. Y. (2009) J. Biol. Chem. 284, 30615–30626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiryaev S. A., Savinov A. Y., Cieplak P., Ratnikov B. I., Motamedchaboki K., Smith J. W., Strongin A. Y. (2009) PLoS One 4, e4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brew K., Nagase H. (2010) Biochim. Biophys. Acta 1803, 55–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark I. M., Swingler T. E., Sampieri C. L., Edwards D. R. (2008) Int. J. Biochem. Cell Biol. 40, 1362–1378 [DOI] [PubMed] [Google Scholar]
- 15.Holmbeck K., Bianco P., Yamada S., Birkedal-Hansen H. (2004) J. Cell. Physiol. 200, 11–19 [DOI] [PubMed] [Google Scholar]
- 16.Murphy G., Knauper V., Lee M. H., Amour A., Worley J. R., Hutton M., Atkinson S., Rapti M., Williamson R. (2003) Biochem. Soc. Symp. 70, 65–80 [DOI] [PubMed] [Google Scholar]
- 17.Murphy G., Stanton H., Cowell S., Butler G., Knäuper V., Atkinson S., Gavrilovic J. (1999) APMIS 107, 38–44 [DOI] [PubMed] [Google Scholar]
- 18.Strongin A. Y., Collier I., Bannikov G., Marmer B. L., Grant G. A., Goldberg G. I. (1995) J. Biol. Chem. 270, 5331–5338 [DOI] [PubMed] [Google Scholar]
- 19.English W. R., Velasco G., Stracke J. O., Knäuper V., Murphy G. (2001) FEBS Lett. 491, 137–142 [DOI] [PubMed] [Google Scholar]
- 20.Nie J., Pei D. (2003) Cancer Res. 63, 6758–6762 [PubMed] [Google Scholar]
- 21.Zhao H., Sohail A., Sun Q., Shi Q., Kim S., Mobashery S., Fridman R. (2008) J. Biol. Chem. 283, 35023–35032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sounni N. E., Rozanov D. V., Remacle A. G., Golubkov V. S., Noel A., Strongin A. Y. (2010) Int. J. Cancer 126, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strongin A. Y., Marmer B. L., Grant G. A., Goldberg G. I. (1993) J. Biol. Chem. 268, 14033–14039 [PubMed] [Google Scholar]
- 24.Chen E. I., Li W., Godzik A., Howard E. W., Smith J. W. (2003) J. Biol. Chem. 278, 17158–17163 [DOI] [PubMed] [Google Scholar]
- 25.Kridel S. J., Sawai H., Ratnikov B. I., Chen E. I., Li W., Godzik A., Strongin A. Y., Smith J. W. (2002) J. Biol. Chem. 277, 23788–23793 [DOI] [PubMed] [Google Scholar]
- 26.Radichev I. A., Remacle A. G., Sounni N. E., Shiryaev S. A., Rozanov D. V., Zhu W., Golubkova N. V., Postnova T. I., Golubkov V. S., Strongin A. Y. (2009) Biochem. J. 420, 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remacle A. G., Chekanov A. V., Golubkov V. S., Savinov A. Y., Rozanov D. V., Strongin A. Y. (2006) J. Biol. Chem. 281, 16897–16905 [DOI] [PubMed] [Google Scholar]
- 28.Remacle A. G., Rozanov D. V., Fugere M., Day R., Strongin A. Y. (2006) Oncogene 25, 5648–5655 [DOI] [PubMed] [Google Scholar]
- 29.Agrawal A., Johnson S. L., Jacobsen J. A., Miller M. T., Chen L. H., Pellecchia M., Cohen S. M. (2010) Chem. Med. Chem. 5, 195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Booher R. N., Kornfeld E. C., Smalstig E. B., Clemens J. A. (1987) J. Med. Chem. 30, 580–583 [DOI] [PubMed] [Google Scholar]
- 31.Hsieh P., Segal R., Chen L. B. (1980) J. Cell Biol. 87, 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowden E. T., Coopman P. J., Mueller S. C. (2001) Methods Cell Biol. 63, 613–627 [DOI] [PubMed] [Google Scholar]
- 33.Angst B. D., Marcozzi C., Magee A. I. (2001) J. Cell Sci. 114, 629–641 [DOI] [PubMed] [Google Scholar]
- 34.Hulpiau P., van Roy F. (2009) Int. J. Biochem. Cell Biol. 41, 349–369 [DOI] [PubMed] [Google Scholar]
- 35.Li W., Savinov A. Y., Rozanov D. V., Golubkov V. S., Hedayat H., Postnova T. I., Golubkova N. V., Linli Y., Krajewski S., Strongin A. Y. (2004) Cancer Res. 64, 8657–8665 [DOI] [PubMed] [Google Scholar]
- 36.Liu Z., Zhou X., Shapiro S. D., Shipley J. M., Twining S. S., Diaz L. A., Senior R. M., Werb Z. (2000) Cell 102, 647–655 [DOI] [PubMed] [Google Scholar]
- 37.Mast A. E., Enghild J. J., Nagase H., Suzuki K., Pizzo S. V., Salvesen G. (1991) J. Biol. Chem. 266, 15810–15816 [PubMed] [Google Scholar]
- 38.Sires U. I., Murphy G., Baragi V. M., Fliszar C. J., Welgus H. G., Senior R. M. (1994) Biochem. Biophys. Res. Commun. 204, 613–620 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z., Winyard P. G., Chidwick K., Murphy G., Wardell M., Carrell R. W., Blake D. R. (1994) Biochim. Biophys. Acta 1199, 224–228 [DOI] [PubMed] [Google Scholar]
- 40.Rozanov D. V., Sikora S., Godzik A., Postnova T. I., Golubkov V., Savinov A., Tomlinson S., Strongin A. Y. (2004) J. Biol. Chem. 279, 50321–50328 [DOI] [PubMed] [Google Scholar]
- 41.Lichte A., Kolkenbrock H., Tschesche H. (1996) FEBS Lett. 397, 277–282 [DOI] [PubMed] [Google Scholar]
- 42.Will H., Atkinson S. J., Butler G. S., Smith B., Murphy G. (1996) J. Biol. Chem. 271, 17119–17123 [DOI] [PubMed] [Google Scholar]
- 43.Rozanov D. V., Deryugina E. I., Monosov E. Z., Marchenko N. D., Strongin A. Y. (2004) Exp. Cell Res. 293, 81–95 [DOI] [PubMed] [Google Scholar]
- 44.Hernandez-Barrantes S., Toth M., Bernardo M. M., Yurkova M., Gervasi D. C., Raz Y., Sang Q. A., Fridman R. (2000) J. Biol. Chem. 275, 12080–12089 [DOI] [PubMed] [Google Scholar]
- 45.Lehti K., Lohi J., Valtanen H., Keski-Oja J. (1998) Biochem. J. 334, 345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toth M., Hernandez-Barrantes S., Osenkowski P., Bernardo M. M., Gervasi D. C., Shimura Y., Meroueh O., Kotra L. P., Gálvez B. G., Arroyo A. G., Mobashery S., Fridman R. (2002) J. Biol. Chem. 277, 26340–26350 [DOI] [PubMed] [Google Scholar]
- 47.Remacle A., Murphy G., Roghi C. (2003) J. Cell Sci. 116, 3905–3916 [DOI] [PubMed] [Google Scholar]
- 48.Belkin A. M., Akimov S. S., Zaritskaya L. S., Ratnikov B. I., Deryugina E. I., Strongin A. Y. (2001) J. Biol. Chem. 276, 18415–18422 [DOI] [PubMed] [Google Scholar]
- 49.Mori H., Tomari T., Koshikawa N., Kajita M., Itoh Y., Sato H., Tojo H., Yana I., Seiki M. (2002) EMBO J. 21, 3949–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. (2001) J. Cell Biol. 153, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie J., Pei J., Blumenthal M., Pei D. (2007) J. Biol. Chem. 282, 6438–6443 [DOI] [PubMed] [Google Scholar]
- 52.Rozanov D. V., Strongin A. Y. (2003) J. Biol. Chem. 278, 8257–8260 [DOI] [PubMed] [Google Scholar]