Abstract
β- and γ-nonmuscle actins differ by 4 amino acids at or near the N terminus and distant from polymerization interfaces. β-Actin contains an Asp1-Asp2-Asp3 and Val10 whereas γ-actin has a Glu1-Glu2-Glu3 and Ile10. Despite these small changes, conserved across mammals, fish, and birds, their differential localization in the same cell suggests they may play different roles reflecting differences in their biochemical properties. To test this hypothesis, we established a baculovirus-driven expression system for producing these actins in isoform-pure populations although contaminated with 20–25% insect actin. Surprisingly, Ca-γ-actin exhibits a slower monomeric nucleotide exchange rate, a much longer nucleation phase, and a somewhat slower elongation rate than β-actin. In the Mg-form, this difference between the two is much smaller. Ca-γ-actin depolymerizes half as fast as does β-actin. Mixing experiments with Ca-actins reveal the two will readily co-polymerize. In the Ca-form, phosphate release from polymerizing β-actin occurs much more rapidly and extensively than polymerization, whereas phosphate release lags behind polymerization with γ-actin. Phosphate release during treadmilling is twice as fast with β- as with γ-actin. With Mg-actin in the initial stages, phosphate release for both actins correlates much more closely with polymerization. Calcium bound in the high affinity binding site of γ-actin may cause a selective energy barrier relative to β-actin that retards the equilibration between G- and F-monomer conformations resulting in a slower polymerizing actin with greater filament stability. This difference may be particularly important in sites such as the γ-actin-rich cochlear hair cell stereocilium where local mm calcium concentrations may exist.
Keywords: Actin, ATPases, Calcium, Cytoskeleton, Microfilaments, Ion Dependence, Isoactins, Nonmuscle Cell, Nucleotide Exchange, Polymerization
Introduction
Birds, fish, and mammals have genes that encode two nonmuscle actin isoforms termed β and γ, based on their migration in an isoelectric focusing gel, with β-actin the more acidic of the two. These two nonmuscle isoforms differ at only four positions out of 375 residues, and the divergent residues are very similar between the two proteins. β-Actin contains an Asp-Asp-Asp N-terminal tripeptide and a Val at position 10 whereas γ-actin has a Glu-Glu-Glu N-terminal tripeptide and an Ile at position 10 (1) (Fig. 1).
FIGURE 1.
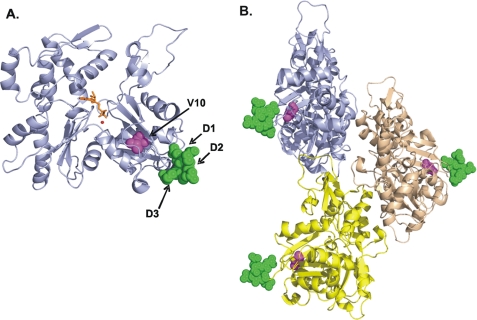
Locations of the structural differences between β- and γ-nonmuscle actin. A, monomer view of the crystal structure of β-actin, modified from Protein Data Bank code 1HLU (22) using Swiss-PdbViewer Version 3.7. The positions of the differing residues are color-highlighted and labeled: D1, D2, D3 green; V10, pink. B, model of the actin trimer based on the filament model of Oda et al. (21), with the positions of the differing residues color-highlighted and labeled as described above. ATP is depicted as an orange stick, and Ca2+ or Mg2+ ion depicted as a red circle.
In nonmuscle cells of mammals, birds, and fish, the β- and γ-nonmuscle actin isoforms exist in varying ratios depending on the cell type, and these ratios are conserved in a cell and tissue-specific way across these three groups of species. Usually they are found in about a 2:1 β- to γ-actin ratio (2). However, there are notable exceptions. For example, in auditory hair cells, the ratio is reversed (3), and in mature skeletal muscle myotubes, γ-nonmuscle actin, which lies in a thin layer just beneath the sarcoplasmic membrane, is the only nonmuscle actin isoform present (4, 5). Despite these small sequence differences, their localization in cultured cells indicates that β- and γ-nonmuscle actins may play very different physiological roles. In motile cells, β-actin tends to accumulate at the membrane-cytoskeleton interface in dynamic ruffling regions near the leading edge of the cell, whereas γ-actin seems to be enriched in stress fibers (6) although this generalization has been challenged in a recent study (7). Spatial and temporal segregation of β- and γ-nonmuscle actins has also been found in gastric parietal cells (8), auditory hair cells (3), osteoblasts (9), and neurons (10, 11).
Despite the apparent differences in physiological behavior of these two isoactins, the molecular basis for this cellular discrimination has been difficult to ascertain. One way for the cell to deal with this problem would be to synthesize the two isoforms in the different parts of the cells where they predominate. Along these lines, it has been demonstrated that some cells can post-transcriptionally sort β-actin mRNA through its isoform-specific 3′-untranslated region to the leading edge resulting in preferential β-actin synthesis in these regions (12–15). However, streaming of actin away from this area (16, 17) would result in an ultimate mixing of isoforms further toward the cell center. Another possibility is differential recognition of the two isoforms by different actin binding proteins. Toward this end, Namba et al. (18) have demonstrated that l-plastin preferentially binds β- over γ-actin, although the reason for this is unclear, especially considering the small differences between the two actins. A third possibility is differential post-translational modifications of the two isoforms. In this vein, it has been reported that β-actin can be N-terminally arginylated following removal of the N-terminal acidic residue (19). Initial estimates based on mass spectroscopic methods suggested that this occurred in 20–40% of the β-actin, although this number may be much lower in different cell types. However, even if a fourth of the β-actin is modified, it is hard to understand how this small fraction could exert its effect over the entire β-actin population. A fourth possibility is that these small inherent structural differences result in propagated allosteric changes that alter distant binding sites for actin-binding proteins. However, based on the monomer crystal structure, the three N-terminal actin residues are in an unstructured finger that reaches out into solution from the surface of the protein making this possibility seemingly less likely (20–22). Position 10, in which the residues differ by only a single methylene carbon, is in the middle of a structural core of subdomain 1, and this difference might result in a conformational change, perhaps via crowding effects.
To date, because of their similar sequences and their presence as mixtures in most cells, a limiting factor in addressing these isoform differences at the biochemical level has been a difficulty in readily obtaining significant amounts of these actins as pure isoform populations. The demonstration that cardiac actin could be successfully expressed in and purified from baculovirus-infected insect cells (23, 24) suggested that this approach might also be a way to obtain pure preparations of human β- and γ-nonmuscle isoactins. In this report, we demonstrate that we are able to do just that although endogenous insect actin is present to the extent of ∼20–25%. We also show that this has little effect on the biochemical properties of the individual isoforms. We have then used these preparations to compare the biochemical properties of these two isoactins in the monomeric state and in their ability to polymerize. Our data show that despite their very similar amino acid sequences, the two isoactins display distinct polymerization characteristics that correlate with their apparently differential roles in motile cells.
EXPERIMENTAL PROCEDURES
Materials
DNase I (grade D) was purchased from Worthington. Affi-Gel 10-activated resin and Micro Bio-Spin P-30 Tris gel filtration chromatography columns were obtained from Bio-Rad. DE52 DEAE-cellulose was acquired from Whatman. N-(1-pyrenyl)maleimide, ATP, ADP, hexokinase, and glucose were acquired from Sigma-Aldrich. Tween-20, enzyme grade, was obtained from Fisher Scientific. Yeast cakes for WT actin preparations were purchased from a local bakery. All other chemicals were reagent-grade quality.
Cells and Cell Culture
Low passage Sf21 cells (BD Biosciences) were cultured in SF900 II medium (Invitrogen) with 1% penicillin/streptomycin. These cells, after being switched to TC-100 Insect Medium with l-glutamine and sodium bicarbonate (Sigma-Aldrich) with 1% penicillin/streptomycin and 10% fetal bovine serum, were used for recombinant virus production and protein expression.
Construction of Baculovirus Transfer Vectors
The coding sequences of human β- and γ-nonmuscle actins were cloned into pDEST8. The AcMNpv 6.9 baculovirus promoter was cloned upstream of the actin gene. The Invitrogen Bac-to-Bac system was then used to generate recombinant bacmids containing either the β- or γ-nonmuscle actin gene. All sequences were confirmed by sequencing of viral DNA from infected cells.
Purification of Recombinant Actin
Infected cells (with a multiplicity of infection equal to 2) were harvested 48 h postinfection by centrifugation, washed in phosphate-buffered saline and lysed in a high Tris buffer (1 m Tris-HCl, pH 7.5, 0.5 mm MgCl2, 0.5 mm ATP, 4% Triton X-100, 1 mg/ml Tween 20, 1 mm dithiothreitol, and a protease inhibitor mixture (benzamidine, leupeptin, aprotinin, antipain, TLCK, TPCK, E-64, each at 1.25 μg/ml)) by sonication. The cell lysate (50 ml from a preparation of ∼9 × 108 cells) was cleared by centrifugation at 40,000K for 1 h using a Beckman L8–70 M ultracentrifuge with a 45Ti rotor. The lysate was then diluted with 1× G-buffer (10 mm Tris-HCl, pH 7.5, 0.2 mm CaCl2, and 0.2 mm ATP) with the same protease inhibitor mixture as above. Control insect actin was also purified from uninfected SF21 cells as described for recombinant actin purification.
Muscle actin was purified from rabbit muscle acetone powder, as described by Spudich and Watt (25). Yeast and nonmuscle actins were purified in the calcium form via a combination of DNase I-agarose affinity chromatography, DEAE-cellulose chromatography, and polymerization/depolymerization cycling as described previously (26). Purity of the actin preparations were assessed by SDS-PAGE analysis (supplemental Fig. S1). The concentration of G-actin was determined from the absorbance at 290 nm using an extinction coefficient of 0.63 m−1 cm−1. The yield was typically 1–2 mg of actin per liter of infected cells. Using mass spectroscopic analysis, we determined there to be ∼20–25% insect actin in our nonmuscle actin protein preparations (data not shown). Mg-actin was generated by diluting actin in G-Buffer with MgCl2 for 10 min and then adding EGTA to chelate calcium as described previously (27).
Actin Thermal Stability
The apparent melting temperatures of yeast and nonmuscle actins were determined using circular dichroism by following the change in ellipticity of the G-actin sample at 222 nm as a function of temperature between 25 and 90 °C as described previously (28). Measurements were made on an Aviv 62 DS spectropolarimeter. Data were fit to a two-state model, and the apparent Tm value was determined by fitting the data to the Gibbs-Helmholtz equation to approximate the temperature at which 50% of the actin was denatured.
Actin ATP Exchange
The ability of G-actin to exchange its bound nucleotide was assessed by first loading the actin with ϵ-ATP2 and then following its displacement from the actin in the presence of a large excess of ATP as described previously (28). Exchange rates were determined by fitting the data to a single exponential expression using BioKine Version 3.1.
Actin Polymerization
Polymerization of G-actin in a total volume of 160 μl was induced by the addition of MgCl2 and KCl to final concentrations of 2 mm and 50 mm respectively (F-salts). Polymerization was monitored at 25 °C by following the increase in light scattering of the sample in a FluoroMax-3 or a Flurolog (model FL3–21) fluorescence spectrometer outfitted with a computer-controlled thermostatted four position multi-sample exchanger (HORIBA Jobin Yvon Inc.). Differences in machines can cause variations in the light scattering values; therefore, data from any given graph were obtained from the same fluorescence spectrometer. Both the excitation and emission wavelengths were set to 360 nm with the slit widths for both set at 1 nm.
For seeded actin polymerization assays, 4.8 μm actin was polymerized as above. Then after polymerization was complete, a 1:1 molar ratio of phalloidin was added to the actin. Filaments were sheared creating phalloidin actin seeds (PAS) and the seeds were added to G-actin to make up 5% of the total actin in the reaction. F-salts then were added, and polymerization was monitored at 10-s intervals as above.
Pi Release from Actin
The rate of Pi release from polymerizing actin samples following ATP hydrolysis was assessed using the commercially available EnzChekTM phosphate assay (Invitrogen) at 25 °C. Briefly, this spectrophotometric assay utilizes the purine nucleoside phosphorylase-dependent phosphorolysis of 6-mercapto-7-methylpurine riboside to ribose 1-phosphate and 2-amino-6-mercapto-7-methylpurine, the latter of which has a characteristic absorbance at 360 nm that is not shared by the nucleotide substrate at pH values greater than 6.5 (29, 30). Following induction of polymerization of 4.8 μm actin, the absorbance was monitored as a function of time, with readings taken automatically at 10-s intervals at 360 nm using a thermostatted cuvette holder set to 25 °C ± 0.1 °C.
Actin Depolymerization
Actin was polymerized to steady-state levels. DNase I was then added in a 1:5 actin to DNase molar ratio. Depolymerization was monitored as a decrease in light scattering over time. Depolymerization rates were determined by fitting the data to a single exponential expression using BioKine Version 3.1.
Electron Microscopy
Actin filaments were visualized by depositing 2 μl of a sample containing 4.8 μm F-actin onto carbon-coated Formvar grids. The grids were negatively stained with 1% uranyl acetate, and observed using a JOEL 1230 transmission electron microscope (University of Iowa Central Electron Microscopy Facility). Image J was used to process the images.
RESULTS
Characterization of G-actin Properties
We first determined if the four amino acid difference between β- and γ-actin causes differences in the physical behavior of the actin monomers. The apparent melting temperatures of the actins with calcium bound at the high affinity binding site in the nucleotide cleft, the form in which they are purified, were determined by following the change in molar ellipticity as a function of increasing temperature in a CD spectropolarimeter. The differing residues between β- and γ-actin have no significant effect on the thermal stabilities (data not shown).
We next determined the ability of the calcium G-actins to exchange a bound fluorescent ATP analog (ϵ-ATP) as a function of time in a solution with a large excess of ATP. We also used yeast and muscle actins as reference points because yeast actin has been shown have one of the fastest exchange rates of all actins and muscle actin one of the slowest. Table 1 shows a slightly but reproducibly faster rate of nucleotide exchange for yeast actin compared with β-actin. The γ-actin nucleotide exchange rate is significantly slower than β-actin but is still much faster than that of muscle actin (31).
TABLE 1.
Effect of isoform residue differences on ATP exchange rates and depolymerization kinetics
ATP exchange rates: The release of actin-bound ϵ-ATP 1 μm actin in G buffer without free ATP was triggered by the addition of 100 μm ATP. The decrease in fluorescence caused by ϵ-ATP exchange was followed over time, and data were fit to a first order reaction mechanism as described under “Experimental Procedures.” Depolymerization Kinetics: The decrease in light scattering of actin filaments was followed as a function of time after the addition of DNase I at 25 °C. The t½ of depolymerization was determined as described under “Experimental Procedures.” The number of experiments performed is indicated in parentheses.
| Cation bound | Actin isoform | Nucleotide exchange t½ | 1/2 Time of depolymerization |
|---|---|---|---|
| S | s | ||
| Ca2+ | Yeast | 35 ± 3 (13) | 70 ± 6 (2) |
| β-Nonmuscle | 66 ± 2 (5) | 127 ± 2 (2) | |
| γ-Nonmuscle | 92 ± 15 (9) | 202 ± 20 (2) | |
| Muscle | 405 ± 41 (3) | NDa | |
| Mg2+ | Yeast | 4 ± 1 (2) | 48 ± 7 (2) |
| β-Nonmuscle | 10 ± 2 (2) | 105 ± 3 (2) | |
| γ-Nonmuscle | 10 ± 4 (2) | 110 ± 5 (2) |
a ND, not determined.
Actin Polymerization
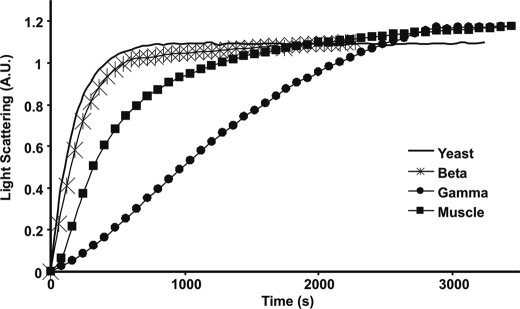
Most actin functions within the cell depend on its ability to form filaments, and isoform differences in polymerization and depolymerization kinetics could account for isoform-specific functions. We therefore wanted to determine if the polymerization rates and filament stabilities of the two isoforms differed from one another. Again, yeast and muscle actins were used as standards. Fig. 2 shows that Ca-β-actin polymerizes nearly as fast as yeast actin. Further, the total light scattering change was almost identical for both β- and γ-actins, indicating little if any difference in critical concentration for the isoactins. Surprising, however, was the extremely slow rate of γ-actin polymerization and the extended nucleation phase, especially because the divergent residues between the two non-muscle isoactins would seem to have little effect on the surfaces of the actin involved in monomer-monomer contacts (Fig. 1). EM analysis of negatively stained samples of filaments of both actins showed no apparent differences between them in filament morphology (supplemental Fig. S2, A and B). Insect actin also showed no apparent differences (data not shown).
FIGURE 2.
Polymerization kinetics of actin isoforms. Polymerization of 4.8 μm actin was initiated by the addition of magnesium and potassium chloride as described under “Experimental Procedures,” and the increase in L.S. was monitored as a function of time at 25 °C. Shown are representative plots of experiments performed at least three times with three independent actin preparations.
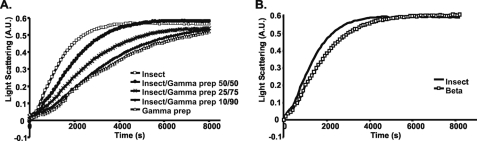
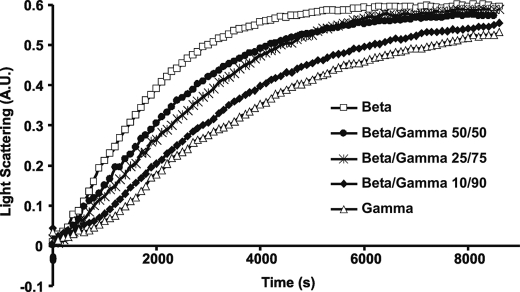
Because of the significant degree to which insect actin was present in our mammalian actin preparations (20–25%) we wished to determine the extent of insect actin influence on γ-actin polymerization kinetics. A titration polymerization assay was performed in which the total actin concentration was kept at 4.8 μm with varying ratios of insect actin to γ-actin (Fig. 3A). The results demonstrated that γ-actin polymerization kinetics remain relatively constant with an additional 10% pure insect actin and were marginally affected by an additional 25%.
FIGURE 3.
Polymerization of insect and γ-actin mixtures. Polymerization of 3.5 μm total actin was initiated by the addition of magnesium and potassium chloride as described under “Experimental Procedures,” the increase in L.S. was monitored as a function of time at 25 °C. The γ-actin preparation used in this work contains 20–25% insect actin. A, γ-actin preparation, mixtures of γ-actin preparation and insect actin, and pure insect actin were polymerized, numbers behind the isoforms represent the relative percentage of each actin preparation within the reaction. B, pure insect and β-actin were polymerized. Shown are representative plots of experiments performed at least three times with three independent actin preparations.
The results from Fig. 2 suggested that insect and β-actin polymerization kinetics were very similar. We thus studied their relative rates of polymerizations at a lower actin concentration where differences might be more apparent. Fig. 3B shows first that both actins polymerize to the same extent and, second, that they do so with almost the same rate although the insect actin repeatedly seems to polymerize slightly faster than the β-actin preparation.
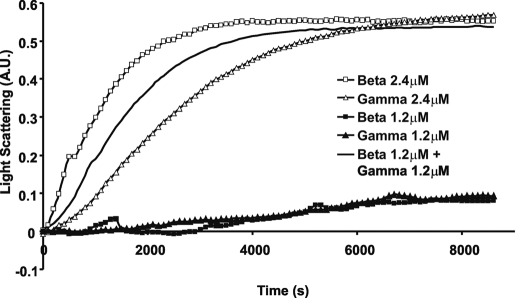
As stated before, despite their high degree of chemical identity, β- and γ-nonmuscle isoactins often differentially localize within the cell. We thus wished to determine whether this asymmetry reflected an inherent inability of the two actins to co-polymerize. We first established the polymerization kinetics for 1.2 μm β- and γ-actins individually. Because this concentration is very near the critical concentration for actin, little if any noticeable polymerization should occur over the course of the experiment as demonstrated in Fig. 4. If the two actins could not co-polymerize, mixing β- and γ-actins with each at 1.2 μm should produce no polymerization. However, Fig. 4 also shows that such a mixture polymerizes to the same extent as either actin does at double the concentration, 2.4 μm, indicating the two actins are totally copolymerizable. The polymerization rate of the mixture is intermediate between the behavior of the two pure samples, reflecting that one isoform is not totally dominant over the other in dictating overall behavior.
FIGURE 4.
Co-polymerization of β- and γ-actin. Polymerization of 2.4 μm or 1.2 μm total actin was initiated by the addition of magnesium and potassium chloride as described under “Experimental Procedures,” and the increase in L.S. was monitored as a function of time at 25 °C. Shown are representative plots of experiments performed four times with two independent actin preparations.
Fig. 2 suggests that the γ-actin preparation has a much more protracted nucleation phase than does the β-actin preparation. We thus assessed the ability of increasing amounts of the β-actin preparation to overcome this apparent γ-actin nucleation barrier. Fig. 5 shows that adding increasing amounts of β-actin to γ-actin while holding total actin constant produced a dose-response curve similar to what we saw with insect actin in overcoming the γ-actin nucleation lag.
FIGURE 5.
Polymerization of β- and γ-actin mixtures. Polymerization of 3.5 μm total actin was initiated by the addition of magnesium and potassium chloride as described under “Experimental Procedures,” and the increase in L.S. was monitored as a function of time at 25 °C. Numbers behind the isoforms represent the relative percentage of each actin isoform within the polymerization reaction. Shown are representative plots of experiments performed four times with two independent actin preparations.
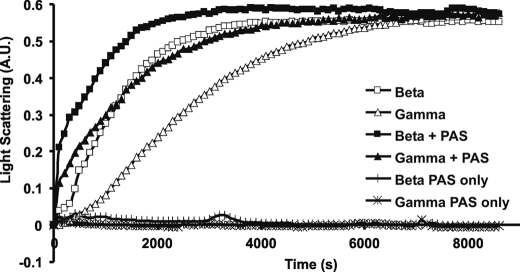
If, in fact, the delay in γ-actin polymerization relative to that of β-actin was caused by a nucleation barrier, the addition of small amounts of F-actin seeds made from γ-actin should largely overcome this observed delay. For this experiment, we generated F-actin seeds by sonicating phalloidin stabilized β- and γ-actin filaments and added them at a molar ratio of 1:20 to samples of G β- and γ-actins, respectively. Fig. 6 shows that, as predicted, the γ-actin seeds largely overcome the delay seen with G γ-actin alone. However, the seeded elongation rate of β-actin is still somewhat faster than that of γ-actin suggesting a small difference in their relative elongation rates as well.
FIGURE 6.
Seeded polymerization of β- and γ-actin. Polymerization of 3.5 μm actin was initiated by the addition of magnesium and potassium chloride as described under “Experimental Procedures,” in the presence or absence of PAS as indicated above and the increase in L.S. was monitored as a function of time at 25 °C. Shown are representative plots of experiments performed four times with two independent actin preparations.
One indication of filament stability is its rate of depolymerization. We thus measured the depolymerization rates of Ca2+ β- and γ-F-actins by following the first-order decrease in light scattering as a function of time following addition of super-stoichiometric amounts of DNase I as a sequestering agent for actin monomers as they are generated. Table 1 shows the t½ values for this process. Both β- and γ-actins depolymerize slower than yeast actin, and the t½ of γ-actin is 1.6 times longer than that of β-actin indicating an enhanced stability for the γ-isoform filament.
Phosphate Release Kinetics
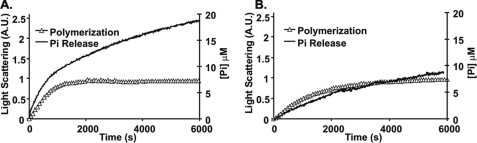
Another determinant of filament stability is the rate at which the inorganic phosphate is released from the actin surface once the ATP is hydrolyzed during polymerization because ADP-Pi F-actin is more stable than ADP F-actin (32). For most polymerizing actins, the Pi release curve is biphasic: an initial rapid release curve concomitant with polymerization and a slower second phase caused by the treadmilling of monomers through the filament at steady state.
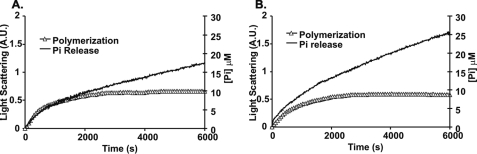
Fig. 7, A and B, show the data obtained in the comparison of Ca2+ β-actin versus γ-actin behavior. For β-actin, the initial Pi release phase appears to actually proceed at a rate noticeably faster than polymerization as determined by light scattering. In comparison, the first phase Pi release with yeast actin is much more synchronized with polymerization. This β-actin result suggests some kind of salt-dependent uncoupling of ATPase and polymerization or abortive filament formation and subsequent cycling of monomers. In contrast, the rate of first-phase Pi release from γ-actin seems to actually lag behind polymerization, similar to what is observed with muscle actin (33, 34). For second-phase treadmilling, the rate of Pi release is approximately twice as fast for β-actin (2.3 × 10−3 ± 0.3 × 10−3 μm Pi/s) than for γ-actin (1.1 × 10−3 ± 0.1 × 10−3 μm/s) similar to the faster depolymerization rate of β- versus γ-actin. Again, this result is suggestive of a more dynamic β-filament.
FIGURE 7.
Pi release associated with polymerization of Ca2+ nonmuscle actin isoforms. 4.8 μm Ca2+ β-actin (A) and γ-actin (B) were polymerized at 25 °C by the addition of salt. Filament formation was monitored by the change in light scattering and Pi release using the Enz Check Assay. The data were normalized and superimposed as described under “Experimental Procedures.” Shown are representative plots of experiments performed at least three times with three independent actin preparations.
Mg-actin Behavior
All of the above assays were performed with calcium initially occupying the high affinity divalent cation binding site on actin. However the predominant actin form in the cell is thought to have bound magnesium due principally to the relative abundance of magnesium versus calcium in the cytosol. Therefore we wanted to determine if the differences in the behavior of Ca2+ β- versus γ-actin were maintained by the actins in their Mg2+ form. As with Ca2+ actins, no differences in thermostabilities of the Mg2+ form of β- and γ-actins were observed (data not shown).
Nucleotide exchange rates for Mg2+ yeast, β-, and γ-actins were all accelerated compared with their Ca2+ counterparts, indicative of a much more dynamic monomer in the Mg2+ state (Table 1). Additionally, the 2-fold difference in exchange rates for β- versus γ-actin in the Ca2+ form virtually disappeared suggesting the release of selective conformational restraints on the calcium form of γ-actin accompanying its conversion from the Ca2+ to the Mg2+ form (Table 1).
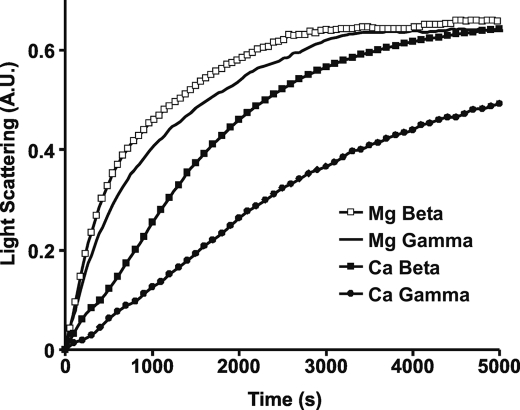
We then repeated the polymerization assay with Mg-actins, using 3.5 μm actin rather than the higher concentration used previously to allow better differentiation between more rapidly polymerizing actins. Fig. 8 shows that the rates of both Mg-actin isoforms are faster than their Ca2+ counterparts. Furthermore, as before, the β-actin polymerizes more rapidly, although the difference between Mg2+ β- and γ-actins has been drastically reduced. This result is consistent with less steric restraints on flexibility of the Mg2+ form of the G-actin monomer allowing it to much more easily assume its polymerization-competent monomer conformation. As with Ca2+ actins, EM examination of the Mg-actin filaments shows similar filament morphologies for β- and γ-actin (supplemental Fig. S2, C and D).
FIGURE 8.
Polymerization kinetics of Ca2+versus Mg2+ nonmuscle actin isoforms. Polymerization of 3.5 μm actin was initiated by the addition of magnesium and potassium chloride as described under “Experimental Procedures,” and the increase in L.S. was monitored as a function of time at 25 °C. Shown are representative plots of experiments performed at least three times with three independent actin preparations.
The relative rates of Pi release for the two isoactins are also much closer in the Mg2+ form than in the Ca2+ form (Fig. 9, A and B). For β-actin, the first phase Pi release rate appears much more closely synchronized with polymerization suggesting the absence of substantial abortive monomer cycling (Fig. 9A). For γ-actin, there is still a lag in Pi release relative to the rate of polymerization, although this discrepancy is smaller than with the Ca2+ actin. Finally, as before, with Mg2+ actin, the treadmilling rate for β-actin (2.8 × 10−3 ± 0.2 × 10−3 μm Pi/s) is still 1.5 times faster than that for γ-actin (1.9 × 10−3 ± 0.1 × 10−3 μm Pi/s).
FIGURE 9.
Pi release associated with polymerization of Mg-nonmuscle actin isoforms. 4.8 μm Mg2+ β-actin (A) and γ-actin (B) were polymerized at 25 °C by the addition of salt. Filament formation was monitored by the change in light scattering and Pi release using the Enz Check Assay. The data were normalized and were superimposed as described under “Experimental Procedures.” Shown are representative plots of experiments performed at least three times with three independent actin preparations.
DISCUSSION
The focus of this work was to determine if, despite their high degree of sequence identity, the two nonmuscle actins exhibit distinct biochemical properties that might explain their different cellular roles (2). However, such an assessment requires ready access to quantities of the individual pure isoforms large enough for biochemical and biophysical experiments, a goal previously difficult to achieve. The closest model of pure nonmuscle β-actin previously established was the scallop adductor muscle β-like actin (35). Although scallop adductor actin is similar in sequence to β-actin, there are seven amino acid substitutions between the two actins, making scallop adductor actin more divergent from β-actin than even γ-actin (35). Based on our results with only a four residue divergence, these differences between the actins could cause differences in their biochemical properties thereby detracting from the usefulness of the scallop actin as a β-actin model system (2, 35).
We have addressed this problem by successfully establishing a baculovirus-driven expression system for each of the individual mammalian nonmuscle isoforms, similar to what had previously been accomplished for the α-cardiac actin by Bookwalter and Trybus (24). Others have also used this system to assess the effect of different myopathic muscle actin mutations in vitro (23, 36). Similarly, our system will allow for the generation in the nonmuscle actins of pathology-producing mutations such as those associated with autosomal dominant non-syndromic hearing loss (37, 38) to try to gain insight into the biochemical alterations that underlie these pathological states. A problem with this system, however, is the significantly higher amounts of contaminating insect actin compared with what was encountered in the production of a non-polymerizing mutant α-cardiac actin (30). Estimates of contaminating insect actin were not provided for the WT α-cardiac actin preparations. We do not understand the reason for this difference. The presence of the insect actin will complicate the ability to use this preparation to study differences in actin-actin binding protein interactions between β- and γ-actins because of the similarities in β- and insect actin behavior. Furthermore, this complication mandates that one include controls with pure insect actin alone in such work. However, the distinct polymerization differences between γ- and insect actins should allow for meaningful comparisons between WT γ-actin and pathology producing mutant γ-actins.
The four residues differentiating β- from γ-actin lie at or near the amino terminus in a position that would not be expected to cause differences in the nucleotide binding behavior or thermostability of the actin monomer or their ability to polymerize. In terms of thermostability, this is what we observed. For nucleotide exchange, monomers of the two actin isoforms exchanged bound nucleotide more slowly than the rapidly exchanging yeast actin but about 4–6-fold more rapidly than muscle actin, previously shown to have one of the slower nucleotide exchange rates (31). We repeatedly observed, though, that the calcium β-isoform exchanged about 50% more rapidly than the γ-isoform suggesting that one or more of the four residue differences between the two actin isoforms could actually influence the dynamics of the protein around the nucleotide cleft, contrary to our original prediction.
Isoform-specific Ion-dependent Polymerization Behavior
Much more surprising, however, was the large difference in polymerization rates for the two actins in the calcium form. The β-actin polymerized much more rapidly than the γ-actin, almost as fast as the rapidly polymerizing yeast actin. The γ-actin polymerized at a rate much slower than even that of muscle actin, one of the slowest polymerizing actins documented to date. This slower polymerization rate of γ-actin versus β-actin seems to result from both slower nucleation and elongation rates, based on our seeded actin studies. In contrast, in the magnesium form generally considered to be the more physiologically relevant due to the Ca/Mg ratios in the cytosol, the difference between the two actin isoforms drastically decreased, although the β-isoactin still polymerized more quickly than the γ-isoform. A similar ion-dependent difference in the polymerization of two actins had previously been observed in comparing Dictyostelium discoideum actin with that from skeletal muscle, but these actins were considerably more divergent from one another than the two under consideration here and from different organisms where they play very different roles (39).
This difference between the two nonmuscle actin isoforms correlates with the different ways they are apparently used in motile cells in which β-isoactin seems to be preferred at the dynamic leading edge while the γ-isoactin tends to be incorporated more in stable stress fibers toward the middle of the cell. Our results show that this spatial separation is not due to an inherent inability of β- and γ-actin to co-polymerize. This difference might be achieved by somehow taking advantage of the apparent inherent difference in filament stability indicated by our DNase I experiment.
Our polymerization-dependent phosphate release assays present a similar picture. Muscle actin retains its phosphate for a substantial time after incorporation of the actin monomer into the filament, whereas with yeast actin, release is essentially simultaneous with polymerization reflecting their relative filament stabilities. In the calcium form, the rapid initial Pi release phase for β-actin appears to occur significantly faster than net polymerization. This observation is consistent with data from Karlsson's group (40), which demonstrated that Pi release precedes polymerization in both calf thymus actin and β-actin expressed and purified from yeast. This relationship may indicate the presence of non-productive or abortive polymerization early in the process. Alternatively in the Ca-induced conformation, addition of F-salts may induce a conformational change, which selectively activates β-actin ATPase without inducing polymerization. In essence, this would cause an uncoupling of these processes. In contrast, γ-actin appears to substantially retain its phosphate in the initial stage and exhibits a much slower treadmilling phase. This observation is consistent with the idea that the γ-filament is an inherently less dynamic and a more stable structure. As with the polymerization studies, conversion of the actin monomers to the magnesium form prior to polymerization reduced the difference between the two proteins although Pi release for β-actin was still faster for both phases than for γ-actin.
Possible Allosteric Interactions Involving the N-terminal Divergent Residues
The picture painted by this overall set of results is one in which the binding of Ca2+ at the high affinity binding site in the nucleotide cleft sets up an energy barrier, intensified by the γ-specific residues, that inhibits the conversion of the monomer conformation from the non-polymerizable to the polymerizable state. In essence there must be allosteric communication between these residues through subdomain 1 to the rest of the protein. It is tempting to speculate that the difference in behavior can be attributed to residue 10 within subdomain 1 where there is a Val/Ile substitution since this is in a core of secondary structural elements that form part of the nucleotide binding cleft. The alternative candidate would be that the other three N-terminal acidic residues that extend from the surface of subdomain 1 on its exterior might interact with other surface residues leading to a propagated change. However, in the monomer, at least, these residues are so unstructured they cannot be observed (22).
Results from previous work with yeast actin not only hint at such allostery but also make the role of the N-terminal acidic residues in this process more likely. In the monomer, the C-terminal peptide is on the opposite face of the planar actin structure in subdomain 1 than is the N-terminal peptide. There can be no direct spatial contact between them. However, we previously demonstrated that the effects of a mutation at residue 372 at the C terminus of yeast actin could be reversed by increasing the number of acidic residues at the N terminus from two to four (41). Second, we demonstrated that substitution of the normal two acidic N-terminal residues of yeast actin with the four found in muscle actin by themselves had no effect in vivo or in vitro (42). Introduction of all but the four N-terminal residues of subdomain 1 and 2 muscle specific residues cumulatively into yeast actin was tolerated well by the cell (31). However, replacement of the two yeast N-terminal acidic residues with the four muscle acidic residues in this subdomain 1/2 hybrid protein resulted in cell death. Replacement with only three acidic residues, on the other hand, was compatible with cell viability (31). Clearly, in this case the effect of the unstructured N-terminal acidic residues is determined by the context of the rest of subdomain 1 in the protein, and this linkage has to work in both directions. Further insight into the nature of this differential allostery between the β- and γ-nonmuscle actins must await the construction of a set of hybrid actins using the baculovirus system in which different combinations of the divergent residues between the two isoforms are switched and the resulting effects on polymerization assessed. However, such experiments are beyond the scope of the present work.
Possible Physiological Relevance of the Ion-dependent Polymerization Differences
One might question the significance of the differential polymerization behaviors of the two nonmuscle actins in the calcium form since the predominant form in the cell should be Mg2+ actin. However, Ca2+ bound in the nucleotide cleft may simply be one of a number of ligands that stabilize an energetically accessible conformation that can significantly differentiate the behavior of the two isoactins. In the cell, actin interacts with a large array of proteins both in the monomeric and polymeric state, and it is entirely possible that the conformational bias observed as a result of calcium binding could also be achieved through the interaction of actin with one or more of its binding partners. The result would be the creation of two populations of dynamically different filaments similar to what is seen in a number of cells.
There is at least one situation where the amount of calcium actin might actually be high enough to directly influence the behavior of these two actin isoforms. The perception of sound depends on a set of cells in the cochlea of the inner ear called hair cells. From these cells extend a highly organized set of deformable membrane-covered actin filament bundles called stereocilia. The bundles are stabilized by a set of actin filament crosslinking proteins. Sound-dependent bending of these bundles opens up potassium and calcium ion channels resulting in the propagation of nerve impulses to the brain (43). It has recently been demonstrated that high concentrations of calcium-binding proteins parvalbumin (44) and calbindin (45), approaching 300 μm and 970 μm respectively, are found in the stereocilia along with significant concentrations of two other calcium-binding proteins (calretinin (46, 47), and oncomodulin (48–50)). Additionally, stereocilia have the Ca2+ ATPase pump PMAC2 at a concentration of ∼2000 molecules/μm2 on the plasma membrane (51). The result could well be locally high persistent calcium concentrations sufficient to create a significant amount of calcium actin because actin actually binds calcium with a Kd in the nanomolar range, tighter than it binds magnesium (52). This is conjecture because to our knowledge it has not yet been possible to actually measure the calcium concentrations in the stereocilia. Another mode of regulation that cannot be dismissed is that the calcium is working directly on actin-binding proteins, which regulate actin dynamics.
A newly published study based on immunolocalization suggests an unequal distribution of β- and γ-actin in the stereocilium such that β-actin is concentrated in the core of the stereociliary bundle while γ-actin forms a shell around the core that will not stain with phalloidin (53). A somewhat analogous situation occurs in Caenorhabditis elegans where intestinal microvilli seem to require the C. elegans Act5 gene product (54). Attempts to substitute the Act1 gene product were not tolerated although the two actins are 99% homologous (54).
A possible trivial explanation for the differential isoform distribution result in the hair cells, which must be considered, is differential accessibility of the two isoform-specific antibodies arising from the method by which the experiment was executed. The lack of phalloidin staining could be due to the masking of phalloidin binding sites in F-actin by an actin-binding protein. Alternatively, it might reflect the sequestering of γ-actin in some type of unorthodox non F-actin state. In response to imposed noise, gaps occur in the bundled filaments which seem to be filled in by the γ-actin in some type of repair process (53). Our results may have direct bearing in this system. If the differential distribution results are valid, the relative inability of the Ca-γ-actin to spontaneously polymerize would help to keep it in this reservoir until directed polymerization within these gaps result in the reconstruction of continuous filaments. Once incorporated into the filament, however, the inherently slower rate of γ-actin depolymerization would work to add strength and stability to the repaired filament.
In summary our baculovirus system-based β- and γ-actin expression system has allowed a rigorous biochemical comparison of these two actins in their isoform-pure states. Our initial demonstration of an unsuspected ability of four small residue differences to allosterically affect filament conformation provides important new insight into the biochemical basis underlying how these proteins might function differently within the cell.
Supplementary Material
Acknowledgments
We thank Dr. Ema Stokasimov for performing the mass spectrometry analysis of insect actin in the β- and γ-actin preparations. We also thank Erin Reinl for work in creating and managing the baculovirus stocks and infected cells used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grants DC008803 (to P. A. R.) and DC004568 (to K. H. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- ϵ-ATP
- 1-N6-ethenoadenosine 5′-triphosphate
- PAS
- phalloidin actin seeds
- WT
- wild type
- L.S.
- light scattering
- A.U.
- arbitrary units.
REFERENCES
- 1.Vandekerckhove J., Weber K. (1978) J. Mol. Biol. 126, 783–802 [DOI] [PubMed] [Google Scholar]
- 2.Khaitlina S. Y. (2001) Int. Rev. Cytol. 202, 35–98 [DOI] [PubMed] [Google Scholar]
- 3.Höfer D., Ness W., Drenckhahn D. (1997) J. Cell Sci. 110, 765–770 [DOI] [PubMed] [Google Scholar]
- 4.Craig S. W., Pardo J. V. (1983) Cell Motil. 3, 449–462 [DOI] [PubMed] [Google Scholar]
- 5.Rybakova I. N., Patel J. R., Ervasti J. M. (2000) J. Cell Biol. 150, 1209–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoock T. C., Newcomb P. M., Herman I. M. (1991) J. Cell Biol. 112, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dugina V., Zwaenepoel I., Gabbiani G., Clément S., Chaponnier C. (2009) J. Cell Sci. 122, 2980–2988 [DOI] [PubMed] [Google Scholar]
- 8.Yao X., Chaponnier C., Gabbiani G., Forte J. G. (1995) Mol. Biol. Cell 6, 541–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watanabe H., Kislauskis E. H., Mackay C. A., Mason-Savas A., Marks S. C., Jr. (1998) J. Cell Sci. 111, 1287–1292 [DOI] [PubMed] [Google Scholar]
- 10.Hannan A. J., Gunning P., Jeffrey P. L., Weinberger R. P. (1998) Mol. Cell Neurosci. 11, 289–304 [DOI] [PubMed] [Google Scholar]
- 11.Micheva K. D., Vallée A., Beaulieu C., Herman I. M., Leclerc N. (1998) Eur. J. Neurosci. 10, 3785–3798 [DOI] [PubMed] [Google Scholar]
- 12.Gu W., Pan F., Zhang H., Bassell G. J., Singer R. H. (2002) J. Cell Biol. 156, 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kislauskis E. H., Zhu X., Singer R. H. (1994) J. Cell Biol. 127, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan F., Hüttelmaier S., Singer R. H., Gu W. (2007) Mol. Cell. Biol. 27, 8340–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross A. F., Oleynikov Y., Kislauskis E. H., Taneja K. L., Singer R. H. (1997) Mol. Cell. Biol. 17, 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallotton P., Gupton S. L., Waterman-Storer C. M., Danuser G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9660–9665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamagishi M., Ishihama Y., Shirasaki Y., Kurama H., Funatsu T. (2009) Exp. Cell Res. 315, 1142–1147 [DOI] [PubMed] [Google Scholar]
- 18.Namba Y., Ito M., Zu Y., Shigesada K., Maruyama K. (1992) J. Biochem. 112, 503–507 [DOI] [PubMed] [Google Scholar]
- 19.Kashina A. S. (2006) Trends Cell Biol. 16, 610–615 [DOI] [PubMed] [Google Scholar]
- 20.Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. (1990) Nature 347, 37–44 [DOI] [PubMed] [Google Scholar]
- 21.Oda T., Iwasa M., Aihara T., Maéda Y., Narita A. (2009) Nature 457, 441–445 [DOI] [PubMed] [Google Scholar]
- 22.Chik J. K., Lindberg U., Schutt C. E. (1996) J. Mol. Biol. 263, 607–623 [DOI] [PubMed] [Google Scholar]
- 23.Rutkevich L. A., Teal D. J., Dawson J. F. (2006) Can J. Physiol. Pharmacol. 84, 111–119 [DOI] [PubMed] [Google Scholar]
- 24.Bookwalter C. S., Trybus K. M. (2006) J. Biol. Chem. 281, 16777–16784 [DOI] [PubMed] [Google Scholar]
- 25.Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 26.Cook R. K., Rubenstein P. A. (1992) in Practical Approaches in Cell Biology (Carraway K., Carraway C. C. ed), pp. 99–122, IRL Press, Oxford [Google Scholar]
- 27.Yao X., Rubenstein P. A. (2001) J. Biol. Chem. 276, 25598–25604 [DOI] [PubMed] [Google Scholar]
- 28.Yao X., Nguyen V., Wriggers W., Rubenstein P. A. (2002) J. Biol. Chem. 277, 22875–22882 [DOI] [PubMed] [Google Scholar]
- 29.Boujemaa-Paterski R., Gouin E., Hansen G., Samarin S., Le Clainche C., Didry D., Dehoux P., Cossart P., Kocks C., Carlier M. F., Pantaloni D. (2001) Biochemistry 40, 11390–11404 [DOI] [PubMed] [Google Scholar]
- 30.Webb M. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4884–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKane M., Wen K. K., Meyer A., Rubenstein P. A. (2006) J. Biol. Chem. 281, 29916–29928 [DOI] [PubMed] [Google Scholar]
- 32.Carlier M. F., Valentin-Ranc C., Combeau C., Fievez S., Pantoloni D. (1994) Adv. Exp. Med. Biol. 358, 71–81 [DOI] [PubMed] [Google Scholar]
- 33.Carlier M. F. (1987) Biochem. Biophys. Res. Commun. 143, 1069–1075 [DOI] [PubMed] [Google Scholar]
- 34.Melki R., Fievez S., Carlier M. F. (1996) Biochemistry 35, 12038–12045 [DOI] [PubMed] [Google Scholar]
- 35.Khaitlina S., Antropova O., Kuznetsova I., Turoverov K., Collins J. H. (1999) Arch. Biochem. Biophys. 368, 105–111 [DOI] [PubMed] [Google Scholar]
- 36.Yates S. P., Otley M. D., Dawson J. F. (2007) Arch. Biochem. Biophys. 466, 58–65 [DOI] [PubMed] [Google Scholar]
- 37.Morin M., Bryan K. E., Mayo-Merino F., Goodyear R., Mencia A., Modamio-Hoybjor S., Del Castillo I., Cabalka J. M., Richardson G., Moreno F., Rubenstein P. A., Moreno-Pelayo M. A. (2009) Hum. Mol. Genet. 18, 3075–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu M., Yang T., Wei S., DeWan A. T., Morell R. J., Elfenbein J. L., Fisher R. A., Leal S. M., Smith R. J., Friderici K. H. (2003) Am. J. Hum. Genet. 73, 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steinmetz M. O., Hoenger A., Stoffler D., Noegel A. A., Aebi U., Schoenenberger C. A. (2000) J. Mol. Biol. 303, 171–184 [DOI] [PubMed] [Google Scholar]
- 40.Nyman T., Schüler H., Korenbaum E., Schutt C. E., Karlsson R., Lindberg U. (2002) J. Mol. Biol. 317, 577–589 [DOI] [PubMed] [Google Scholar]
- 41.McKane M., Wen K. K., Boldogh I. R., Ramcharan S., Pon L. A., Rubenstein P. A. (2005) J. Biol. Chem. 280, 36494–36501 [DOI] [PubMed] [Google Scholar]
- 42.Cook R. K., Root D., Miller C., Reisler E., Rubenstein P. A. (1993) J. Biol. Chem. 268, 2410–2415 [PubMed] [Google Scholar]
- 43.Hudspeth A. J. (1989) Nature 341, 397–404 [DOI] [PubMed] [Google Scholar]
- 44.Heller S., Bell A. M., Denis C. S., Choe Y., Hudspeth A. J. (2002) J. Assoc. Res. Otolaryngol. 3, 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sans A., Etchecopar B., Brehier A., Thomasset M. (1986) Brain Res. 364, 190–194 [DOI] [PubMed] [Google Scholar]
- 46.Dechesne C. J., Winsky L., Kim H. N., Goping G., Vu T. D., Wenthold R. J., Jacobowitz D. M. (1991) Brain Res. 560, 139–148 [DOI] [PubMed] [Google Scholar]
- 47.Edmonds B., Reyes R., Schwaller B., Roberts W. M. (2000) Nat. Neurosci. 3, 786–790 [DOI] [PubMed] [Google Scholar]
- 48.Sakaguchi N., Henzl M. T., Thalmann I., Thalmann R., Schulte B. A. (1998) J. Histochem. Cytochem. 46, 29–40 [DOI] [PubMed] [Google Scholar]
- 49.Henzl M. T., Shibasaki O., Comegys T. H., Thalmann I., Thalmann R. (1997) Hear Res. 106, 105–111 [DOI] [PubMed] [Google Scholar]
- 50.Hackney C. M., Mahendrasingam S., Penn A., Fettiplace R. (2005) J. Neurosci. 25, 7867–7875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vollrath M. A., Kwan K. Y., Corey D. P. (2007) Annu. Rev. Neurosci. 30, 339–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carlier M. F., Pantaloni D., Korn E. D. (1986) J. Biol. Chem. 261, 10778–10784 [PubMed] [Google Scholar]
- 53.Belyantseva I. A., Perrin B. J., Sonnemann K. J., Zhu M., Stepanyan R., McGee J., Frolenkov G. I., Walsh E. J., Friderici K. H., Friedman T. B., Ervasti J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacQueen A. J., Baggett J. J., Perumov N., Bauer R. A., Januszewski T., Schriefer L., Waddle J. A. (2005) Mol. Biol. Cell 16, 3247–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.