Abstract
The proteasome, a key component of the ubiquitin-proteasome pathway, has emerged as an important cancer therapeutic target. PS-341 (also called Bortezomib or Velcade) is the first proteasome inhibitor approved for newly diagnosed and relapsed multiple myeloma and is currently being tested in many clinical trials against other types of cancers. One proposed mechanism by which PS-341 exerts its anticancer effect is inactivation of nuclear factor-κB (NF-κB) through prevention of IκBα degradation. In this study, we show that PS-341 at concentrations that effectively inhibited the growth of human cancer cells, instead of increasing IκBα stability, paradoxically induced IκBα degradation. As a result, PS-341 facilitated p65 nuclear translocation and increased NF-κB activity. Moreover, IκBα degradation by PS-341 occurred early before induction of apoptosis and could not be inhibited by a pan-caspase inhibitor or caspase-8 silencing; however, it could be prevented with calpain inhibitors, calcium-chelating agents, calpain knockdown, or calpastatin overexpression. In agreement, PS-341 increased calpain activity. These data together indicate that PS-341 induces a calpain-mediated IκBα degradation independent of caspases. In the presence of a calpain inhibitor, the apoptosis-inducing activity of PS-341 was dramatically enhanced. Collectively, these unexpected findings suggest not only a novel paradigm regarding the relationship between proteasome inhibition and NF-κB activity but also a strategy to enhance the anticancer efficacy of PS-341.
Keywords: Apoptosis, Cancer, Protease/Calpain, Proteases/Proteosomes, Protein/Degradation, IkappaBalpha, NF-kappaB, PS-341
Introduction
The proteasome, a key component of the ubiquitin-proteasome pathway, has emerged as an important cancer therapeutic target (1–3). PS-341 (also called Bortezomib or Velcade), a dipeptidyl boronic acid, is a highly specific proteasome inhibitor with potent anticancer activity (4, 5). It is an approved drug for treatment of patients with multiple myeloma and mantle cell lymphoma. Many preclinical studies have demonstrated that PS-341 alone or in combination with other cancer therapeutic agents induces apoptosis in a variety of human cancer cells in vitro, including both hematologic and solid tumor malignancies, and inhibits the growth of tumor xenografts in vivo. Currently, there are many ongoing clinical trials that test the anticancer efficacy of PS-341 or its combinations with other agents in several types of cancers, including lung cancer (2, 6, 7).
Nuclear factor-κB (NF-κB)5 is a transcription factor that regulates the expression of a large number of genes involved in multiple biological processes, including apoptosis (8, 9). Its activation is often associated with cell resistance to apoptosis via up-regulation of the expression of multiple anti-apoptotic genes such as Bcl-2 and c-FLIP (9, 10). Therefore, NF-κB has long been an attractive target for cancer therapy. It is well known that IκBα ubiquitination and its subsequent degradation by the 26 S proteasome leading to nuclear translocation of p65 and an increase in NF-κB DNA binding are the primary mechanisms of NF-κB activation in cells upon stimulation (9, 10). Thus, a putative mechanism by which PS-341 induces apoptosis and exerts its anticancer activity is to inhibit NF-κB activation via prevention of IκBα degradation (1, 5).
PS-341 either as a single drug or in combination with other agents has been demonstrated to be active in inducing growth arrest and apoptosis in lung cancer cells in vitro and in inhibiting the growth of lung tumors in animal models (6, 7). To date, PS-341 is the only proteasome inhibitor to be evaluated in clinical trials against lung cancer and to show activity when used both as a single agent and in combination regimens (7). However, the mechanisms by which PS-341 induces apoptosis or growth arrest in lung cancer, particularly the role of NF-κB in these events, remain unclear.
Therefore, we have been interested in studying how PS-341 induces apoptosis in human cancer cells, particularly lung cancer cells. This study particularly focused on the role of NF-κB inhibition in PS-341-induced apoptosis in human lung cancer cells. Unexpectedly, we found that PS-341, instead of preventing IκBα degradation, induces calpain-dependent IκBα degradation in human lung cancer cells and other types of cancer cells. Moreover, we demonstrate that PS-341-induced IκBα degradation results in NF-κB activation and impacts cell responses to PS-341-induced apoptosis. Thus, this study suggests a novel mechanism underlying PS-341-mediated biological functions and a novel strategy to enhance the anticancer efficacy of PS-341.
EXPERIMENTAL PROCEDURES
Reagents
PS-341 was provided by Millennium Pharmaceuticals (Cambridge, MA). It was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 10 mm, and aliquots were stored at −80 °C. Stock solutions were diluted to the desired final concentrations with growth medium just before use. The pan-caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (Z-VAD-FMK) was purchased from Enzyme System Products (Livermore, CA). Ac-Leu-Leu-Nle-al (ALLN), leupeptin, and 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA) were purchased from Biomol (Plymouth Meeting, PA). Epoxomycin, [l-trans-3-ethoxycarbonyloxirane-2-carbonyl]-l-Leu-(3-methylbutyl)amide (EST), calpeptin, and PD150606 were purchased from Calbiochem. MG132, EGTA, and cisplatin were purchased from Sigma. The membrane-permeable calpain-specific fluorogenic substrate, t-butoxycarbonyl-Leu-Met-chloromethylcoumarin, and calpastatin expression plasmid (3878564) were purchased from Invitrogen. Docetaxel was obtained from Sanofi Aventis (Bridgewater, NJ).
Cell Lines and Cell Culture
All cell lines used in this study were originally purchased from the American Type Culture Collection (ATCC; Manassas, VA) and kept in our laboratory. They were grown in monolayer culture in RPMI 1640 medium supplemented with 5% fetal bovine serum at 37 °C in a humidified atmosphere consisting of 5% CO2 and 95% air.
Western Blot Analysis
The procedures for preparation of whole-cell protein lysates and Western blotting were described previously (11). Rabbit polyclonal antibodies against caspase-8, caspase-9, calpastatin, and PARP and mouse monoclonal phospho-IκBα (Ser-32/36) antibody were purchased from Cell Signaling Technology, Inc. (Beverly, MA). Mouse monoclonal anti-caspase-3 antibody was purchased from Imgenex (San Diego). Mouse monoclonal anti-IκBα and anti-calpain regulatory subunit antibodies were purchased from Santa Cruz Biotechnology. Rabbit polyclonal anti-actin and anti-glyceraldehyde-3-phosphate dehydrogenase antibodies were purchased from Sigma and Trevigen, Inc. (Gaithersburg, MD), respectively.
Gene Silencing with Small Interfering RNA
The nonsilencing control and caspase-8 small interfering RNA duplexes were described previously (12). Calpain regulatory subunit (SC-29887) and calpastatin (SC-29889) siRNAs were purchased from Santa Cruz Biotechnology. Transfection of these siRNA duplexes was conducted in 6-mm dishes using the HiPerFect transfection reagent (Qiagen, Valencia, CA) following the manufacturer's fast forward protocol. Briefly, 1 × 106 cells were seeded into a 60-mm culture dish 1 h prior to transfection. The given siRNA at 20 nm was mixed with 40 μl of HiPerFect transfection reagent, diluted with Opti-MEM (Invitrogen) medium, and then added to the cells. After 38 h post-transfection, the cells were treated with 50 nm PS-341 for an additional 10 h before collecting the cells for Western blotting. Gene silencing effects and other protein expression were evaluated by Western blot analysis as described above.
Cell Survival Assay
Cells were seeded in 96-well cell culture plates and treated the next day with the agents indicated. The viable cell number was determined using the sulforhodamine B assay, as described previously (13).
Detection of Apoptosis
Apoptosis was evaluated by annexin V staining using annexin V-PE apoptosis detection kit purchased from BD Biosciences following the manufacturer's instructions.
Proteasome Activity Assay
Fluorometric measurement of 20 S proteasome activity was the same as we described previously (14).
Calpain Activity Assay
Intracellular calpain activity was measured as described previously (15, 16). Cells in a 96-well plate or in chamber slides were exposed to PS-341 for a given time and then loaded with 10 μm t-butoxycarbonyl-Leu-Met-chloromethylcoumarin. After a 30–60-min incubation, cellular fluorescence was then quantified with a SPECTRAmax dual-scanning microplate spectrofluorometer (Molecular Devices Co., Sunnyvale, CA) with 351-nm excitation and 430-nm emission filters or recorded under a fluorescent microscope.
Indirect Immunofluorescent Staining
The procedure for p65 staining using rabbit anti-human Rel/p65 antibody (Santa Cruz Biotechnology, Inc.) was described previously (17).
Transient Transfection and Reporter Activity Assay
The pNF-κB-luc reporter plasmid, which contains the luciferase reporter gene driven by a basic promoter element (TATA box) joined to seven repeats of an NF-κB-response element, was purchased from Stratagene (La Jolla, CA). A pCH110 plasmid encoding β-galactosidase was purchased from GE Healthcare and used for normalization. The procedures for transfection, luciferase activity assay, and β-galactosidase assay were described previously (18).
Adenoviral Infection
Adenovirus harboring an empty vector (Ad-CMV) or a super repressor of IκBα (Ad-SRIκBα) was generously provided by Dr. L. W. Chung (Emory University, Atlanta, GA). The amplification, purification, and infection of adenovirus were described previously (17).
RESULTS
PS-341 Reduces IκBα Levels by Induction of Its Proteolysis while Inhibiting Proteasome Activity
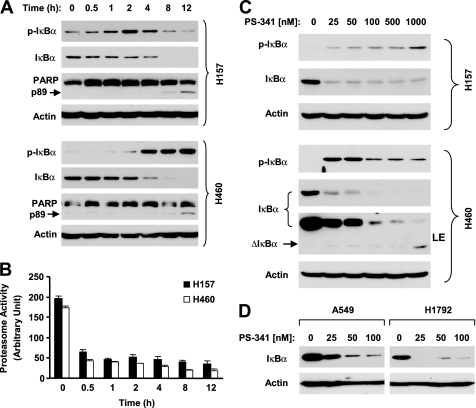
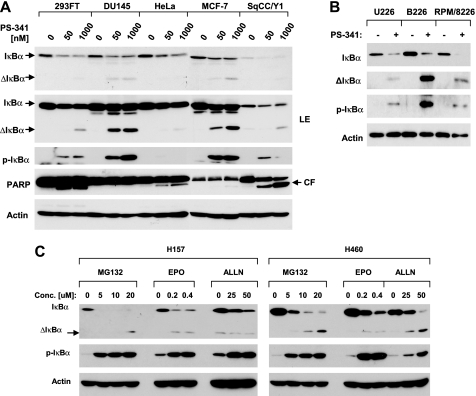
IκBα is known to be degraded via a ubiquitin-proteasome mechanism (9). PS-341 as a proteasome inhibitor theoretically should stabilize IκBα protein through inhibition of proteasome activity. When examining the effects of PS-341 on IκBα degradation in two human lung cancer cell lines, we surprisingly found that PS-341 decreased IκBα levels while increasing p-IκBα levels in a time-dependent fashion. In H157 cells, PS-341 substantially increased p-IκBα levels at 1 h followed by a great decrease in IκBα levels after 2 h. In H460 cells, a drastic p-IκBα increase was accompanied by a substantial reduction of IκBα levels after a 2-h treatment. In both cell lines, the IκBα reduction was sustained up to 12 h (Fig. 1A). By measuring the 20 S proteasome activity, we found that PS-341, under the same experimental conditions, effectively inhibited the 20 S proteasome activity starting from 0.5 h post-treatment in both tested cell lines (Fig. 1B). Moreover, we examined the effects of different concentrations of PS-341 on modulation of IκBα levels. As presented in Fig. 1C, PS-341 at all the tested concentrations (i.e. 25–1000 nm) decreased IκBα levels in both cell lines albeit without an apparent dose-dependent relationship in H157 cells. Accordingly, we detected increased levels of p-IκBα in both cell lines. In the other two lung cancer cell lines, PS-341 also effectively reduced the levels of IκBα (Fig. 1D). Moreover, we examined the effects of PS-341 on IκBα degradation in other types of solid tumor cell lines, including prostate (DU145), cervical (HeLa), breast (MCF-7), and head and neck cancer (SqCC/Y1), to determine whether IκBα reduction by PS-341 is a common phenomenon. In agreement with the findings in lung cancer cell lines, PS-341 at both low (50 nm) and high (1000 nm) concentrations decreased IκBα levels while increasing p-IκBα levels in all of the tested cell lines (Fig. 2A). Similar results were also obtained in several myeloma (U226, B226, and RPM/8226) cell lines (Fig. 2B).
FIGURE 1.
PS-341 induces IκBα degradation (A, C, and D) and inhibits proteasome activity (B) in human lung cancer cells. The given cells lines were treated with 50 nm PS-341 for the indicated time (A) or with the indicated concentrations of PS-341 for 8 h (C and D). After the aforementioned treatments, the cells were then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis. B, given cell lines were treated with 50 nm PS-341 for the indicated time and then subjected to proteasome activity assay. Columns, means of triplicate treatments; bars, ±S.D. LE, longer exposure.
FIGURE 2.
Induction of IκBα degradation by PS-341 in other types of cancer cells lines (A and B) and by other proteasome inhibitors in human lung cancer cells (C). A, given cell lines were treated with low (50 nm) or high (1000 nm) concentrations of PS-341 for 10 h. B, given cell lines were treated with 50 nm PS-341 for 8 h. C, given cell lines were treated with the indicated concentrations of MG132, epoxomycin (EPO), or ALLN for 8 h. After the aforementioned treatments, the cells were then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis. ΔIκBα, cleaved IκBα; LE, longer exposure; CF, cleaved fragment.
Furthermore, we determined whether other proteasome inhibitors exerted similar effects as PS-341 in modulation of IκBα levels. We used three proteasome inhibitors MG132, epoxomycin, and ALLN in this experiment and found that all these agents increased p-IκBα levels and decreased IκBα levels in both H157 and H460 cells (Fig. 2C). Thus, it appears that reduction of IκBα levels is a common phenomenon for proteasome inhibitors.
We noted that a short form of a protein band could be detected in the Western blot analysis for IκBα from cells exposed to either PS-341 (Fig. 1C and Fig. 2, A and B) or other proteasome inhibitors (Fig. 2C). This short form of protein appeared or was increased as the IκBα levels were decreased. Given that IκBα can be cleaved by a calpain-dependent mechanism (19, 20), it is very likely that this short form of protein is a cleaved form of IκBα. Thus, it appears that inhibition of proteasome with either PS-341 or other inhibitors reduced IκBα through promotion of its cleavage.
We also examined the effects of cisplatin and docetaxel, two cancer chemotherapeutic agents, on IκBα levels in our cell systems in comparison with PS-341. Under the tested conditions, we found that only PS-341, but not cisplatin and docetaxel, increased p-IκBα levels and decreased IκBα levels with increased levels of cleaved fragment (supplemental Fig. S1), suggesting that IκBα degradation is unique to PS-341 treatment.
PS-341 Induces Calpain-dependent IκBα Proteolysis Independent of Caspases
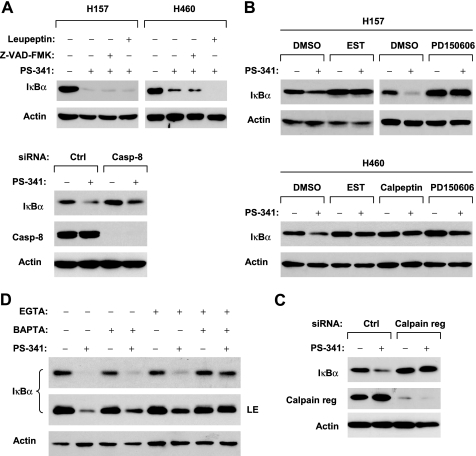
IκBα has been shown to be degraded through proteasome-independent mechanisms (e.g. caspase or calpain protease) (19–22). In our study, we noted that cleavage of PARP, a hallmark of apoptosis, was detected only at 8 h after PS-341 treatment, which was far later than IκBα reduction (after 1 h) (Fig. 1A). Moreover, strong cleavage of IκBα was detected in DU145 and MCF-7 cells, in which PARP cleavage was minimally detected or not detected, whereas relatively weak IκBα cleavage was detected in 293FT, HeLa, and SqCC/Y1 cells in which PARP was strongly cleaved (Fig. 2A). These results together suggest that the observed IκBα cleavage is unlikely due to caspase activation. Indeed, in the presence of the pan-caspase inhibitor, Z-VAD-FMK, which at the tested concentration abolished PS-341-induced caspase-3 activation (supplemental Fig. S2), PS-341 still effectively induced IκBα reduction in both H157 and H460 cells (Fig. 3A). A previous study suggested that caspase-8 is involved in IκBα cleavage (23). Thus, we also specifically addressed whether caspase-8 is involved in mediating PS-341-induced IκBα degradation through inactivating caspase-8 by knocking down caspase-8 expression. As presented in Fig. 3A (lower panel), knockdown of caspase-8 failed to prevent PS-341-induced IκBα reduction, indicating that caspase 8 is not involved in IκBα degradation induced by PS-341. Collectively, these data strongly suggest that PS-341 induces a caspase-independent IκBα proteolysis.
FIGURE 3.
PS-341 induces caspase-independent (A), calpain-mediated (B–D) IκBα proteolysis. A, top panel, the indicated cell lines were treated with 50 nm PS-341 alone or 50 nm PS-341 plus 50 μm Z-VAD-FMK, and 50 μm leupeptin, respectively, for 12 h. Bottom panel, H460 cells were transfected with control (Ctrl) or caspase-8 (Casp-8) siRNA. Forty eight hours later, the cells were treated with DMSO or 50 nm PS-341 for an additional 12 h. B, given cell lines were treated with 50 nm PS-341 alone or PS-341 combined with 200 μm EST, PD150606, and calpeptin, respectively, for 8 h. C, H460 cells were transfected with 20 nm control (Ctrl) or calpain regulatory (reg) siRNA. Thirty eight hours later, the cells were treated with DMSO or 50 nm PS-341 for an additional 10 h. D, given cell lines were treated with 50 nm PS-341 alone or PS-341 combined with 1.25 mm EGTA alone, 30 μm BAPTA alone, and EGTA plus BAPTA, respectively, for 8 h. After the aforementioned treatments, the cells were then subjected to preparation of whole-cell protein lysates and subsequent Western blot analysis. LE, longer exposure.
In addition, we determined whether the trypsin-like protease and cysteine protease inhibitor, leupeptin, could prevent PS-341-induced IκBα degradation. As presented in Fig. 3A, PS-341-induced IκBα reduction could not be prevented by leupeptin in either H157 or H460 cells, indicating that PS-341 decreases IκBα levels independent of leupeptin-sensitive proteases as well.
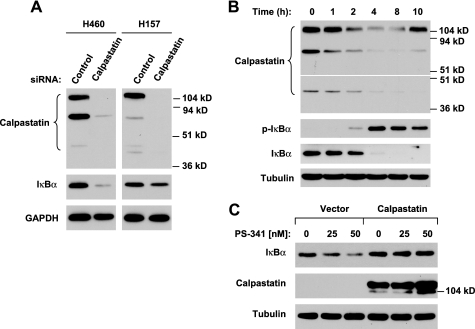
Finally, we investigated whether PS-341 induces IκBα degradation through a calpain-mediated mechanism. To this end, we first examined whether the presence of calpain inhibitors abolished the ability of PS-341 to induce IκBα degradation. As presented in Fig. 3B, the presence of either EST or PD150506 prevented IκBα from reduction induced by PS-341 in both H157 and H460 cells. In agreement, another calpain inhibitor calpeptin inhibited PS-341-induced IκBα reduction in H460 cells as well. Furthermore, we silenced the expression of the calpain regulatory subunit, which is a common regulatory subunit for both μ-calpain (calpain 1) and m-calpain (calpain 2) (24), and we examined its effect on PS-341-induced IκBα degradation. We assumed that knockdown of the calpain regulatory subunit would inhibit both μ-calpain and m-calpain activity. In agreement with the effects of the calpain inhibitors, as discussed above, knockdown of calpain regulatory subunit abolished the ability of PS-341 to reduce IκBα levels (Fig. 3C). These results clearly indicate that PS-341 induces calpain-dependent IκBα proteolysis.
It is known that calpains are calcium-dependent cysteine proteases (24). Thus, we further looked at the impact of calcium depletion on PS-341-induced IκBα degradation. The presence of the calcium-chelating agent BAPTA or EGTA alone in part prevented IκBα reduction by PS-341; however, the co-presence of both EGTA and BAPTA abolished the ability of PS-341 to decrease IκBα levels (Fig. 3D). These results again support the critical role of calpains in mediating PS-341-induced IκBα degradation.
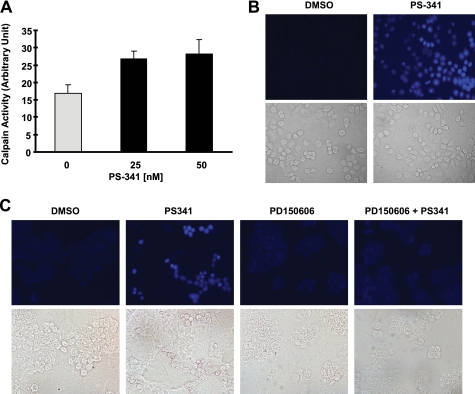
Moreover, we determined whether PS-341 indeed increases calpain activity. As presented in Fig. 4, A and B, PS-341 significantly increased the proteolytic hydrolysis of the calpain-specific fluorogenic substrate, t-butoxycarbonyl-Leu-Met-chloromethylcoumarin, exhibiting increased fluorescence. These results indicate that PS-341 increases intracellular calpain activity. In the presence of the calpain inhibitor PD150606, PS-341 did not increase calpain activity (i.e. fluorescence intensity) (Fig. 4C), indicating the specificity of the assay for calpain activity. This result provides another piece of evidence supporting involvement of calpain in PS-341-induced IκBα degradation.
FIGURE 4.
PS-341 increases calpain activity (A and B), which can be blocked by a calpain inhibitor (C). A and B, H460 cells were exposed to the indicated concentrations of PS-341 for 4 h (A) or 25 nm PS-341 for 8 h (B) and then subjected to calpain activity assay with either a microplate spectrofluorometer (A) or fluorescent microscopy (B). Columns, means of triplicate determinations; bars, ± S.D. C, H460 cells were exposed to DMSO, 50 nm PS-341, 100 μm PD150606, and the combination of PD150606 and PS-341 for 8 h and then subjected to calpain activity assay with fluorescent microscopy.
PS-341 Induces IκBα Proteolysis Involving Down-regulation of Calpastatin
To gain insight into the mechanism underlying proteasome inhibition-induced calpain activation, we further examined whether PS-341 modulates the levels of calpastatin, a well known endogenous calpain inhibitor (25), because proteasome inhibition was reported to reduce calpastatin levels (26). In Western blotting, we detected multiple sizes (e.g. ∼110, 70, and 40 kDa) of calpastatin, which are identical to published observations (27, 28). siRNA-mediated silencing of calpastatin substantially reduced the levels of these proteins, indicating that they are all specific calpastatin proteins. Meanwhile, IκBα levels were drastically reduced in calpastatin-silenced cells (Fig. 5A). PS-341 treatment decreased the levels of calpastatin accompanied by an increase in p-IκBα levels and reduction of IκBα in a time-dependent manner (Fig. 5B). Enforced expression of ectopic calpastatin increased basal levels of IκBα and prevented IκBα from degradation by PS-341 (Fig. 5C). Collectively, these results indicate that calpastatin inhibition is involved in mediating IκBα proteolysis by PS-341.
FIGURE 5.
Calpastatin inhibition decrease IκBα levels (A) and mediates PS-341-induced IκBα degradation (B and C). A, indicated cell lines were transfected with control or calpastatin siRNA for ∼48 h. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, H460 cells were treated with 50 nm PS-341 for the indicated times. C, HEK293T cells were transiently transfected with empty vector or calpastatin plasmid for ∼36 h and then treated with the indicated concentrations of PS-341 for an additional 16 h. After the aforementioned treatments, the cells were subjected to preparation of whole-cell protein lysates for Western blot analysis.
IκBα Phosphorylation Is Required for PS-341-induced IκBα Degradation
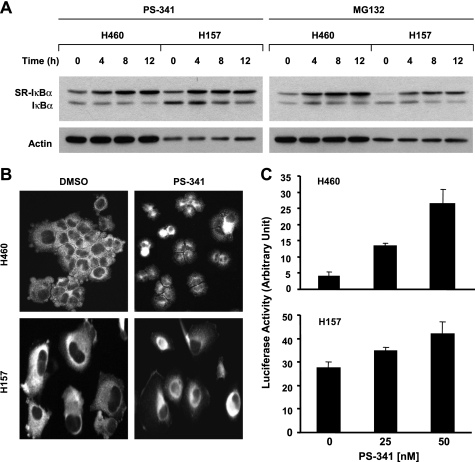
Given that inhibition of the proteasome with different proteasome inhibitors increases p-IκBα levels, we then asked whether IκBα phosphorylation is required for proteasome inhibition-initiated IκBα degradation. To address this issue, we took advantage of SRIκBα, which carries serine-to-alanine mutations at residues 32 and 36 and thus cannot be phosphorylated at Ser-32 and Ser-36. We first infected two lung cancer cell lines H460 and H157 with Ad-SRIκBα and then treated cells with either PS-341 or MG132 to determine whether PS-341 or MG132 could still induce SRIκBα degradation. For up to 12 h of treatment, neither PS-341 nor MG132 decreased the levels of SRIκBα; instead, both agents increased the levels of SRIκBα (Fig. 6A). Accordingly, we noted that the endogenous IκBα levels were not reduced or only slightly reduced (Fig. 6A). Thus, these results clearly indicate that IκBα phosphorylation at Ser-32 and Ser-36 is required for proteasome inhibition-initiated IκBα degradation.
FIGURE 6.
Proteasome inhibitors do not induce degradation of SRIκBα that carries serine to alanine mutations (A) and do induce p65 nuclear translocation (B) and NF-κB activity (C). The given cell lines were infected with 30 multiplicities of infection of Ad-SRIκBα for 48 h and then treated with 50 nm PS-341 or 10 μm MG132 for the indicated times. The cells were lysed for preparation of whole-cell protein lysates and subsequent Western blot analysis. The migration difference between the endogenous IκBα and SR-IκBα is likely due to mutation (i.e. amino acid change) and phosphorylation inability. B, given cell lines were treated with DMSO or 50 nm PS-341 for 4 h and then subjected to immunofluorescent staining of p65. C, indicated cell lines were transfected with pNF-κB-luc plasmid together with β-galactosidase plasmid as an internal control using FuGENE 6 transfection reagent. On the 2nd day, the cells were treated with the indicated concentrations of PS-341 for 8 h and then subjected to cell lysis and luciferase activity assay. Columns, means of triplicate determinations; Bars, ±S.D.
PS-341 Activates NF-κB
Since IκBα degradation is an essential step for p65 nuclear translocation and subsequent NF-κB activation (9, 21), we then questioned whether IκBα reduction by PS-341 resulted in NF-κB activation. To this end, we analyzed p65 localization in cells treated with PS-341 using immunofluorescent staining. As shown in Fig. 6B, we primarily detected p65 staining in the cytoplasm in DMSO-treated H460 or H157 cells. However, a strong nuclear staining of p65 was detected in PS-341-treated H460 or H157 cells, indicating that PS-341 induces p65 nuclear translocation in these cell lines. Moreover, we examined NF-κB activity in both H460 and H157 cell lines treated with PS-341 and found that PS-341 increased NF-κB luciferase activity in a dose-dependent manner (Fig. 6C).
c-FLIP, survivin, and COX-2 are NF-κB-regulated genes (9, 29, 30). PS-341 increased the levels of these proteins; however, the effects of PS-341 on increasing the levels of these proteins were abolished or attenuated in cells infected with Ad-SRIκBα (supplemental Fig. S3). These results clearly indicate that PS-341 modulates the expression of these proteins through an IκBα degradation- or NF-κB activation-dependent mechanism. Taken together with the above findings, we conclude that PS-341 increases NF-κB activity.
Moreover, we further analyzed the effects of PS-341 on p65 nuclear translocation in more cancer cell lines. Consistently, PS-341 induces p65 nuclear translocation in every tested cell line (supplemental Fig. S4). These data again support the notion that PS-341 activates NF-κB.
Inhibition of Calpain Augments the Effect of PS-341 on Induction of Apoptosis
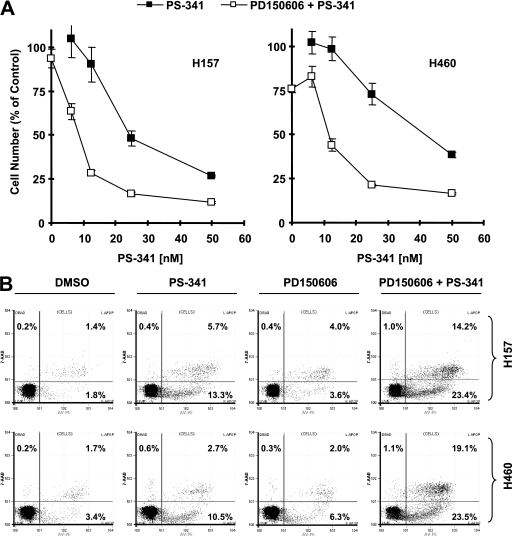
Activation of NF-κB is generally associated with apoptosis resistance, although it can be pro-apoptotic under certain conditions (8, 9, 22, 31, 32). Therefore, we investigated the impact of IκBα degradation on PS-341-induced apoptosis. To do this, we compared the effects of PS-341 on cell survival and apoptosis in the absence and presence of a calpain inhibitor. As presented in Fig. 7A, the calpain inhibitor PD150606 itself had a very weak effect on decreasing cell numbers. The combination of PS-341 with PD150606 was much more potent than PS-341 alone in decreasing cell numbers in both tested cell lines. Consistently, the combination of PS-341 with PD150606 caused the most apoptotic cells compared with PS-341 alone in both cell lines (Fig. 7B). For example, in H460 cells, PS-341 alone induced 13% apoptosis; however, the combination of PS-341 and PD150606 caused more than 40% apoptosis (Fig. 7B). Thus, it is clear that the presence of PD150606 substantially enhances the effect of PS-341 on induction of apoptosis, implying that inhibition of calpain activation may augment the anticancer activity of PS-341.
FIGURE 7.
Calpain inhibitor PD150606 augments effects of PS-341 on decreasing cell survival (A) and inducing apoptosis (B). A, H157 or H460 cells were treated with the given concentrations of PS-341 alone, 100 μm PD150606 alone, and the combination of PS-341 and PD150606. After 3 days, the cell numbers were estimated with the sulforhodamine B assay. Points, means of four replicate determinations; bars, ±S.D. B, given cell lines were treated with DMSO, 20 nm (H157) or 30 nm (H460) PS-341 alone, 100 μm PD150606 alone, or the combination of PS-341 and PD150606. After 48 h, the cells were harvested for estimating apoptotic cells with the annexin V assay. The percent positive cells in the upper right and lower right quadrants were added to yield the total of apoptotic cells.
DISCUSSION
Although inhibition of IκBα degradation and NF-κB activation is a putative mechanism for action of PS-341, no study has been done to demonstrate this mechanism in lung cancer cells. Our efforts in this regard have generated surprising results that instead of inhibiting IκBα degradation PS-341 actually potently decreases IκBα levels and results in NF-κB activation. These findings are consistent with three previous studies demonstrating that PS-341 as well other proteasome inhibitors induce IκBα degradation and activate NF-κB in endometrial carcinoma cell lines (33), the HepG2 liver cancer cell line (23), and multiple myeloma cells (34). We also found that PS-341 decreased IκBα levels in other types of cancer cell lines, including myeloma, prostate, cervical, breast, and head and neck cancer cells. One previous study shows that PS-341 indeed increases IκBα levels in human glioblastoma multiforme cells (35). Nevertheless, it seems that induction of IκBα degradation by PS-341 is likely to be a common event in many types of cancer cells.
It is known that activated IκB kinase (IKK) complex phosphorylates IκBα on serines 32 and 36, leading to subsequent ubiquitination and proteasome-mediated degradation of IκBα. Our results clearly show that PS-341 induces IκBα phosphorylation on serines 32 and 36 accompanied with reduction of IκBα levels. Moreover, we found that PS-341 failed to induce degradation of the IκBα mutant, SR-IκBα, in which serines 32 and 36 are mutated to alanines; rather, it increased the levels of SR-IκBα (Fig. 6A). Thus, we conclude that PS-341 induces phosphorylation-dependent IκBα degradation. This finding is in agreement with previous work demonstrating that PS-341 and other proteasome inhibitors induce IKK-dependent IκBα phosphorylation and degradation in other types of cancer cell lines through detection of phosphorylated IKKs and inhibition of IKKs with small molecules or siRNA (33, 34, 36). We assume that PS-341 may induce an IKK-dependent IκBα phosphorylation and degradation in lung cancer cells as well.
Before this study, it was unclear why PS-341 as a proteasome inhibitor induces IκBα degradation. It is worth mentioning that other proteasome inhibitors such as MG132 were also shown to induce IκBα degradation in colon cancer (36) and endometrial carcinoma cells (33). Our own data demonstrate that the proteasome inhibitors MG132, epoxomycin, and ALLN increase IκBα phosphorylation and decrease IκBα levels in human lung cancer cells as well. Thus, it is likely that proteasome inhibition is associated with PS-341-induced IκBα phosphorylation and degradation.
IκBα can also be degraded by proteases (e.g. calpain) or caspases (21, 22) in addition to proteasome. A recent study shows that PS-341 induces IκBα degradation involving caspase-8 in HepG2 cells (23). Our study demonstrates that PS-341 induces IκBα degradation or proteolysis independent of caspase activation, including caspase-8 activation based on the following facts. First, IκBα phosphorylation and degradation apparently occurred much earlier (at 2 h) than PARP cleavage (at 8 h) (Fig. 1A). Second, strong IκBα cleavage occurred in the cell lines where PARP cleavage was not detected or only minimally detected (Fig. 2A). Third, the presence of the pan-caspase inhibitor Z-VAD-FMK failed to prevent PS-341-induced IκBα degradation (Fig. 3A). Finally, inactivation of caspase-8 by siRNA-mediated knockdown of caspase-8 did not inhibit PS-341-induced IκBα degradation (Fig. 3A, lower panel).
It has been documented that IκBα can undergo calpain-mediated proteolysis, which is parallel to the ubiquitination-proteasome mechanism (19). Calpain-mediated IκBα proteolysis generates an ∼30-kDa cleaved IκBα intermediate in a previous report (19). In our study, we also detected an ∼30-kDa cleaved band of IκBα in cells exposed to PS-341 or other proteasome inhibitors (Figs. 1 and 2). The presence of the calpain inhibitor EST, calpeptin, or PD150606 abolished the ability of PS-341 to induce IκBα degradation (Fig. 3B). In addition, knockdown of the calpain regulatory subunit with a calpain regulatory siRNA abolished the ability of PS-341 to decrease IκBα levels (Fig. 3C). Consistently, the calcium chelators EGTA and BAPTA inhibited PS-341-induced IκBα degradation as well (Fig. 3D) because calpains are calcium-dependent proteases. Moreover, we detected increased calpain activity in cells exposed to PS-341 (Fig. 4). Together, these results clearly indicate that PS-341 induces IκBα proteolysis through a calpain-dependent mechanism. Thus, it is likely that inhibition of the proteasome may favor calpain activation, leading to IκBα degradation.
It is well known that calpain activity is negatively regulated by its endogenous inhibitor calpastatin (25). It was previously shown that proteasome inhibition induced calpastatin degradation, leading to calpain activation and subsequent Tau protein degradation in neuroblastoma cells (26). Similarly, we found that knockdown of calpastatin substantially decreased IκBα levels, further supporting the notion that calpain can cause IκBα degradation. Importantly, PS-341 decreased the levels of calpastatin accompanied with IκBα degradation (Fig. 5B). Consistently, overexpression of an exogenous calpastatin elevated basal IκBα levels and prevented IκBα from degradation by PS-341 (Fig. 5C). Thus, it appears that proteasome inhibition down-regulates or inhibits calpastatin, leading to calpain activation and subsequent IκBα degradation.
Because NF-κB regulates the expression of multiple anti-apoptotic genes, its activation is often associated with apoptosis resistance (9, 10, 32). In agreement with the finding in endometrial carcinoma cells (33) and multiple myeloma cells (34), we have also shown that PS-341 induces p65 nuclear translocation and increases NF-κB activity (Fig. 5) in our cell systems. Moreover, PS-341 increased the expression of survivin, c-FLIP, and COX-2, all of which are NF-κB-regulated genes (9, 29, 30). Importantly, suppression of IκBα degradation by expressing SR-IκBα abrogated the increase of these proteins by PS-341 (supplemental Fig. S3), furthering the notion that PS-341 activates NF-κB in our cell systems. In endometrial carcinoma cells, NF-κB activation does not mediate cytotoxic effects of proteasome inhibitors because neither IKKα nor IKKβ small hairpin RNA protected cells from death induced by these inhibitors (33). In multiple myeloma cells, inhibition of IKKβ enhances PS-341-induced cytotoxicity (34). In human lung cancer cells, we found that the presence of a calpain inhibitor (e.g. PD150606), which inhibits PS-341-induced IκBα degradation, substantially enhanced the effects of PS-341 on decreasing cell survival and inducing apoptosis (Fig. 7), implying that IκBα degradation consequently confers cell resistance to undergo apoptosis, presumably through activation of NF-κB. Thus, our findings in this regard may suggest a novel therapeutic strategy for enhancing the anticancer activity of PS-341 by preventing calpain activation.
In summary, we report a novel finding that PS-341 induces calpain-dependent IκBα degradation in human lung cancer as well other types of cancer cells. As a result, PS-341 activates NF-κB, which can confer apoptotic resistance and counteract the apoptosis inducing activity of PS-341. Thus, prevention of calpain-dependent IκBα degradation will substantially augment the apoptosis inducing activity of PS-341.
Supplementary Material
Acknowledgments
We are grateful to Dr. L. W. Chung for providing Ad-SRIκBα and to Dr. H. A. Elrod for editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant CA128613 (to S.-Y. S. and F. R. K. for Project 2). This work was also supported by Department of Defense VITAL Grant W81XWH-04-1-0142 (to S.-Y. S. for Project 4).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- NF-κB
- nuclear factor-κB
- siRNA
- small interfering RNA
- IKK
- IκB kinase
- BAPTA
- 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone
- EST
- [l-trans-3-ethoxycarbonyloxirane-2-carbonyl]-l-Leu-(3-methylbutyl)amide
- PARP
- poly(ADP-ribose) polymerase.
REFERENCES
- 1.Adams J. (2004) Nat. Rev. Cancer 4, 349–360 [DOI] [PubMed] [Google Scholar]
- 2.Rajkumar S. V., Richardson P. G., Hideshima T., Anderson K. C. (2005) J. Clin. Oncol. 23, 630–639 [DOI] [PubMed] [Google Scholar]
- 3.Zavrski I., Jakob C., Schmid P., Krebbel H., Kaiser M., Fleissner C., Rosche M., Possinger K., Sezer O. (2005) Anticancer Drugs 16, 475–481 [DOI] [PubMed] [Google Scholar]
- 4.Adams J., Kauffman M. (2004) Cancer Invest. 22, 304–311 [DOI] [PubMed] [Google Scholar]
- 5.Richardson P. G., Mitsiades C., Hideshima T., Anderson K. C. (2006) Annu. Rev. Med. 57, 33–47 [DOI] [PubMed] [Google Scholar]
- 6.Schenkein D. P. (2005) Clin. Lung Cancer 7, S49–S55 [DOI] [PubMed] [Google Scholar]
- 7.Scagliotti G. (2006) Crit. Rev. Oncol. Hematol. 58, 177–189 [DOI] [PubMed] [Google Scholar]
- 8.Karin M. (2006) Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 9.Aggarwal B. B. (2004) Cancer Cell 6, 203–208 [DOI] [PubMed] [Google Scholar]
- 10.Kucharczak J., Simmons M. J., Fan Y., Gélinas C. (2003) Oncogene 22, 8961–8982 [DOI] [PubMed] [Google Scholar]
- 11.Sun S. Y., Yue P., Wu G. S., El-Deiry W. S., Shroot B., Hong W. K., Lotan R. (1999) Oncogene 18, 2357–2365 [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Yue P., Zhou Z., Khuri F. R., Sun S. Y. (2004) J. Natl. Cancer Inst. 96, 1769–1780 [DOI] [PubMed] [Google Scholar]
- 13.Sun S. Y., Yue P., Dawson M. I., Shroot B., Michel S., Lamph W. W., Heyman R. A., Teng M., Chandraratna R. A., Shudo K., Hong W. K., Lotan R. (1997) Cancer Res. 57, 4931–4939 [PubMed] [Google Scholar]
- 14.Zou W., Yue P., Lin N., He M., Zhou Z., Lonial S., Khuri F. R., Wang B., Sun S. Y. (2006) Clin. Cancer Res. 12, 273–280 [DOI] [PubMed] [Google Scholar]
- 15.Rosser B. G., Powers S. P., Gores G. J. (1993) J. Biol. Chem. 268, 23593–23600 [PubMed] [Google Scholar]
- 16.Xu L., Deng X. (2004) J. Biol. Chem. 279, 53683–53690 [DOI] [PubMed] [Google Scholar]
- 17.Jin F., Liu X., Zhou Z., Yue P., Lotan R., Khuri F. R., Chung L. W., Sun S. Y. (2005) Cancer Res. 65, 6354–6363 [DOI] [PubMed] [Google Scholar]
- 18.Liu X., Yue P., Khuri F. R., Sun S. Y. (2004) Cancer Res. 64, 5078–5083 [DOI] [PubMed] [Google Scholar]
- 19.Han Y., Weinman S., Boldogh I., Walker R. K., Brasier A. R. (1999) J. Biol. Chem. 274, 787–794 [DOI] [PubMed] [Google Scholar]
- 20.Schaecher K., Goust J. M., Banik N. L. (2004) Neurochem. Res. 29, 1443–1451 [DOI] [PubMed] [Google Scholar]
- 21.Hu X. (2003) Cytokine 21, 286–294 [DOI] [PubMed] [Google Scholar]
- 22.Lamkanfi M., Declercq W., Vanden Berghe T., Vandenabeele P. (2006) J. Cell Biol. 173, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calvaruso G., Giuliano M., Portanova P., De Blasio A., Vento R., Tesoriere G. (2006) Mol. Cell. Biochem. 287, 13–19 [DOI] [PubMed] [Google Scholar]
- 24.Saez M. E., Ramirez-Lorca R., Moron F. J., Ruiz A. (2006) Drug Discov. Today 11, 917–923 [DOI] [PubMed] [Google Scholar]
- 25.Wendt A., Thompson V. F., Goll D. E. (2004) Biol. Chem. 385, 465–472 [DOI] [PubMed] [Google Scholar]
- 26.Delobel P., Leroy O., Hamdane M., Sambo A. V., Delacourte A., Buée L. (2005) FEBS Lett. 579, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grynspan F., Griffin W. B., Mohan P. S., Shea T. B., Nixon R. A. (1997) J. Neurosci. Res. 48, 181–191 [PubMed] [Google Scholar]
- 28.Wang K. K., Posmantur R., Nadimpalli R., Nath R., Mohan P., Nixon R. A., Talanian R. V., Keegan M., Herzog L., Allen H. (1998) Arch. Biochem. Biophys. 356, 187–196 [DOI] [PubMed] [Google Scholar]
- 29.Kawakami H., Tomita M., Matsuda T., Ohta T., Tanaka Y., Fujii M., Hatano M., Tokuhisa T., Mori N. (2005) Int. J. Cancer 115, 967–974 [DOI] [PubMed] [Google Scholar]
- 30.Shishodia S., Sethi G., Konopleva M., Andreeff M., Aggarwal B. B. (2006) Clin. Cancer Res. 12, 1828–1838 [DOI] [PubMed] [Google Scholar]
- 31.Perkins N. D., Gilmore T. D. (2006) Cell Death Differ. 13, 759–772 [DOI] [PubMed] [Google Scholar]
- 32.Radhakrishnan S. K., Kamalakaran S. (2006) Biochim. Biophys. Acta 1766, 53–62 [DOI] [PubMed] [Google Scholar]
- 33.Dolcet X., Llobet D., Encinas M., Pallares J., Cabero A., Schoenenberger J. A., Comella J. X., Matias-Guiu X. (2006) J. Biol. Chem. 281, 22118–22130 [DOI] [PubMed] [Google Scholar]
- 34.Hideshima T., Ikeda H., Chauhan D., Okawa Y., Raje N., Podar K., Mitsiades C., Munshi N. C., Richardson P. G., Carrasco R. D., Anderson K. C. (2009) Blood 114, 1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin D., Zhou H., Kumagai T., Liu G., Ong J. M., Black K. L., Koeffler H. P. (2005) Oncogene 24, 344–354 [DOI] [PubMed] [Google Scholar]
- 36.Németh Z. H., Wong H. R., Odoms K., Deitch E. A., Szabó C., Vizi E. S., Haskó G. (2004) Mol. Pharmacol. 65, 342–349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.