Abstract
Antibacterial peptides of the innate immune system combat pathogenic microbes, but often have additional roles in promoting inflammation and as growth factors during tissue repair. Midkine (MK) and pleiotrophin (PTN) are the only two members of a family of heparin-binding growth factors. They show restricted expression during embryogenesis and are up-regulated in neoplasia. In addition, MK shows constitutive and inflammation-dependent expression in some non-transformed tissues of the adult. In the present study, we show that both MK and PTN display strong antibacterial activity, present at physiological salt concentrations. Electron microscopy of bacteria and experiments using artificial lipid bilayers suggest that MK and PTN exert their antibacterial action via a membrane disruption mechanism. The predicted structure of PTN, employing the previously solved MK structure as a template, indicates that both molecules consist of two domains, each containing three antiparallel β-sheets. The antibacterial activity was mapped to the unordered C-terminal tails of both molecules and the last β-sheets of the N-terminals. Analysis of the highly conserved MK and PTN orthologues from the amphibian Xenopus laevis and the fish Danio rerio suggests that they also harbor antibacterial activity in the corresponding domains. In support of an evolutionary conserved function it was found that the more distant orthologue, insect Miple2 from Drosophila melanogaster, also displays strong antibacterial activity. Taken together, the findings suggest that MK and PTN, in addition to their earlier described activities, may have previously unrealized important roles as innate antibiotics.
Keywords: Antibiotics, Evolution/Protein, Immunology/Defensins, Immunology/Innate Immunity, Organisms/Human, Peptides/Conformation, Protein/Domains
Introduction
The innate immune system is an ancient part of host defense and antimicrobial peptides (AMPs)2 are critical to execute its functions. The ability to produce AMPs is broadly distributed among all plants and animals, enhancing survival in an environment where pathogenic microbes are a constant threat (1). Some AMPs are constitutively produced, while the expression of others is induced during the inflammatory response to infection. Despite considerable diversity among AMPs and increasing evidence that they affect bacteria in a multitude of ways, rupture of bacterial integrity plays a key role in the antibacterial activity of most AMPs (2).
In most cases, AMPs of the innate immune system serve additional purposes. They can act as growth factors to promote tissue repair (e.g. promoting angiogenesis) and they can recruit and activate inflammatory cells (3). This pluripotency is characteristic of important AMPs such as the cathelicidins, the antibacterial chemokines, and the defensins (4–8).
Midkine (MK) and pleiotrophin (PTN) constitute a structurally distinct, highly conserved family, of small heparin-binding growth factors (9). MK is a 13.4-kDa protein consisting of 123 amino acids, whereas PTN is a 15.4-kDa protein of 136 amino acids (10, 11). MK was first identified as a gene activated by retinoic acid in murine carcinoma cells (12, 13). Independently, a chicken homologue was purified from basement membranes at the same time (14). PTN was originally described as a factor promoting the outgrowth of neurites and as a mitogenic factor for murine fibroblasts (14, 15). Both MK and PTN show restricted expression during embryogenesis, being mainly expressed in the central nervous system, the respiratory system and the gastrointestinal tract. They are up-regulated in several neoplastic diseases (16). In humans, MK is up-regulated during inflammation in some tissues of the adult and can be detected in plasma of healthy individuals (17–19). In addition, MK recruits and activates neutrophils, suggesting important roles for this growth factor during inflammation (20, 21).
MK and PTN bind and activate several receptors, for example the anaplastic lymphoma kinase (ALK) receptor, the receptor protein-tyrosine phosphatase β/ζ (RPTPβ/ζ), N-syndecan, whereas MK can also bind LDL-related protein (LRP) and the α4β1- and α6β1-integrins. Ligand binding results in diverse cellular effects including mitogenic responses and differentiation (22).
Both MK and PTN are found in many species, from Drosophila to humans. They have not, however, been found in the Caenorhabditis elegans genome, suggesting their origin among insects. The existence of two genes would appear to be the result of an early gene duplication (23). MK and PTN have a structure with β-sheets, held together by disulfide bridges, a structure also found in one class of AMP, namely the defensins (24). In addition, MK and PTN are highly cationic and contain heparin-binding sites, a common feature of AMPs (25). Taken together, MK and PTN share many functional and structural features with AMPs. The results presented here clearly show that human MK and PTN have antibacterial properties and that antibacterial domains are conserved in MK and PTN orthologues.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Recombinant human PTN and MK were from PeproTech (London, UK). According to the manufacturer, the purity of MK and PTN was greater than 98% as determined by SDS-PAGE and HPLC analyses. Determination of disulfide bonds in human recombinant MK and PTN was performed essentially as described (26, 27). In short, the proteins were digested using trypsin (1:20 (w/w) enzyme:substrate ratio) at 37 °C for 16 h. Digestions were performed both with and without a preceding reduction/alkylation procedure (dithiothreitol and iodoacetamide). Analysis was performed using HPLC-MS/MS on a QTOF Ultima API (ESI-MS/MS) (Waters, Manchester, UK) coupled to a CapLC (Waters). In addition, non-reduced samples were subject to MALDI-TOF MS on a MALDI micro MX (Waters). Injected samples were trapped on a C18 pre-column (300 μm × 5 mm, 5 μm, 100 Å; LC-packings/Dionex, Stockholm, Sweden) and separated on an Atlantis C18 reversed phase analytical column (75 μm × 150 mm, 3 μm, 100 Å) (Waters). All experimentally obtained peptide and peptide conglomerate masses were compared with the expected masses obtained after trypsin cleavage, using the FindPept, PeptideMass, and the PeptideCutter tools (59). When MK and PTN were digested with trypsin, without prior reduction and alkylation, masses corresponding to peptide complexes with retained sulfide bridges were identified using ESI-MS/MS and MALDI-TOF MS (26, 27). Sequence coverage spanning the positions of all cysteines, thus directly representing the primary amino acid sequence of both chemokines, was obtained from MS/MS of samples only when trypsin digestion was performed after dithiothreitol and iodoacetamide treatment. These experiments therefore confirm that both MK and PTN had proper tertiary conformations, i.e. contained the correct set of disulfide bonds. On a quantitative basis, no thiol groups were detected in MK and PTN, respectively, using 5, 5′-dithiobis(2-nitrobenzoate) (DTNB), further demonstrating that all cysteines are engaged in disulfide bonds (28). 20-mer peptides corresponding to various regions of human MK, PTN, and corresponding orthologues were from Sigma (PEP screen®).
To analyze antibacterial activity of Miple2 protein, residues 21–279 of Miple2 were subcloned into the pGEX-T2 GST fusion protein expression vector (details available upon request). The resulting construct, pGEX-T2:Miple2, was confirmed by sequence analysis. GST-Miple2 fusion protein was induced and purified from Escherichia coli BL21 (DE3) bacterial lysates by standard protocols using glutathione Sepharose beads. GST-Miple2 recombinant protein was proteolytically cleaved with thrombin (4 units/ml). After cleavage, the thrombin was removed with p-aminobenzamidine agarose. The sample was dialyzed overnight with a molecular mass cut-off of 15 kDa, followed by concentration. On an SDS-PAGE gel stained with Coomassie Blue, Miple2 was the only visible protein. To rule out that contaminating GST or thrombin contributed to the antibacterial activity of Miple2, these proteins were used in the viable count assay with Staphylococcus aureus. Neither GST (10 μm) nor thrombin (4 units/ml) showed any antibacterial activity.
Bacterial Strains and Growth Conditions
The Streptococcus pyogenes strain AP1 (40/58) of M1 serotype was from the World Health Organization Collaborating Centre for Reference and Research on Streptococci, Prague, Czech Republic. Staphylococcus aureus (strain 5120), E. coli (strain 37.4), and Pseudomonas aeruginosa (strain 27-1) were obtained from the Division of Infection Medicine, Department of Clinical Sciences, Lund University, Sweden. All bacteria were routinely grown in Todd-Hewitt (TH; Difco/Becton & Dickinson, Franklin Lakes, NJ) liquid medium at 37 °C and 5% CO2.
Viable Count Assay
All bacteria were cultivated to mid-logarithmic phase (optical density at 620 nm was 0.4) in TH medium, washed, and diluted (1:1,000) in incubation buffer (10 mm Tris-HCl, containing 5 mm glucose; pH 7.4). P. aeruginosa were grown for 18 h at 37 °C. 50 μl of bacteria (106 colony forming units (cfu)/ml) were incubated in the absence or presence of peptides at various concentrations, (0.003–1 μm) for 1 h at 37 °C. To quantify antibacterial activity, serial dilutions of the incubation mixtures were plated on TH agar plates, incubated overnight at 37 °C, and the number of cfu determined.
To investigate the activity of MK and PTN in the presence of salt, the viable counts assay was also performed after addition of different concentrations of sodium chloride to the incubation buffer (5–150 mm). The assay was also performed in the presence of human plasma (pooled citrated plasma from at least 12 donors; Department of Transfusion Medicine, University Hospital, Lund, Sweden) and sodium chloride (150 mm) at plasma concentrations ranging from 1 to 30%. To investigate the importance of heparin-binding regions of MK and PTN in antibacterial activity, the proteins were incubated with low molecular mass heparin (5 kDa; FragminTM; Pfizer, Stockholm, Sweden) at a MK/PTN: heparin ratio of 1:1 for 20 min before exposure to bacteria in the viable counts assay.
Liposome Leakage Assay
The liposomes investigated were anionic (DOPE/DOPG 75/25 mol/mol). DOPG (1,2-dioleoyl-sn-glycero-3-phosphoglycerol, monosodium salt) and DOPE (1,2-dioleoyl-sn-glycero-3-phoshoetanol-amine) were from Avanti Polar Lipids (Alabaster, AL) and of >99% purity. The lipid mixtures were dissolved in chloroform, after which the solvent was removed by evaporation under vacuum overnight. Subsequently, 10 mm Tris buffer, pH 7.4, with or without additional 150 mm NaCl, was added together with 0.1 m carboxyfluorescein (CF) (Sigma). After hydration, the lipid mixture was subjected to eight freeze-thaw cycles consisting of freezing in liquid nitrogen and heating to 60 °C. Unilamellar liposomes of about Ø 140 nm were generated by multiple extrusions through polycarbonate filters (pore size, 100 nm) mounted in a LipoFast mini-extruder (Avestin, Ottawa, Ontario, Canada) at 22 °C. Untrapped CF was removed by two subsequent gel filtrations (Sephadex G-50, GE Healthcare, Uppsala, Sweden) at 22 °C, with Tris buffer (or Tris plus 150 mm NaCl) as eluent. CF release from the liposomes was determined by monitoring the emitted fluorescence at 520 nm from liposome dispersions (10 mm lipid in 10 mm Tris with 5 mm glucose, pH 7.5, with or without 150 mm additional NaCl). An absolute leakage scale was obtained by disrupting the liposomes at the end of each experiment by addition of 0.8 mm Triton X-100 (Sigma), causing 100% release and dequenching of CF. A SPEX-fluorolog 1650 0.22-m double spectrometer (SPEX Industries, Edison, NJ) was used for the liposome leakage assay. Measurements were performed in, at least, duplicates at 37 °C.
Transmission Electron Microscopy
Bacteria were cultured to mid-logarithmic phase as described above. S. pyogenes and E. coli were incubated for 1 h at 37 °C with 0.15 μm PTN or MK and 0.4 μm PTN or MK, respectively. To verify that the bacteria were killed, part of the sample was plated on TH agar plates, incubated at 37 °C overnight, and the cfu counted the following day. The remaining parts of the samples were processed for electron microscopy as described (29). In short, samples (5 μl) were absorbed for 45 s onto the grids followed by two washes with distilled water. The samples were stained for 3 s with 0.75% uranyl formate droplets, followed by staining for an additional 15–20 s with 0.75% uranyl formate. The samples were investigated using a Jeol JEM 1230 EX electron microscope operated at 60 kV accelerating voltage.
Phylogenetic Analysis
MK and PTN amino acid sequences were retrieved from GenBank at the NCBI site (supplemental Table S1). Each sequence was analyzed with the Psi-Blast (NCBI) (30) to find orthologue and paralogue sequences. Sequences showing structural homologies of more than 70% were selected. These sequences were aligned using ClustalW (31) with Blosum 80 protein weight matrix settings (32). Internal adjustments were made, taking the structural alignment into account, utilizing the ClustalW interface. The level of consistency of each position within the alignment was estimated by alignment evaluating software Tcoffee (33). MK and PTN amino acid sequences were used to construct a phylogenetic tree applying the neighbor-joining method (34). The generated tree was rooted with human PTN, and the reliability of each branch was assessed using 1,000 bootstrap replications.
Molecular Modeling
A model structure of full-length human PTN (GenBankTM accession number P21246) was created by coupling generated comparative homology models of PTN N- and C-terminal domains, respectively. The theoretical model of PTN containing amino acid residues 33–96 (N-terminal domain) and residues 97–168 (C-terminal domain) was built using the NMR solution structures of human MK with Protein Data Bank codes 1MKN and 1MKC, respectively, as templates (35). The sequence of PTN was aligned against the sequence of MK using the alignment displayed in Fig. 4A. The modeling was performed using the program MODELLER version 9v7 (36). When modeling the N-terminal domain of PTN, a restraint defining the disulfide bond between cysteines 47 and 76 was added. Ten models for each domain were generated using the automodel class, and the model with the lowest DOPE assessment score was selected. The final model of full-length PTN was built in MODELLER using the selected models for the N- and C-terminal domains as templates. A model structure of full-length human MK (GenBankTM accession number P21741) was created in a similar manner as PTN (described above). The model of MK contains amino acid residues 21–143. Putative signal peptides of Miple1 and Miple2 were determined using neural networks and hidden Markov models as described (37).
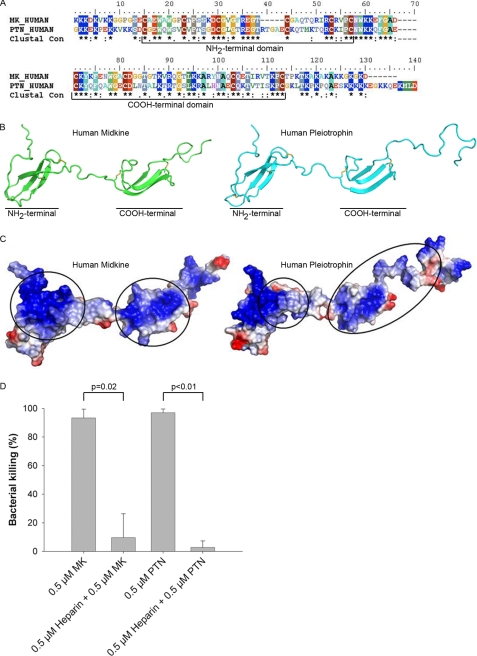
FIGURE 4.
Alignment of the human MK and PTN sequences, and predicted structure of PTN. A, alignment of the MK and PTN sequences show high homology (45%) and similarity (61%). Ten cysteine residues are present, six in the N-terminal domain and four in the C-terminal domain. B, structure of MK was used as a template to predict the structure of PTN. Both molecules contain two domains consisting of three antiparallel β-sheets, held together by a hinge region. PTN has a longer unordered C-terminal tail than MK. C, models of MK and PTN showing the distribution of positively charged (blue) and negatively charged residues (red). The heparin-binding sites are indicated (circles). D, to investigate the importance of heparin-binding domains in antibacterial activity of the holoproteins, MK and PTN were preincubated with low molecular weight heparin prior to incubation with S. aureus using the viable count assay. A strong inhibition of the antibacterial activity was seen at equimolar concentrations of heparin and MK/PTN. Heparin alone did not affect the viability of bacteria. The data shown represent mean ± S.D. from three separate experiments. Statistical significance was determined based on the Student's t test for paired observations.
Radial Diffusion Assay
Radial diffusion assay (RDA) was performed essentially as described earlier (38). E. coli was grown to mid-logarithmic phase (optical density at 620 nm was 0.4) in 10 ml of full strength (3%, w/v) trypticase soy broth (TSB) (BD Biosciences, San Jose, CA). The bacteria were washed once with 10 mm Tris-HCl (containing 5 mm glucose; pH 7.4). 4 × 106 cfu were added to 5 ml of the underlay gel (0.03% TSB, 1% low electro endosmosis-type agarose (Low-EEO; Sigma), 0.02% Tween-20 (Sigma) in 10 mm Tris-HCl), and poured onto a Ø 90 mm Petri dish. After agarose solidification, Ø 4 mm wells were punched, and 6 μl of buffer alone or peptide (100 μm) were added to each well. Plates were incubated at 37 °C for 3 h to allow the peptides to diffuse. 5 ml of the overlay gel (6% TSB, 1% low-EEO agarose in 10 mm Tris-HCl) was used to cover the underlay gel. Antimicrobial activity was seen as a clearing zone around each well after incubating 18–24 h at 37 °C. The assay was performed in triplicates with either the recombinant human MK and PTN holoprotein or, overlapping peptides, (comprised of 20 amino acids) derived from the mature holoprotein sequences of MK and PTN (GenBankTM accession numbers NP_001012333 and CAA37121). Similar overlapping peptides from the MK and PTN orthologues from X. laevis (GenBankTM accession number P48530; P48533) D. rerio (GenBankTM accession numbers Midkine-related growth factor A NP_571145; Midkine-related growth factor B NP_571791; PTN NP_001003981) and D. melanogaster (GenBankTM accession number miple1 NP_612022; miple2 NP_651996) were also used. All peptides used were water-soluble. The peptides were dissolved in water and no other solvents were used.
Calculations
Theoretical pI were computed using the EXPASy primary structure analysis tool. Statistical significance was determined based on the Student's t test for paired observations.
RESULTS
Human MK and PTN Possess Antibacterial Activity
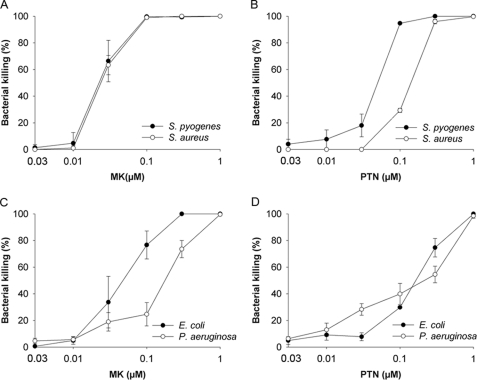
MK and PTN were investigated for possible antibacterial activity against a panel of pathogenic bacteria, i.e. the Gram-positive bacteria S. pyogenes and S. aureus as well as the Gram-negative bacteria, P. aeruginosa, and E. coli. The bacteria were incubated in buffer alone or with recombinant MK or PTN at concentrations ranging from 0.003 μm to 1 μm followed by a viable count assay. Both MK and PTN showed high antibacterial activity against Gram-positive bacteria (Fig. 1, A & B) and slightly lower activity against Gram-negative bacteria (Fig. 1, C & D).
FIGURE 1.
Antibacterial activity of human midkine and pleiotrophin. A–D, MK and PTN were investigated for antibacterial activity using a viable count assay. The Gram-positive pathogens S. pyogenes and S. aureus, and the Gram-negative pathogens E. coli and P. aeruginosa, were grown to mid-logarithmic phase followed by incubation with proteins at the indicated concentrations or in buffer alone, for 1 h at 37 °C. Serial dilutions were made and after culture overnight on agar plates the number of cfu were counted. The number of colonies remaining after exposure to MK and PTN, respectively, was compared with the number of cfu obtained after incubation in buffer alone, to calculate % killing. The data shown represent mean ± S.D. from three separate experiments.
Antibacterial Activity of MK and PTN in the Presence of Salt and Plasma
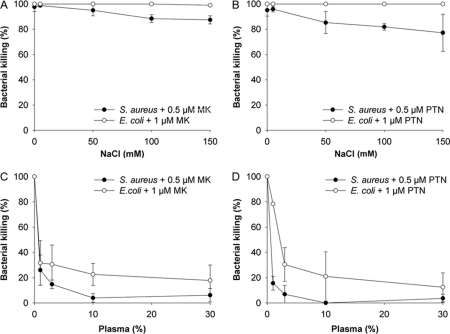
Physiological concentrations of sodium chloride impair the antimicrobial activity of many AMPs, such as defensins (39). Saliva, during fasting conditions, contains 5 mm sodium chloride; plasma has a concentration slightly below 150 mm, whereas epithelial surfaces have concentrations in between (40). Therefore, we chose to study the antibacterial activity of MK and PTN in the presence of sodium chloride at concentrations ranging from 5–150 mm. Because S. pyogenes itself is sensitive to salt, S. aureus and E. coli were investigated. The antibacterial activity was largely unaffected (Fig. 2A), except in the case of S. aureus incubated with PTN, where a dose-dependent decrease of the antibacterial activity by 23% was seen at 150 mm sodium chloride (Fig. 2B). The presence of plasma is also known to inhibit the activity of AMPs. The influence of human citrated plasma on the killing capacity of MK and PTN was therefore investigated using S. aureus and E. coli in the viable count assay. Already at low concentrations of plasma, a strong inhibition of both MK- and PTN-induced killing was seen, although less pronounced in the case of E. coli (Fig. 2, C & D).
FIGURE 2.
Antibacterial activity of human MK and PTN in the presence of salt and plasma. To investigate if the presence of sodium chloride and plasma proteins interfere with the antibacterial activity of MK and PTN, respectively, the viable count assay was used. S. aureus and E. coli were grown to mid-logarithmic phase followed by incubation with MK and PTN, respectively (0.5 μm in the case of S. aureus and 1 μm in the case of E. coli) for 1 h at 37 °C. Serial dilutions were plated and the resulting cfu counted. Decrease of antibacterial activity was seen in the case of E. coli for both MK and PTN at higher concentrations of sodium chloride. Addition of plasma showed a strong inhibition of the antibacterial activity. The data shown represent mean ± S.D. from three separate experiments.
MK and PTN Have Membrane-disrupting Properties
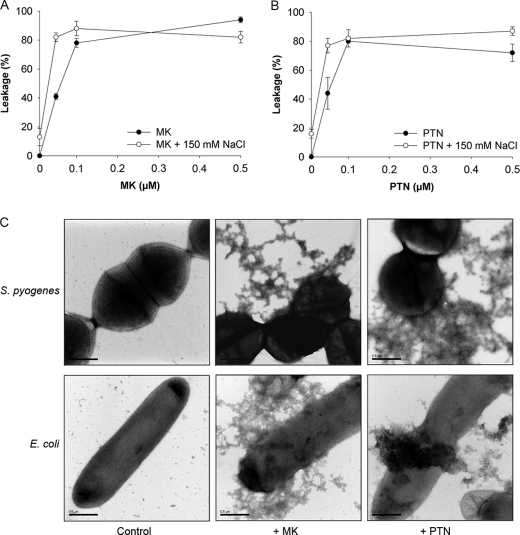
Many AMPs have a mode of action that includes disruption of the bacterial plasma membrane. To explore if this is the case for MK and PTN, model lipid bilayers were investigated in liposome leakage experiments. Addition of human recombinant MK and PTN caused leakage of fluorescent dye from micelles even at low concentrations of the proteins. Addition of sodium chloride at 150 mm did not affect the interaction among MK, PTN, and the lipid bilayers (Fig. 3, A & B). Electron microscopy was used to investigate effects on the bacterial morphology and integrity during the killing process. S. pyogenes and E. coli were incubated in either buffer alone, with MK or PTN (Fig. 3C). In analogy to the liposome results, bacteria incubated with MK and PTN, showed membrane protrusions and leakage of intracellular content, indicating damage to the bacterial membranes.
FIGURE 3.
Membrane-disrupting properties of human MK and PTN. A and B, membrane-disruptive effects of MK (A) and PTN (B) were analyzed by measuring release of carboxyfluorescein from liposomes in the absence or presence of sodium chloride (150 mm). A dose-dependent leakage, which was not affected by the presence of sodium chloride, was seen. C, ultrastructural changes of bacterial morphology after incubation with MK or PTN was investigated using electron microscopy. S. pyogenes and E. coli were incubated in buffer alone (control), and in the presence of MK or PTN (both proteins at concentrations of 0.15 μm in the case of S. pyogenes and 0.4 μm in the case of E. coli), respectively. The samples were fixed and processed for negative staining. The electron micrographs show intact bacteria after incubation in buffer (control) while incubation in the presence of MK and PTN caused bacterial disintegration with membrane blebbing and leakage of intracellular contents. Bar, 0.5 μm.
Homology and Structure
A comparison of full-length human MK and PTN proteins displayed an identity of 45% and a similarity of 61% (Fig. 4A). Considering that the cysteines are engaged in disulfide bonds, pI values of 10.4 and 10.2 were calculated for MK and PTN, respectively. The structures of MK and PTN have been characterized by nuclear magnetic resonance (NMR) and consist of two domains, each made up of three antiparallel β-sheets. However, final structure calculations were only made for MK (35, 41). A comparative homology model of PTN was made using MK as a template (Fig. 4B). The tail portions outside of the domains do not seem to form stable structures and thus move freely. The domains are stabilized by five disulfide bonds that, in the case of MK, probably provide the resistance to high temperature and acidic conditions (9, 42).
The secondary structures of MK and PTN are very similar, and it is apparent that the β-sheets of the N-terminal regions are very coherent (Fig. 4B). The structure of the C-terminal parts is more flexible than the N terminus, but still similar. Many AMPs have heparin binding properties, and heparin-binding sites of both MK and PTN have been determined (Fig. 4C) (43, 35). The critical heparin-binding site of both MK and PTN is found in the C-terminal region. However, parts of both the N and C termini are needed to form the complete binding site (43). The charge distribution is shown in the same figure (Fig. 4C), demonstrating that MK and PTN are quite similar in this respect, although PTN have more negatively charged residues in the tail than MK.
Antibacterial Domains of Human MK and PTN
The MK and PTN holoproteins are antibacterial (Fig. 1) but some regions within the molecules may be of particular importance. To determine whether the heparin-binding sites are involved in exerting the antibacterial activity, low molecular weight heparin was used to block the heparin-binding sites in MK and PTN. After preincubation with heparin at equimolar concentrations, both MK and PTN lost most of their antibacterial activity (Fig. 4D) demonstrating crucial importance for the heparin-binding sites in the antibacterial activity. To further map regions of the proteins that contain antibacterial activity, overlapping peptides, 20 amino acids in length, were synthesized. These were used to screen for antibacterial activity in other parts of the proteins by radial diffusion assay with E. coli (supplemental Fig. S1). The regions with the highest antibacterial activity were localized in the last β-strand in the N-terminal domain (peptides 5 and 6) in the middle β-strand of the C-terminal domain (peptide 9) coherent with the heparin-binding site. But activity was also found in two peptides derived from the C-terminal tail (peptides 11 and 12). The location of the regions with the highest antibacterial activity of MK and PTN are shown in Fig. 5A. The peptides displayed a similar pattern of activity with S. aureus using viable count assay (data not shown).
FIGURE 5.
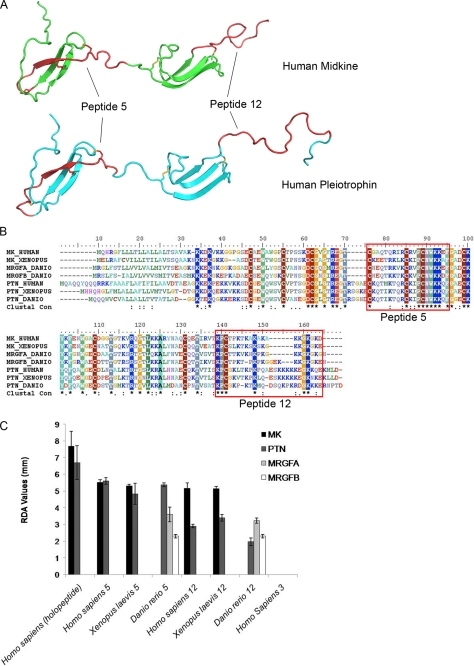
Antibacterial regions of human MK and PTN compared with orthologues from X. laevis and D. rerio. A, two regions corresponding to the highest antibacterial activity of human MK and PTN (supplemental Fig. S1) are indicated in the structural models (peptide 5: red in MK (upper) and PTN (lower); peptide 12: red in MK (upper) and in PTN (lower)). B, alignment of human MK and PTN and the corresponding orthologues from X. laevis and D. rerio. The location of peptides 5 and 12 is indicated. C, possible differences in antibacterial activity of peptides 5 and 12 was compared between species using a radial diffusion assay. Peptide 3 derived from MK and PTN, respectively, showed no activity in initial screening and thus served as negative controls. E. coli was grown to mid-logarithmic phase, dispersed in agarose that was allowed to solidify on Petri dishes. Wells were punched, and buffer alone or peptides (100 μm) were added to each well. Plates were incubated at 37 °C for 3 h to allow the peptides to diffuse. Antibacterial activity was seen as a clearing zone around each well after incubation for 18–24 h at 37 °C. Baseline value of a non-active peptide in the RDA is 0 mm. Experiments were done in triplicates, and results are shown as mean ± S.D.
Alignment and Phylogenetic Tree
To investigate homologies and possible conservation of antibacterial domains during evolution, sequence alignment of MK and PTN orthologues in the amphibian X. laevis and the fish D. rerio were compared with their human counterparts (Fig. 5B; identities and similarities are presented in supplemental Table S2). As a result of gene duplication, D. rerio has two closely related MK genes, Midkine-Related Factor A and B (MRGFA and MRGFB) that are distinct from PTN (44). Upon alignment of the mature proteins, the ten cysteine residues are perfectly conserved, suggesting that the overall structure is conserved among the species investigated. In addition, the highly cationic regions, with a content of lysines and arginines, also show a high degree of similarity.
Antibacterial Domains in Orthologues
MK and PTN orthologues were subsequently investigated for the presence of antibacterial activity to test whether this feature has been conserved evolutionarily. Peptides of 20 amino acids, with an overlap of 10 amino acids, from the different orthologues were synthesized. A distribution of antibacterial activity, similar to that observed in human MK and PTN, was found in the corresponding peptides of the orthologues proteins (Fig. 5C, supplemental Fig. S1). High antibacterial activity was seen in a lysine-rich region including cysteines 4–6 (Peptide 5) and in the C-terminal tail including cysteine 10 (Peptide 12) (Fig. 5, B & C). Synthetic peptides, corresponding to the regions of Peptides 5 and 12 of the MK and PTN orthologues were examined to compare their antibacterial activity using RDA. The observed antibacterial activity was almost equally high, when comparing peptide 5 and 12 of human and X. laevis whereas the corresponding peptides derived from the fish D. rerio were less active (Fig. 5C). In addition, antibacterial activity of the human peptides was assessed in the presence of sodium chloride at physiological concentrations (50–150 mm). PTN5 (10 μm) was almost unaffected by salt while the activity of MK12 decreased by 35% at 150 mm sodium chloride. The antibacterial activity of the other peptides decreased by ∼60% in the presence of 150 mm sodium chloride (supplemental Fig. S2, A, B, & D, E). The antibacterial activity of all peptides was abolished in the presence of plasma (1–30% plasma; data not shown). Using the liposome leakage assay, all peptides (MK12, PTN5, and PTN12) except the MK5-peptide, exerted strong membrane-disruptive activity also in the presence of sodium chloride (150 mm) (supplemental Fig. S2, C & F).
Phylogenetic Tree
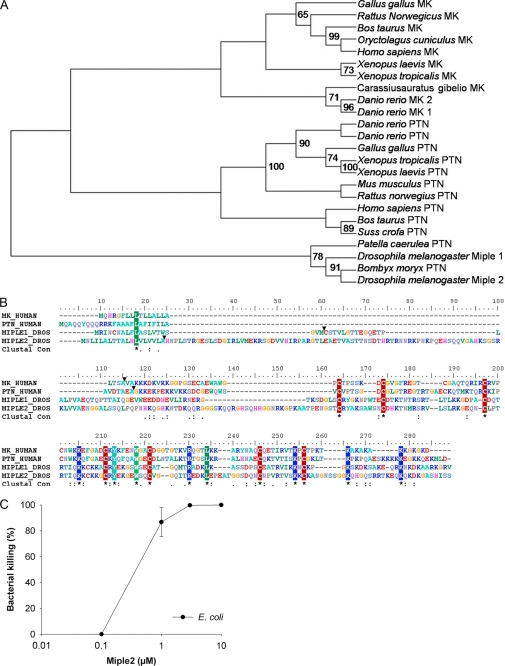
The most distant orthologues of MK and PTN are Miple1 and Miple2 of D. melanogaster. A phylogenetic tree was constructed using representative MK and PTN orthologues of invertebrates and vertebrates, yielding information about a common ancestor (Fig. 6A). During the evolution of vertebrates, MK and PTN are separated and belong to two different clades. The high degree of similarity between the two proteins suggests that they have originated from the same gene by gene duplication, but it is not evident which one has diverged and which one is the original (23).
FIGURE 6.
Evolution of MK and PTN. A, phylogenetic tree showing the evolutionary relationship of selected MK and PTN orthologues using the neighbor-joining method. Numbers on the branches indicate the reliability of each branch using 1,000 boot-strap replications. B, alignment of human MK and PTN sequences with Miple1 and Miple2 of D. melanogaster show complete conservation of the ten cysteine residues. Arrowheads indicate cleavage sites of signal peptides. C, antibacterial activity of Miple2 assessed by viable count assay. The number of cfu remaining after exposure to MK and PTN, respectively, were compared with the number of cfu obtained after incubation in buffer alone, to calculate % killing. The data shown represent mean ± S.D. from three separate experiments.
Antibacterial Activity of Drosophila Miple2
Investigation of sequence alignments of human MK and PTN with Miple1 and Miple2 of D. melanogaster, highlights conservation of all cysteine residues (Fig. 6B). To see if the early predecessors also display antibacterial activity, viable count assays using recombinant Miple2 against E. coli and S. aureus, were performed. These assays clearly demonstrate antibacterial activity of Miple2 against E. coli bacteria (Fig. 6C) but not against S. aureus at the same concentration range (data not shown). Thus, antibacterial activity of the MK/PTN family of heparin-binding growth factors is clearly evolutionarily preserved.
DISCUSSION
This study shows that the heparin-binding growth factors MK and PTN exert strong antibacterial activity against a panel of Gram-positive and Gram-negative bacteria, by disrupting the integrity of their plasma membranes. Interestingly, the activity was largely unaffected by the presence of 150 mm sodium chloride, a concentration equal to that of human plasma. Strong antibacterial activity was mapped to the unordered C-terminal tail and to the last β-sheet of the N termini of both molecules. We also demonstrate that the antibacterial activity is conserved in corresponding regions of MK and PTN orthologues, and that the holoprotein of the most distant orthologue, Drosophila Miple2 exerts strong antibacterial activity.
MK and PTN both contain two domains consisting of three antiparallel β-sheets that are held together by a hinge region. The β-defensins are an important family of AMPs that share structural similarities with MK and PTN. The β-defensins are cationic and have three anti-parallel β-sheets that are stabilized by disulfide-bonds between six cysteine residues, similar to the N-terminal domain of MK and PTN (45, 46). Additionally, the β-defensins have a short C-terminal amphipathic α-helix, containing antibacterial activity, which does not seem to be the case for neither MK nor PTN (47). Instead, the latter molecules have an unordered C termini with a high content of lysines that may attain a α-helical structure when inserted into a lipid bilayer, for example the bacterial plasma membrane. Another property in common with β-defensins is that, at least MK, dimerize in solution (46). Oligomerization can provide a more efficient exposure of antibacterial residues to the target organism, as demonstrated for β-defensins (48). Heparin binding properties, depending on the presence of specifically arranged lysines in Cardin-Weintraub motifs, is a common theme among antibacterial peptides (25). In the case of MK and PTN, the mapping experiments suggest that the heparin-binding region of the molecules has a high antibacterial activity. In this context, it should also be noted that the overlapping peptides obviously do not exactly reflect the complete three-dimensional structure and activity of the holoprotein. Indeed, as seen in the molecular models (Fig. 4C), MK and PTN both display an amphipathic character with distinct and separated cationic and hydrophobic “faces”, features typical of most AMPs. Taken together, the positioning of cationic amino acid residues is likely to promote the membrane-disrupting properties of AMPs, including MK and PTN.
Many AMPs can both act as growth factors and proinflammatory mediators, one example being the β-defensins (49, 7). Another example is the antibacterial fragment LL-37, derived from the human cathelicidin, which activates the receptor FPRL-1 on neutrophils and promotes angiogenesis (4, 5). Conversely, growth factors, such as HB-EGF, possess antibacterial properties, further illustrating this multifunctionality (50). MK has neutrophil-activating properties, although the receptor on neutrophils has not been characterized (8). Antibacterial chemokines constitute another family of proteins that both structurally and functionally resemble MK and PTN. Generally, these molecules consist of an N-terminal region with three antiparallel β-sheets and a C-terminal amphipathic α-helix (51). For example, the CC chemokine CCL20 and the CXC chemokine CXCL6, both have mitogenic- and leukocyte-activating properties, in addition to being antibacterial themselves (52–55).
Looking at the amino acid sequences, MK and PTN are highly conserved among vertebrates. The conserved positions of cysteines suggest that the structures of these molecules are very similar among vertebrates. Not surprisingly, mapping of the antimicrobial domains in orthologues of higher species (X. laevis and D. rerio) showed the same pattern, with the highest antibacterial activities in the last strand of the N-terminal β-sheet and in the unordered C-terminal regions. The C-terminal region is also the most conserved region among the orthologues.
Moreover, the antibacterial feature clearly exists in Miple2, an early MK/PTN orthologue of Drosophila, suggesting that the antibacterial properties of this family of molecules are ancient and highly conserved. Intriguingly, Miple2 is strongly expressed in the endodermal epithelium of the Drosophila gut, in agreement with a role as a potential AMP in vivo (23).
In D. rerio the function of MK differs from the function in humans, suggesting that there has been a functional diversification after a gene duplication resulting in two MK genes (44). In adult humans, MK expression is found in the small intestine, colon, kidney, and lung (56), whereas in the adult D. rerio, it is only found in the brain (44). In X. laevis, the expression (liver, kidney, and brain) is more similar to humans (57).
Keratinocytes of the human epidermis constitutively express MK, and it is possible that both MK and PTN accumulate in the limited interstitial volume present between cells, where high and antibacterial concentrations may be reached (9). In addition, the heparin binding properties of MK and PTN can provide an immobilized antibacterial gradient on the surface of epithelial cells.
A few inflammatory conditions have been investigated for MK expression in mice and humans. In humans, MK is present in plasma and increase during inflammatory bowel disease (17–19). An increased production of MK is also seen in meningitis where monocytes and other leukocytes may contribute to the accumulation of MK at sites of inflammation (18).
MK may play key roles in the pathogenesis of rheumatoid arthritis where increased levels are present in both serum and synovial fluid of patients suffering from the disease. In an arthritis model, using MK knock-out mice, mice seldom developed disease and these animals have a decreased recruitment of leukocytes to the inflammatory site (21). Thus, MK has inflammation responsive elements in its promoter-region, including NFκB, explaining expression at sites of inflammation (58).
In conclusion, these findings suggest that MK and PTN, in addition to their previously described activities, can serve important roles as innate antibiotics. Future investigation should reveal whether these properties of MK and PTN are functionally relevant in vivo.
Supplementary Material
Acknowledgments
We thank Pia Andersson, Maria Baumgarten, and Lise-Britt Wahlberg for excellent technical assistance and Fredrik Hugosson for providing recombinant Miple2.
This work was supported by grants from the Swedish Research Council (Project 20674), the Swedish Heart and Lung Foundation (Project 20080246), the Medical Faculty at Lund University, Swedish Government Funds for Clinical Research (ALF), and the Foundations of Bergh, Crafoord, Ihre, Hedberg, Kock, Marianne & Marcus Wallenberg, and Österlund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- AMP
- antimicrobial peptide
- MALDI-TOF
- matrix-assisted laser desorption/ionization-time of flight
- MK
- midkine
- PTN
- pleiotrophin
- TH
- Todd-Hewitt broth
- cfu
- colony forming units
- CF
- carboxyfluorescein
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phosphoglycerol, monosodium salt
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phoshoethanol-amine
- GST
- glutathione S-transferase
- RDA
- radial diffusion assay.
REFERENCES
- 1.Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 2.Brogden K. A. (2005) Nat. Rev. Microbiol. 3, 238–250 [DOI] [PubMed] [Google Scholar]
- 3.Elsbach P. (2003) J. Clin. Invest. 111, 1643–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Yang Chen Q., Schmidt A. P., Anderson G. M., Wang J. M., Wooters J., Oppenheim J. J., Chertov O. (2000) J. Exp. Med. 192, 1069–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koczulla R., von Degenfeld G., Kupatt C., Krötz F., Zahler S., Gloe T., Issbrücker K., Unterberger P., Zaiou M., Lebherz C., Karl A., Raake P., Pfosser A., Boekstegers P., Welsch U., Hiemstra P. S., Vogelmeier C., Gallo R. L., Clauss M., Bals R. (2003) J. Clin. Invest. 111, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole A. M., Ganz T., Liese A. M., Burdick M. D., Liu L., Strieter R. M. (2001) J. Immunol. 167, 623–627 [DOI] [PubMed] [Google Scholar]
- 7.Yang D., Chertov O., Bykovskaia S. N., Chen Q., Buffo M. J., Shogan J., Anderson M., Schröder J. M., Wang J. M., Howard O. M., Oppenheim J. J. (1999) Science 286, 525–528 [DOI] [PubMed] [Google Scholar]
- 8.Murphy C. J., Foster B. A., Mannis M. J., Selsted M. E., Reid T. W. (1993) J. Cell. Physiol. 155, 408–413 [DOI] [PubMed] [Google Scholar]
- 9.Muramatsu T. (2002) J. Biochem. 132, 359–371 [DOI] [PubMed] [Google Scholar]
- 10.Li Y. S., Milner P. G., Chauhan A. K., Watson M. A., Hoffman R. M., Kodner C. M., Milbrandt J., Deuel T. F. (1990) Science 250, 1690–1694 [DOI] [PubMed] [Google Scholar]
- 11.Deuel T. F., Zhang N., Yeh H. J., Silos-Santiago I., Wang Z. Y. (2002) Arch. Biochem. Biophys. 397, 162–171 [DOI] [PubMed] [Google Scholar]
- 12.Kadomatsu K., Tomomura M., Muramatsu T. (1988) Biochem. Biophys. Res. Commun. 151, 1312–1318 [DOI] [PubMed] [Google Scholar]
- 13.Tomomura M., Kadomatsu K., Matsubara S., Muramatsu T. (1990) J. Biol. Chem. 265, 10765–10770 [PubMed] [Google Scholar]
- 14.Rauvala H. (1989) EMBO J. 8, 2933–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milner P. G., Li Y. S., Hoffman R. M., Kodner C. M., Siegel N. R., Deuel T. F. (1989) Biochem. Biophys. Res. Commun. 165, 1096–1103 [DOI] [PubMed] [Google Scholar]
- 16.Muramatsu H., Zou P., Suzuki H., Oda Y., Chen G. Y., Sakaguchi N., Sakuma S., Maeda N., Noda M., Takada Y., Muramatsu T. (2004) J. Cell Sci. 117, 5405–5415 [DOI] [PubMed] [Google Scholar]
- 17.Lucas S., Henze G., Schnabel D., Barthlen W., Sakuma S., Kurtz A., Driever P. H. (2010) Pediatr Int. 52, 75–79 [DOI] [PubMed] [Google Scholar]
- 18.Yoshida Y., Ikematsu S., Muramatsu H., Sakakima H., Mizuma N., Matsuda F., Sonoda K., Umehara F., Ohkubo R., Matsuura E., Goto M., Osame M., Muramatsu T. (2008) Intern Med. 47, 83–89 [DOI] [PubMed] [Google Scholar]
- 19.Krzystek-Korpacka M., Neubauer K., Matusiewicz M. (2010) Inflamm. Bowel Dis. 16, 208–215 [DOI] [PubMed] [Google Scholar]
- 20.Takada T., Toriyama K., Muramatsu H., Song X. J., Torii S., Muramatsu T. (1997) J. Biochem. 122, 453–458 [DOI] [PubMed] [Google Scholar]
- 21.Maruyama K., Muramatsu H., Ishiguro N., Muramatsu T. (2004) Arthritis Rheum 50, 1420–1429 [DOI] [PubMed] [Google Scholar]
- 22.Palmer R. H., Vernersson E., Grabbe C., Hallberg B. (2009) Biochem. J. 420, 345–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Englund C., Birve A., Falileeva L., Grabbe C., Palmer R. H. (2006) Dev. Genes Evol. 216, 10–18 [DOI] [PubMed] [Google Scholar]
- 24.Lai Y., Gallo R. L. (2009) Trends Immunol. 30, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersson E., Rydengård V., Sonesson A., Mörgelin M., Björck L., Schmidtchen A. (2004) Eur. J. Biochem. 271, 1219–1226 [DOI] [PubMed] [Google Scholar]
- 26.Fabri L., Maruta H., Muramatsu H., Muramatsu T., Simpson R. J., Burgess A. W., Nice E. C. (1993) J. Chromatogr 646, 213–225 [DOI] [PubMed] [Google Scholar]
- 27.Yan W. K., Goette M., Hofmann G., Zaror I., Sim J. (2010) Protein Expr. Purif. 70, 270–276 [DOI] [PubMed] [Google Scholar]
- 28.Riddles P. W., Blakeley R. L., Zerner B. (1983) Methods Enzymol. 91, 49–60 [DOI] [PubMed] [Google Scholar]
- 29.Bengtson S. H., Sandén C., Morgelin M., Marx P. F., Olin A. I., Leeb-Lundberg L. M. F., Meijers J. C. M., Herwald H. (2009) J. Innate Immun. 1, 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 32.Henikoff S., Henikoff J. G. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poirot O., Suhre K., Abergel C., O'Toole E., Notredame C. (2004) Nucleic Acids Res. 32, W37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 35.Iwasaki W., Nagata K., Hatanaka H., Inui T., Kimura T., Muramatsu T., Yoshida K., Tasumi M., Inagaki F. (1997) EMBO J. 16, 6936–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Current Protocols in Bioinformatics, Chapter 5, Unit 56, John Wiley & Sons, Hoboken, NJ: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 38.Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. (1991) J. Immunol. Methods 137, 167–173 [DOI] [PubMed] [Google Scholar]
- 39.Ganz T. (2003) Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 40.Aps J. K., Martens L. C. (2005) Forensic Sci. Int. 150, 119–131 [DOI] [PubMed] [Google Scholar]
- 41.Kilpelainen I., Kaksonen M., Kinnunen T., Avikainen H., Fath M., Linhardt R. J., Raulo E., Rauvala H. (2000) J. Biol. Chem. 275, 13564–13570 [DOI] [PubMed] [Google Scholar]
- 42.Kojima S., Inui T., Kimura T., Sakakibara S., Muramatsu H., Amanuma H., Maruta H., Muramatsu T. (1995) Biochem. Biophys. Res. Commun. 206, 468–473 [DOI] [PubMed] [Google Scholar]
- 43.Raulo E., Tumova S., Pavlov I., Pekkanen M., Hienola A., Klankki E., Kalkkinen N., Taira T., Kilpelaïnen I., Rauvala H. (2005) J. Biol. Chem. 280, 41576–41583 [DOI] [PubMed] [Google Scholar]
- 44.Winkler C., Schafer M., Duschl J., Schartl M., Volff J. N. (2003) Genome Res. 13, 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoover D. M., Chertov O., Lubkowski J. (2001) J. Biol. Chem. 276, 39021–39026 [DOI] [PubMed] [Google Scholar]
- 46.Schibli D. J., Hunter H. N., Aseyev V., Starner T. D., Wiencek J. M., McCray P. B., Jr., Tack B. F., Vogel H. J. (2002) J. Biol. Chem. 277, 8279–8289 [DOI] [PubMed] [Google Scholar]
- 47.Krishnakumari V., Nagaraj R. (2008) Peptides 29, 7–14 [DOI] [PubMed] [Google Scholar]
- 48.Bai Y., Liu S., Jiang P., Zhou L., Li J., Tang C., Verma C., Mu Y., Beuerman R. W., Pervushin K. (2009) Biochemistry 48, 7229–7239 [DOI] [PubMed] [Google Scholar]
- 49.Niyonsaba F., Ushio H., Nakano N., Ng W., Sayama K., Hashimoto K., Nagaoka I., Okumura K., Ogawa H. (2007) J. Invest Dermatol. 127, 594–604 [DOI] [PubMed] [Google Scholar]
- 50.Malmsten M., Davoudi M., Walse B., Rydengård V., Pasupuleti M., Mörgelin M., Schmidtchen A. (2007) Growth Factors 25, 60–70 [DOI] [PubMed] [Google Scholar]
- 51.Viola A., Luster A. D. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 171–197 [DOI] [PubMed] [Google Scholar]
- 52.Keates S., Han X., Kelly C. P., Keates A. C. (2007) J. Immunol. 178, 8013–8021 [DOI] [PubMed] [Google Scholar]
- 53.Hoover D. M., Boulegue C., Yang D., Oppenheim J. J., Tucker K., Lu W., Lubkowski J. (2002) J. Biol. Chem. 277, 37647–37654 [DOI] [PubMed] [Google Scholar]
- 54.Gijsbers K., Gouwy M., Struyf S., Wuyts A., Proost P., Opdenakker G., Penninckx F., Ectors N., Geboes K., Van Damme J. (2005) Exp. Cell Res. 303, 331–342 [DOI] [PubMed] [Google Scholar]
- 55.Linge H. M., Collin M., Nordenfelt P., Mörgelin M., Malmsten M., Egesten A. (2008) Antimicrob. Agents Chemother. 52, 2599–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsutsui J., Kadomatsu K., Matsubara S., Nakagawara A., Hamanoue M., Takao S., Shimazu H., Ohi Y., Muramatsu T. (1993) Cancer Res. 53, 1281–1285 [PubMed] [Google Scholar]
- 57.Sekiguchi K., Yokota C., Asashima M., Kaname T., Fan Q. W., Muramatsu T., Kadomatsu K. (1995) J. Biochem. 118, 94–100 [DOI] [PubMed] [Google Scholar]
- 58.You Z., Dong Y., Kong X., Beckett L. A., Gandour-Edwards R., Melamed J. (2008) BMC Med. Genomics 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) The Proteomics Protocols Handbook, pp. 571–607, Humana Press, Totawa, NJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.