Abstract
The two estrogen receptor (ER) subforms, ERα and ERβ, are capable of forming DNA-binding homodimers and heterodimers. Although binding to DNA is thought to stabilize ER dimers, how ERα/α, ERβ/β, and ERα/β dimerization is regulated by DNA and the chaperone protein Hsp90 is poorly understood. Using our highly optimized bioluminescence resonance energy transfer assays in conjunction with assays for transcriptional activation of ERs, we determined that DNA binding appears to play a minor role in the stabilization of ER dimers, especially in the case of ERβ/β homodimers. These findings suggest that ER dimers form before they associate with chromatin and that DNA binding plays a minor role in stabilizing ER dimers. Additionally, although Hsp90 is essential for the proper dimerization of ERα/α and ERα/β, it is not required for the proper dimerization of ERβ/β. Despite this, Hsp90 is critical for the estrogen-dependent transcriptional activity of the ERβ/β homodimer. Thus, Hsp90 is implicated as an important regulator of distinct aspects of ERα and ERβ action.
Keywords: DNA-binding Protein, Estrogen, Nuclear Receptors, Transcription Factors, Transcriptional Regulation, Hsp90, Estrogen Receptor Alpha, Estrogen Receptor Beta, Heterodimers, Homodimers
Introduction
The biological functions of estrogens are transduced by two estrogen receptors (ERs),2 ERα and ERβ, which are members of the nuclear receptor (NR) superfamily. These two ER isoforms contain conserved domain structures. The ligand-binding domain (LBD) mediates ligand binding, receptor dimerization, and the receptor-mediated transactivation function of target genes upon ligand binding to the C-terminal activation function (AF-2) domain. The LBD of ERβ is only 60% homologous to that of ERα; these differences allow for the existence of subtype-specific ligands (1–5). Ligand binding to the LBDs of ERα and ERβ induces conformational changes within the receptor that influence its ability to homodimerize or heterodimerize and to recognize distinct DNA sequences in the promoter regions of target genes. The DNA-binding domains (DBDs) of ERα and ERβ also contain weak dimerization modules and are 96% conserved. Of the residues composing the zinc finger domains that directly recognize DNA, 100% of the residues are conserved. Thus, although ERα and ERβ homodimers have been found to regulate common target genes, their abilities to regulate distinct target genes likely account for their opposing cellular functions: ERα promotes cell proliferation, whereas ERβ inhibits it (6, 7). The ability of these receptors to regulate distinct gene sets despite the similarities in their DBDs is likely due to a multitude of factors, including interaction with cofactors or transcription factors and the chromatin architecture of the target gene.
Transcriptional regulation by ERs is tightly controlled by a variety of interacting partners, including chaperone proteins belonging to the heat shock protein family. Hsp90 is a molecular chaperone protein that regulates signal transduction by NRs and protein kinases. This molecular chaperone associates with the unliganded form of ERs as well as the androgen receptor, progesterone receptor, glucocorticoid receptor, and mineralocorticoid receptor (8–10). Ligand binding to each NR induces a conformational change within the receptor, causing its dissociation from Hsp90, leading to receptor dimerization, interaction with cofactors, DNA binding, and target gene activation. Thus, molecular chaperoning is an essential initial step in the tightly regulated process of ligand-dependent transcriptional control of ERs. In the ligand-free form, ERα is sequestered and held inactive by a large molecular complex organized around Hsp90, a p23 protein, and one immunophilin (9). This molecular complex stabilizes ERα, and inhibition of Hsp90 by chemical ligands targets the client ERα to ubiquitination and its subsequent 26 S proteasome-mediated degradation (11, 12). Whereas the interaction of ERα and other NRs with Hsp90 molecular chaperone complexes is well documented, substantially less data are available on the role of this molecular chaperone in transcriptional regulation by ERβ. Nevertheless, both ERα and ERβ have been shown to interact with Hsp90 in the absence of endogenous and exogenous estrogens, and exposure of cells to Hsp90 inhibitors results in proteasome-mediated degradation of both receptor isoforms (13). However, different Hsp90 ligands differentially affect the degradation of ERα and ERβ, suggesting that the stability of the receptor subtypes is differentially regulated by Hsp90 (13).
Upon ligand binding and dissociation from the Hsp90 molecular complex, ERα and ERβ may form DNA-binding homodimers or heterodimers depending on the context of the bound ligand. These estrogen-responsive DNA-binding sequences may be canonical estrogen response elements (EREs) with which ERs directly interact; however, ERs also activate the transcription of other DNA sequences via tethering to other transcription factors such as Sp1 and AP-1. Direct DNA binding to EREs is thought to stabilize ER dimers via the dimerization interface located within the DBD, although this dimerization interface is thought to be substantially weaker than the LBD dimerization interface (14–17). This weaker dimerization interface located within the DBD has been proposed to be a constitutive dimerization interface, and DNA binding in response to estrogen is thought to stabilize ER dimers via interactions between the two dimerization interfaces located in the DBD and LBD (17).
Because of the fact that the co-presence of ERα and ERβ results in a heterogeneous population of homodimers and heterodimers, many aspects of ERα/β heterodimer signaling and physiological effects have remained elusive. Using highly optimized bioluminescence resonance energy transfer (BRET) assays, we have shown previously that ERα is the dominant partner within a heterodimer in that ligand binding to ERα alone, and not ERβ, can induce heterodimerization (18). However, the role of Hsp90 in ERα/β heterodimer action has not been explored. Furthermore, despite the well recognized mechanism of ER signaling via interaction with the Hsp90 molecular chaperone complex, ligand binding, interaction with coactivators, and recognition of EREs to activate the transcription of target genes, the order in which these events occur in the cellular context remains controversial.
Another controversial area of study in the field of ER biology is whether ER monomers are preloaded on DNA with the Hsp90 complex in the absence of ligand or whether ERs associate with DNA as fully formed dimers. Previous studies have shown that when ER DBDs recognize half-site EREs in the promoter regions of target genes, the dimerization interface within the DBD stabilizes dimerized ERs on the DNA (19–21); however, the widely accepted notion that ER DBDs do not associate with DNA as monomers but rather only as dimers has been challenged by a few in vitro studies (17, 22) and thus remains ambiguous. Moreover, the role of molecular chaperoning by Hsp90 in ERβ estrogen-regulated transcriptional activity is poorly understood, and the sequential process in which estrogenic ligand binding to ERs causes their dissociation from Hsp90, homo- or heterodimerization, and the initiation of target gene transcription is completely unexplored in the case of the ERα/β heterodimer. The purpose of this study was to elucidate the role of the Hsp90 complex in ER homodimer and heterodimer formation as well as to decipher the role of DNA binding in the crucial initial step of homo- or heterodimerization prior to estrogen-dependent transcriptional activation. We have found that, although functional interaction with Hsp90 is essential for the transcriptional activity of both ERα and ERβ, the requirement of Hsp90 for dimerization is markedly different for different dimer pairs. Specifically, whereas ERα dimerization requires functional Hsp90, this molecular chaperone is not critical for the dimerization potential of ERβ. Hsp90 appears to play a less critical role in ERα/β heterodimerization than in ERα/α homodimerization, although the transcription of non-degraded ERα/α homodimers is able to remain active when Hsp90 is inhibited. Furthermore, ERs appear to associate with DNA after they are dimerized, and DNA recognition appears to play a minor role in stabilizing all three dimer pairs, especially in the case of the ERβ/β homodimer. This is in keeping with previous findings that ERβ/β homodimers maintain a high level of ligand-independent dimerization and transcriptional activation (23, 24). These results suggest that the mechanism of ligand-dependent transcriptional regulation by ERs for all three dimer pairs is shared at some steps but differs at other crucial steps.
EXPERIMENTAL PROCEDURES
Drugs and Inhibitors
17β-Estradiol (E2) was obtained from Sigma. 17-Dimethylaminoethylamino-17-demethoxygeldanamycin (17-DMAG) was a kind gift from the laboratory of Dr. Shannon Kenney (University of Wisconsin, Madison). 8-[(Benzylthio)methyl]-(7CI,8CI) (TPBM) was identified in a high throughput screen (25) performed at the University of Illinois using a library developed by K. Putt and P. Hergenrother (26–28) as well as the National Institutes of Health NCI Diversity Set.
Cell Culture and Transfection
HEK293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cell cultures were split 1:12 when they reached confluency (∼3 days). One day before transfection, HEK293 cells were seeded at a confluency of ∼50% in phenol red-free Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum stripped six times with charcoal and dextran (stripped fetal serum). For BRET assays, cells were transfected with 435 ng of total construct DNA using TurboFect transfection reagent (Fermentas) according to the manufacturer's instructions. For reporter gene assays, cells were transfected in batches with 2.5 ng of ERα alone, ERβ alone, or ERα + ERβ along with 50 ng of pTK-ERE-Luc plasmid and 15 ng of pCMX-β-gal per well and simultaneously seeded in phenol red-free Dulbecco's modified Eagle's medium + 5% stripped fetal serum in 48-well plates. For Western blots, HEK293 cells were transfected with 625 ng of each ER and treated with the indicated ligands in phenol red-free Dulbecco's modified Eagle's medium + 5% stripped fetal serum.
BRET Assays
HEK293 cells were either transfected with a single BRET fusion plasmid (pCMX-ERα-RLuc or pCMX-RLuc-ERβ) or cotransfected with Renilla luciferase (RLuc) and yellow fluorescent protein (YFP) BRET fusion plasmids (pCMX-ERα-RLuc + pCMX-YFP-ERβ for ERα/ERβ heterodimers, pCMX-ERα-RLuc + pCMX-ERα-YFP for ERα homodimers, or pCMX-RLuc-ERβ + pCMX-YFP-ERβ for ERβ homodimers) as described above. Empty expression vector pCMX-pL2 was used to keep the total amount of transfected DNA constant. Twenty-four hours after transfection, cells were trypsinized, counted, and resuspended in phosphate-buffered saline in quadruplicate at ≈50,000 cells/well of a 96-well white-bottom microplate. Cells were incubated with ligand for 1 h in the 96-well format unless alternative treatment times are indicated. For BRET assays that were treated with ligands for >1 h, ligands were diluted in medium, and cells were treated in batches in 6-well plates. The amount of dimethyl sulfoxide (DMSO) vehicle was held constant at 0.6%/well for 1-h treatments and 0.1% for longer time points treated in batches in medium. Cells transfected with pCMX-pL2, pCMX-ERα-RLuc, or pCMX-RLuc-ERβ alone were used as controls and incubated with DMSO under the same experimental conditions as the cotransfected conditions. Coelenterazine h (Promega) was added in phosphate-buffered saline at a final concentration of 5 μm, and 460- and 530-nm emission detection measurements were immediately taken at 0.1 s/wavelength read/well on a PerkinElmer Life Sciences VICTOR3 V plate reader. Although RLuc peaks in the presence of coelenterazine h at an emission wavelength of 470 nm, the closest filter available on the plate reader detected emission at 460 nm. The BRET ratio was calculated as described (29) (also see Fig. 1b).
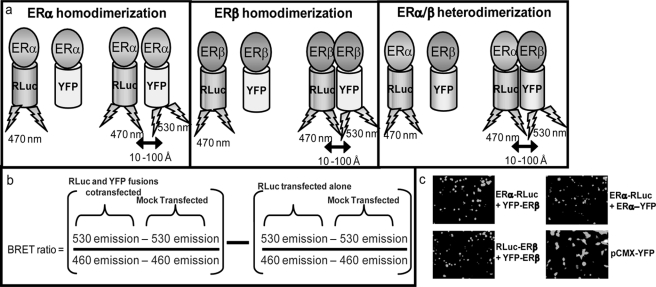
FIGURE 1.
BRET methodology. a, shown is a schematic representing ligand-dependent dimerization and resonance energy transfer between RLuc and YFP fusions via BRET. b, the BRET ratio is calculated as the ratio of YFP emission to RLuc emission with subtraction of the background and a correction factor for emission spectrum isolation. c, ERα and ERβ BRET fusions localize to the nucleus, in contrast with the cytoplasmic presence of pCMX-YFP.
Firefly Luciferase Assays
HEK293 cells were transfected in batches in 48-well plates using 2.5 ng of each indicated ER and 50 ng of pTK-ERE-Luc vector per well as described above. After allowing 48 h for protein expression and incubating with the indicated ligands for 24 h, cells were lysed, and firefly luciferase emission was detected upon the addition of the firefly luciferase substrate (Promega) on the VICTOR3 V plate reader using a luminescence detection setting. β-Galactosidase was analyzed using the Tropix β-galactosidase detection kit, and emission was detected on the VICTOR3 V plate reader using a luminescence detection setting. Luciferase counts were normalized to β-galactosidase counts in each well. For the side-by-side BRET/firefly luciferase reporter assays, increasing titrations (1, 0.1, 0.2, and 0.5 μg per well of 6-well plates) of the pERE-Luc reporter were cotransfected along with BRET fusions following the BRET transfection protocol described above. The total amount of DNA was kept constant per well using the pCMX-pL2 empty vector.
Western Blotting
HEK293 cells were transfected in 6-well plates as described above. After allowing 48 h for protein expression and treatment with the indicated ligands for the indicated times, cells were lysed, and total protein lysates was run on an 8% SDS-polyacrylamide gel. Protein was transferred to a nitrocellulose membrane, and ERα, ERβ, or β-actin (loading control) expression was detected using antibodies from Santa Cruz Biotechnology (HC-20 and H-150, for ERα and ERβ, respectively) and Sigma (A5441 for β-actin).
Co-immunoprecipitations
HEK293 cells were transiently transfected with ERα or ERβ (3 μg/well of each 10-cm dish). Twenty-four hours post-transfection, cells were treated with the indicated ligands by replacing the medium, and cells were lysed 24 h after treatment. Lysate was immunoprecipitated with antibody to endogenous Hsp90 (H-114, Santa Cruz Biotechnology). Immunoprecipitated lysates were then run on an SDS-PAGE system, and Western blotting was performed using an antibody to ERα (HC-20) or ERβ (H-150).
RESULTS
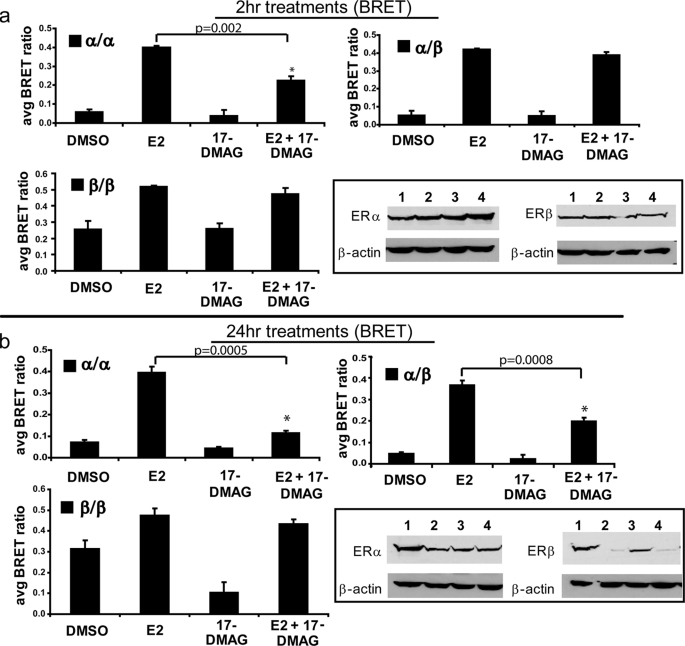
To elucidate the role of Hsp90 in the ligand-dependent dimerization of ERα and ERβ, we employed our highly optimized BRET assays (Fig. 1a) (18). This assay detects proximity between two proteins of interest fused to a donor (RLuc) and acceptor (YFP) protein (30–33). To eliminate interference dimerization of fusion constructs with endogenous ERs, the ER-negative cell line HEK293 was utilized. The high transfection efficiency (>90%) and low doubling time of this cell line also made it an attractive candidate for use in these studies. HEK293 cells were transfected with either ERα or ERβ fused to either RLuc or YFP. Thus, if a ligand was able to induce ER homodimerization or heterodimerization, depending on the fusion proteins cotransfected, RLuc would be brought into proximity with YFP. Upon the addition of the RLuc substrate coelenterazine h, which causes RLuc to emit at a peak wavelength of 470 nm, energy was transferred to YFP, exciting it at 515 nm and causing it to emit at a peak wavelength of 530 nm. Thus, YFP emission was indicative of dimerization, and the wavelength emissions of RLuc and YFP were used to calculate a BRET ratio to quantify the signal (Fig. 1b) (see Ref. 29 for a more detailed explanation). We have previously characterized and optimized this assay and all BRET fusion constructs used (18); nuclear localization of constructs is shown in Fig. 1c. HEK293 cells transfected with ERα and ERβ fusions to YFP and RLuc were exposed to the Hsp90 inhibitor 17-DMAG, which is a synthetic analog of the Hsp90 inhibitor geldanamycin. Geldanamycin and its synthetic analogs bind with high affinity to the ATP-binding pocket of Hsp90, inhibiting its function as a molecular chaperone for NRs. A 2-h treatment with 17-DMAG has been shown previously to be sufficient to release ERs from the Hsp90 complex, but ERs are not yet degraded at this time point (Fig. 2a, inset) (34). Western blots were confirmed quantitatively by detecting emission of the RLuc and YFP BRET fusions to ER in the presence of coelenterazine h (for RLuc emission) or YFP excitation (for YFP emission) (data not shown). BRET assays were used to directly examine the dimerization of ERα/α, ERβ/β, and ERα/β (Fig. 2, a and b); 10 nm E2 induced all three dimer pairs, whereas 212 nm 17-DMAG alone did not influence the background BRET ratio for any dimer pair at this time point. The E2-induced dimerization of ERα/α, ERβ/β, and ERα/β was statically significant in both the absence (compare DMSO versus E2) and presence (compare 17-DMAG versus E2 + 17-DMAG) of the Hsp90 inhibitor. However, the combination of 212 nm 17-DMAG and 10 nm E2 resulted in a statistically significant decrease (p = 0.002) in E2-mediated ERα/α homodimerization after 2 h of treatment (Fig. 2a, upper panel), which was not a sufficient amount of time for 17-DMAG-mediated ER degradation (Fig. 2a, inset). We therefore conclude that ERα/α homodimerization is heavily dependent upon the interaction of ERα monomers with the Hsp90 molecular chaperone complex. In contrast, ERα/β heterodimerization is influenced to a lesser, statistically non-significant extent by Hsp90 inhibition (p = 0.09) than is ERα homodimerization, suggesting that these heterodimers may be less dependent on molecular chaperoning by Hsp90 for their dimerization potential. Similar to ERα/β heterodimers, ERβ/β homodimers were minimally influenced by Hsp90 disruption with a 2-h co-treatment of 212 nm 17-DMAG and 10 nm E2, and this decrease was not statistically significant (p = 0.12). These data suggest that there are inherent differences in the influence of Hsp90 on ER dimer pairs, with ERα/α homodimers being the most heavily dependent on Hsp90 for its dimerization potential.
FIGURE 2.
Hsp90 is important to the function of all three dimer pairs but plays a minor role in ERβ homodimerization. a, co-treatment with the Hsp90 inhibitor 17-DMAG for 2 h, which did not induce ER degradation (inset), reduced E2-dependent dimerization of ERα/α homodimers, although ERβ/β homodimers and ERα/β heterodimers were minimally affected. b, co-treatment with 17-DMAG overnight, which induced ER degradation (inset), reduced E2-dependent dimerization of ERα/α homodimers and ERα/β heterodimers, although ERβ/β homodimers were minimally affected. Error bars represent mean ± S.D. *, statistically significant decrease in dimerization compared with E2 alone. Inset, lane 1, DMSO; lane 2, 10 nm E2; lane 3, 212 nm 17-DMAG; lane 4, 10 nm E2 + 212 nm 17-DMAG.
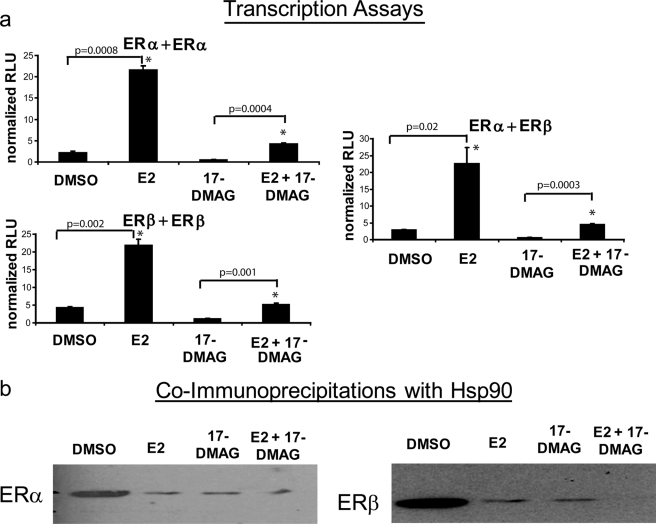
Transfection of HEK293 cells with BRET fusions to ER and co-treatment with E2, 17-DMAG, or the combination of E2 + 17-DMAG for 24 h led to ER degradation (Fig. 2b, inset) (13). Degradation of ERs via Western blotting was confirmed quantitatively by detecting emission of the RLuc and YFP BRET fusions to ER in the presence of coelenterazine h (for RLuc emission) or YFP excitation (for YFP emission) (data not shown). As shown in Fig. 2b, a statistically significant decrease in ERα/α homodimerization (p = 0.0005) occurred with a 24-h co-treatment of E2 + 17-DMAG compared with E2 alone, suggesting that the critical Hsp90 transcriptional regulation of ERα homodimers occurs at the initial dimerization step, and this is likely due to the combination of initial ERα/α homodimer abrogation and proteasomal degradation of ERα at this time point. In contrast, a 24-h co-treatment did not drastically reduce ERβ/β homodimerization (p = 0.06), despite the fact that ERβ was partially degraded at this time point (Fig. 2b, inset) (13). This is in keeping with previous findings that ERβ maintains a high level of ligand-independent dimerization (18) and transcriptional activity (23, 24). Because these ligand-independent ERβ/β homodimers are not predicted to be associated with Hsp90, the total population of ERβ/β homodimers is less influenced by Hsp90 inhibition, despite degradation of the ERβ subpopulation that is associated with the chaperone complex. The intermediate statistically significant decrease in ERα/β heterodimerization (p = 0.0008) upon a 24-h co-treatment with E2 and 17-DMAG is likely due to ERα degradation at this time point, which is in keeping with our previous finding that ERα is the dominant heterodimeric partner (18). The decrease in the BRET signal in the presence of 17-DMAG alone compared with the vehicle DMSO is likely due to degradation of these receptors. The transcriptional activity of ERs was significantly decreased by a 24-h treatment with 17-DMAG (p = 0.001, 0.002, and 0.02 for ERα + ERα, ERβ + ERβ, and ERα + ERβ, respectively) (Fig. 3a). However, when comparing DMSO to E2 as well as 17-DMAG to 17-DMAG + E2, the increases in transcriptional activity were statistically significant (p < 0.05), and we thus cannot exclude the possibility that the reduced signal was due to ERα/α degradation resulting from 17-DMAG addition. Indeed, the fold induction of DMSO to E2 and 17-DMAG to 17-DMAG + E2 was similar (9.5-fold versus 7.4-fold, respectively, for ERα/α homodimer transcriptional activity). This is likely the result of transcriptional activity of non-degraded ERs in the presence of the Hsp90 inhibitor. Co-immunoprecipitation experiments showed that E2 and the Hsp90 inhibitor 17-DMAG, as well as the combination of these ligands, disrupted the ER-Hsp90 interaction (Fig. 3b). Taken together, these results indicate that the contribution of the Hsp90 molecular chaperone complex differentially regulates ERs at the initial dimerization step.
FIGURE 3.
a, Hsp90 inhibition ablated the transcriptional activity of ERα and ERβ. Overnight 17-DMAG treatment inhibited E2-mediated activation of a pERE-Luc reporter in the presence of all three dimer pairs. Error bars represent mean ± S.D. *, statistically significant increase in transcriptional activity compared with DMSO (left bars) or 17-DMAG alone (right bars). RLU, relative luciferase units. b, co-immunoprecipitations showed that Hsp90 interacted with ERα (left panel) and ERβ (right panel), and this interaction was disrupted by treatment with 10 nm E2, 250 nm 17-DMAG, and the combination of both ligands.
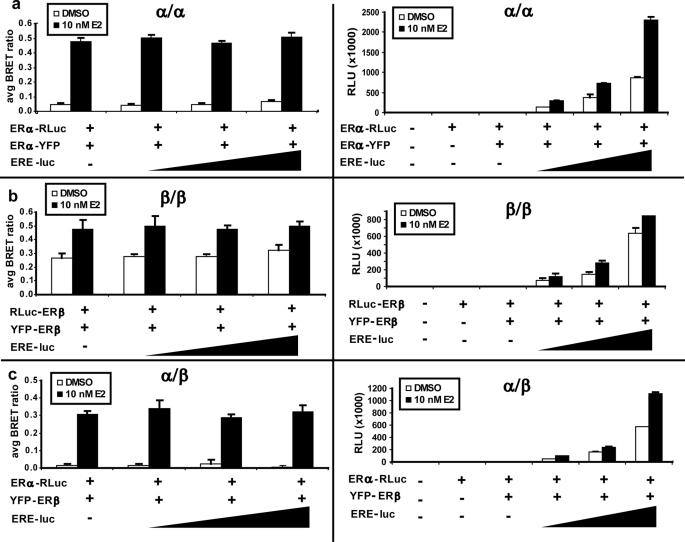
This initial upstream dimerization step is critical to the transcriptional activity of ERs on the regulatory regions of target genes. To elucidate the contribution of DNA binding to this critical process, we first performed BRET assays using a cotransfected pERE-Luc plasmid (Fig. 4). Fig. 4 (right panels) shows that the ERE-luciferase element was transcriptionally activated by cotransfected ER BRET fusions in an estrogen-dependent manner (p < 0.05). Fig. 4 (left panels) shows that the presence of the ERE had a negligible effect on the BRET signal (p > 0.05), indicating that DNA binding has a minor effect on dimerization. However, it is possible that the transfected EREs do not compete with binding of ER fusion proteins to chromatinized EREs of target genes.
FIGURE 4.
DNA binding minimally influences the BRET ratio. ERα homodimer (a), ERβ homodimer (b), and ERα/β heterodimer (c) BRET ratios were minimally increased by the addition of ERE-luciferase elements by transient transfection in 6-well plates. Left panels, dimerization via BRET assays; right panels, estrogen-dependent transcriptional activation of a pERE-luciferase reporter gene by BRET fusion constructs. Error bars represent mean ± S.D. RLU, relative luciferase units.
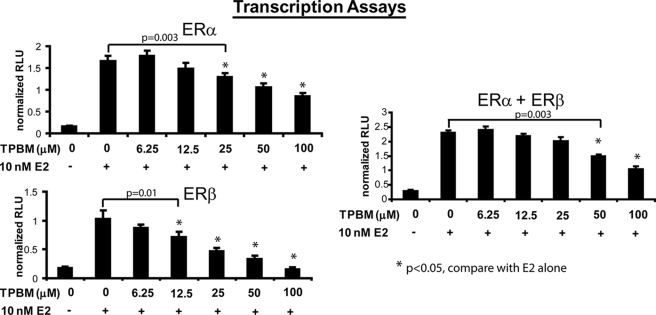
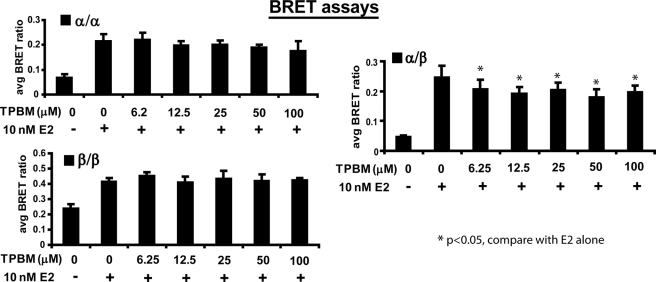
Therefore, to further examine the effect of DNA binding to dimerization, we treated cells with the compound theophylline (TPBM), which disrupts the ability of ERs to bind DNA (25) and hence their transcriptional activity (Fig. 5). Using this assay, concentrations of TPBM gave statistically significant decreases in transcriptional activity at a lower limit of 25 μm for ERα + ERα, 12.5 μm for ERβ + ERβ, and 50 μm for ERα + ERβ. TPBM was identified in a high throughput screen for the identification of inhibitors of ERα transcriptional activity. TPBM inhibits the growth of estrogen-dependent cell lines, does not bind to the ligand-binding pocket of ERs, and does not chelate ERα zinc fingers (25). As shown in Fig. 6, disruption of DNA binding by TPBM treatment minimally destabilized ER dimer pairs, as all treatments and conditions yielded a statistically significant increase in dimerization compared with the vehicle DMSO for all dimer pairs (p < 0.05). However, when comparing each co-treatment of TPBM with E2, statistically significant decreases were observed in ERα/β heterodimerization compared with E2 alone (p < 0.05), whereas miniscule decreases in ERα/α and ERβ/β homodimerization were not statistically significant (p > 0.05). This was particularly pronounced in the case of ERβ/β homodimers, which appeared to be completely unaffected, despite the finding that the transcriptional activity of ERβ was maximally disrupted by TPBM treatment (compare Figs. 5 and 6). From these data, it appears that disruption of the ability of ERα and ERβ to bind DNA does not drastically influence their dimerization potential.
FIGURE 5.
TPBM abrogates the transcriptional activity of ERs. Increasing concentrations of TPBM ablated the ability of ERα and ERβ to activate a pERE-luciferase reporter gene. Error bars represent mean ± S.D. *, statistically significant decrease in transcriptional activity compared with E2 alone. RLU, relative luciferase units.
FIGURE 6.
Disruption of DNA binding with TPBM slightly impairs dimerization for all three ER dimer combinations. ERβ/β homodimers were minimally disrupted by TPBM despite its marked ability to abrogate ERβ activation of an ERE-luciferase element. Error bars represent mean ± S.D. *, statistically significant decrease in dimerization compared with E2 alone.
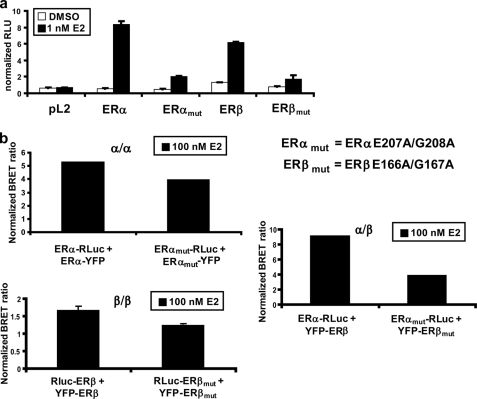
To examine more directly the contribution of DNA binding to dimerization, we constructed mutants of the DBD within ERα and ERβ. Specifically, residues 207 and 208 of ERα and the corresponding residues of ERβ (residues 166 and 167) were mutated to alanine to give the respective mutants ERα(E207A/G208A) and ERβ(E166A/G167A). These mutations have been characterized previously to eliminate recognition by ERs of an ERE (35, 36). In our experiments, these mutations reduced the ability of ERα and ERβ to bind DNA and activate an ERE-luciferase element to <25% of the wild type (Fig. 7a). To ensure equal expression levels of these mutants compared with wild-type receptors, equal amounts of DNA encoding each fusion protein were transfected alone. Relative receptor levels were determined by measuring RLuc emission in the presence of coelenterazine h or YFP emission upon excitation at 515 nm (data not shown). BRET assays revealed that ERα/α homodimers and ERβ/β homodimers were minimally influenced by the reduced ability of ERs to bind to DNA, and this reduction was not statistically significant (p > 0.05); in contrast, the ERα/β heterodimers were affected by 50%, which is a statistically significant difference (p = 0.042) (Fig. 7b). This significant decrease in ERα/β heterodimerization agrees with the findings in Fig. 6, which showed that ERα/β heterodimerization was significantly disrupted by treatment with TPBM. However, the decreases in heterodimerization observed in Fig. 7 with the DBD mutants were more pronounced than the decreases observed in Fig. 6 with TPBM. Although statistically congruent, these differences may be explained by inherent characteristics of the BRET assay, which detects conformational changes within a dimerized unit. The DBDs of ERα and ERβ contain a dimerization interface, and changes in dimer conformations can alter the placement of the RLuc and YFP fusions within the dimer, thus influencing the BRET ratio. Perhaps mutation of residues that mediate ERE recognition influences the dimerized conformations of ERα/β heterodimers differently than ER homodimers. For ER homodimers, the BRET signal is minimally reduced compared with the drastically reduced ability of ERs to bind DNA, which suggests that DNA binding does not strongly influence homodimerization. However, the stronger effects of mutations on ERα/β heterodimers detected in BRET assays imply that the dimerization interfaces of ER homodimers may be different from those of heterodimers, in which case the DNA-binding residues could be directly involved in heterodimerization.
FIGURE 7.
Disruption of DNA binding minimally influences the dimerization potential of ERs. a, mutation of specific residues within the DBD nearly completely ablated the ability of ERs to bind DNA. b, mutational ablation of DNA binding slightly decreased the BRET ratio for all three dimer pairs but had a greater impact on ERα/β heterodimerization. Error bars represent mean ± S.D. *, statistically significant decrease in dimerization compared with both wild-type receptors. RLU, relative luciferase units.
DISCUSSION
The three ER dimer pairs exhibit diverse biological profiles in the presence of different ligands and DNA sequence elements; thus, both dimerization and DNA binding are critical steps determining the transcriptional outcome and cellular response to endogenous and exogenous estrogenic ligands. This highly regulated process is kept silent in the absence of ligand via the interaction of ERs with the molecular chaperone protein Hsp90, which holds ERs poised for ligand binding but prevents both their activation and degradation. The influence of molecular chaperoning by Hsp90 on ERβ is poorly understood, and the stepwise process in which ligands bind to ERs to cause their dissociation from Hsp90, homo- or heterodimerization, and initiation of gene transcription on select DNA sequences is completely unexplored in the case of the ERα/β heterodimer. This study sought to analyze the upstream contribution of Hsp90, as well as the downstream involvement of DNA binding, to ER dimerization and transcriptional output. Using our highly optimized BRET assays, we determined that Hsp90 is intimately involved in the formation of the ERα/α homodimer and plays a lesser but still significant role in the formation of the ERα/β heterodimer. Disruption of the Hsp90 molecular chaperone complex is minimally disruptive to ERβ/β homodimers, which is in keeping with previous findings that ERβ/β homodimers maintain a high level of ligand-independent dimerization and transcriptional activity (23, 24). Unliganded ERβ appears to exist in two pools: a homodimerized pool and a separate subset that is associated with Hsp90. We therefore hypothesize that transient dissociations of ligand-independent ERβ/β homodimers may be able to donate an ERβ molecule to the ERα/β heterodimeric complex. Indeed, the formation of ERα/β heterodimers is predicted to be favored over the formation of either homodimer (37). The intermediate effect of Hsp90 on ERα/β heterodimerization is likely reflective of each partner's contribution to the dimer. The finding that decreases in heterodimerization are most drastically observed when ERα is degraded is consistent with our previous finding that ERα is the dominant heterodimeric partner; likewise, the fact that heterodimerization is not abrogated to the same extent as is ERα/α homodimerization is consistent with our previous finding that ERβ also plays an important role in heterodimerization. Taken together, these findings indicate that Hsp90 is required for the proper physiological response of all three ER dimer pairs. However, the effect of Hsp90 regulation of ERs appears to differ among the three dimer pairs at the initial upstream dimerization step. The differential requirement for Hsp90 in the formation of ER homodimers and heterodimers implicates it as an important regulator for cellular programs controlled by both ERα and ERβ.
Thus far, hormonal therapies have been directed at blocking ERα with selective ER modulators such as tamoxifen or reducing the synthesis of endogenous estrogens in postmenopausal women with aromatase inhibitors such as Anastrozole. However, these hormonal strategies are thwarted by the resistance that is often acquired against tamoxifen as well as the undesirable side effects of down-regulating the opposing cellular functions of both ERα and ERβ in all estrogen-responsive tissues. A more recent therapeutic approach has been to target the Hsp90 molecular chaperone complex as a means of disrupting ER stability and function; this approach addresses the problem of tamoxifen resistance and tissue specificity with aromatase inhibitors. The geldanamycin analog allylamino-17-demethoxygeldanamycin has been shown to be effective at micromolar concentrations without overt toxicity in Phase I clinical trials (38, 39). This drug is now in Phase II clinical trials (40), and 17-DMAG has entered into a Phase I clinical trial (41). Thus, because of its regulation of ERs, Hsp90 is a promising new therapeutic target for the management of hormonally regulated tumors such as breast cancer. We speculate that these Hsp90 inhibitors may be especially advantageous for therapeutically targeting breast cancers that express ERβ, as the subpopulation of ligand-independent ERβ homodimers will likely not be targeted by this drug. Thus, these ligand-independent ERβ homodimers will remain active to mediate their growth inhibitory anti-apoptotic effects.
Furthermore, using increasing titrations of ERE DNA sequences, the chemical compound TPBM (which disrupts ER-DNA interactions), and ER DBD mutants, we found that DNA binding appears to play a minor role in the stabilization of ER dimers, and the BRET assays therefore allow the detection of both “free” and DNA-bound dimers. Because DNA binding has a minor effect on ER dimers, we predict that the majority of dimers detected by BRET assays are not bound by DNA and that these dimers form prior to their association with chromatin. Among the three dimer pairs, ERβ/β homodimers appear to be least dependent upon DNA binding for dimer formation, which is consistent with their ability to form dimers independently of ligand. Thus, we surmise that ERβ binding to DNA makes no significant contribution to ERβ/β homodimerization, and ERβ might maintain a high level of free dimers that are not necessarily transcriptionally active on EREs. Whereas ERα/β heterodimers are similar to homodimers in that DNA binding has a minimal impact on their stabilization, it appears from our BRET assays with mutagenized ERs that the ERα/β heterodimer conformation induced upon ligand binding may utilize different residues from those employed by either homodimer. Indeed, the recent co-crystallization of full-length retinoid X receptor/peroxisome proliferator-activated receptor heterodimers has shown that, in addition to the LBD and DBD dimerization interfaces, a third dimerization interface exists within the hinge region (42). Given the similarities in NR structures, it is likely that ER dimers exhibit dimerization structures similar to this. We therefore speculate that subtle differences in the interactions among these three putative dimerization interfaces in the presence of bound ligand could elicit different conformations in the three dimer pairs. Our finding that mutation of the residues that mediate contacts with DNA changes the BRET ratio for ERα/β heterodimers compared with either homodimer suggests that these residues may be more directly involved in ERα/β heterodimerization than in homodimerization. Therefore, we speculate that subtle differences in ERα/β heterodimer conformation may influence differential interaction with cofactors, ultimately resulting in a biological response that is unique from that of either homodimer.
Previous studies using chromatin immunoprecipitation have shown that unliganded ER is not prebound to EREs; however, this technology does not address whether E2-bound ER is released from the Hsp90 complex to rapidly bind DNA as monomers or whether ERs dimerize prior to binding DNA and associate with DNA as stable dimers. Chromatin immunoprecipitation is limited by the ability of an antibody to recognize its target epitope, and these inherent limitations could therefore prevent antibody recognition of ER within the Hsp90 complex. Because high quality chromatin immunoprecipitation-grade ERβ antibodies are currently unavailable, this technology is not useful for accurately examining whether Hsp90 is recruited with ERβ at the promoters of target genes. Our BRET assays lend new insight into the controversial stepwise process of ligand binding to ER, its dissociation from the Hsp90 complex, dimerization, and recognition of DNA in that this novel technology circumvents the need for conventional antibodies used in chromatin immunoprecipitation and instead directly examines ER dimer formation in live cells in real time. We have found that ER dimer pairs are minimally influenced by DNA binding, and this is especially true in the case of the ERβ/β homodimer. Taken together, these results suggest that the critical initial step of ER dimerization occurs prior to DNA binding, and the majority of the dimers observed in BRET assays are not associated with chromatin. Therefore, DNA binding appears to play a minor role in the stabilization of dimers. This observation is consistent with the notion that the DBD serves as a weak dimerization module.
Acknowledgments
We thank Janet Mertz for suggestions, the Shannon Kenney laboratory for providing Hsp90 inhibitors, and Elaine Alarid for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA125387 and RO3 MH089442 (to W. X.), RO1 DK071909 (to D. J. S.), and T32 CA009135 (to E. P.).
- ER
- estrogen receptor
- NR
- nuclear receptor
- LBD
- ligand-binding domain
- DBD
- DNA-binding domain
- ERE
- estrogen response element
- BRET
- bioluminescence resonance energy transfer
- E2
- 17β-estradiol
- 17-DMAG
- 17-dimethylaminoethylamino-17-demethoxygeldanamycin
- TPBM
- 8-[(benzylthio)methyl]-(7CI,8CI)
- RLuc
- Renilla luciferase
- YFP
- yellow fluorescent protein
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Shiau A. K., Barstad D., Radek J. T., Meyers M. J., Nettles K. W., Katzenellenbogen B. S., Katzenellenbogen J. A., Agard D. A., Greene G. L. (2002) Nat. Struct. Biol. 9, 359–364 [DOI] [PubMed] [Google Scholar]
- 2.Sun J., Meyers M. J., Fink B. E., Rajendran R., Katzenellenbogen J. A., Katzenellenbogen B. S. (1999) Endocrinology 140, 800–804 [DOI] [PubMed] [Google Scholar]
- 3.Saji S., Hirose M., Toi M. (2005) Cancer Chemother. Pharmacol. 56, Suppl. 1, 21–26 [DOI] [PubMed] [Google Scholar]
- 4.Stauffer S. R., Coletta C. J., Tedesco R., Nishiguchi G., Carlson K., Sun J., Katzenellenbogen B. S., Katzenellenbogen J. A. (2000) J. Med. Chem. 43, 4934–4947 [DOI] [PubMed] [Google Scholar]
- 5.Meyers M. J., Sun J., Carlson K. E., Marriner G. A., Katzenellenbogen B. S., Katzenellenbogen J. A. (2001) J. Med. Chem. 44, 4230–4251 [DOI] [PubMed] [Google Scholar]
- 6.Helguero L. A., Faulds M. H., Gustafsson J. A., Haldosén L. A. (2005) Oncogene 24, 6605–6616 [DOI] [PubMed] [Google Scholar]
- 7.Chang E. C., Frasor J., Komm B., Katzenellenbogen B. S. (2006) Endocrinology 147, 4831–4842 [DOI] [PubMed] [Google Scholar]
- 8.Fang Y., Fliss A. E., Robins D. M., Caplan A. J. (1996) J. Biol. Chem. 271, 28697–28702 [DOI] [PubMed] [Google Scholar]
- 9.Johnson J. L., Toft D. O. (1994) J. Biol. Chem. 269, 24989–24993 [PubMed] [Google Scholar]
- 10.Drysdale M. J., Brough P. A., Massey A., Jensen M. R., Schoepfer J. (2006) Curr. Opin. Drug Discov. Dev. 9, 483–495 [PubMed] [Google Scholar]
- 11.Roe S. M.., Prodromou C., O'Brien R., Ladbury J. E., Piper P. W., Pearl L. H. (1999) J. Med. Chem. 42, 260–266 [DOI] [PubMed] [Google Scholar]
- 12.Maloney A., Workman P. (2002) Expert Opin. Biol. Ther. 2, 3–24 [DOI] [PubMed] [Google Scholar]
- 13.Gougelet A., Bouclier C., Marsaud V., Maillard S., Mueller S. O., Korach K. S., Renoir J. M. (2005) J. Steroid Biochem. Mol. Biol. 94, 71–81 [DOI] [PubMed] [Google Scholar]
- 14.Kuntz M. A., Shapiro D. J. (1997) J. Biol. Chem. 272, 27949–27956 [DOI] [PubMed] [Google Scholar]
- 15.White R., Fawell S. E., Parker M. G. (1991) J. Steroid Biochem. Mol. Biol. 40, 333–341 [DOI] [PubMed] [Google Scholar]
- 16.Schwabe J. W., Chapman L., Finch J. T., Rhodes D. (1993) Cell 75, 567–578 [DOI] [PubMed] [Google Scholar]
- 17.Kumar V., Chambon P. (1988) Cell 55, 145–156 [DOI] [PubMed] [Google Scholar]
- 18.Powell E., Xu W. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19012–19017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green S., Kumar V., Theulaz I., Wahli W., Chambon P. (1988) EMBO J. 7, 3037–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green S., Chambon P. (1989) Cancer Res. 49, 2282s–2285s [PubMed] [Google Scholar]
- 21.Hirst M. A., Hinck L., Danielsen M., Ringold G. M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5527–5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murdoch F. E., Byrne L. M., Ariazi E. A., Furlow J. D., Meier D. A., Gorski J. (1995) Biochemistry 34, 9144–9150 [DOI] [PubMed] [Google Scholar]
- 23.Tremblay A., Tremblay G. B., Labrie F., Giguère V. (1999) Mol. Cell 3, 513–519 [DOI] [PubMed] [Google Scholar]
- 24.Tremblay G. B., Tremblay A., Copeland N. G., Gilbert D. J., Jenkins N. A., Labrie F., Giguère V. (1997) Mol. Endocrinol. 11, 353–365 [DOI] [PubMed] [Google Scholar]
- 25.Mao C., Patterson N. M., Cherian M. T., Aninye I. O., Zhang C., Montoya J. B., Cheng J., Putt K. S., Hergenrother P. J., Wilson E. M., Nardulli A. M., Nordeen S. K., Shapiro D. J. (2008) J. Biol. Chem. 283, 12819–12830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Putt K. S., Chen G. W., Pearson J. M., Sandhorst J. S., Hoagland M. S., Kwon J. T., Hwang S. K., Jin H., Churchwell M. I., Cho M. H., Doerge D. R., Helferich W. G., Hergenrother P. J. (2006) Nat. Chem. Biol. 2, 543–550 [DOI] [PubMed] [Google Scholar]
- 27.Putt K. S., Hergenrother P. J. (2004) Anal. Biochem. 333, 256–264 [DOI] [PubMed] [Google Scholar]
- 28.Hergenrother P. J. (2006) Curr. Opin. Chem. Biol. 10, 213–218 [DOI] [PubMed] [Google Scholar]
- 29.Pfleger K. D., Eidne K. A. (2006) Nat. Methods 3, 165–174 [DOI] [PubMed] [Google Scholar]
- 30.Bacart J., Corbel C., Jockers R., Bach S., Couturier C. (2008) Biotechnol. J. 3, 311–324 [DOI] [PubMed] [Google Scholar]
- 31.Koterba K. L., Rowan B. G. (2006) Nuclear Recept. Signal. 4, e021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michelini E., Mirasoli M., Karp M., Virta M., Roda A. (2004) Anal. Chem. 76, 7069–7076 [DOI] [PubMed] [Google Scholar]
- 33.Xu Y., Piston D. W., Johnson C. H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beliakoff J., Bagatell R., Paine-Murrieta G., Taylor C. W., Lykkesfeldt A. E., Whitesell L. (2003) Clin. Cancer Res. 9, 4961–4971 [PubMed] [Google Scholar]
- 35.Jakacka M., Ito M., Weiss J., Chien P. Y., Gehm B. D., Jameson J. L. (2001) J. Biol. Chem. 276, 13615–13621 [DOI] [PubMed] [Google Scholar]
- 36.Jakacka M., Ito M., Martinson F., Ishikawa T., Lee E. J., Jameson J. L. (2002) Mol. Endocrinol. 16, 2188–2201 [DOI] [PubMed] [Google Scholar]
- 37.Pace P., Taylor J., Suntharalingam S., Coombes R. C., Ali S. (1997) J. Biol. Chem. 272, 25832–25838 [DOI] [PubMed] [Google Scholar]
- 38.Bagatell R., Khan O., Paine-Murrieta G., Taylor C. W., Akinaga S., Whitesell L. (2001) Clin. Cancer Res. 7, 2076–2084 [PubMed] [Google Scholar]
- 39.Beliakoff J., Whitesell L. (2004) Anticancer Drugs 15, 651–662 [DOI] [PubMed] [Google Scholar]
- 40.Sausville E. A., Tomaszewski J. E., Ivy P. (2003) Curr. Cancer Drug Targets 3, 377–383 [DOI] [PubMed] [Google Scholar]
- 41.Egorin M. J., Lagattuta T. F., Hamburger D. R., Covey J. M., White K. D., Musser S. M., Eiseman J. L. (2002) Cancer Chemother. Pharmacol. 49, 7–19 [DOI] [PubMed] [Google Scholar]
- 42.Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T. P., Rastinejad F. (2008) Nature 456, 350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]