Abstract
Pmel17 is a melanocyte/melanoma-specific protein that subcellularly localizes to melanosomes, where it forms a fibrillar matrix that serves for the sequestration of potentially toxic reaction intermediates of melanin synthesis and deposition of the pigment. As a key factor in melanosomal biogenesis, understanding intracellular trafficking and processing of Pmel17 is of central importance to comprehend how these organelles are formed, how they mature, and how they function in the cell. Using a series of deletion and missense mutants of Pmel17, we are able to show that the integrity of the junction between the N-terminal region and the polycystic kidney disease-like domain is highly crucial for endoplasmic reticulum export, subcellular targeting, and fibril formation by Pmel17 and thus for establishing functional melanosomes.
Keywords: Endoplasmic Reticulum, Lysosomes, Protein Domains, Protein Export, Protein Sorting, Trafficking, Fibril, Melanosomes, Pmel17, gp100
Introduction
Pmel17 (also called gp100, silver, or ME20) is an extensively glycosylated type I membrane protein specifically expressed in melanocytes and melanoma. It forms a fibrillar sheet structure in melanosomes, on which precursors of melanin synthesis are deposited and concentrated (1–3). This concentration might serve to accelerate the rate of melanin production (1, 3, 4) as well as to sequester toxic reaction intermediates, a function deduced from the finding that Pmel17-defective melanocytes display reduced viability in vivo (5).
Following co-translational translocation into the endoplasmic reticulum (ER)2 membrane as a so-called P1 precursor form (100 kDa), Pmel17 gets transported through the Golgi apparatus where it becomes a substrate for sialyltransferases (6). Through maturation of its oligosaccharides, P1 is converted into the larger (120 kDa) P2 form that enters the trans-Golgi network and from there migrates (directly or via the plasma membrane) to stage I melanosomes (7). These are a specialized set of multivesicular bodies (MVBs) destined to later develop into the mature pigment-storing organelles (8). Therein Pmel17 is transferred to intralumenal vesicles (ILVs) via budding into the interior of the MVB (9), a step thought to be essential for subsequent cleavage of P2 into an N-terminal 90-kDa Mα fragment and a C-terminal membrane-standing 28-kDa Mβ fragment (10). This cleavage, carried out by a yet unidentified furin-like proprotein convertase (pPC) is essential for fibril formation (11). However, Mα and Mβ remain linked to each other by a disulfide bridge. A subsequent membrane-proximal cleavage of Mβ by a metalloprotease of the a disintegrin and metalloprotease (ADAM) family eventually releases the lumenal Mα fragment from the ILV surface (12). This is followed by a sequence of poorly characterized further cleavages and finally the assembly of some of the resulting fragments into mature fibrils.
The organization of domains within Pmel17 has recently been described, and the first steps have been undertaken to investigate their function in the molecule. In particular, all three large domains within Mα (namely the N-terminal region (NTR, ∼aa 22–210), the polycystic kidney disease-like domain (PKD, ∼aa 211–293), and the repeat domain (RPT, ∼aa 314–424) (see Fig. 1A)) are essential for fibril formation (10, 13). Although all these domains (or fragments thereof) may directly contribute to the fibrillar core (1, 4, 14), the NTR and PKD domains seem to be additionally crucial for subcellular targeting of the polypeptide. Pmel17 lacking either of these domains fails to accumulate in LAMP1/LAMP2-containing organelles when transiently overexpressed in HeLa cells but rather localizes to early endosomes (10, 13). Interestingly, although targeting of Pmel17 to MVBs is normal, its delivery from the limiting membrane to ILVs appears to be impaired when either the NTR or the PKD domain is missing (10). This may underlie the observed subcellular sorting defect of the respective mutants.
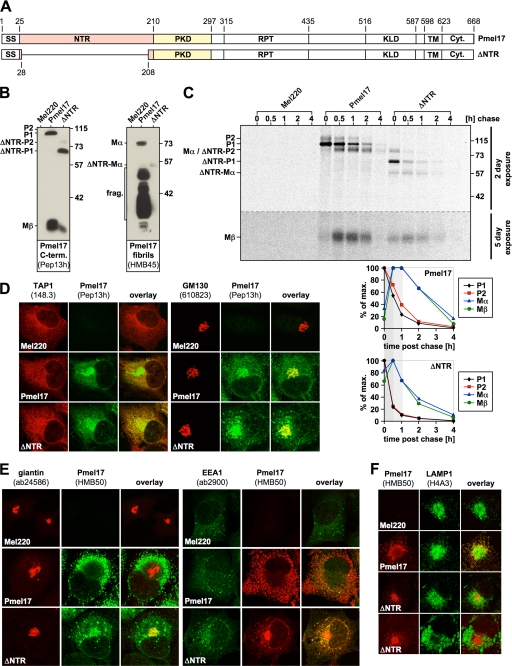
FIGURE 1.
The NTR is required for normal subcellular targeting of Pmel17. A, schematic representation of the ΔNTR construct. B, ΔNTR can be processed by pPCs. A total membrane fraction derived from the indicated stable Mel220 transfectants was lysed in 1% SDS/1% β-mercaptoethanol plus protease inhibitors (Complete, Roche Applied Science) and analyzed by Western blot using Pmel17-specific antibodies. C, ΔNTR shows both accelerated pPC processing and turnover. Cells from Fig. 1B, were pulse-labeled for 30 min with 35S and subsequently chased for the indicated times. 2% Triton X-100 lysates were immunoprecipitated with Pmel17-specific antibody HMB50, eluted with 0.5% SDS under vigorous vortexing for 30 min, and analyzed by autoradiography (top panel). A 2-day exposure is shown for the upper part of the gel separated by the dashed line from a 5-day exposure for the lower part of the same gel. Quantitative PhosphorImager analysis of the pulse-chase data with maximal levels for each band set to 100% is shown (bottom panel). D, newly synthesized ΔNTR localizes to the ER and Golgi. Cells from B were analyzed by immunofluorescence using antibodies against newly synthesized Pmel17 (Pep13h) and organelle markers TAP1 (148.3) (ER) or GM130 (610823) (Golgi). E, folded HMB50-reactive ΔNTR co-localizes with Golgi and early endosomal markers. Cells from B were analyzed by immunofluorescence using antibodies against folded Pmel17 (HMB50) and organelle markers giantin (ab24586) (Golgi) or EEA1 (ab2900) (early endosomes). F, ΔNTR fails to migrate into LAMP1-positive compartments. Cells from B were analyzed by immunofluorescence using antibodies against folded Pmel17 (HMB50) and organelle marker LAMP1 (H4A3) (lysosomes/melanosomes).
To allow a more detailed mapping of the sorting requirements, we constructed a series of deletion and missense mutants of Pmel17 where either the whole NTR or only a small region at the NTR-PKD domain boundary is affected. These were stably expressed in the Pmel17-negative melanoma cell line LG2-MEL-220 (Mel220) (15). Consistent with earlier reports (10, 13) our data show that Pmel17 lacking the whole NTR (Δ28–208) fails to migrate into LAMP1-containing organelles in melanoma cells but, rather, gets largely retained in early secretory compartments (and to some extent in early endosomes) where it undergoes rapid degradation. Interestingly, a mutant with a smaller deletion (Δ190–208) completely fails to get released from the ER and a very similar, albeit slightly milder defect is also observed with three Pmel17 missense mutants, all affected in the same region close to the NTR-PKD domain boundary. The subset of these mutants with the slightly milder phenotype can eventually leave the ER, albeit at an extremely slow rate and only to a very low extent. Interestingly, these mutants nevertheless largely accumulate in LAMP1-positive compartments over time and they do so to levels comparable with wild-type Pmel17 (wt-Pmel17). We use these mutants to demonstrate that an intact NTR-PKD domain boundary is also necessary beyond efficient ER release for proper subcellular targeting and fibril formation. Hence, this region within Pmel17 is crucial for the function of the polypeptide and thus for melanosome biogenesis.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
LG2-MEL-220 (Mel220), a human Pmel17-deficient melanoma cell line (15), was grown in Iscove's modified Dulbecco's medium (Sigma)/10% fetal calf serum (HyClone) containing non-essential amino acids (Invitrogen), GlutaMax (Invitrogen), and penicillin/streptomycin (Invitrogen). Mel220 cells expressing wild-type or mutant Pmel17-i (6, 16) were grown in medium additionally containing 2 mg/ml G418 (Invitrogen).
Antibodies
Pep13h (9) and Pmel-N (17) are rabbit antisera recognizing the C and N termini of Pmel17, respectively. I51 is a rabbit antiserum raised against residues 206–220 within the PKD domain of Pmel17 (14). HMB50 (18), NKI-beteb (Abcam), and HMB45 (NeoMarkers) are mouse monoclonal antibodies (IgG2a, IgG2b, and IgG1, respectively) recognizing the folded PKD domain (HMB50 and NKI-beteb) (19) or a sialylated epitope within the RPT domain (HMB45) (20) of Pmel17. We consider HMB50 and NKI-beteb to be conformation-specific antibodies, because they only work in applications where folded protein is to be detected (immunofluorescence, immunohistochemistry, flow cytometry, and immunoprecipitation), whereas denatured protein in a Western blot format is not recognized (21). Moreover, thermal denaturation completely abrogates reactivity of Pmel17 with either of these antibodies (data not shown). Supplemental Table 1 provides an overview of the features of the Pmel17-specific antibodies used in this study. The monoclonal antibodies 148.3 (22), 610823 (BD), and H4A3 (IgG1, Abcam) recognize the ER marker TAP1 (transporter associated with antigen processing 1), the Golgi marker GM130, and the lysosomal marker LAMP1, respectively. The rabbit polyclonal antibodies Clyde (23), S-7696 (Sigma), E-1031 (Sigma), ab24586 (Abcam), ab16052 (Abcam), 555798 (BD), and ab18528 (Abcam) recognize the ER marker calnexin, the ER exit site marker Sec23, the ERGIC marker ERGIC-53, the Golgi marker giantin, the trans-Golgi network marker TGN46, the early endosomal marker EEA1, and the lysosomal marker LAMP2a, respectively. Horseradish peroxidase- or fluorophore-labeled isotype-specific or conventional goat-anti-mouse and goat-anti-rabbit antibodies were purchased from Molecular Probes or Jackson ImmunoResearch.
Vector Constructs and Pmel17 Expression
Pmel17-i (also termed gp100) (16) was cloned into vector pEGFP-N1 (Clontech) as an EcoRI/SalI fragment and subjected to the standard QuikChange mutagenesis protocol (Stratagene) using primer pair 5′-GTGACTGTCTACCCTAGCCGGAAACGCCGATCGTATGTGCCTCTTGC-3′/5′-GCAAGAGGCACATACGATCGGCGTTTCCGGCTAGGGTAGACAGTCAC-3′ to generate a construct encoding IR-wt. From the resulting vectors mutant or wild-type Pmel17 was amplified in a Pfu-driven PCR (annealing temperature, 52 °C/36 cycles) using primer pair 5′-AATGAATTCCACCATGGATCTGGTGCTAAAAAGATGCCTTCTTCAT-3′/5′-TTGAATTCGCCGCTCAGACCTGCTGCCC-3′ and cloned into the expression vector pBMN-IRES-neo as an EcoRI/EcoRI fragment. The respective vector containing wild-type Pmel17 then served as a template for a standard QuikChange mutagenesis using primer pairs 5′-GGGGGCTACAAAAGTACCCATTACTGACAGGTGCC-3′/5′-GGCACCTGGTCAGTAATGGGTACTTTTGTAGCCCCC-3′, 5′-GGAAGTGACTGTATACATTACTGACCAGGTGCC-3′/5′-GGCACCTGGTCAGTAATGTATACAGTCACTTCC-3′, 5′-GTGACTGTCTACCCTAGGCGGGGATCCCGG-3′/5′-CCGGGATCCCCGCCTAGGGTAGACAGTCAC-3′, 5′-GTGACTGTCTACCATTCGCGAGGATCCCGGAGC-3′/5′-GCTCCGGGATCCTCGCGAATGGTAGACAGTCAC-3′, 5′-CTACCATCGCCGGAAATCCCGGAGCTATGTG-3′/5′-CACATAGCTCCGGGATTTCCGGCGATGGTAG-3′, and 5′-CCATCGCCGGGGACGCCGGAGCTATGTG-3′/5′-CACATAGCTCCGGCGTCCCCGGCGATGG-3′ to generate constructs encoding ΔNTR (Δ28–208), Δ190–208, H190P, R191S, G193K, and S194R, respectively. All pBMN vectors containing mutant or wild-type Pmel17 were sequenced from both directions before retroviral transduction into Mel220 cells (24). Mel220 transfectants expressing wild-type or mutant Pmel17 were selected in medium containing 2 mg/ml G418 (Invitrogen) for 3 weeks, and expression of Pmel17 was assessed by Western blot. Levels of expression of wt-Pmel17 in Mel220 transfectants were compared with endogenous Pmel17 expression in a derivative of the melanoma cell line buf1280 (25) and found to be ∼2-fold higher (supplemental Fig. S1).
Immunofluorescence and Flow Cytometry
Mel220 transfectants were seeded overnight on glass coverslips. The next day, cells were washed with PBS containing 0.9 mm CaCl2 and 0.5 mm MgCl2 (PBS+/+) and fixed with 2% formaldehyde (15 min at room temperature). After quenching with PBS+/+/10 mm glycine followed by a wash with PBS+/+/0.5% bovine serum albumin, cells were permeabilized for 1h in staining buffer (PBS+/+/0.5% bovine serum albumin/0.5% saponin). Staining was performed in a humidity chamber for 1 h with the indicated primary antibodies at concentrations recommended by the manufacturer or 1:50 for 148.3, 1:100 for HMB50, and 1:35 for Clyde. After three washes with staining buffer, Alexa647- and Alexa488-conjugated secondary antibodies (Molecular Probes) were applied at a 1:100 dilution in the same buffer, before cells were washed again three times, mounted in ProLong Gold reagent (Invitrogen), and analyzed by confocal fluorescence microscopy using a TCS SP2 confocal microscope (Leica Microsystems).
For quantification of differences in the subcellular distribution patterns of LAMP1- and Pmel17 we first rotated the respective immunofluorescence pictures in a way that the nucleus of the shown cell points to one side (right or left), whereas the bulk of perinuclear LAMP1-fluorescence points to the other side as schematically depicted in supplemental Fig. S3A. We then determined the vertically averaged pixel intensity for every position along the x axis using the Plot Profile function of the image processing software ImageJ 1.41o (National Institutes of Health) and exported the respective histogram data to Microsoft Excel 2004 (Microsoft). There, the histograms for LAMP1 and Pmel17 staining were independently normalized to fit a range from 0 (lowest observed fluorescence intensity) to 100 (highest observed fluorescence intensity) before they were subtracted from each other in every position along the x axis. The absolute values of the respective differences were added and finally divided by the total number of pixels along the x axis. We named the resulting value “average difference” between the LAMP1 and Pmel17 distributions.
Flow cytometry using surface or intracellularly labeled cells was performed as described previously (26, 27) using antibodies NKI-beteb, HMB50, or Pep13h at concentrations 1:10, 1:100, and 1:100, respectively, followed by Alexa647- or Alexa488-conjugated secondary antibodies. All data were acquired on a FACSCalibur flow cytometer and analyzed using FlowJo 6.4.7 software (Tree Star).
Electron Microscopy
For conventional Epon embedding of cell samples, Mel220 transfectants were fixed in 2.5% glutaraldehyde/2% sucrose in 0.1 m sodium cacodylate buffer, pH 7.4 (NaCaCo buffer), for 30 min at room temperature followed by another 30 min in the same fixative solution at 4 °C. Subsequently, cells were rinsed with NaCaCo buffer and further processed as described (28).
For cryo-immunoelectron microscopy, samples were fixed in 2% paraformaldehyde/0.1% glutaraldehyde in PBS for 15 min at room temperature followed by another 15 min in the same fixative solution at 4 °C. Subsequently, cells were rinsed with PBS and further processed as described (28). For immunolabeling cells were stained with Pmel17-specific antibody HMB50 at 1:25 followed by gold-anti-mouse conjugate (Jackson ImmunoResearch Laboratories).
For embedding of cell samples in LR-gold resin (London Resin Gold), cells were fixed in 4% paraformaldehyde/0.1% glutaraldehyde/2% sucrose in 0.1 m HEPES buffer for 30 min at room temperature followed by another 90 min in the same fixative solution, but lacking glutaraldehyde at 4 °C. Subsequently, cells were rinsed with PBS and 50 mm NH4Cl/100 mm glycine/3% sucrose for 15 min to quench free aldehydes, scraped in 1% gelatin, and transferred to 5% agar. Once set, samples were placed in 0.5% tannic acid in 0.1 m HEPES for 30 min, rinsed twice in Tris/50 mm maleate/3% sucrose (sucrose-maleate buffer), and stained with 2% uranyl acetate in sucrose-maleate buffer. Samples were dehydrated through a graded series of ethanol (50% to 95%) at −20 °C and embedded in LR-gold resin (EMS) at −20 °C. For immunolabeling, cells were stained with Pmel17-specific antibody HMB50 at 1:10 followed by gold-anti-mouse conjugate (Jackson ImmunoResearch Laboratories).
Samples were viewed using an FEI Tencai Biotwin transmission electron microscope at 80 Kv. Images were taken using Morada CCD and iTEM (Olympus) software.
Determination of UPR Induction by Reverse Transcription-PCR
Mel220 cells expressing wild-type Pmel17 or construct Δ190–208 were treated overnight with 5 μg/ml tunicamycin (Sigma) or were left untreated, before mRNA was prepared using the RNeasy Mini kit (Qiagen) and reverse-transcribed into cDNA using the First Strand Synthesis kit (Stratagene) in combination with Oligo(dT) primers. XBP-1 was amplified using primer pair 5′-CCTTGTAGTTGAGAACCAGG-3′/5′-GGGGCTTGGTATATATGTGG-3′ (29) in a standard Taq-driven PCR (annealing temperature, 50 °C/40 cycles), and the activation of the unfolded protein response (UPR) was assessed by monitoring the presence of the spliced, PstI-resistant form (indicating UPR induction) versus the unspliced, PstI-sensitive form (indicating absence of UPR induction) of XBP-1.
Pulse-chase Analysis, Immunoprecipitation, and Western Blotting
Radiolabeling was performed as described (26). Briefly, 1.5 × 107 starved Mel220 cells expressing Pmel17 derivatives were pulse-labeled at 37 °C with [35S]methionine/cysteine (PerkinElmer Life Sciences) at 0.5 mCi/ml in 1.5 ml for 30 min and subsequently chased in Iscove's modified Dulbecco's medium/10% fetal calf serum containing an excess of cold l-methionine/l-cysteine (both at 0.45 mg/ml) for up to 4 h. Following this, cells were harvested and frozen at −80 °C until the next day.
For immunoprecipitation antibody HMB50 was first covalently coupled to protein A-Sepharose and stored at 4 °C (26). Cell pellets were thawed, lysed in 2% Triton X-100 (containing protease inhibitor mixture (Roche Applied Science)) at 107 cells/ml, and precleared using protein A-Sepharose beads. Subsequently, the supernatant was applied to the HMB50-coupled beads, and immunoprecipitation was carried out as described (26). After separation of immunoprecipitates by SDS-PAGE, gels were dried, exposed to PhosphorImager screens, and analyzed with ImageQuant 5.2 (Amersham Biosciences). Western blotting was carried out as described (26).
We used the prestained marker mix Seablue Plus 2 (Invitrogen) for estimation of protein molecular weights in gels. Because this prestained marker mix displayed an inaccurate running behavior in the big gel system employed (PROTEAN II xi cell with 20 cm long gels, Bio-Rad), we recalibrated the bands using proteins of known molecular mass as standards (TPP2, 135 kDa; Grp94, 94 kDa; TAP1, 70 kDa; calreticulin, 57 kDa; and HLA-A2, 42 kDa). The recalibrated marker bands are shown in the figures.
RESULTS
The NTR Domain Is Required for Normal Subcellular Targeting and Function of Pmel17
Both the NTR and the PKD domain of Pmel17 have been implicated in subcellular targeting of the polypeptide to ILVs in stage I melanosomes (10, 13). However, which of the one or more subregions within the ∼270-aa stretch encompassing the two domains mediates this effect is unknown. Yet, because this sorting defect has so far only been described in transient overexpression systems employing the cervical carcinoma cell line HeLa, we first decided to examine whether it would also be observed in melanoma cells. These cells may provide a more native environment (e.g. with regard to repertoires of ER chaperones or pPCs present), in which Pmel17 is normally expressed and functional.
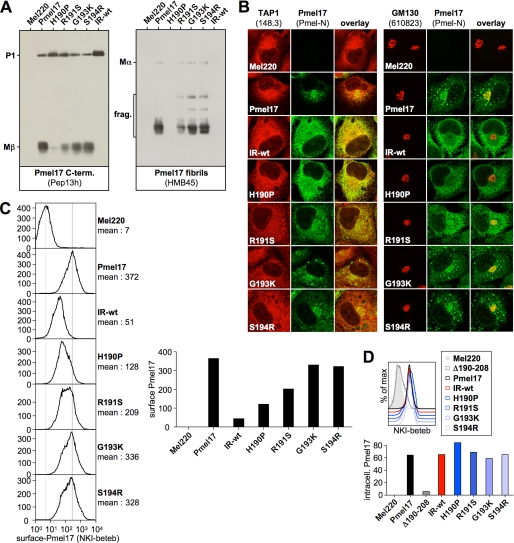
To this end, we constructed a Pmel17 deletion mutant lacking the NTR (Δ28–208) (Fig. 1A) (hereinafter ΔNTR) and stably expressed it in the Pmel17-deficient melanoma cell line LG2-MEL-220 (Mel220) (15). Expression was confirmed by Western blot using antibody Pep13h (Fig. 1B, left panel), which recognizes newly synthesized Pmel17, but not mature fibrils (19, 30). ΔNTR appeared to undergo pPC-mediated cleavage as judged by the detection of Mβ (Fig. 1B, left panel) and Mα (Fig. 1B, right panel) fragments, but did not give rise to the set of fibrillogenic fragments reactive with antibody HMB45 (Fig. 1B, right panel), suggesting that it represents a loss-of-function mutant. Levels of Mβ and Mα were lower at steady state when compared with wt-Pmel17, indicating either reduced rate of cleavage or enhanced degradation of the ΔNTR-derived cleavage products. To distinguish between these two possibilities we performed a pulse-chase experiment (Fig. 1C). If anything, conversion of P2 into Mα and Mβ occurred with more rapid kinetics for ΔNTR than for wt-Pmel17 as shown by both faster decay of P2 and earlier appearance of its cleavage products (Fig. 1C, see bottom panels for quantification). This suggests that the NTR may have a possible function in protecting Pmel17 from premature cleavage early during secretion. However, although generated earlier, both Mα and Mβ also disappeared faster than their counterparts in wt-Pmel17-expressing cells, thus providing an explanation for the lower steady-state levels of these fragments in ΔNTR-transfected Mel220 cells. These results are consistent with an earlier report showing that the NTR is not required for pPC-mediated cleavage of Pmel17 (13, 31) but disagree with data by Theos and co-workers, who had described an NTR-deletion mutant (Δ29–200) that appeared to be resistant to pPC-processing unless furin was overexpressed (10). In the light of our findings and the findings by Hoashi and colleagues (13, 31) we speculate that the latter construct may be structurally affected, thus sterically impairing its association with a pPC until overexpression of a convertase drives the equilibrium toward sufficient interaction (to allow cleavage). We additionally note that the fact that ΔNTR gets cleaved efficiently in Mel220 cells (Fig. 1C), while displaying a substantially altered subcellular distribution when compared with wt-Pmel17 (see below), is surprising in the light of the current understanding of Pmel17 maturation. One possible explanation for this phenomenon may be that inward budding of the protein within MVBs may in fact not be essential for pPC-mediated cleavage of Pmel17, or at least of its N-terminal deletion mutant.
Like wt-Pmel17, newly synthesized (Pep13h-reactive) ΔNTR was found in both the ER and the Golgi apparatus and sometimes additionally in a punctate, vesicular pattern probably reflecting early endosomes (Fig. 1D). We also assessed the subcellular localization of wt-Pmel17 and ΔNTR with HMB50, an antibody detecting folded Pmel17 (see “Experimental Procedures”) that reportedly stains only stage II melanosomes by immunofluorescence (IF) (19). However, the same antibody at least weakly labels earlier secretory and endocytic compartments by immunoelectron microscopy (Ref. 30 and data not shown) and recognizes ER and Golgi forms of Pmel17 by immunoprecipitation (see Fig. 4C) (9, 18, 19, 21). As expected, no HMB50-reactive wt-Pmel17 was found in the Golgi by IF, whereas ΔNTR extensively overlapped with the Golgi marker giantin (Fig. 1E, left panel). Consistent with the report by Theos and co-workers (10) ΔNTR was additionally found to some minor degree in EEA1-co-labeled early endosomes (Fig. 1E, right panel). Moreover, ΔNTR was also detected in a reticular pattern surrounding the nucleus, indicating that a substantial portion of the folded protein is localized to the ER (Fig. 1E). In line with this data and consistent with previous reports (10, 13) we did not observe co-localization of ΔNTR with the lysosomal/melanosomal marker LAMP1 (Fig. 1F), indicating that the NTR is essential to route the protein to the appropriate compartments within the cell. Taken together, these data show that the NTR is essential for proper subcellular migration but not for pPC-mediated cleavage of Pmel17.
FIGURE 4.
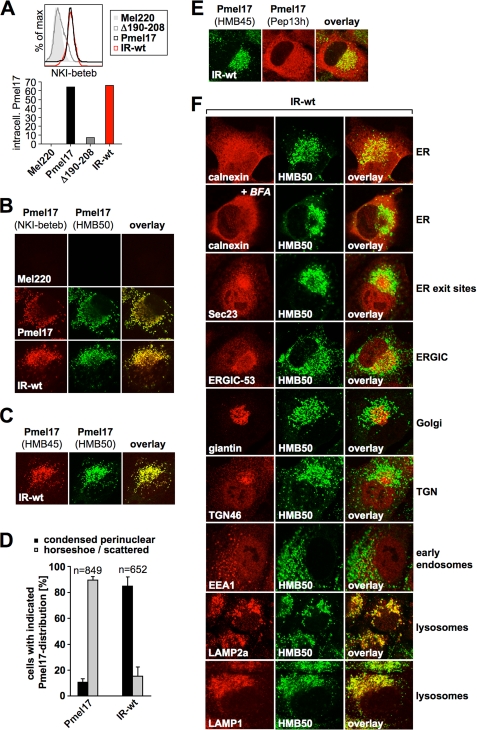
Mel220 transfectants stably expressing IR-wt build up a large post-ER pool of this mutant that localizes to lysosomes. A, high levels of folded IR-wt can be detected intracellularly. Mel220 transfectants stably expressing IR-wt or Δ190–208 were fixed in 2% formaldehyde, permeabilized, and stained with antibodies reactive with folded Pmel17 (NKI-beteb) and analyzed by flow cytometry (top panel). After background subtraction (untransfected Mel220 cells) data are represented as a bar diagram (bottom panel). B, NKI-beteb- and HMB50-reactive IR-wt displays a distinct subcellular pattern as wt-Pmel17. Mel220 transfectants expressing wt-Pmel17 or IR-wt were analyzed by immunofluorescence using antibodies against folded Pmel17 (NKI-beteb and HMB50). C, HMB50-reactive IR-wt localizes to a post-Golgi compartment. Mel220 transfectants expressing IR-wt were analyzed by immunofluorescence using antibodies against sialylated (post-Golgi- or Golgi-localized) Pmel17 (HMB45) and folded Pmel17 (HMB50). D, in contrast to wt-Pmel17, HMB50-reactive IR-wt is mostly present in a condensed perinuclear pattern. Mel220 transfectants expressing wt-Pmel17 or IR-wt were analyzed by immunofluorescence using antibodies against folded Pmel17 (HMB50). Cells displaying either a condensed perinuclear pattern or a horseshoe-shaped/scattered pattern of fluorescence were blind-counted, and the results from two slides are displayed as a bar diagram. E, antibodies Pep13h and HMB45 recognize distinct populations of IR-wt. Cells from Fig. 4C were analyzed by immunofluorescence using antibodies against sialylated (post-Golgi- or Golgi-localized) Pmel17 (HMB45) and newly synthesized Pmel17 (Pep13h). F, folded HMB50-reactive post-ER IR-wt localizes to lysosomes. Cells from Fig. 4C were analyzed by immunofluorescence using antibodies against folded Pmel17 (HMB50) and organelle markers calnexin (Clyde) (ER), Sec23 (S-7696) (ER-exit sites), ERGIC-53 (E-1031) (ERGIC), giantin (ab24586) (Golgi), trans-Golgi network46 (ab16052) (TGN), EEA1 (555798) (early endosomes), LAMP2a (ab18528) (lysosomes), and LAMP1 (H4A3) (lysosomes).
The Integrity of a Small Region in the Vicinity of the NTR-PKD Domain Boundary Is Crucial for ER Exit of Pmel17
Our data (Fig. 1, E and F) and the data of others (10, 13) demonstrate that the presence of the NTR is essential for normal trafficking of Pmel17. We next wanted to assess whether a smaller subregion within the N terminus could be identified that is required for normal subcellular distribution of the protein. To this end, we introduced an 18-aa deletion into Pmel17 comprising only residues 190–208 (Fig. 2A) (hereinafter Δ190–208). Thus, Δ190–208 and ΔNTR differ only by the N-terminal extension of the respective deletion but share its C-terminal border. This border is located only 2 aa before the junction of exons 6 and 7. We note that exon boundaries are well known to correlate with domain boundaries in many polypeptides (32), and within Pmel17 the respective area likely corresponds to the border between the NTR and the PKD domains (Fig. 2A). Further supporting this, domain linker prediction using domain linker prediction-support vector machine (DLP-SVM) (33) identifies region 200–217 within a Pmel17 sample sequence ranging from aa 160 to 280 as the most likely area to represent a loop separating two independent structural domains.
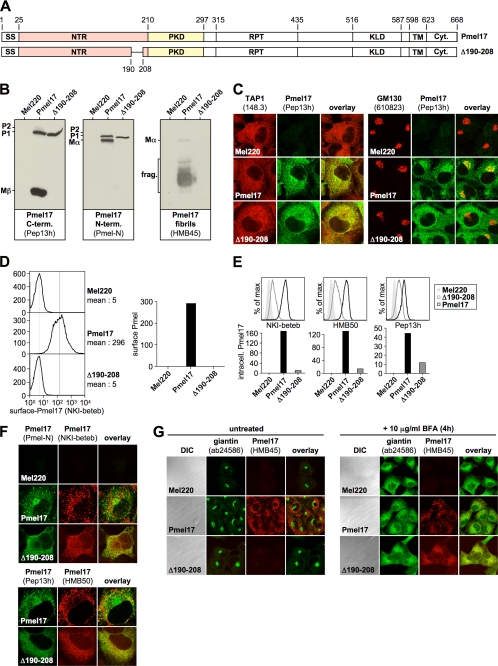
FIGURE 2.
The integrity of a small region in the vicinity of the NTR-PKD domain boundary is crucial for ER exit of Pmel17. A, schematic representation of the Δ190–208 construct. B, only the ER-associated P1 form can be detected for Δ190–208 at steady state. Membrane lysates of Mel220 transfectants stably expressing Δ190–208 were prepared as in Fig. 1B and analyzed by Western blot using Pmel17-specific antibodies. C, newly synthesized Δ190–208 is completely retained in the ER. Cells from Fig. 2B were analyzed by immunofluorescence using antibodies against newly synthesized Pmel17 (Pep13h) and organelle markers TAP1 (148.3) (ER) or GM130 (610823) (Golgi). D, folded NKI-beteb-reactive Δ190–208 is completely absent from the cell surface. Cells from Fig. 2B were surface-labeled with antibody NKI-beteb against folded Pmel17 and analyzed by flow cytometry (left panel). After background subtraction (untransfected Mel220 cells) data are represented as a bar diagram (right panel). E, low levels of folded Δ190–208 can be detected intracellularly. Cells from Fig. 2B were fixed in 2% formaldehyde, permeabilized, and stained intracellularly with antibodies reactive with folded (NKI-beteb or HMB50) or newly synthesized Pmel17 (Pep13h) and analyzed by flow cytometry (top panel). After background subtraction (untransfected Mel220 cells) data are represented as a bar diagram (bottom panel). F, folded HMB50- and NKI-beteb-reactive Δ190–208 localizes to the ER. Cells from Fig. 2B were analyzed by immunofluorescence using antibodies against folded Pmel17 (NKI-beteb (top panel) or HMB50 (bottom panel)) and newly synthesized Pmel17 (Pmel-N (top panel) or Pep13h (bottom panel)). G, Δ190–208 does not react with antibody HMB45, which specifically recognizes sialylated (post-Golgi- or Golgi-localized) Pmel17. Cells from Fig. 2B were treated or not with 10 μg/ml brefeldin A (BFA) for 4 h to shift Golgi-associated proteins back into the ER and analyzed by immunofluorescence using antibodies against sialylated (post-Golgi or Golgi-localized) Pmel17 (HMB45) and organelle marker giantin (ab24586) (Golgi).
Δ190–208 could be stably expressed in Mel220 cells, but strikingly only the ER-associated P1 form could be detected by Western blot, whereas the (post-)Golgi form P2 and its cleavage products Mα and Mβ were completely absent (Fig. 2B, left and middle panels). As expected, P1 was fully sensitive to endoglycosidase H indicating that the protein had not yet passed through the Golgi (data not shown). HMB45-reactive fibrillogenic fragments were also not detected for this mutant (Fig. 2B, right panel) and neither were fibrils found by immunoelectron microscopy (data not shown), demonstrating that the respective mutant had a loss-of-function phenotype. Consistent with this, we found newly synthesized (Pep13h-reactive) Δ190–208 completely retained in the ER as judged by full overlap with ER marker TAP1 and absence of co-localization with Golgi marker GM130 by IF (Fig. 2C). ER retention of the protein was also supported by the lack of any detectable surface levels of folded Δ190–208 (Fig. 2D).
Interestingly, however, we were able to detect low intracellular levels of folded Δ190–208 using two different conformation-sensitive antibodies (HMB50 and NKI-beteb) (Fig. 2E, left and middle panels), and the respective staining completely overlapped with the newly synthesized protein in the ER (Fig. 2F, top and bottom panels). This suggests that there is no significant pool of post-ER Δ190–208 forms, and consequently ER retention of this mutant is complete. We note that much lower labeling for Δ190–208 than for wt-Pmel17 was not surprising, because by IF these antibodies only efficiently detect stage II melanosomes in wt-Pmel17-expressing cells (as discussed above) and these striated organelles are absent in Δ190–208-transfected cells. Finally, to demonstrate ER retention of Δ190–208, we stained Mel220 transfectants with antibody HMB45, a reagent that recognizes an epitope within Pmel17 only if it has been sialylated in the Golgi (20). As expected HMB45 easily detected wt-Pmel17, but failed to detect Δ190–208 (Fig. 2G, left panel). This is consistent with the finding that the latter never gets exposed to Golgi enzymes. However, when its exposure to Golgi enzymes was enforced by the retrotransfer of Golgi proteins back into the ER using brefeldin A, HMB45 detected Δ190–208 in the ER (Fig. 2G, right panel). Thus deletion of residues 190–208 did not in principle destroy the HMB45-reactive epitope.
Interestingly, ER retention of Δ190–208 did not appear to be caused by a complete inability to acquire a native conformation, because it neither induced the UPR (supplemental Fig. S2A) nor was degraded rapidly, which would be expected for an excessively misfolded ERAD substrate (supplemental Fig. S2B). This, combined with its reactivity with two conformation-sensitive antibodies (Fig. 2F), suggests that Δ190–208 behaves more like a “normal ER-resident protein” rather than a polypeptide folded into a largely non-native structure. Although Δ190–208 may contain local alterations to the “normal” conformational folding, any such alterations appear not to result in significant exposure of hydrophobic patches recognizable by the ERAD machinery. Certainly there do not appear to be detrimental effects on folding of the PKD domain, which reacts with the HMB50 and NKI-beteb monoclonal antibodies in a conformation-sensitive fashion. Consequently, the inability of Δ190–208 to get released from the ER seems more likely to reflect a requirement of an intact NTR-PKD domain boundary for proper trafficking of Pmel17 rather than for global folding of the molecule. Altogether, this suggests that the integrity of the NTR-PKD domain junction is crucial for ER exit and normal trafficking of Pmel17.
Amino Acid Exchanges Targeting the NTR-PKD Domain Boundary within Pmel17 Dramatically Affect ER Export
We noted that the ER export defect observed with Δ190–208 was very reminiscent of the behavior of another Pmel17 mutant that we had generated. In this mutant (hereinafter IR-wt) we had inserted at the NTR-PKD domain boundary (aa 190–196) an optimal pPC-cleavage site derived from the human proinsulin receptor (Fig. 3A). Thus, this construct is affected in the same area as the deletion mutant Δ190–208.
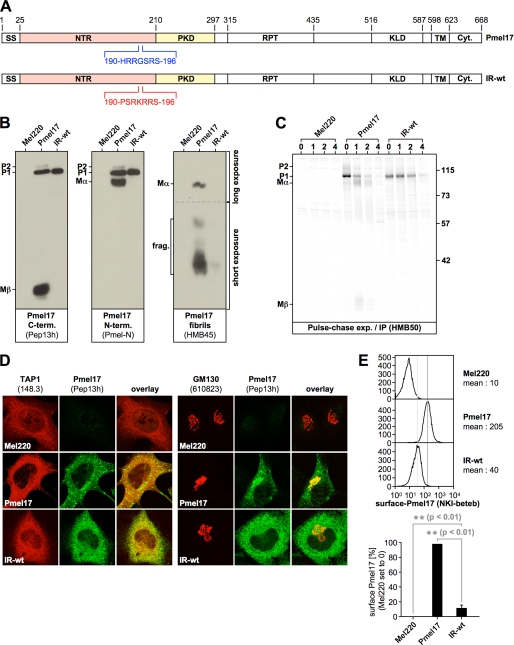
FIGURE 3.
Amino acid exchanges targeting the NTR-PKD domain boundary dramatically affect ER export. A, schematic representation of the IR-wt construct. B, only the ER-associated P1 form can be detected for IR-wt at steady state, when assessed with antibodies recognizing newly synthesized Pmel17. Membrane lysates of Mel220 transfectants stably expressing IR-wt were prepared as in Fig. 1B and analyzed by Western blot using Pmel17-specific antibodies. To visualize Mα better, a longer exposure of the upper part (separated by a dashed line from the lower part) of the same HMB45-blot (right panel) is shown. C, newly synthesized IR-wt shows almost no export from the ER or proprotein convertase-mediated processing. Mel220 transfectants stably expressing IR-wt were pulse-labeled for 30 min with 35S and subsequently chased for the indicated times. 2% Triton X-100 lysates were immunoprecipitated with Pmel17-specific antibody HMB50, eluted with 0.5% SDS under vigorous vortexing for 30 min, and analyzed by autoradiography. One representative out of two independent experiments is shown. D, newly synthesized IR-wt is largely retained in the ER. Cells from Fig. 3B were analyzed by immunofluorescence using antibodies against newly synthesized Pmel17 (Pep13h) and organelle markers TAP1 (148.3) (ER) or GM130 (610823) (Golgi). E, IR-wt is present at the cell surface only at minute levels. Cells from Fig. 3B were surface-labeled with antibody NKI-beteb against folded Pmel17 and analyzed by flow cytometry (top panel). After background subtraction (untransfected Mel220 cells) the data from three independent experiments are represented as a bar diagram (bottom panel). The difference in surface expression between wt-Pmel17 and IR-wt (**, p < 0.01) is statistically significant, as assessed by a one-way analysis of variance test with the Dunnett posttest.
Interestingly, IR-wt showed a dramatic ER retention defect very similar to that of Δ190–208. Antibodies recognizing newly synthesized protein in Western blot detected at steady-state practically only P1 (Fig. 3B, left and middle panels), and the amount of HMB45-reactive fibrillogenic fragments was extremely low when compared with wt-Pmel17 (Fig. 3B, right panel). Surprisingly, no fragments were detected with antibodies directed against the N or C termini of Pmel17 that would correspond to products generated by cleavage at the newly introduced pPC-cleavage site (Fig. 3B), and cleavage was also not detectable in a pulse-chase experiment (Fig. 3C). This indicates that the respective motif is not recognized in the sequence context into which it had been inserted. IR-wt-P1 was fully sensitive to endoglycosidase H (data not shown), suggesting that this protein was localized to the ER and also pulse-chase analysis confirmed that processing of IR-wt was almost completely abolished (Fig. 3C). In line with this, IF demonstrated the newly synthesized (Pep13h-reactive) protein to be confined to the ER, where it fully overlapped with ER marker TAP1 (Fig. 3D, left panel), whereas no significant co-localization with the Golgi marker GM130 was observed (Fig. 3D, right panel). Furthermore, IR-wt reached only very low (albeit reproducibly detectable) levels at the cell surface of Mel220 cells (Fig. 3E).
Altogether, this indicates that the introduction of a few amino acid changes close to the NTR-PKD domain boundary dramatically affects ER export of Pmel17 and underscores once more that the integrity of this region in the protein is essential for its normal trafficking.
Despite Only Minimal Release from the ER, over Time IR-wt Builds up a Large Lysosomal Population
The experiments shown in Fig. 3 demonstrate that newly synthesized IR-wt is largely retained in the ER. Surprisingly, however, unlike Δ190–208, when assessed by flow cytometry, intracellular IR-wt was extensively labeled with conformation-sensitive antibody NKI-beteb to a level identical as wt-Pmel17 (Fig. 4A). This indicates that a large pool of folded IR-wt was abundant somewhere in the cell. To determine which subcellular compartment this folded population of IR-wt was associated with, we employed IF and surprisingly found the NKI-beteb-reactive protein distributed in the juxtanuclear area showing a vesicular pattern typical for endosomes or lysosomes (Fig. 4B). The same pattern was observed with another conformation-sensitive antibody (HMB50) (Fig. 4B) and strikingly also with antibody HMB45 (Fig. 4C), which recognizes only Pmel17 that has already traversed the Golgi. This demonstrates that the respective population of IR-wt was localized to a post-ER compartment. Thus, all three antibodies, which for wt-Pmel17 recognize only the mature protein by IF, specifically detect a post-ER pool of IR-wt. This pool is clearly different from the bulk of newly synthesized and largely ER-retained (Pep13h-reactive) IR-wt as judged by the absence of overlap between the two populations (Fig. 4E). In fact, it seems that this post-ER pool of IR-wt builds up very slowly over time, because, in contrast to mature wt-Pmel17, it was not detectable immediately after transfection, but only after prolonged expression of the construct (data not shown).
However, we noted that this condensed perinuclear pattern typical of NKI-beteb-/HMB50-/HMB45-reactive IR-wt differed substantially from the subcellular pattern detected for wt-Pmel17 (Fig. 4B). In particular, wt-Pmel17 seemed to distribute into a broad band surrounding the perinuclear area and resembling a “horseshoe profile” (and in some cases of very high expression Pmel17 was scattered all over the cell) (Fig. 4, B and D; see also Fig. 5C). This suggests that, although both wt-Pmel17 and IR-wt efficiently label with conformation-sensitive antibodies, their respective post-ER populations reside in different locations in the cell.
FIGURE 5.
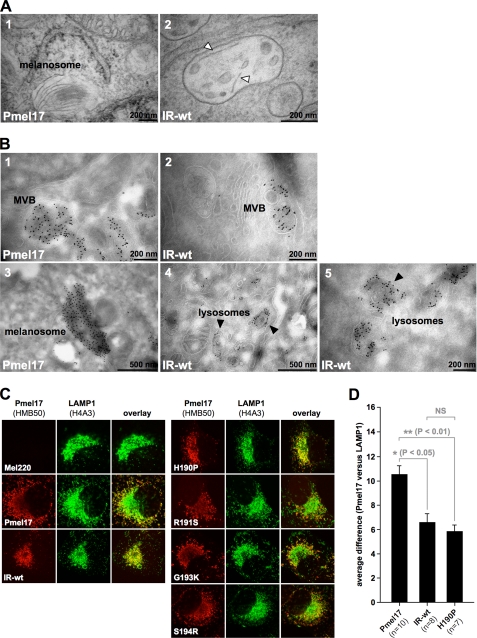
The post-ER populations of IR-wt and H190P are non-functional and largely route to lysosomes. A, IR-wt is severely impaired or blocked in fibril formation. Electron microscopic analysis of Epon-embedded Mel220 transfectants stably expressing wt-Pmel17 or IR-wt. Panel 1 shows a typical melanosome frequently found for wt-Pmel17. Panel 2 shows an example of the very rare occurrence of immature organelles that contain individual striae-like structures (white arrowheads). Almost always such structures could be clearly identified as membrane segments as judged by a visible double layer structure. However, in a few remaining cases image quality did not allow us to fully exclude that they represent an immature fibril in the formation process. Hence, IR-wt is at least severely impaired, but more likely completely blocked in fibril formation. B, folded HMB50-reactive IR-wt gets mostly delivered to compartments with lysosomal morphology. Cells expressing wt-Pmel17 or IR-wt were fixed and examined by cryo-immunoelectron microscopy (panels 1–5) using antibody HMB50. Panels 1 and 2 display MVBs with extensive immunolabeling over intralumenal vesicles. Panel 3 shows a typical melanosome in wt-Pmel17-expressing cells. Black arrowheads in panels 4 and 5 point to lysosomal, often multilamellar compartments densely labeled with gold particles in IR-wt-expressing cells. C, wt-Pmel17 mostly distributes to LAMP1low melanosomes, whereas IR-wt and H190P extensively co-localize with LAMP1 in LAMP1high lysosomes. Mel220 cells stably expressing the indicated Pmel17 mutants were analyzed by immunofluorescence using antibodies against folded Pmel17 (HMB50) and LAMP1 (H4A3). D, quantification of the average difference (see “Experimental Procedures”) between the Pmel17 and the LAMP1 profiles in the indicated Mel220 transfectants in a statistically relevant number of cells. The pattern differences between wt-Pmel17 and either IR-wt (*, p < 0.05) or H190P (**, p < 0.01) are statistically significant, as assessed by a one-way analysis of variance test with the Dunnett post-test. Pattern differences between IR-wt and H190P are not statistically significant (NS).
Next, we decided to determine in which compartment the IR-wt protein, reactive with conformation-sensitive antibodies, resides. Consistent with its reactivity with antibody HMB45, this population did not co-localize with ER marker calnexin, ER-exit site marker Sec23, ERGIC marker ERGIC-53, or Golgi marker giantin (Fig. 4F, rows 1 and 3–5). Moreover, brefeldin A treatment did not redistribute the protein back into the ER (as expected for proteins in a pre-Golgi or Golgi compartment) (Fig. 4F, row 2). This confirmed once again that HMB50-reactive IR-wt had already traversed and migrated beyond the Golgi. Interestingly, no overlap of this population with markers of the trans-Golgi network (TGN) or early endosomes was observed (Fig. 4F, rows 6 and 7). Instead, HMB50-reactive IR-wt fully co-localized with markers of lysosomes and melanosomes, LAMP1 and LAMP2a (Fig. 4F, rows 8 and 9), indicating that post-Golgi IR-wt resides in one (or both) of these compartments.
Altogether, this suggests that at steady state there are two distinct populations of IR-wt in the cell. The vast majority of newly synthesized protein fails to leave the ER as does the related mutant Δ190–208, which is affected in a similar region. However, unlike Δ190–208, a very small fraction of IR-wt does eventually escape and is targeted to LAMP1/LAMP2a-positive compartments, wherein it extensively accumulates over time. Hence, this post-Golgi pool of IR-wt may be used to assess whether an intact NTR-PKD domain boundary is, beyond proper trafficking, also necessary for fibril formation in acidic lysosome(like) compartments.
An Intact NTR-PKD Domain Boundary Is Necessary for Normal Fibril Formation
To determine whether an intact NTR-PKD domain boundary is necessary for fibril formation we performed electron microscopy (EM) using IR-wt-expressing cells. Interestingly, despite equal levels of NKI-beteb-reactive Pmel17 in Mel220 cells transfected with wt-Pmel17 or IR-wt (Fig. 4A), we detected striated stage II melanosomes only in those cells expressing the wild-type protein (Fig. 5A, panel 1). In stark contrast, we could not find any morphologically normal, fibril-containing stage II melanosomes in IR-wt-expressing Mel220 cells. At most, we saw a few immature-looking MVBs that occasionally contained some individual striae, most of which could be identified as membranes judged by a visible double layer structure (data not shown). However, in a low number of cases the resolution of the picture did not allow us to fully exclude that these striae represent fibrils in the process of formation (Fig. 5A, panel 2). Nevertheless, such cases were rare and the respective organelles were clearly distinguishable from bona fide stage II melanosomes observed in wt-Pmel17-expressing cells (Fig. 5A, panel 1) by both organelle shape and density of striations. From this we conclude that IR-wt is at least dramatically impaired, but more likely completely blocked in fibril formation. However, because this mutant gives rise to an abundant post-ER population localizing to LAMP1/LAMP2a-positive compartments, yet does not significantly contribute to fibrils, the obvious question is, in which organelles is it localized on an ultrastructural level? Interestingly, as for wt-Pmel17 (Fig. 5B, panel 1) HMB50-associated immunolabeling of IR-wt was to some extent observed in MVBs, but within the MVBs it was not restricted to the limiting membrane. In fact, it clearly extended over the ILVs (Fig. 5B, panel 2). This suggests that the defect of IR-wt in fibril formation is not caused by an impairment in the budding step as suggested for some N-terminal deletion mutants reported by Theos and co-workers (10). However, the vast majority of IR-wt-associated immunolabeling seemed to be concentrated in small perinuclear and often multilamellar structures morphologically resembling lysosomes (Fig. 5B, panels 4 and 5). In contrast, in wt-Pmel17-expressing cells most of the labeling was detected in ellipsoid, fibril containing organelles with the typical characteristics of stage II melanosomes (Fig. 5B, panel 3).
Therefore, and in line with our IF data (Fig. 4, B and D), EM analysis confirms that the post-ER pools of IR-wt and wt-Pmel17 reside in different subcellular compartments. Moreover, our results suggest that the integrity of the NTR-PKD domain boundary is necessary for appropriate fibril formation in melanoma cells.
Loss of Integrity at the NTR-PKD Domain Boundary Causes Pmel17 Misrouting to Lysosomes
The EM data strongly suggested that the post-ER populations of (non-functional) IR-wt and (functional) wt-Pmel17 localize to lysosomes and melanosomes, respectively (Fig. 5B, panels 3–5). To examine this further, we co-stained Mel220 transfectants with antibodies against folded Pmel17 (HMB50) or LAMP1 and analyzed the cells by IF. Consistent with our data shown in Fig. 4 (B and D), we found most of the wt-Pmel17 protein distributed in the characteristic “horseshoe” pattern in the majority of cells, while only relatively low levels of LAMP1 were associated with this broad U-shaped band surrounding the perinuclear area (see supplemental Fig. S3A for a schematic representation and Fig. 5C, left panel, row 2). In contrast, only relatively little Pmel17-associated fluorescence was found in the immediate juxtanuclear region, where in turn the LAMP1 staining peaked. Thus, for the wild-type protein we observed a high degree of complementarity for the distribution of Pmel17 and LAMP1 in the cell. This was also confirmed in a quantitative approach measuring differences in Pmel17 and LAMP1 fluorescence patterns along the body of a cell (supplemental Fig. S3A for a schematic representation and supplemental Fig. S3B and Fig. 5D). In the light of an earlier EM-based study, which demonstrated that in melanoma cells lysosomes contain high levels of LAMP1, but low levels of Pmel17, while the reverse is true for melanosomes (30), this strongly supports the view that functional Pmel17 is mainly routed to melanosomes (LAMP1low compartments), but not to lysosomes in Mel220 cells. In contrast, IR-wt-associated fluorescence was almost completely restricted to the perinuclear area, where the protein fully overlapped with LAMP1 in an identical pattern (Fig. 5C, left panel, row 3, supplemental Fig. S3B and Fig. 5D for quantification). Differences in the subcellular distribution of wt-Pmel17 and IR-wt relative to LAMP1 were substantial and statistically significant (Fig. 5D). This indicates that IR-wt migrates to lysosomes (LAMP1high compartments) rather than melanosomes, an observation that is also strongly supported by our EM results shown in Fig. 5B. Interestingly, experiments co-expressing wt-Pmel17 and IR-wt in Mel220 cells did not provide evidence that the two constructs (wild-type and mutant) affect each other's subcellular distribution and maturation to a significant extent (supplemental Figs. S4 and S5). Rather strong Pmel17-specific labeling (HMB50) was found in both juxtanuclear LAMP1high compartments as well as in horseshoe-shaped LAMP1low compartments in these cells.
Taken together, by both morphological criteria on the ultrastructural level, as well as by its subcellular distribution pattern and co-localization characteristics with LAMP1, IR-wt is demonstrated to misroute into lysosomal compartments. Because this mutant is affected at the NTR-PKD domain boundary, the integrity of this region may also be necessary for an appropriate routing of the polypeptide after export from the ER.
The Pmel17 Point Mutants H190P and R191S Are Affected in ER Export
The results in Figs. 3–5 showed that the quadruple amino acid exchange mutant IR-wt (190HR191/193GS194→PS/KR) is severely compromised in ER export, subcellular targeting, and fibril formation. Although our data strongly suggest that this construct is not cleaved in the ER by pPCs at the newly introduced pPC-cleavage motif generated by these exchanges (Fig. 3, A–C), we cannot exclude that the very low, but steady flow of IR-wt that appears to slowly build up the lysosomal protein pool eventually undergoes cleavage in an acidic compartment, where pPCs are normally active (i.e. we know that Pep13h-reactive, ER-localized IR-wt is not cleaved at position 195 (Fig. 3B), but we cannot make a statement about the lysosomal population detected by antibody HMB50). Thus, theoretically the behavior of this mutant may at least in part depend on the presence of the additional pPC-recognition motif rather than on an altered NTR-PKD domain boundary per se.
To address this issue we individually introduced into Pmel17 the four single amino acid substitutions that together constitute the IR-wt mutation and analyzed whether any of these single missense mutations would recapitulate the IR-wt phenotype. We underline that, with the exception of S194R, none of these single mutations (H190P, R191S, and G193K) would de novo generate a consensus pPC-cleavage motif (RX(R/K)R↓) (34). All four mutants could be stably expressed in Mel220 cells (Fig. 6A). Strikingly, H190P almost completely mimicked the processing and trafficking defect of IR-wt as judged by virtual absence of Mβ at steady state (Fig. 6A, left panel), dramatically lower levels of HMB45-reactive fibrillogenic fragments (Fig. 6A, right panel), almost complete restriction of the newly synthesized protein to the ER (Fig. 6B), and very low surface levels (Fig. 6C). Moreover, like IR-wt, H190P reactive with conformation-sensitive antibodies also accumulated to a large extent (Fig. 6D) in lysosomal compartments, as assessed by a comparison of its subcellular distribution pattern with that of LAMP1 in the cell (Fig. 5, C and D, and supplemental Fig. S3B). In contrast, mutants G193K and S194R behaved like wt-Pmel17 in all assays (Figs. 5C and 6, A–C), suggesting that they are functionally normal. Interestingly, mutant R191S displayed an intermediate phenotype with reduced levels of Mβ (Fig. 6A, left panel), reduced levels of HMB45-reactive fibrillogenic fragments (Fig. 6A, right panel), and substantially lower surface levels (Fig. 6C) when compared with wt-Pmel17. Also, when the distribution of newly synthesized R191S was examined by IF, ER staining clearly dominated over Golgi labeling (Fig. 6B), although, in contrast to H190P and IR-wt, a significant fraction of the protein weakly co-localized with Golgi marker GM130 (Fig. 6B, right panel). This is consistent with a somewhat milder ER retention defect for R191S. However, when the patterns of folded R191S and LAMP1 were compared, most cells showed the horseshoe profile typical for wt-Pmel17 (Fig. 5C), indicating that R191S was only moderately (if at all) compromised in post-ER trafficking.
FIGURE 6.
Pmel17 missense mutants H190P and R191S display a strong and a weak ER export defect, respectively. A, H190P and R191S generate lower steady-state levels of proprotein convertase-produced fragments as well as lower levels of HMB45-reactive fibrillogenic fragments. Membrane lysates of Mel220 transfectants stably expressing Pmel17 mutants H190P, R191S, G193K, and S194R were prepared as in Fig. 1B and analyzed by Western blot using Pmel17-specific antibodies. B, newly synthesized H190P is largely retained in the ER and also R191S seems to be exported slower from this compartment. Cells from Fig. 6A were analyzed by immunofluorescence using antibodies against newly synthesized Pmel17 (Pmel-N) and organelle markers TAP1 (148.3) (ER) or GM130 (610823) (Golgi). C, H190P and R191S are present at the cell surface at abnormally low levels. Cells from Fig. 6A were surface-labeled with antibody NKI-beteb against folded Pmel17 and analyzed by flow cytometry (left panel). After background subtraction (untransfected Mel220 cells) data are represented as a bar diagram (right panel). D, high levels of folded H190P and R191S can be detected intracellularly with antibody NKI-beteb. Cells from Fig. 6A were fixed in 2% formaldehyde, permeabilized, and stained with antibodies reactive with folded Pmel17 (NKI-beteb) and analyzed by flow cytometry (top panel). After background subtraction (untransfected Mel220 cells) data are represented as a bar diagram (bottom panel).
Taken together, our results show that a single amino acid substitution at the NTR-PKD domain boundary can be sufficient to recapitulate the dramatic ER export and sorting defect displayed by IR-wt. This once again underlines the importance of the integrity of this region for Pmel17 function.
Point Mutations H190P and R191S Abrogate or Severely Impair Fibril Formation by Pmel17, Respectively
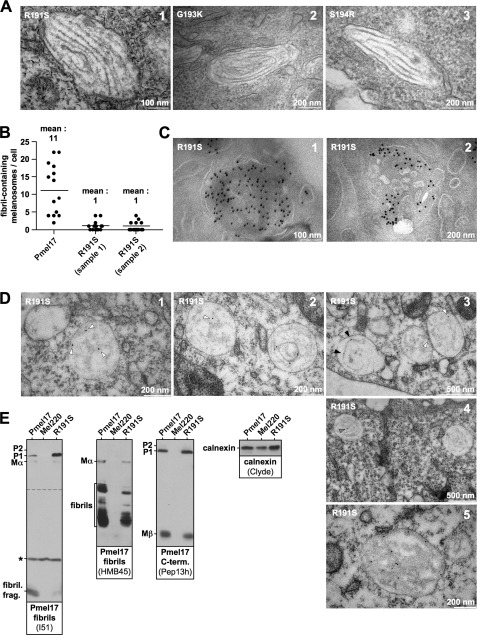
Our data in Fig. 6 show that two point mutations in Pmel17, H190P and R191S, substantially affect ER export of the protein, although to a different extent. Next, we wanted to test by EM, whether any of these mutations would also interfere with fibril formation in vivo. As expected, the two Pmel17 mutants G193K and S194R, which behaved normally in terms of ER export, gave rise to normal looking fibril-containing stage II melanosomes (Fig. 7A, panels 2 and 3) and also the R191S mutant, which was only mildly impaired in release from the ER was not absolutely blocked in formation of fibrils (Fig. 7A, panel 1). However, in two independently prepared samples we observed an 11-fold lower number of fibril-containing melanosomes for this mutant compared with wt-Pmel17 (Fig. 7B). Thus with R191S, fibril formation was clearly and dramatically affected. Consistent with the results in Fig. 5 (C and D), H190P did not form any apparent fibrils when assessed by EM, but rather represented a loss-of-function mutant.
FIGURE 7.
Pmel17 mutant R191S is substantially affected in fibril formation. A, fibril formation by Pmel17 mutants. Electron microscopic analysis of Epon-embedded Mel220 transfectants stably expressing R191S, G193K, and S914R. Panels 2 and 3 show typical melanosomes found for G193K and S194R. Panel 1 shows one of the very rare examples of a morphologically normal melanosome in R191S-expressing cells (see also B). B, mutant R191S is quantitatively impaired in fibril formation. Fibril-containing melanosomes were counted in Epon-embedded samples of Mel220 cells stably expressing wt-Pmel17 or mutant R191S. To confirm the results with mutant R191S a second sample was independently prepared and quantitated in the same way. C, mutant R191S is frequently found in MVBs and localizes to intralumenal vesicles therein. Mel220 cells expressing Pmel17 mutant R191S were fixed and examined by cryo-immunoelectron microscopy (panels 1 and 2) using antibody HMB50. D, Pmel17 mutant R191S frequently forms somewhat amorphous structures in immature looking MVB-like organelles. Mel220 cells expressing Pmel17 mutant R191S were fixed and examined by immunolabeling of LR-gold-embedded samples with antibody HMB50 (panels 1–5). White arrowheads point to gold-labeled, somewhat amorphous structures frequently found in R191S-expressing cells. These structures may represent Pmel17 protofibrils. Black arrowheads point to individual linear filaments sometimes seen in these cells. Gray arrowheads point to gold-labeled bundles of fibrils. E, mutant R191S produces lower steady-state levels of fibrillogenic fragments. Membrane lysates of Mel220 transfectants stably expressing wt-Pmel17 or R191S were prepared as in Fig. 1B and analyzed by Western blot using Pmel17-specific antibodies. Antibody I51 detects a recently described fibril-associated fragment containing the PKD domain of Pmel17. Antibody HMB45 detects a set of fibril-associated fragments containing the RPT domain of Pmel17. Calnexin-staining was used as a loading control. To visualize the P1-band better, a longer exposure of the upper part (separated by a dashed line from the lower part) of the same I51 blot (left panel) is shown. *, a nonspecific protein recognized by I51.
The slightly milder fibril formation defect associated with R191S raised the question of why this mutant was functionally affected, because at least the post-ER population of this Pmel17 derivative showed no obvious alterations in trafficking (Fig. 5C). To address this issue, we first examined where the bulk of folded HMB50-reactive R191S resided in the cell. Interestingly, cryo-immunoelectron microscopy revealed considerable Pmel17-specific staining in MVBs, where gold-particles were distributed all over the organelle, including extensive labeling of the ILVs (Fig. 7C, panels 1 and 2). This suggests that the R191S mutation does not cause an apparent defect in budding into the interior of the MVB, but rather that a later step in fibril formation is affected. Consistent with this, immunolabeling of LR-gold-embedded EM samples, an approach that in our hands strongly improves the preservation of fibrillar structures (data not shown), showed almost all of the Pmel17-specific staining to be associated with immature looking, large, roundish organelles (Fig. 7D, panels 1–3). Most of the gold particles therein labeled loose, somewhat amorphous, moderately electron-dense structures that may correspond to abnormal aggregates that have not (or not yet) assembled into mature fibrils (Fig. 7D, panels 1–3, white arrowheads). We note, however, that these structures morphologically resemble Pmel17-protofibrils that had recently been described to emanate from the ILV surface in stage I melanosomes (2). In the case of wild-type protein these would presumably later assemble into mature fibrils. Thus, an attractive alternative interpretation of these fibrous networks observed in R191S-expressing cells is that they represent protofibrils incapable of associating laterally, rather than merely reflecting malformed aggregates. In this case, assembly of fibrils into sheets rather than fibril formation per se may be impaired with the R191S mutant. Consistent with this, individual filamentous structures decorated with gold were sporadically seen in otherwise still immature looking organelles (Fig. 7D, panel 3, black arrowheads). Very rarely these filaments were organized in bundles that resemble mature fibrils (Fig. 7D, panel 4, gray arrowheads), and not surprisingly immunogold labeling could be dense along this fabric in these few cases (Fig. 7D, panel 5). These more conventional looking organelles likely correspond to the few fibril-harboring melanosomes of normal morphology that are occasionally observed in R191S-expressing cells (Fig. 7, A and B). Interestingly, we found significantly lower levels of HMB45-reactive fibrillogenic fragments in R191S-expressing cells (Figs. 6A, right panel, and 7E, second panel), and a very recently described 7-kDa PKD-derived fragment reactive with antibody I51 (14) was present to a substantially lower extent in these cells (Fig. 7E, first panel). Thus, it is possible that fibril formation by R191S is impaired, because in most cells the key fibril-forming fragments rarely reach a particular threshold density required for efficient aggregation.
Taken together, our results on the Pmel17 point mutants H190P and R191S once more demonstrate the functional importance of an intact NTR-PKD domain boundary for fibril formation of the polypeptide. Moreover, the distinct phenotype of the two mutants suggests that integrity of this junction is crucial to multiple steps in the processing of Pmel17, including subcellular targeting, maturation, and eventually assembly of submolecular fragments into fibrils.
DISCUSSION
Pmel17 is a molecule localized to melanosomes that exerts a critical function within these organelles by assembling into a fibrillar sheet structure, onto which potentially toxic reaction intermediates of the melanin synthesis pathway as well as the mature pigment get deposited and concentrated (1–3). At present there is good evidence that at least fragments containing the PKD domain as well as fragments containing the RPT domain are an integral part of these fibrillar aggregates. First, antibodies targeting these regions within Pmel17 directly recognize the fibrils (9, 14, 30), and second, fibril-enriched subcellular fractions contain high amounts of both a set of 35- to 45-kDa HMB45-reactive RPT-derived fragments (see Fig. 7E, second panel) (13, 14) and a very prominent 7-kDa PKD-derived fragment reactive with antibody I51 (see Fig. 7E, first panel) (14). A contribution of the NTR to the fibrils is also possible, but much less clear (14).
This raises the question of whether the region at the NTR-PKD domain boundary that is affected in most of the constructs used in this study is actually part of the fibrils or whether it is positioned slightly upstream of (or at) the N-terminal border of the 7-kDa PKD-containing fragment. We note that the reactivity with antibody I51 demands a minimal N-terminal extension of this amyloidogenic fragment to about residue 206. Furthermore, if antibody HMB50 reacts with the fibrils via recognition of that same fragment (which the study of Watt and colleagues (14) strongly suggests) its C-terminal border should lie within residues 234–293 according to epitope-mapping studies (19). Moreover, Watt and co-workers identified a 7-kDa protease-resistant fragment derived from in vitro-assembled PKD-containing amyloid fibrils that according to mass-spectrometric analysis contained peptide-segments roughly covering regions from 226 to 267. Together, this strongly suggests that the PKD-derived fibrillogenic fragment found in cells covers at least residues 206–267, which would result in a peptide of 6.8 kDa, thus closely matching the 7-kDa size observed in gels (14). Interestingly, we detect this fragment only by Western blotting (Fig. 7E, first panel), but not in pulse-chase experiments employing antibody HMB50 (Fig. 1C). This would suggest that the respective fragment cannot be labeled by 35S and consequently that methionines and cysteines are absent. Indeed, as the stretch between residues 185–300 of Pmel17 does not contain either of these amino acids, the N terminus of the PKD-derived fragment is likely to lie downstream of residue 184. Unfortunately, a more accurate mapping is not possible on the basis of the current data, and the NTR-PKD domain boundary is equally likely to be “just inside” or “just outside” the fibrils.
In either case it is clear that this region is very close to a critical key site of Pmel17 processing, and it is therefore plausible to assume that mutations specifically targeting this area might interfere with the efficient generation of fibrils. This would be particularly true if the PKD-derived fragment forms the amyloid core as suggested in the recent study by Watt and colleagues (14). In fact, besides the reduced delivery of R191S out of the ER per se, we speculate that this scenario may additionally contribute to the highly reduced levels of the PKD-derived fibrillogenic fragment in cells expressing this mutant (Fig. 7E, first panel). This may explain why R191S fails to be efficient in the formation of fibrils despite its high abundance in LAMP1low compartments and thus relatively normal subcellular trafficking of at least its post-ER population (Figs. 5C and 7C, panels 1 and 2). Surprisingly, we did not observe substantial differences in NKI-beteb-associated fluorescence between wt-Pmel17- or R191S-expressing cells in flow cytometry-based assays (Fig. 6D), although this antibody very likely recognizes the PKD-derived fragment in this setting. This suggests that the antibody concentrations used in the experiment may have been saturating. Alternatively, it is possible that NKI-beteb predominantly recognizes a distinct set of fragments generated from R191S (but not from wt-Pmel17), which does not react with I51. If so, detection of such fragments would probably pose a substantial challenge, because the absence of sulfur-containing amino acids in the respective region would likely render these fragments also undetectable by pulse-chase-based techniques.
We further speculate that, for fibril formation by Pmel17 to be efficient, certain local threshold concentrations of the amyloidogenic fragments may have to be exceeded. This might explain why a small subset of R191S-expressing cells nevertheless contains a few melanosomes with fibrils (Figs. 7, A, panel 1, and B), because among the bulk transfectants a fraction of cells with very high Pmel17 expression may succeed in generating enough fibrillogenic material even from a relatively unfavorable precursor. Indeed, concentration dependence of amyloid formation has been observed in other systems and the local availability of potentially aggregating peptide species may not only determine whether or not fibrils are formed, but also regulate the particular type of fibril that appears (35). We note that this phenomenon might explain the somewhat amorphous, HMB50-reactive fabric frequently observed in immature-looking organelles of R191S-expressing cells (Fig. 7D, panels 1–3, white arrowheads) if it arose as a consequence of abnormally low presence of fibrillogenic peptides. Alternatively, these somewhat loose, fibrous structures may actually represent protofibrils that have failed to assemble properly into the characteristic amyloid sheets observed in wt-Pmel17-expressing cells (2). If this is true, arginine 191 may be part of an interaction interface required for lateral assembly of nascent protofibrils into a planar arrangement. Replacement of this residue with serine may hence substantially decrease the rate (and/or stability) of sheet formation, thus causing protofibrils to largely accumulate in an unassembled state. In such a scenario, fibril formation per se would not be affected in R191S, but rather a later step of higher order assembly of these fibrils would be impaired.
Interestingly, two other mutations close to the NTR-PKD domain boundary (IR-wt and H190P) also strongly interfere with fibril formation, although large post-ER pools can be built up for both constructs (Fig. 6D). This once again confirms the importance of integrity in this key region for the fibril formation process. Strikingly, we observe drastically reduced levels of fibrillogenic fragments in both these cases (Figs. 3B, right panel, and 6A, right panel), strongly suggesting that defective processing is causing the impairment. Interestingly, once released from the ER, IR-wt seems to traffic relatively normally in the cell as it successfully accesses MVBs and buds into their interior (Fig. 5B, panel 2). However, the fibril formation process downstream of this trafficking step appears to be affected as IR-wt ultimately acquires a lysosomal distribution very similar to that reported for ΔRPT-mutants expressed in HeLa cells (10, 13). As in all these cases (including the ΔRPT-mutants) RPT-based fragments are either absent or only present at minute levels (Figs. 3B, right panel, and 6A, right panel), this might indicate a role for these peptides in regulating the fate of early stage melanosomes to develop into mature, pigment storing organelles instead of developing into lysosomes. However, the loss-of-function phenotype associated with IR-wt would also be consistent with a direct role for RPT-derived fragments in fibril formation (4). Interestingly, like IR-wt, Pmel17 mutant H190P is also highly deficient in fibril formation as assessed by EM. Conformational effects caused by the introduction of a structurally “inflexible” proline at the NTR-PKD domain junction might underlie this phenotype, although both H190P and IR-wt interact efficiently with antibodies recognizing only folded Pmel17, namely HMB50 and NKI-beteb (Figs. 5C, 6C, and 6D). This suggests that at least neither mutant is globally misfolded. Alternatively, the identity of histidine at position 190 may be required for fibril formation. We note that, in acidic organelles like melanosomes, histidine through its side-chain pK value of 6.0 would be positively charged and thus of dramatically different character than the hydrophobic proline present in its position in the H190P mutant. Indeed, the drastic effect on fibril formation of even a mild exchange of arginine (positively charged) to serine (polar) in position 191 (Fig. 7B) and thus in the immediate vicinity of H190 makes this a plausible scenario as well.
Very surprising are the drastic ER export defects associated with Δ190–208, IR-wt, and H190P (and to a lesser extent also with R191S) (Figs. 2 (B–G), 3 (B–E), and 6 (A–C)). Experiments with Δ190–208, which is completely ER-retained, suggest that excessive global misfolding of the construct does not underlie the phenotype. First, the UPR was not induced in cells stably expressing Δ190–208 (supplemental Fig. S2A). Second, its half-life of >3 h was not consistent with Δ190–208 being an unstable, rapidly turned over ERAD substrate (supplemental Fig. S2B). Thirdly, Δ190–208 reacted with both conformation-sensitive antibodies used in this study (Fig. 2F), as did IR-wt and H190P (Figs. 4A, 4B, 5C, and 6D). Our results yet do not exclude that the structure of Δ190–208 (as well as IR-wt and H190P) may be locally disturbed when compared with wt-Pmel17. However, any such potential alteration does not appear to result in a destructive effect on the conformation-sensitive epitopes that antibodies HMB50 and NKI-beteb recognize. This suggests that at least the PKD domain can be folded in these mutants, and consequently the structure of these molecules is not globally disrupted. Also, in whatever structural conformation Δ190–208 might exist in the cell, it does not appear to be in a conformation where hydrophobic patches are excessively exposed at the polypeptide surface, because this would be predicted to induce its rapid degradation. Overall, the data suggest that Δ190–208 behaves more like a normal ER-resident protein rather than an excessively misfolded polypeptide, which would be recognized by the cellular quality control machinery upstream of the UPR and ERAD pathways.
In principle, there are two plausible scenarios that might explain the observed ER retention of the constructs. Either the respective mutants are actively retained in the ER by factors that do not (or do not persistently) interact with wt-Pmel17, or these mutants fail to participate in an interaction that is under normal circumstances required for efficient ER exit of the wild-type protein. Indeed, cases have been described where an ER-lumenal interaction with a “tethering factor” is necessary to recruit otherwise ER-retained molecules into coat protein complex II (COPII)-coated vesicles and allow for their efficient export (36). However, most ER export motifs identified so far are located rather in the cytosolic tails of cargo proteins, which is not surprising, because this arrangement allows for a direct interaction with the COPII coat (37). Indeed, Pmel17 has been shown to contain such a cytosolic export motif at the C terminus, and its deletion as found in the silver mutation causes a drastically decreased efficiency of release from the ER (38). Therefore, it is also a possible scenario that mutations targeting the NTR-PKD domain boundary drive the polypeptide into a conformational state that has a far reaching structural effect regulating accessibility of the cytosolic tail. Such an effect may cause masking of the C-terminal export motif, resulting in retention, because the signal for incorporation into COPII vesicles and thus forward trafficking is lacking. Indeed, examples where mutant proteins fail to leave the ER, because the COPII machinery cannot access a critical export motif due to permanent masking, have been described (39). Moreover, keeping cytosolic export signals inaccessible via a conformational switch is a mechanism used by some polypeptides to regulate their ER-to-Golgi transport (40). It is thus possible that the NTR-PKD domain boundary is a sensitive structure that, when affected, permanently locks mutants like Δ190–208, IR-wt, or H190P in a conformation that does not allow the COPII coat to efficiently access the ER export motif. This would then trap the mutants in the ER. Indeed, we note that a recent report on the Dominant White mutant of Pmel17 supports a possible structural cross-talk between the transmembrane domain and the lumenal domain of the molecule (41).
Interestingly, Pmel17 lacking the whole NTR (Δ28–208) is efficiently exported into the Golgi (Fig. 1, C and D), showing that, at least in the context of this truncated protein, ER release does not essentially require the residues comprising the NTR-PKD domain boundary (190–208). Moreover, as assessed by pulse-chase experiments, at least in the HeLa/overexpression system almost any lumenal region, including the NTR and the PKD (in the respective study (10) defined as residues 29–201 and 235–292, respectively) could be deleted within Pmel17 without dramatically affecting ER export anywhere near to the extent that what we observe for Δ190–208 or IR-wt (10) (see e.g. Fig. 3C). Consequently, the particular residues at the NTR-PKD junction are unlikely to directly interact with an essential ER-exit-promoting factor.
Finally, another possible scenario that could explain the export defect of mutants like Δ190–208 is active ER retention. Chaperone networks in the ER detecting subtle local alterations to the native Pmel17 structure may serve such a purpose, although the absence of UPR induction and lack of rapid delivery of the polypeptide to the ERAD pathway do not support such an argument. Alternatively, mutant Pmel17 may self-associate or associate with unknown retention factors other than chaperones. However, by EM we did not observe visible aggregate formation in the ER by Δ190–208, IR-wt, and H190P, suggesting that ER retention is at least not caused by uncontrolled initiation of the aggregation process.
Taken together, in this report we identify a novel segment within Pmel17 that is of critical importance for ER export, subcellular targeting, and fibril formation by the polypeptide. Hence, our study provides novel insights into how Pmel17 establishes the fibrillar network that is of crucial importance for the biology of melanosomes.
Supplementary Material
Acknowledgments
We are indebted to Drs. M. Marks, A. Gown, and M. Skelly for the kind donation of Pmel17-specific antibodies Pep13h, Pmel-N, and HMB50, respectively. We further thank Dr. M. Marks for very helpful discussions and communication of unpublished results. We are grateful to Dr. M. Graham for help with the electron microscopic analysis. Dr. J. Grotzke is acknowledged for critically reading the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R37-AI23081 (to P. C.). This work was also supported by the Howard Hughes Medical Institute and Yale SPORE in Skin Cancer Grant 5P50 CA121974 (to P. C.). Furthermore, support was provided by a Cancer Research Institute Fellowship (to R. M. L.) and a Marie Curie Outgoing International Fellowship from the European Union and the Fonds National de la Recherche Scientifique, Belgium (to N. V.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, additional references, and Figs. S1–S5.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated protein degradation
- aa
- amino acid(s)
- ILV
- intralumenal vesicle
- MVB
- multivesicular body
- NTR
- N-terminal region
- PKD
- polycystic kidney disease-like domain
- pPC
- proprotein convertase
- RPT
- repeat domain
- UPR
- unfolded protein response
- wt
- wild type
- PBS
- phosphate-buffered saline
- IF
- immunofluorescence
- EM
- electron microscopy
- COPII
- coat protein complex II.
REFERENCES
- 1.Fowler D. M., Koulov A. V., Alory-Jost C., Marks M. S., Balch W. E., Kelly J. W. (2006) PLoS Biol. 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurbain I., Geerts W. J., Boudier T., Marco S., Verkleij A. J., Marks M. S., Raposo G. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19726–19731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Z. H., Hou L., Moellmann G., Kuklinska E., Antol K., Fraser M., Halaban R., Kwon B. S. (1996) J. Invest. Dermatol. 106, 605–610 [DOI] [PubMed] [Google Scholar]
- 4.McGlinchey R. P., Shewmaker F., McPhie P., Monterroso B., Thurber K., Wickner R. B. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 13731–13736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quevedo W. C., Fleischmann R. D., Dyckman J. (1981) in Phenotypic Expression in Pigment Cells (Seiji M. ed) pp. 177–184, Tokyo University Press, Tokyo [Google Scholar]
- 6.Theos A. C., Truschel S. T., Raposo G., Marks M. S. (2005) Pigment Cell Res. 18, 322–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raposo G., Marks M. S. (2007) Nat. Rev. Mol. Cell Biol. 8, 786–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo G., Marks M. S. (2002) Traffic 3, 237–248 [DOI] [PubMed] [Google Scholar]
- 9.Berson J. F., Harper D. C., Tenza D., Raposo G., Marks M. S. (2001) Mol. Biol. Cell 12, 3451–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Theos A. C., Truschel S. T., Tenza D., Hurbain I., Harper D. C., Berson J. F., Thomas P. C., Raposo G., Marks M. S. (2006) Dev. Cell 10, 343–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berson J. F., Theos A. C., Harper D. C., Tenza D., Raposo G., Marks M. S. (2003) J. Cell Biol. 161, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kummer M. P., Maruyama H., Huelsmann C., Baches S., Weggen S., Koo E. H. (2009) J. Biol. Chem. 284, 2296–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoashi T., Muller J., Vieira W. D., Rouzaud F., Kikuchi K., Tamaki K., Hearing V. J. (2006) J. Biol. Chem. 281, 21198–21208 [DOI] [PubMed] [Google Scholar]
- 14.Watt B., van Niel G., Fowler D. M., Hurbain I., Luk K. C., Stayrook S. E., Lemmon M. A., Raposo G., Shorter J., Kelly J. W., Marks M. S. (2009) J. Biol. Chem. 284, 35543–35555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigneron N., Ooms A., Morel S., Ma W., Degiovanni G., Van den Eynde B. J. (2005) Tissue Antigens 65, 156–162 [DOI] [PubMed] [Google Scholar]
- 16.Adema G. J., de Boer A. J., Vogel A. M., Loenen W. A., Figdor C. G. (1994) J. Biol. Chem. 269, 20126–20133 [PubMed] [Google Scholar]
- 17.Nichols S. E., Harper D. C., Berson J. F., Marks M. S. (2003) J. Invest. Dermatol. 121, 821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esclamado R. M., Gown A. M., Vogel A. M. (1986) Am. J. Surg. 152, 376–385 [DOI] [PubMed] [Google Scholar]
- 19.Harper D. C., Theos A. C., Herman K. E., Tenza D., Raposo G., Marks M. S. (2008) J. Biol. Chem. 283, 2307–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiamenti A. M., Vella F., Bonetti F., Pea M., Ferrari S., Martignoni G., Benedetti A., Suzuki H. (1996) Melanoma Res. 6, 291–298 [DOI] [PubMed] [Google Scholar]
- 21.Yasumoto K., Watabe H., Valencia J. C., Kushimoto T., Kobayashi T., Appella E., Hearing V. J. (2004) J. Biol. Chem. 279, 28330–28338 [DOI] [PubMed] [Google Scholar]
- 22.Meyer T. H., van Endert P. M., Uebel S., Ehring B., Tampé R. (1994) FEBS Lett. 351, 443–447 [DOI] [PubMed] [Google Scholar]
- 23.Hammond C., Helenius A. (1994) J. Cell Biol. 126, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton G. M., Medzhitov R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14943–14945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seliger B., Ritz U., Abele R., Bock M., Tampé R., Sutter G., Drexler I., Huber C., Ferrone S. (2001) Cancer Res. 61, 8647–8650 [PubMed] [Google Scholar]
- 26.Leonhardt R. M., Keusekotten K., Bekpen C., Knittler M. R. (2005) J. Immunol. 175, 5104–5114 [DOI] [PubMed] [Google Scholar]
- 27.Leonhardt R. M., Lee S. J., Kavathas P. B., Cresswell P. (2007) Infect Immun 75, 5105–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrithers M. D., Chatterjee G., Carrithers L. M., Offoha R., Iheagwara U., Rahner C., Graham M., Waxman S. G. (2009) J. Biol. Chem. 284, 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y., Gao B., Ye L., Kong L., Jing W., Yang X., Wu Z., Ye L. (2005) J. Microbiol. 43, 529–536 [PubMed] [Google Scholar]
- 30.Raposo G., Tenza D., Murphy D. M., Berson J. F., Marks M. S. (2001) J. Cell Biol. 152, 809–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoashi T., Tamaki K., Hearing V. J. (2009) FASEB J. 24, 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M., Grigoriev A. (2004) Trends Genet. 20, 399–403 [DOI] [PubMed] [Google Scholar]
- 33.Tanaka T., Yokoyama S., Kuroda Y. (2006) Biopolymers 84, 161–168 [DOI] [PubMed] [Google Scholar]
- 34.Remacle A. G., Shiryaev S. A., Oh E. S., Cieplak P., Srinivasan A., Wei G., Liddington R. C., Ratnikov B. I., Parent A., Desjardins R., Day R., Smith J. W., Lebl M., Strongin A. Y. (2008) J. Biol. Chem. 283, 20897–20906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen J. S., Otzen D. E. (2008) Protein Sci. 17, 2–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bökel C., Dass S., Wilsch-Bräuninger M., Roth S. (2006) Development 133, 459–470 [DOI] [PubMed] [Google Scholar]
- 37.Barlowe C. (2003) Trends Cell Biol. 13, 295–300 [DOI] [PubMed] [Google Scholar]
- 38.Theos A. C., Berson J. F., Theos S. C., Herman K. E., Harper D. C., Tenza D., Sviderskaya E. V., Lamoreux M. L., Bennett D. C., Raposo G., Marks M. S. (2006) Mol. Biol. Cell 17, 3598–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Matteson J., An Y., Moyer B., Yoo J. S., Bannykh S., Wilson I. A., Riordan J. R., Balch W. E. (2004) J. Cell Biol. 167, 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun L. P., Seemann J., Goldstein J. L., Brown M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6519–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuliawat R., Santambrogio L. (2009) Eur J. Cell Biol. 88, 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.