Abstract
APOBEC3G (Apo3G) is a single-stranded DNA-dependent deoxycytidine deaminase, which, in the absence of the human immunodeficiency virus (HIV) viral infectivity factor, is encapsulated into HIV virions. Subsequently, Apo3G triggers viral inactivation by processively deaminating C→U, with 3′→5′ polarity, on nascent minus-strand cDNA. Apo3G has a catalytically inactive N-terminal CD1 domain and an active C-terminal CD2 domain. Apo3G exists as monomers, dimers, tetramers, and higher order oligomers whose distributions depend on DNA substrate and salt. Here we use multiangle light scattering and atomic force microscopy to identify oligomerization states of Apo3G. A double mutant (F126A/W127A), designed to disrupt dimerization at the predicted CD1-CD1 dimer interface, predominantly converts Apo3G to a monomer that binds single-stranded DNA, Alu RNA, and catalyzes processive C→U deaminations with 3′→5′ deamination polarity, similar to native Apo3G. The CD1 domain is essential for both processivity and polarity. We propose a structure-based model to explain the scanning and catalytic behavior of Apo3G.
Keywords: Enzyme Mechanisms, Human Immunodeficiency Virus, Mutagenesis Mechanisms, Protein-DNA Interaction, Viral Immunology, DNA Deaminase, HIV Restriction Factor
Introduction
Human APOBEC3G (Apo3G)3 is a single-stranded DNA (ssDNA) deoxycytidine deaminase that can restrict replication of HIV in strains lacking the virus infectivity factor protein (1). Virus infectivity factor facilitates polyubiquitination of Apo3G, resulting in proteasomal degradation (2–6). When virus infectivity factor is absent, however, Apo3G is encapsulated in the HIV virus particle, most likely as a multimer (7), through interactions with HIV RNA or 7SL RNA and the nucleocapsid portion of Gag (7–15). Following release from the HIV virion into T cells, Apo3G generates large numbers of deaminations, typically 5′-CCC→CCU, in viral (−) single-stranded cDNA during reverse transcription (16–18). Subsequent replication of the cDNA generates G→A mutations in HIV (+) DNA, causing proviral inactivation (16–18). As a counterpart to its role as a C-deaminase, Apo3G also has a non-enzymatic cellular function. It restricts retrotransposition by binding Alu RNA transcripts in the cytoplasm, thereby blocking the transposition cycle (19).
In this study, we address the relationship of Apo3G structure to biochemical function. An Apo3G monomer has two homologous deaminase domains, an active C-terminal domain (CD2) required for catalysis and motif specificity and a catalytically inactive N-terminal domain (CD1) needed for DNA and RNA binding (20, 21). The CD1 domain is believed to be required for the incorporation of Apo3G into virions (21).
Apo3G deaminates ssDNA processively, with a strongly polar 3′→5′ deamination bias (22, 23). There is no obvious energy source, e.g. ATP or GTP (22). Therefore, the processive sliding and jumping/hopping motions must be random, yet the deaminations are decidedly non-random (22, 23). Therefore, the spatial deamination bias must be a catalytic bias, not a motional bias. The question is: “What is the biochemical/structural mechanism?”
There was an overriding serious impediment in the way of addressing the mechanism because highly purified native Apo3G is composed of monomers, dimers, trimers, tetramers, and higher order oligomers in the absence of DNA (23, 24), whose relative amounts change dramatically when bound to ssDNA and show additional changes in composition depending on salt concentration (23). Apo3G may be present as an oligomer bound to RNA when released from the HIV virion into a T cell (7, 25), but most, if not all, of the RNA must be degraded to activate Apo3G for catalysis (25, 26). Thereafter, the function of the oligomer is uncertain. Does Apo3G work solely as a dimer, as is currently thought (24, 27), or alternatively, as a combination of monomers and oligomers (23)?
Here we investigate the biochemical properties of an Sf9-expressed Apo3G mutant, F126A/W127A. This mutant was designed to disrupt dimerization based on structures of APOBEC2 (Apo2) (28) and the Apo3G-CD2 catalytic domain (29). We analyze the ssDNA scanning and C-deamination properties of the monomer and the Apo3G-CD2 catalytic domain (in the absence of CD1) relative to those of the naturally occurring heterogeneous assortment of Apo3G monomers and oligomers (23, 24). We propose a model to account for the processive and polar characteristics of Apo3G-catalyzed deamination on ssDNA based on biochemical comparisons of the native and Apo3G monomer, in conjunction with structural studies on Apo2 (28) and Apo3G-CD2 (29).
EXPERIMENTAL PROCEDURES
Apo2 and Apo3G Structural Modeling
The predicted model of Apo3G CD1 was obtained by using the interactive SWISS-MODEL program using the homologous Apo2 (Protein Data Bank (PDB): 2nyt) structure as a template (see Fig. 1B). For the Apo3G dimer head-head configuration (see Fig. 1D), the Apo3G-CD1 model was aligned with each Apo2 monomer in the Apo2 tetrameric interface. For the Apo3G dimer tail-tail configuration (see Fig. 1E), the Apo3G-CD2 (PDB: 3e1u) structures were aligned with each Apo2 monomer in the tetrameric interface. All figures were made using PyMOL.
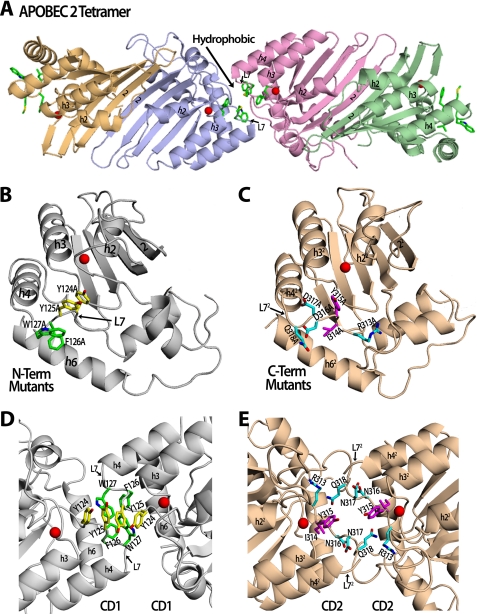
FIGURE 1.
Predicted structural models of Apo3G dimerization based on the Apo2 structure. A, the solved structure of the Apo2 tetramer containing hydrophobic residues (green sticks) on loop 7 that are at both the interface and the outside surface. Each monomer is displayed as a different color, and the zinc atom is a red sphere. B, the model of N-terminal (N-Term) Apo3G-CD1 (gray) shows the residues that were mutated to alanine. Tyr-124/Tyr-125 residues are shown as yellow sticks, and Phe-126/Trp-127 residues are shown as green sticks. C, the structure of C-terminal (C-Term) Apo3G-CD2 (wheat) shows the residues that were mutated to alanine. Arg-313/Asp-316/Asp-317/Gln-318 residues are shown as cyan sticks, and Ile-314/Tyr-315 mutant residues are shown as magenta sticks. D, a zoomed in view of the proposed Apo3G dimer interface in a head-head (CD1-CD1) configuration. Hydrophobic residues Tyr-124/Tyr-125 (yellow) and Phe-126/Trp-127 (green) on loop 7 (L7) were mutated to alanine and may be involved in forming a hydrophobic core at the dimer interface, similar to hydrophobic residues found in the equivalent loop in the Apo2 tetramer interface. E, a zoomed in view of the proposed Apo3G dimer interface in a tail-tail (CD2-CD2) configuration. Charged residues Arg-313/Asp-316/Asp-317/Gln-318 (cyan) and Ile-314/Tyr-315 (purple) were mutated to alanine and may be involved in forming a dimer interface using loop 7 (L72), which has previously been shown to interact with the DNA substrate (29, 36, 39).
Substrates
DNA oligonucleotides were synthesized on an Applied Biosystems 3400 DNA/RNA synthesizer and are listed in supplemental Table S1. Fluorescein-dT was purchased from Glen Research. Fluorescently labeled RNA was produced by transcribing the DNA sequence, cloned into the pSP72 vector (Promega) using XhoI and HindIII sites, with SP6 RNA polymerase with a pool of nucleotides containing fluorescein-12-UTP (Roche Applied Science).
Apo3G Expression and Purification
A baculovirus-expressed GST-Apo3G protein was constructed as described previously (22). Apo3G-CD2 domain (amino acids 197–380) was cloned into XmaI and EcoRI sites in the pAcG2T vector (BD Biosciences). Apo3G recombinant virus was obtained as described previously (30). Sf9 cells were infected with recombinant Apo3G virus at a multiplicity of infection of 1 and harvested after 72 h. Cells were lysed as described previously (22) in the presence of 50 μg ml−1 of RNase A. Cleared lysates were then incubated with glutathione-Sepharose resin (GE Healthcare) and subjected to a series of salt washes (0.25–1 m NaCl) (23) before on-column cleavage of the glutathione S-transferase tag with 40 units of thrombin (Calbiochem) for 16 h at 21 °C. Cleaved Apo3G fractions were collected and stored at −70 °C. Apo3G is ∼95% pure.
Site-directed Mutagenesis
Mutant Apo3G proteins were constructed by site-directed mutagenesis (QuikChange site-directed mutagenesis protocol, Stratagene) with modifications by Wang and Malcom (31) using the pAcG2T-Apo3G vector as the template (22). The primers that were used are listed in supplemental Table S2.
Multiangle Light Scattering (MALS)
Apo3G (50–100 μg in 30 μl) was subjected to size-exclusion chromatography using a silica-based column (8-nm diameter, 300-mm length, KW-803 Shodex). A solution containing 50 mm HEPES, pH 6.7, and 200 mm NaSO4 was used as the eluent buffer. Chromatography was performed at 0.5 ml min−1, and the column effluent was passed directly on-line into a MALS detector (Dawn Heleos, Wyatt Technology) and refractometer (Optilab rEX, Wyatt Technology). Data analysis to determine molecular masses was performed with ASTRA software. Experiments were conducted at the University of Southern California (USC) NanoBiophysics Core Facility.
Atomic Force Microscopy (AFM)
Apo3G imaging buffer (25 mm HEPES, pH 7.3, 5 mm MgCl2, 0.1 mm dithiothreitol) and sample were prepared as described previously (23). For experiments done in the presence of DNA substrate, 400 nm (final concentration) of ssDNA substrate was added to the Apo3G sample (100 nm final concentration) before depositing as for the protein alone. All AFM images were captured in air using a Nanoscope IIIa (Digital Instruments, Santa Barbara, CA) microscope in tapping mode. Experiments were conducted at the USC NanoBiophysics Core Facility. Pointprobe Plus tapping mode silicon probes (Agilent, Tempe, AZ) with resonance frequencies of ∼170 kHz were used for imaging. Images were collected at a speed of 2–3 Hz with an image size of 1 × 1 μm at 512 × 512-pixel resolution. Volume analysis was done as described previously (32, 33). The Sigma Plot program was used to generate statistical plots.
Deamination Assays
Apo3G activity on ssDNA substrates was measured in reactions containing 50 nm 32P-labeled DNA, 5 or 1000 nm Apo3G in buffer (50 mm HEPES, pH 7.3, 5 mm MgCl2, 1 mm dithiothreitol) and incubated for 2.5–30 min at 37 °C. ssDNA substrates (0.5 μm) with an internal fluorescein label were incubated with Apo3G (15–1000 nm), and deaminations were carried out as described above. Reactions were terminated by an extraction with phenol/chloroform/isoamyl alcohol (25:24:1), and deaminations were detected by the uracil DNA glycosylase assay, as described previously (22). Gel band intensities were visualized by phosphorimaging or with an FX fluorescence scanner (Bio-Rad) and then measured with ImageQuant software (GE Healthcare).
Pre-steady state reactions were mechanically sampled and quenched using a KinTek quench-flow instrument as described previously (23). Samples were then treated as described above.
Steady State Rotational Anisotropy Assays
Apo3G-RNA or -DNA binding was monitored by changes in steady state fluorescence depolarization (rotational anisotropy). Reaction mixtures (70 μl), containing fluorescein-labeled RNA or DNA (50 nm) in buffer (50 mm HEPES, pH 7.3, 150 mm NaCl, 10 mm MgCl2, 1 mm dithiothreitol) and varying concentrations of 0–2400 nm Apo3G, were incubated at 37 °C. Rotational anisotropy was measured using a QuantaMaster QM-1 fluorometer (Photon Technology International) with a single emission channel. Samples were excited with vertically polarized light at 494 nm, and both vertical and horizontal emission was monitored at 520 nm (4 nm band pass). The fractions of RNA or DNA bound to protein were determined as described previously (34).
Pre-steady State Anisotropy Assays
Reactions were at 21 °C in buffer containing 50 mm HEPES, pH 7.3, 5 mm MgCl2, and 1 mm dithiothreitol. Stopped-flow anisotropy measurements were performed using an Applied Photophysics Π*180 spectrofluorometer equipped for anisotropy as described previously (23).
RESULTS
Apo3G exists as a combination of monomers, dimers, tetramers, and higher oligomeric forms (23, 24) whose relative distributions change significantly when bound to ssDNA (23). Oligomerization appears to be essential for cellular functions such as virion incorporation (7) and Alu retrotransposon restriction (9, 27); however, the molecular interactions responsible for forming the different oligomers are poorly defined. Our principal objectives are: to determine the scanning, substrate binding, and deamination properties of monomeric Apo3G; to compare the biochemical mechanisms of monomeric Apo3G with CD2 and with native Apo3G; and to determine the role of CD1 in the scanning mechanism. Therefore, it is necessary to identify the amino acids responsible for oligomerization and introduce strategic mutations that interfere with oligomer assembly and to verify the composition of Apo3G native and mutant forms.
Generating Apo3G Oligomerization Mutants Based on the Apo2 Crystal Structure
We performed comparative homology modeling and structure-guided mutagenesis, based on the previously determined crystal structures of the homologous Apo2 tetramer (28) and the Apo3G CD2 domain (29), to design Apo3G mutants that are predicted to disrupt oligomerization. The double-domain Apo3G monomer is equivalent to the single-domain Apo2 dimer. Furthermore, the dimerization interface of Apo3G is predicted to be equivalent to the Apo2 tetramer interface. Apo2 tetramer oligomerization occurs through a head-head (i.e. N-N) dimer that forms a hydrophobic dimer-dimer interface created by residues found on loop 7 of each dimer (28) (Fig. 1A). Through sequence alignment of Apo2, Apo3G CD1 domain (head), and Apo3G CD2 domain (tail), the loop 7 cluster of residues was used to identify homologous residues on the equivalent loop 7 of the Apo3G CD1 domain and Apo3G CD2 domain (supplemental Fig. S1). We mutated these residues to alanine to determine whether we could disrupt oligomerization at the CD1 domain (Y124A/Y125A; F126A/W127A; Fig. 1B) or CD2 domain (R313A/D316A/D317A/Q318A;I314A/Y315A; Fig. 1C). Apo3G dimerization could presumably occur in a head-head (CD1-CD1, Fig. 1D), tail-tail (CD2-CD2, Fig. 1E), or head-tail (CD1-CD2, not shown) orientation. The hydrophobic residues located on loop 7 of Apo2 are highly conserved in the Apo3G CD1 domain (Fig. 1, A and D, supplemental Fig. S1). This homology is in contrast to the Apo3G CD2 domain loop 72, which contains mainly charged residues that were previously shown to be involved in DNA binding and specificity (29) (Fig. 1E, supplemental Fig. S1). Based on these observations, we predicted that Apo3G dimerization occurs in a CD1-CD1 head-head orientation (Fig. 1D).
MALS Analysis of the Oligomerization State of Native and Mutant Apo3G Proteins
The oligomerization state of Apo3G in native and mutant forms was determined using MALS (Fig. 2). Purified protein is injected into a high pressure liquid chromatography size-exclusion column followed in series by detectors that measure UV absorbance, MALS, and refractive index (35) (see “Experimental Procedures”). The light-scattering chromatogram (Fig. 2A, Raleigh ratio, right y axis) in combination with the molecular mass line plot (Fig. 2A, left y axis) shows that native Apo3G (predicted molecular mass 46.4 kDa) is present primarily as a dimer (∼80%, molecular mass 113 kDa) along with a small but significant population of monomers (∼18%) and ∼2% higher order oligomers with an average molecular mass of ∼650 kDa (Fig. 2A) (35). The molecular mass line plots, which plot the mass throughout the peak, each have a steep slope, demonstrating that native Apo3G is polydisperse in aqueous solution in the presence of salt (200 mm Na2SO4). There appear to be unresolved monomers, trimers, and tetramers contained in the dimer peak (Fig. 2A, line plot, left y axis). This result is in contrast to the constant mass plotted throughout the peak of a monodisperse protein, i.e. bovine serum albumin (supplemental Fig. S2). Similar results for native Apo3G were found over a 4-fold concentration range, suggesting that dilution of the protein during gel filtration is unlikely to be causing dissociation of multisubunit structures (data not shown).
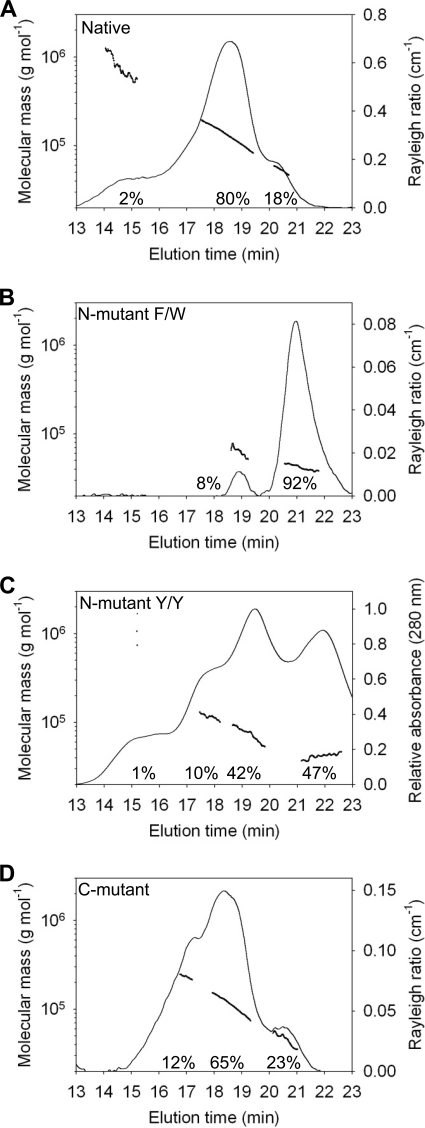
FIGURE 2.
Determination of the molecular mass of Apo3G native and mutants by MALS. Apo3G native and mutants were resolved by size-exclusion chromatography in running buffer with 200 mm NaSO4 and 50 mm HEPES, pH 6.7. The molecular mass is plotted throughout the eluted peaks (line plot, left y axis). The Rayleigh light scattering (35) or UV absorbance chromatograms showing the protein distributions are plotted on the right y axis. A, the median molecular masses and distributions for Apo3G native are 18% monomers (51 kDa), 80% dimers (113 kDa), and 2% large molecular mass species (650 kDa). B, the median molecular masses and distributions for F126A/W127A mutant (N-mutant F/W) are 92% monomers (43 kDa) and 8% dimers (63 kDa). C, the median molecular masses and distributions for Y124A/Y125A mutant (N-mutant Y/Y) are 47% monomers (42 kDa), 42% dimers (67 kDa), 9% tetramers (117 kDa), and 1% large molecular mass species (650 kDa). D, the median molecular masses and distributions for R313A/D316A/D317A/Q318A (C-mutant) are 23% monomers (46 kDa), 65% dimers (99 kDa), and 12% tetramers (216 kDa). The chromatograms for A, B, and D correspond to the relative intensity of light scattering (right y axis). The chromatogram for C corresponds to the relative intensity of absorption at 280 nm (right y axis).
We performed a MALS analysis on four Apo3G mutants predicted to interfere with oligomerization, based on crystal contacts in the Apo2 tetramer structure (28) (Fig. 1). Two mutants are located in the N-terminal CD1 domain (F126A/W127A and Y124A/Y125A) and two are in the C-terminal CD2 domain (R313A/D316A/D317A/Q318A and I314A/Y315A).
Mutant F126A/W127A causes a severe disruption in oligomer formation resulting in about 92% monomers and 8% dimers, with no larger oligomer forms detected (Fig. 2B, N-mutant F/W). The small population of dimers may result from residual CD1-CD1 interactions, perhaps from relatively weak tail-tail CD2-CD2 or head-tail CD1-CD2 interactions. It appears that the oligomer forms of Apo3G are stabilized in the presence of salt via hydrophobic interactions involving CD1, with just the two adjacent residues, Phe-126 and Trp-127, playing a dominant role. In contrast, the CD1 residues Tyr-124 and Tyr-125 have a far less significant role in oligomer formation. The mutant Y124A/Y125A is composed of roughly 47% monomers, 42% dimers, 10% tetramers, and 1% much larger molecular mass species (∼650 kDa) (Fig. 2C, N-mutant Y/Y). The absorbance chromatogram is used in place of the Rayleigh light-scattering ratio (35) to depict the forms for the Apo3G N-mutant Y124A/Y125A (Fig. 2C, chromatogram, right y axis) because of a stronger signal, but the two profiles are similar (data not shown). The molecular mass values associated with each absorbance peak are obtained from an analysis of MALS (35) (Fig. 2C, line plot, left y axis).
The quadruple mutant, R313A/D316A/D317A/Q318A, in the CD2 domain of Apo3G (Fig. 2D, C-mutant), corresponds to the region at the Apo2 tetrameric interface and predicted CD1 oligomerization region (residues 124–127), based on sequence alignment (supplemental Fig. S1). However, the CD2 domain does not contain the hydrophobic residues conserved in CD1 and Apo2; these residues have previously been shown to be involved in substrate specificity (29, 36, 37). The Apo3G C-mutant (23% monomer, 65% dimer, Fig. 2D) and native Apo3G (18% monomer, 80% dimer, (Fig. 2A) are roughly comparable. However, the C-mutant contains about 12% tetramers with no larger oligomeric forms (Fig. 2D), whereas the native protein contains some tetramers (molecular mass ∼190 kDa), but they cannot be resolved in the dimer peak (Fig. 2A). Native Apo3G contains a small amount (∼2%) of 14 subunit oligomers, molecular mass ∼ 650 kDa (compare Fig. 2D with Fig. 2A). Apparently, these C-terminal residues are needed to form large oligomers, but because there is no substantial effect on dimer formation, perhaps these amino acids are restricted to tail-tail interactions (Fig. 1E). If this supposition is valid, then because the effects of mutating these C-terminal residues are relatively minor, it suggests that tail-tail interactions do not establish a major dimer interface, whereas the N-terminal CD1 residues do establish a major dimer interface via head-head interactions. Similar results were found with the C-terminal mutant I314A/Y315A (supplemental Fig. S3).
AFM Comparison of Apo3G Native and N-mutant F/W Oligomeric Forms
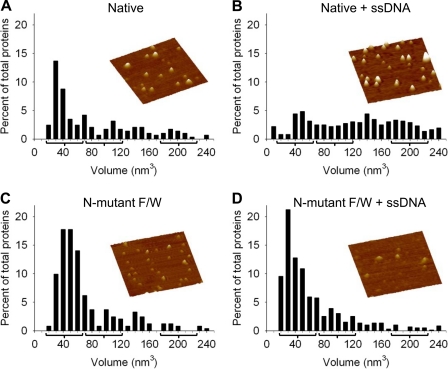
The MALS analysis provides a characterization of native and mutant forms of Apo3G in solution in the absence of ssDNA (Fig. 2). The oligomeric forms of native Apo3G change significantly in the presence of ssDNA (23). We used AFM to image the native and monomer N-mutant F/W forms of Apo3G with and without DNA (Fig. 3, supplemental Fig. S4). The volume of the deposited protein determined from the AFM images can be used to examine the distribution of oligomeric states because the AFM volume of proteins exhibits a linear relationship with molecular weight (32, 33), with volume = −14.7 + 1.2 (molecular weight). In the absence of ssDNA and presence of Mg2+, which is used to facilitate AFM imaging, both native Apo3G (23) (Fig. 3A) and the N-mutant F/W (Fig. 3C) have large monomer populations. In contrast, in the presence of ssDNA, native Apo3G contains a broad distribution of monomers, dimers, tetramers, and higher order oligomers (Fig. 3B), whereas the monomeric N-mutant F/W remains principally monomeric whether or not ssDNA is present (Fig. 3D).
FIGURE 3.
Oligomerization state of Apo3G native and N-mutant F/W in the absence and presence of ssDNA. AFM imaging was done in the presence of 5 mm MgCl2. The oligomeric state of native Apo3G alone (A) and in the presence of ssDNA (B) demonstrated that the Apo3G native composition shifts from a distribution of mostly monomers (A) to a broad distribution of oligomers in the presence of ssDNA (B). The oligomer interface N-mutant F/W imaged by AFM in the absence (C) and presence (D) of ssDNA did not demonstrate a significant shift in composition upon the addition of ssDNA. Volume distributions of Apo3G are plotted against percentage of total proteins. The predicted peaks for a monomer (42 nm3), dimer (97 nm3), and tetramer (208 nm3) are denoted with a bracket. Total volume distribution is to 1000 nm3 (see supplemental Fig. S4). Partial volume distribution is shown here to give a focused view of the major monomer, dimer, and tetramer populations. For Apo3G native and Apo3G N-mutant F/W, the population that distributes from 0 to 240 nm3 is ∼60 and ∼90% of the total proteins, respectively. The total proteins counted were: A, 285; B, 722; C, 242; and D, 588. Representative AFM images of Apo3G native and the N-mutant F/W are shown for each condition. Images are 300 × 300 nm with a height scale of 5.5 nm.
Apo3G-catalyzed 3′→5′ Deamination Polarity and Processivity Are Determined by the Non-catalytic N-terminal CD1 Domain
Native Apo3G, composed of a mix of monomers and oligomers, has been shown to catalyze processive C-deamination with a strong bias favoring deamination in target motifs located toward the 5′-region of linear ssDNA in the presence of salt (23) (Fig. 4A, Native). The native enzyme deaminates the 5′-CCC target motif 5-fold more rapidly than the 3′-CCC motif (Fig. 4A, Deamination ratio, 5′-C/3′-C = 5). The N-mutant F/W, which is predominantly a monomer, shows an 11-fold bias favoring the 5′-deamination motif (Fig. 4A, N-mutant F/W). The reactions are performed under “single-hit” conditions (< 15% substrate usage), which, by Poisson statistics (38), ensures that any given ssDNA substrate is acted on by at most one form of Apo3G (monomer or oligomer). The deamination band at 5′-C and 3′-C, observed for the native and N-mutant F/W enzymes, results from processive double deaminations carried out principally by a single enzyme (22, 23). Thus, the Apo3G monomeric mutant deaminates C→U, processively on ssDNA with a strong 3′→5′ directional bias. Apo3G does not need to oligomerize to carry out either processive or directional deamination.
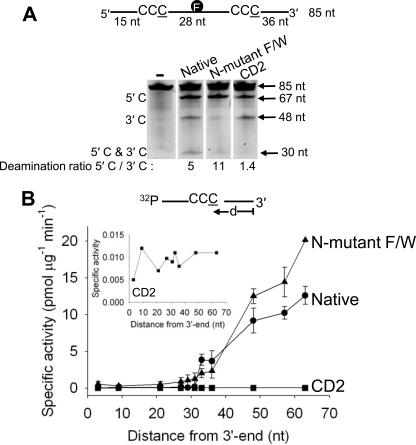
FIGURE 4.
Processivity, polarity, and specific activity of the Apo3G native, N-mutant F/W, and CD2 domain. A, native and mutant Apo3G were incubated with an 85-nt internally fluorescein-labeled ssDNA substrate with two CCC motifs 28 nt apart. Single deaminations of the 5′-C and 3′-C are detected as the appearance of labeled 67- and 48-nt fragments, respectively; double deamination of both C residues on the same molecule results in 30-nt labeled fragment (5′-C and 3′-C). Deamination ratios of the 5′-C and 3′-C deaminations are shown below each gel. The enzyme to DNA ratio varied from 1:20 to 1:1, depending on the specific activity, to obtain a substrate usage of up to 15% in a timed reaction. B, specific activity and polarity of Apo3G native (●), N-mutant F/W (▴), and CD2 domain (■) plotted as a function of the distance (d) of the CCC motif from the 3′-end of a 69-nt ssDNA substrate (50 nm). Inset, an expanded view of the specific activity of the CD2 domain. Error bars indicate S.E.
The C-terminal CD2 catalytic domain alone, which does not oligomerize, can catalyze ssDNA-dependent C→U deamination in the absence of the non-catalytic CD1 (29, 36, 39). The 5′-C and 3′-C deamination bands have similar integrated intensities (Fig. 4A, CD2, Deamination ratio, 5′-C/3′-C = 1.4), and there is no detectable 30-nt double deamination band. Therefore, unlike either the Apo3G native or the N-mutant F/W, CD2 acts non-processively with essentially no directional bias.
The 3′→5′ deamination bias can be examined directly by measuring the specific activity of Apo3G acting on a single 5′-CCC motif located at different distances (d) from the 3′-ssDNA end. Deamination is strongly favored at the 5′-end (Fig. 4B), where the specific activity for the monomeric N-mutant F/W (∼20 pmol μg−1 min−1) is ∼2-fold higher than native enzyme (∼12 pmol μg−1 min−1). There is a 20-fold monotonic increase in specific activity for both enzyme forms as the target C residue in the 5′-CCC motif (underlined C is deaminated) is moved in a 3′ to 5′ direction, from nucleotide position 33 to position 66 (Fig. 4B). In contrast, CD2 has ∼1000-fold lower specific activity (∼0.010 pmol μg−1 min−1), which does not depend on the location of the CCC motif (Fig. 4B, inset).
The 30-nt region at the 3′-end constitutes a deamination “dead zone” (23). Within the dead zone, the Apo3G native and N-mutant F/W deamination activities show a steep, i.e. discontinuous, loss of deamination activity, which remains at a constant depressed level irrespective of motif location (Fig. 4B). Native Apo3G has a specific activity ∼0.07 pmol μg−1 min−1 in the 30-nt dead zone, which shows a stepwise jump of ∼50-fold to 3.8 pmol μg−1 min−1 at CCC (nt 33), located 3 nt out of the dead zone. The N-mutant F/W has a specific activity of ∼0.5 pmol μg−1 min−1 in the dead zone, which jumps by 4-fold to 2.0 pmol μg−1 min−1 at nt 33. The deamination activity of CD2 stays constant outside and inside the dead zone (Fig. 4B, inset), in contrast to the monomeric N-mutant F/W, which favors 3′→5′ deamination outside the dead zone (Fig. 4B). These data suggest that when CD1 is present with CD2, the non-catalytic CD1 domain imposes an asymmetric 3′→5′ catalytic constraint.
Comparison of Apo3G Native and Monomeric N-mutant F/W ssDNA Substrate Binding and Catalysis
A quench-flow analysis was used to determine the single-turnover C-deamination rates for the native and monomeric N-mutant F/W at two locations, one close to the 5′-ssDNA end (Fig. 5A) and the other nearer to the 3′-end but outside the 30-nt dead zone (Fig. 5B). A 10-fold molar excess of Apo3G (500 nm) over ssDNA (50 nm) is sufficient to saturate the substrate and allows a measurement of the maximum rate to convert C→U, at the two CCC motif positions.
FIGURE 5.
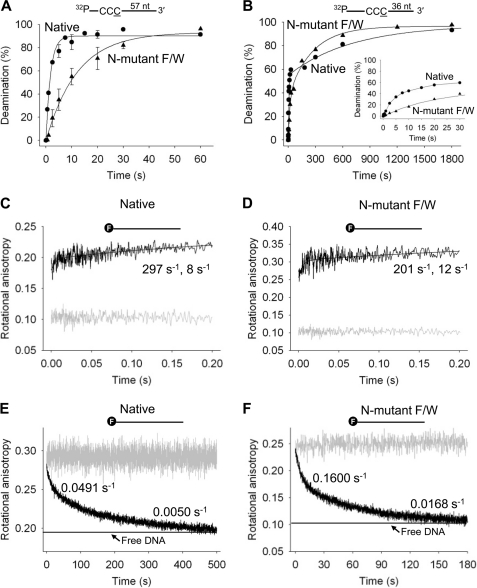
Pre-steady state analysis of deamination and binding of Apo3G native and N-mutant F/W on ssDNA. A and B, pre-steady state analysis of deamination rates of Apo3G native and N-mutant F/W on ssDNA. Single turnover reaction rates for Apo3G native (●) and N-mutant F/W (▴) were determined on ssDNA substrates with CCC motifs 57 (A) or 36 nt (B) from the 3′-end of a 69-nt substrate under enzyme saturating conditions (500 nm Apo3G, 50 nm DNA). Error bars indicate S.E. C and D, stopped-flow fluorescence pre-steady state interactions of Apo3G native and N-mutant F/W. The on-rate of Apo3G native (C) and N-mutant F/W (D) was determined when 1 μm enzyme was added to a 69-nt fluorescein-labeled (denoted with an F) ssDNA substrate (100 nm) by monitoring changes in rotational anisotropy of free ssDNA (gray line) following the addition of Apo3G as a function of time. E and F, the off-rate of 0.2 μm of Apo3G native (E) and N-mutant F/W (F) dissociating from fluorescein-labeled DNA (50 nm) was determined by preincubating Apo3G with ssDNA (gray line) and monitoring changes in rotational anisotropy following the addition of 100-fold excess unlabeled 69-nt ssDNA as trap (black line). The anisotropy of free fluorescein-labeled ssDNA mixed with unlabeled ssDNA trap is denoted with a horizontal line. Data were fit to either a double or a single exponential to obtain rates. Kinetic rates are discussed under “Results” and listed in supplemental Table S3.
Native Apo3G converts CCC→CCU ∼2.7-fold faster when it is 57 nt (Fig. 5A, 0.52 s−1) versus 36 nt from the 3′-end (Fig. 5B, 0.19 s−1). The more rapid turnover at the 5′-motif is consistent with the favored 3′→5′ deamination polarity (Fig. 4A, Native, deamination ratio 5′-C/3′-C = 5). The data for the 5′-motif fit well to a single exponential, whereas the data for the 3′-motif are biphasic with 56% of the turnovers occurring at a rate of 0.19 s−1 and 44% occurring at a 200-fold slower rate (0.001 s−1).
The monomeric N-mutant F/W deaminates the 5′-CCC motif at a maximum rate of 0.11 s−1 (Fig. 5A) when compared with 0.06 s−1 for the 3′-motif (Fig. 5B). Similar to the native enzyme, the kinetic data for the N-mutant F/W fit best to a single exponential for the 5′-motif and to a double exponential for the 3′-motif, 39% of the turnovers occurring at 0.06 s−1 and 61% occurring at a 15-fold slower rate (0.004 s−1). In comparison with native Apo3G, the N-mutant F/W has an ∼5-fold slower maximum C→U turnover rate at the 5′-motif (0.11 s−1 versus 0.52 s−1) and an ∼3-fold slower rate at the 3′-motif (0.06 s−1 versus 0.19 s−1).
Stopped-flow fluorescence measurements were used to determine the kinetics of binding by Apo3G native and N-mutant F/W enzymes (Fig. 5, C and D). The on-rates, measured using 20-fold excess enzyme over DNA, are rapid; about half the binding occurs within the instrument dead time (1.1 ms) (Fig. 5, C and D). The kinetics are biphasic, characterized by a faster rate of 297 s−1 and slower rate of 8 s−1, for native Apo3G (Fig. 5C) when compared with 201 and 12 s−1 for the N-mutant F/W (Fig. 5D). The heterogeneity in binding for the native enzyme is probably caused by the heterogeneous forms of the enzyme in solution (Fig. 2A), consistent with previous binding studies (23). In the case of the N-mutant F/W, which is predominantly a monomer (Fig. 2B), the small fraction of oligomer forms could assemble at high protein concentrations. Consistent with this supposition, a 5-fold reduction in Apo3G concentration (4-fold excess enzyme over DNA) causes the N-mutant F/W binding to become monophasic (supplemental Fig. S5B; rate = 208 s−1), with a single on-rate that is approximately the same as measured at the high enzyme to DNA ratio (Fig. 5D; faster rate = 201 s−1). Therefore, there may be a reduction in oligomerization occurring at the lower enzyme/DNA ratio (4:1) for the N-mutant F/W. Biphasic binding is retained for the native enzyme (supplemental Fig. S5A; faster rate = 79 s−1, slower rate = 8 s−1).
The off-rates of the native and N-mutant F/W enzymes are biphasic (Fig. 5, E and F). Roughly 40% of the native Apo3G remains bound for 20 s, and 60% remains bound for 200 s (Fig. 5E). About 43% of the Apo3G N-mutant F/W remains bound for 6 s, and 57% stays bound for 60 s (Fig. 5F). The N-mutant Apo3G dissociates from ssDNA 3–5-fold faster than the native form(s) of the enzyme. Taken together with the rapid kinetic C→U turnover data (Fig. 5, A and B), the enzymes remain bound in their various forms for sufficient time to catalyze from as little as 8 to as many as 80 deaminations for the native enzyme and from 0.5 to 5 deaminations for the N-mutant F/W.
Interaction of Apo3G N-mutant F/W with Alu RNA
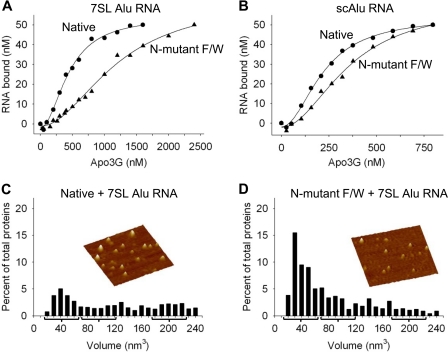
The Apo3G residue Trp-127 appears to be required for two cellular functions that involve RNA binding, virion incorporation (9, 27, 40, 41), and Alu retrotransposon restriction (9). Furthermore, oligomerization has been considered necessary for Apo3G to bind RNA (27). We have determined the binding affinity of Apo3G native and N-mutant F/W for two Alu RNAs; one is a 91-nt Alu sequence derived from 7SL RNA (9), and the other is a small cytoplasmic Alu RNA, which is a product of RNA processing (42, 43).
Native Apo3G binds 7SL Alu RNA with ∼2.5-fold higher affinity than the N-mutant F/W with apparent dissociation constants (Kd) of 446 and 1200 nm, respectively (Fig. 6A). The Apo3G native and N-mutant F/W bind a 108-nt small cytoplasmic Alu (scAlu) (native Kd of 220 nm; N-mutant F/W Kd of 357 nm, Fig. 6B) or 40-nt random RNA sequence (native Kd of 298 nm; N-mutant F/W Kd of 211 nm, data not shown) with approximately the same affinity. AFM was used to determine whether Apo3G native and N-mutant F/W form oligomers on 7SL-derived Alu RNA. In the presence of 91-nt Alu RNA, the Apo3G native volume distribution shifts from a relatively narrow distribution of mostly monomers (Fig. 3A) to a broad distribution of oligomeric forms (Fig. 6C, supplemental Fig. S6A), similar to when DNA is present (Fig. 3B). The N-mutant F/W is present mostly as a monomer alone (Fig. 3C), when bound to RNA (Fig. 6D, supplemental Fig. S6B), and when bound to DNA (Fig. 3D). These data demonstrate that Apo3G monomers are able to bind RNA.
FIGURE 6.
Apo3G native and N-mutant F/W interactions with Alu RNAs. A and B, the apparent dissociation constant (Kd) of Apo3G native and mutants from Alu RNAs was determined by monitoring changes in rotational anisotropy of free fluorescein-labeled RNA (50 nm) following the addition of enzyme as a function of concentration. A, Apo3G native (●) binds to a 91-nt Alu RNA sequence derived from 7SL RNA with an apparent Kd of 446 nm, which is a 2.5-fold higher affinity interaction than for the N-mutant F/W (▴, Kd of 1200 nm). B, the Apo3G native (●) and N-mutant F/W (▴) bind a 108-nt small cytoplasmic Alu (scAlu) RNA with approximately the same affinity, having an apparent Kd of 220 and 357 nm, respectively. C and D, AFM was used to determine the oligomeric state of Apo3G native (C) and N-mutant F/W (D) in the presence of a 91-nt Alu RNA and 5 mm MgCl2. The apparent dissociation constants for native and mutant Apo3G in the AFM buffer were 374 and 742 nm, respectively. Volume distributions of Apo3G are plotted against a percentage of total proteins. The predicted peaks for a monomer (42 nm3), dimer (97 nm3), and tetramer (208 nm3) are denoted with a bracket. Total volume distribution is to 1000 nm3 (see supplemental Fig. S6). Partial volume distribution is shown here to give a focused view of the monomer, dimer, and tetramer populations. For Apo3G native and N-mutant F/W, the population that distributes from 0 to 240 nm3 is ∼50 and ∼80%, respectively. Total proteins counted were: C, 361; and D, 211. Representative AFM images of Apo3G native and N-mutant F/W are shown for each condition. Images are 300 × 300 nm with a height scale of 5.5 nm.
DISCUSSION
There are questions concerning the composition of Apo3G prior to its encapsulation in an HIV virion as a “Trojan horse” and after its entry in T cells. It is generally agreed that Apo3G is encapsulated as a multimer, perhaps by interacting with the Alu portion of 7SL RNA or HIV RNA, but presumably not as a monomer (7, 14). However, in contrast to a currently favored model for Apo3G encapsulation (27), which implies that multimerization is necessary for RNA binding, our data show that Apo3G monomers are also able to bind RNA (Fig. 6). Although Apo3G may enter the virion as an RNA-induced multimer (7, 25) that is disrupted by RNase H to release Apo3G monomers and dimers (25), it is not certain what form Apo3G takes when catalyzing deaminations of HIV proviral cDNA.
Exploring the Composition of Apo3G
The uncertainty in the composition of native Apo3G stems from its ability to assume several different forms. Low angle x-ray-scattering and cross-linking experiments show Apo3G dimers (27, 44) that appear to be arranged in either a head-to-head configuration with the two N-terminal CD1 domains bound (27) or a tail-to-tail configuration with the two C-terminal CD2 domains bound (45). Using MALS, we find that Apo3G in solution has a polydisperse light-scattering peak containing mostly dimers (Fig. 2A, ∼80%), along with monomers present at the low molecular mass side of the main peak and unresolved trimers and tetramers at the high molecular mass side of the peak (Fig. 2A), in agreement with recent analytical ultracentrifugation data (24). An analysis of Apo3G composition using AFM further reveals that although in the absence of ssDNA, Apo3G exists mainly as monomers and dimers, with a minority of larger species (Fig. 3A), there is significant increase in the percentage of trimers, tetramers, and large oligomers that appear in the population in the presence of ssDNA (Fig. 3B). Therefore, it seems plausible that native Apo3G may act on ssDNA in a variety of different forms (23).
We have used the crystal structure of Apo2 (28) (Fig. 1A) to identify homologous amino acids to mutate in the N-terminal CD1 domain of Apo3G that corresponds to the Apo2 hydrophobic interface (Fig. 1B, supplemental Fig. S1). Disruption of this predicted interface should interfere with N-terminal head-to-head dimerization, thereby generating Apo3G monomers. One such mutant, F126A/W127A (N-mutant F/W), resulted in predominantly monomers (∼92%) (Fig. 2B) that remain as monomers in the presence of ssDNA (compare Fig. 3C with Fig. 3D). In contrast, the native Apo3G exhibits a major change in composition in the presence of ssDNA (compare Fig. 3A with Fig. 3B). Because native Apo3G can form oligomers larger than dimers, either in solution (Fig. 2A) or when bound to ssDNA (Fig. 3B), an additional oligomer interface apart from CD1-CD1 is likely to exist. The C-terminal mutants R313A/D316A/D317A/Q318A and I314A/Y315A have fewer larger molecular mass species when compared with native Apo3G (Fig. 2, A and D, supplemental Fig. S3) suggesting that large complexes may form through the CD2-CD2 interface.
Comparing Polarity and Processivity of Native Apo3G with Monomeric Apo3G and CD2
A side-by-side comparison of native, N-mutant F/W, and catalytic CD2 Apo3G shows that the monomer mutant retains the biochemical characteristics of the native enzyme, such as 3′→5′ deamination polarity (22, 23) and processivity (22, 23), whereas CD2 (lacking CD1) does not (Fig. 4A). The monomer mutant deaminates both 5′-CCC and 3′-CCC target motifs processively, with an 11-fold directional bias favoring deamination of the 5′-motif over the 3′-motif (Fig. 4A, Native and N-mutant F/W lanes). There is a 30-nt dead zone region at the ssDNA 3′-end characterized by a stepwise drop in specific activity, 50-fold for native Apo3G and 4-fold for the N-mutant F/W monomer (Fig. 4B).
Unlike native Apo3G and the monomeric N-mutant F/W, CD2 specific activity is roughly constant over the entire ssDNA molecule (Fig. 4B); it is not processive (Fig. 4A, CD2 lane), shows no deamination polarity (Fig. 4, A and B), and has no dead zone (Fig. 4B, inset). The catalytically active CD2 domain has a net negative charge (−4.5), whereas the inactive CD1 domain has a large net positive charge (+11). In the absence of CD1, the apparent Kd for ssDNA of the CD2 domain increases from 0.15 to 12 μm (data not shown), which probably accounts for the loss of processive deaminations. Mutating two residues, Phe-126 and Trp-127 to Ala, causes a 5-fold decrease in the catalytic rate (Fig. 5, A and B) and a 3–5-fold increase in the off-rate (Fig. 5, E and F). Thus, the non-catalytic CD1 plays an indispensable role in stabilizing ssDNA binding, enhancing catalysis, and establishing 3′→5′ deamination polarity and processivity. Our data indicate that the functions of the CD1 and CD2 domains are not nearly as segregated as suggested previously, i.e. with CD1 responsible for encapsidation of Apo3G into virions and CD2 for catalysis (21, 46, 47). Although it is true that the chemistry of the deamination reaction occurs solely in the CD2 domain, CD1 clearly has a major influence on catalysis.
Structure-based Model for Apo3G Polarity and Processivity
All previous studies with purified native Apo3G were performed using a heterogeneous mix of monomers, dimers, and higher order oligomers in solution, which exhibit severe changes in composition ratio in the presence of ssDNA (Fig. 3, A and B) and salt (23). It is seemingly impossible to determine a mechanism of scanning and deamination using data from a complex and variable mix of native forms. In contrast, our present study, showing that the N-mutant F/W monomer retains all of the salient catalytic properties of the mix of species of the native enzyme (Fig. 4), enables us to propose a structure-based model for Apo3G ssDNA scanning and deamination (Fig. 7).
FIGURE 7.
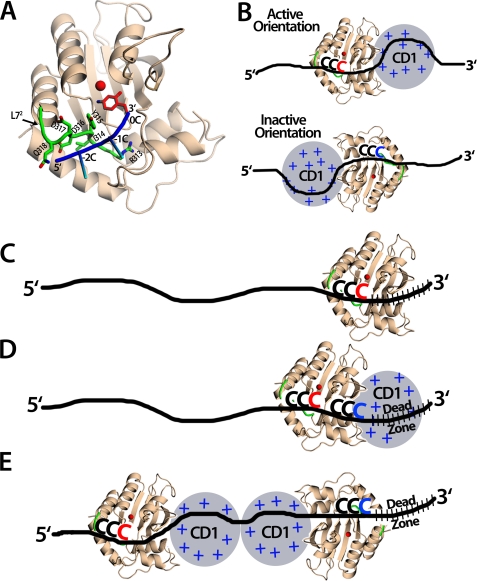
Models of Apo3G with ssDNA. A, the Apo3G crystal structure was modeled with a short piece of ssDNA containing the CCC hot spot. The Apo3G-CD2 crystal structure (shown in wheat) was superimposed with the mouse cytidine deaminase co-crystal structure with cytidine in the active site (PDB 2fr6) to place the cytosine base in this position (shown in red). Residues on loop 7 (L72) have previously been shown to be important for Apo3G substrate specificity (29, 36, 39) (shown in green), specifically, Asp-316 and Asp-317 (29). Other residues on L72 may contact the bases neighboring the target cytosine through electrostatic, hydrophobic, or base-stacking interactions. It is predicted that for Apo3G to efficiently deaminate the target cytosine at position 0, the DNA (shown in blue, with the −2 and −1 cytosines shown as sticks) should be positioned in a 5′→3′ orientation across L72 to allow the hot spot cytosines to interact with the residues on L72. Exactly how these residues interact with the cytosines remains unknown. B, an Apo3G monomer can bind ssDNA in either orientation, but only the active orientation is efficient for cytosine deamination. Top, Apo3G is shown in the active orientation that allows for the Apo3G-CD2 (crystal structure shown in wheat) to bind DNA so that the hot spot CCC motif (red) binds L72 in a 5′→3′ orientation. Apo3G-CD1 (gray circle) is more positively charged and binds DNA (black line) with a higher affinity than Apo3G-CD2. Bottom, Apo3G is shown in the inactive orientation where the Apo3G-CD2 residues on L72 are predicted not to contact the hot spot motif. Because this interaction is important for recognizing the target cytosine, in this orientation, Apo3G-CD2 would not be able to efficiently deaminate the CCC motif (blue). C, Apo3G-CD2 is capable of deaminating in the dead zone. Apo3G-CD2 (wheat) is not restricted and can deaminate the target cytosine (red) in the dead zone (slashed black lines). D, the monomer Apo3G, although positioned in the active orientation, does not deaminate cytosine in the dead zone. Apo3G-CD1 (gray circle) binds DNA tightly and may restrict deamination in the dead zone (slashed black lines) by blocking Apo3G-CD2 (wheat) access to the target cytosine (blue). However, CCC motifs upstream of the dead zone can be deaminated (red). E, a predicted model of the Apo3G dimer in a head-head orientation capable of deaminating the ssDNA 5′-proximal CCC motif. The Apo3G-CD2 near the 5′-end of the DNA is positioned in the active orientation to deaminate the 5′-CCC motif (red). The Apo3G-CD2 positioned near the 3′-end of the DNA is in the inactive orientation and inefficiently deaminates the 3′-CCC motif (blue) in the dead zone (slashed black lines).
The presence of 3′→5′ catalytic directionality appears to be imposed by a constraint within the active site of the enzyme such that a 5′-CCC motif, situated anywhere along ssDNA, is in a favorable orientation for deamination when bound with the first (−2) and second (−1) C in the 5′-CCC motif extending away from the zinc atom within the active center toward loop 72 (Fig. 7A). Residues on loop 72 of the CD2 domain are required for deamination (29, 36, 39) and important for deamination specificity (29). The pyrimidine rings of the −2 and −1 C residues of the CCC motif are essential for motif recognition and deamination of the third (0) C residue, thus corroborating a favored binding orientation within the active site, one that is most efficient for deamination (48). The biochemical data (Fig. 4, A and B) and structural modeling (Fig. 7A) suggest that Apo3G catalyzes deamination most efficiently when approaching the deamination target with the CD2 domain oriented toward the 5′-ssDNA end (Fig. 7B, Active Orientation). The reverse orientation is unlikely to allow contact between residues of loop 72 and the −2 and −1 C residues in the CCC motif, precluding the proper geometry for the C at position 0 in the motif to orient with the zinc molecule as required for deamination (Fig. 7B, Inactive Orientation). The model clearly suggests why it is the C at position 0 in any given 5′-CCC motif that is deaminated in strong preference to the other two C residues.
After binding randomly to DNA (22), because there is no apparent energy source present, Apo3G translocates randomly in small steps by sliding (22, 23) and in larger steps by jumping/hopping (22, 23) or intersegmental transfer (49). The deaminations occur processively, i.e. one Apo3G molecule (monomer, dimer, tetramer) catalyzes multiple deaminations on the same ssDNA prior to attacking a different substrate (22, 23). Although random movements in the 5′- and 3′-directions must on average be the same, the catalytic orientational specificity always favors deamination toward the 5′-side (Fig. 7B, Active Orientation). Thus, Apo3G is predisposed to exhibit a deamination polarity. The presence of a dead zone located at the 3′-ssDNA end (Fig. 4B) also suggests the presence of a favored catalytic orientation. Because CD2-catalyzed deamination does not depend on motif location, i.e. it has no dead zone (Figs. 4B and 7C), we speculate that CD1, because of its large overall positive charge and high affinity for DNA, creates a dead zone whenever it binds randomly in the last 30 nt of the 3′-ssDNA end, thereby shielding targets from being deaminated by CD2 (Fig. 7D). CD2 acts mainly as a catalyst, which is active when oriented facing the 5′-end of the ssDNA but much less so when facing the 3′-end (Fig. 7B). The same general model can be used to explain directional deamination for an Apo3G monomer (Fig. 7D), for a dimer (Fig. 7E), and for larger oligomers.
One prediction of the model is that an enhanced mobility of the enzyme on the ssDNA would enable increased sampling of all CCC motifs, located either 5′ or 3′ to the original binding site. Once a favored 5′-motif is deaminated, a more mobile enzyme can slide and jump back to sample a 3′-motif more efficiently than a less mobile enzyme, resulting in a reduction in the 5′-C/3′-C deamination ratio. Examination of data on Apo3G binding and deamination polarity in the presence and absence of salt (Mg2+) supports this prediction (supplemental Fig. S7). Both the native and the monomer mutant of Apo3G bind less tightly to the ssDNA in the absence of salt (Kd of 418 and 320 nm, respectively) than in its presence (Kd of 153 and 107 nm, respectively), and both show a substantial reduction in the deamination polarity in the absence of salt (compare supplemental Fig. S7A with Fig. 4A). Further evidence suggesting that weaker interactions lead to greater mobility comes from examination of the fluorescence of ssDNA containing pyrrolo-dC (see supplemental “Experimental Procedures”), which is a “marker” of DNA conformational alterations (50). Changes in pyrrolo-dC fluorescence occur only after Apo3G binds to the ssDNA (23) and therefore reflect DNA conformational alterations during enzyme translocation (supplemental Fig. S7, D and E). Notably, both native and monomeric Apo3G show smaller intensity changes when they scan the ssDNA backbone in the absence versus the presence of salt (supplemental Fig. S7, B and C), consistent with their decreased binding affinities. In addition, in the presence of salt, the Apo3G monomer causes greater structural changes when compared with the native enzyme (supplemental Fig. S7, B and C), and the monomer has a 2-fold higher 3′→5′ deamination bias (Fig. 4A, Deamination ratio, 5′-C/3′-C = 11 and 5, for the monomer and native enzyme forms, respectively). These data suggest that the positively charged CD1 domain, by binding the DNA with either low or high affinity (low or high salt conditions, respectively), can either enhance or restrict the movement of Apo3G on the DNA.
The structure-based model (Fig. 7) explains why deamination is favored in a 5′-direction. The monomer data are essential to explain the properties of the native forms of Apo3G. Key to the model is the observation that the Apo3G monomer mutant retains the biochemical features of processivity and deamination polarity exhibited by the native mix of monomers, dimers, and larger oligomer forms. The features of processivity and polarity are determined by the CD1 domain of the monomer, thus revealing the overriding importance of the non-catalytic CD1 in the spatial properties of deamination and in imposing an asymmetric catalytic constraint on full-length Apo3G. These properties are responsible for generating 3′→5′ deamination gradients on ssDNA in vitro, which correspond to mutational gradients in HIV RNA observed in vivo (16, 22, 23, 51).
Acknowledgment
We thank N. Chelyapov at the USC NanoBiophysics Core Facility for assistance with MALS and AFM data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants ESO13192 and R37GM21422 (to M. F. G.), GM080294 and GM079480 (to D. A. E.), and GM087986 (to X. S. C.). This work was also supported by a California HIV/AIDS Research Program grant (to X. S. C.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Tables S1–S3, and Figs. S1–S7.
- Apo3G
- APOBEC3G
- ssDNA
- single-stranded DNA
- HIV
- human immunodeficiency virus
- CD2
- C-terminal domain
- CD1
- N-terminal domain
- MALS
- multiangle light scattering
- AFM
- atomic force microscopy
- nt
- nucleotides
- F/W
- F126A/W127A.
REFERENCES
- 1.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 2.Kao S., Khan M. A., Miyagi E., Plishka R., Buckler-White A., Strebel K. (2003) J. Virol. 77, 11398–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marin M., Rose K. M., Kozak S. L., Kabat D. (2003) Nat. Med. 9, 1398–1403 [DOI] [PubMed] [Google Scholar]
- 4.Sheehy A. M., Gaddis N. C., Malim M. H. (2003) Nat. Med. 9, 1404–1407 [DOI] [PubMed] [Google Scholar]
- 5.Stopak K., de Noronha C., Yonemoto W., Greene W. C. (2003) Mol. Cell 12, 591–601 [DOI] [PubMed] [Google Scholar]
- 6.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 7.Burnett A., Spearman P. (2007) J. Virol. 81, 5000–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alce T. M., Popik W. (2004) J. Biol. Chem. 279, 34083–34086 [DOI] [PubMed] [Google Scholar]
- 9.Bach D., Peddi S., Mangeat B., Lakkaraju A., Strub K., Trono D. (2008) Retrovirology 5, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogerd H. P., Cullen B. R. (2008) RNA 14, 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cen S., Guo F., Niu M., Saadatmand J., Deflassieux J., Kleiman L. (2004) J. Biol. Chem. 279, 33177–33184 [DOI] [PubMed] [Google Scholar]
- 12.Douaisi M., Dussart S., Courcoul M., Bessou G., Vigne R., Decroly E. (2004) Biochem. Biophys. Res. Commun. 321, 566–573 [DOI] [PubMed] [Google Scholar]
- 13.Khan M. A., Goila-Gaur R., Opi S., Miyagi E., Takeuchi H., Kao S., Strebel K. (2007) Retrovirology 4, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strebel K., Khan M. A. (2008) Retrovirology 5, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svarovskaia E. S., Xu H., Mbisa J. L., Barr R., Gorelick R. J., Ono A., Freed E. O., Hu W. S., Pathak V. K. (2004) J. Biol. Chem. 279, 35822–35828 [DOI] [PubMed] [Google Scholar]
- 16.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004) Nat. Struct. Mol. Biol. 11, 435–442 [DOI] [PubMed] [Google Scholar]
- 17.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003) Nature 424, 99–103 [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003) Nature 424, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu Y. L., Witkowska H. E., Hall S. C., Santiago M., Soros V. B., Esnault C., Heidmann T., Greene W. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 15588–15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haché G., Liddament M. T., Harris R. S. (2005) J. Biol. Chem. 280, 10920–10924 [DOI] [PubMed] [Google Scholar]
- 21.Navarro F., Bollman B., Chen H., König R., Yu Q., Chiles K., Landau N. R. (2005) Virology 333, 374–386 [DOI] [PubMed] [Google Scholar]
- 22.Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 23.Chelico L., Sacho E. J., Erie D. A., Goodman M. F. (2008) J. Biol. Chem. 283, 13780–13791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salter J. D., Krucinska J., Raina J., Smith H. C., Wedekind J. E. (2009) Biochemistry 48, 10685–10687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soros V. B., Yonemoto W., Greene W. C. (2007) PLoS Pathog. 3, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu Y. L., Soros V. B., Kreisberg J. F., Stopak K., Yonemoto W., Greene W. C. (2005) Nature 435, 108–114 [DOI] [PubMed] [Google Scholar]
- 27.Huthoff H., Autore F., Gallois-Montbrun S., Fraternali F., Malim M. H. (2009) PLoS Pathog. 5, e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prochnow C., Bransteitter R., Klein M. G., Goodman M. F., Chen X. S. (2007) Nature 445, 447–451 [DOI] [PubMed] [Google Scholar]
- 29.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008) Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bransteitter R., Pham P., Calabrese P., Goodman M. F. (2004) J. Biol. Chem. 279, 51612–51621 [DOI] [PubMed] [Google Scholar]
- 31.Wang W., Malcolm B. A. (1999) BioTechniques 26, 680–682 [DOI] [PubMed] [Google Scholar]
- 32.Ratcliff G. C., Erie D. A. (2001) J. Am. Chem. Soc. 123, 5632–5635 [DOI] [PubMed] [Google Scholar]
- 33.Yang Y., Wang H., Erie D. A. (2003) Methods 29, 175–187 [DOI] [PubMed] [Google Scholar]
- 34.Bertram J. G., Bloom L. B., O'Donnell M., Goodman M. F. (2004) J. Biol. Chem. 279, 33047–33050 [DOI] [PubMed] [Google Scholar]
- 35.Wyatt P. J. (1993) Anal. Chim. Acta 272, 1–40 [Google Scholar]
- 36.Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008) Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
- 37.Furukawa A., Nagata T., Matsugami A., Habu Y., Sugiyama R., Hayashi F., Kobayashi N., Yokoyama S., Takaku H., Katahira M. (2009) EMBO J. 28, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Creighton S., Bloom L. B., Goodman M. F. (1995) Methods Enzymol. 262, 232–256 [DOI] [PubMed] [Google Scholar]
- 39.Chen K. M., Martemyanova N., Lu Y., Shindo K., Matsuo H., Harris R. S. (2007) FEBS Lett. 581, 4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huthoff H., Malim M. H. (2007) J. Virol. 81, 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang T., Tian C., Zhang W., Luo K., Sarkis P. T., Yu L., Liu B., Yu Y., Yu X. F. (2007) J. Virol. 81, 13112–13124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Batzer M. A., Deininger P. L. (2002) Nat. Rev. Genet. 3, 370–379 [DOI] [PubMed] [Google Scholar]
- 43.Shaikh T. H., Roy A. M., Kim J., Batzer M. A., Deininger P. L. (1997) J. Mol. Biol. 271, 222–234 [DOI] [PubMed] [Google Scholar]
- 44.Wedekind J. E., Gillilan R., Janda A., Krucinska J., Salter J. D., Bennett R. P., Raina J., Smith H. C. (2006) J. Biol. Chem. 281, 38122–38126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett R. P., Salter J. D., Liu X., Wedekind J. E., Smith H. C. (2008) J. Biol. Chem. 283, 33329–33336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goila-Gaur R., Strebel K. (2008) Retrovirology 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith J. L., Bu W., Burdick R. C., Pathak V. K. (2009) Trends Pharmacol. Sci. 30, 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rausch J. W., Chelico L., Goodman M. F., Le Grice S. F. (2009) J. Biol. Chem. 284, 7047–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. (2008) Nat. Struct. Mol. Biol. 15, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 50.Zhang X., Wadkins R. M. (2009) Biophys. J. 96, 1884–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suspène R., Rusniok C., Vartanian J. P., Wain-Hobson S. (2006) Nucleic Acids Res. 34, 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]