Abstract
The expansive family of metazoan ADP-ribosylation factor and ADP-ribosylation factor-like small GTPases is known to play essential roles in modulating membrane trafficking and cytoskeletal functions. Here, we present the crystal structure of ARL6, mutations in which cause Bardet-Biedl syndrome (BBS3), and reveal its unique ring-like localization at the distal end of basal bodies, in proximity to the so-called ciliary gate where vesicles carrying ciliary cargo fuse with the membrane. Overproduction of GDP- or GTP-locked variants of ARL6/BBS3 in vivo influences primary cilium length and abundance. ARL6/BBS3 also modulates Wnt signaling, a signal transduction pathway whose association with cilia in vertebrates is just emerging. Importantly, this signaling function is lost in ARL6 variants containing BBS-associated point mutations. By determining the structure of GTP-bound ARL6/BBS3, coupled with functional assays, we provide a mechanistic explanation for such pathogenic alterations, namely altered nucleotide binding. Our findings therefore establish a previously unknown role for ARL6/BBS3 in mammalian ciliary (dis)assembly and Wnt signaling and provide the first structural information for a BBS protein.
Keywords: Diseases, Protein/Structure, Centriole, Signal Transduction, Subcellular Organelles, ARL6, BBS3, Bardet-Biedl Syndrome, Cilia, Small GTPase
Introduction
Bardet-Biedl syndrome (BBS)4 is a genetically heterogeneous human disorder characterized by a wide array of phenotypes, including obesity, polycystic kidney disease, retinal degeneration, polydactyly, and sensory impairments (1–4). It represents a paradigm for ciliopathies, a class of genetic disorders sharing the common etiology of basal body and/or cilia dysfunction (5–9). The physiological relevance of cilia, the motile and/or sensory organelles present on most vertebrate cell types, is emphasized by the growing number of pleiotropic diseases belonging to this class, which in addition to BBS includes the closely related Alström syndrome, as well Meckel syndrome, Joubert syndrome, and Senior-Løken syndrome.
Once thought to be a vestigial organelle, the nonmotile (primary) cilium is now well established as being essential for the transduction of chemical, visual, and mechanical sensory stimuli, and its receptor-studded membrane orchestrates a number of important signaling cascades necessary for vertebrate development, including Hedgehog, Wnt, and PDGFRαα signaling (6, 10–12).
The microtubule-based ciliary axoneme is assembled at the distal end of a basal body, a structure derived from the mother centriole that docks at the plasma membrane prior to ciliogenesis (13, 14). Nearly all cilia are built and maintained by a multiprotein intraflagellar transport (IFT) machinery that mobilizes ciliary cargo, for example structural components, receptors, and signaling molecules, into the organelle via an anterograde kinesin motor and transports components back to the base by way of a retrograde dynein motor (15–18). The IFT machinery is thought to dock near the tips of the transitional fibers, which are structures of unknown composition that form a nine-membered pinwheel-like radial array that joins the distal end of the basal body to the ciliary membrane (19, 20). Near this site, effectively a “ciliary gate,” post-Golgi trafficking of vesicular cargoes destined for the cilium terminates by fusion with the membrane; the transport of the released cargoes into the organelle may then be facilitated by the IFT machinery (21–23).
Few cellular factors are known to participate in the functional transition from vesicular trafficking to ciliary trafficking (24) or, potentially, in the reverse process where ciliary components are trafficked out of the organelle. Although ciliary trafficking is nonvesicular, it relies on IFT protein subunits that share β-propeller/solenoid topologies (and ostensibly functions) similar to those of coat vesicle-associated proteins (23, 25, 26). Our group previously discovered a member of the ARF-like family of small GTPases, ARL6, that could conceivably possess such a trafficking function. ARL6 is associated with Bardet-Biedl syndrome type 3 (BBS3) (27, 28), presently making it 1 of 14 BBS-associated proteins (4). In Caenorhabditis elegans sensory cilia, ARL6 was shown to undergo IFT (28). Moreover, ARF-like proteins, which are generally implicated in membrane trafficking and regulation of microtubule-associated processes (29–32), have at least two members that possess established ciliary functions. ARL3 orthologues from Leishmania, C. elegans, and mice are associated with cilia/ciliary photoreceptor functions (33–35); similarly, ARL13B specifically localizes to cilia in C. elegans, and in the mouse it participates in cilium formation and Hedgehog signaling (34, 36). The exclusive presence of ARL6 in organisms that possess cilia and its association with IFT (28) therefore suggests a potential role for this small GTPase in coordinating or facilitating the functional bridging of intraciliary trafficking with vesicular trafficking.
Although all BBS proteins studied to date appear to play important roles in either trafficking cargo to the basal body/centrosome or within cilia via IFT (2), ARL6/BBS3 is suggested to have a potentially distinct function from that of a core group of BBS proteins that assemble into a complex, termed the BBSome (37). This notion arises from the finding that the BBSome, which consists of a biochemically separable protein complex containing BBS1, BBS2, BBS4, BBS5, BBS7, BBS8, and BBS9 proteins, does not co-fractionate with BBS3 in a sucrose gradient (37). Interestingly, one component of the BBSome (BBS1) interacts with Rabin8, a GDP/GTP exchange factor for the small GTPase RAB8. Rabin8 co-localizes with the BBSome at the centrosome/basal body and affects the nucleotide cycling of RAB8, which enters the cilium in its GTP-bound state and is necessary for ciliogenesis (37). RAB8 was also recently shown to interact with Rabaptin5, which localizes at the basal body, and associates with an IFT protein termed Elipsa/DYF-11 (38, 39). Given the well established role of Rabaptin5 as a RAB5 effector in endocytosis, it was suggested that this Rabaptin5-RAB8-Elipsa/DYF-11 trio may provide critical functional interactions between the ciliary membrane (or vesicles associated with cilia) and the IFT machinery (39). In C. elegans, RAB-5 itself was shown to concentrate at the base of cilia together with an early endosome-associated STAM-Hrs complex that regulates polycystin-1/polycystin-2 protein removal from cilia (40). How BBSome proteins, which are themselves associated with both the basal body and IFT, participate in this basal body-ciliary sorting-trafficking process remains elusive, and the role of BBS3, potentially distinct from core BBSome components, is presently unknown.
To shed light on the molecular and cellular functions of ARL6/BBS3, we first determined the crystal structure of the GTP-bound form of the human protein. The ARL6/BBS3 structure represents the first to be solved for a BBS or IFT protein and displays unique characteristics not found in other small GTPases. In combination with GTP binding functional assays, the structure demonstrates how single-residue mutations uncovered in BBS patients abrogate nucleotide binding and thus protein function. Furthermore, we now demonstrate that ARL6 in ciliated mammalian cells localizes to the distal end basal bodies, near or at transition fibers, and thus in close proximity to the site of vesicle docking/fusion at the base of cilia. Consistent with a role for ARL6 at this subcellular location, we show that disrupting its cellular function, via overproduction of GDP- or GTP-locked forms, leads to ciliary anomalies in vivo. Furthermore, excess cellular wild-type ARL6 causes an up-regulation of Wnt signaling. Importantly, this effect is not observed with pathogenic variants of ARL6/BBS3. Together, our findings reveal that a BBS protein not directly (or tightly) associated with the BBSome localizes to a novel site within the basal body region, where it may regulate cilia (dis)assembly and, perhaps as a result, subsequent ciliary signaling events. We propose that ARL6/BBS3 may perform this role by modulating membrane trafficking at the base of the ciliary organelle.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
For the crystallography studies, amino acid residues 16–186 of ARL6 from the Mammalian Genome Collection clone (Geneservice Coordinate AT30-A7) were amplified by PCR and subcloned into the pET28a-LIC vector (GI:145307000), using the In-Fusion PCR cloning system (Clontech). The construct was transformed into Escherichia coli strain BL21-CodonPlus(DE3)-RIL (Stratagene), and the cells were grown at 37 °C in Terrific Broth to an A600 of 3.0 using the LEX bubbling system. The water bath was lowered to 15 °C for 30 min prior to induction using 1 mm isopropyl 1-thio-β-d-galactopyranoside and then left at 15 °C overnight. To harvest, cells were centrifuged, flash-frozen in liquid nitrogen, and stored at −80 °C. The thawed cell pellets from 1.8 liters of culture were resuspended in 100 ml of lysis buffer (10 mm Tris-HCl, pH 7.5, 0.5 m NaCl, 5 mm imidazole) with 1 mm phenylmethylsulfonyl fluoride, 0.5% CHAPS and lysed with microfluidizer at 20,000 p.s.i. Cleared supernatant was passed through an anion exchange column DE52 (Whatman) pre-equilibrated with the lysis buffer and then loaded onto a 5-ml HisTrap HP (GE Healthcare) column also pre-equilibrated with the lysis buffer. The target protein was eluted with an imidazole gradient from 50 to 500 mm in 50 ml. The eluted protein fractions were collected, pooled, and further purified by a gel filtration (Superdex 75) column pre-equilibrated with a buffer containing 20 mm HEPES, pH 8.0, 500 mm NaCl, and 1 mm dithiothreitol. Collected protein fractions were adjusted to contain 5 mm MgCl2 and then incubated with 5× protein molarity concentration of GDP for 1 h. The protein was then concentrated using an Amicon Ultra-15 centrifugal filter to a final concentration of 79.2 mg/ml. The purified protein had a mass of 21,354.8 Da.
Crystallization and Structure Determination
Diffracting crystals were grown using sitting drop vaporization at room temperature. The mother liquor contained 2 m (NH4)2SO4, 0.2 m NaCl, 0.1 m sodium cacodylate, pH 5.5. Mountable crystal appeared after 2 days. Diffraction data were collected at a copper rotating anode source (RIGAKU FR-E) and processed using the HKL2000 (41) software suite. Molecular replacement was performed using the program PHASER (42) with an ensemble of the two structures (PDB codes 1ZD9 and 2H16) as a search model. An initial ARL6 model was automatically built in ARP/wARP (43), followed by several rounds of restrained coordinate and temperature factor refinements in REFMAC (44), geometry validation in MOLPROBITY (45) server, and manual adjustment in COOT (46). Diffraction data and model statistics are summarized in Table 1. Coordinates and structure factor amplitudes have been deposited in the PDB as code 2H57 (47).
TABLE 1.
Diffraction data and crystallographic model statistics
r.m.s.d., root mean square deviation.
| Wavelength | 1.5418 Å |
| Unit cell | a = 80.06 Å, b = 134.88 Å, c = 63.88 Å |
| Space group | P21212 |
| Resolution range | 30.00 to 2.00 Å (2.07 to 2.00 Å)a |
| No. of unique reflections | 46,561 (4483) |
| Redundancy | 5.3/5.2 |
| Completeness | 97.9% (96.2%) |
| 〈I/σI〉 | 39.5 (3.0) |
| Rsymb | 0.043% (0.599%) |
| Rwork/Rfreec | 20.6% (24.5%) |
| Average B-factor | 34.2 Å2 |
| r.m.s.d.d from ideal geometry | |
| Bond length | 0.017 Å |
| Bond angle | 1.393° |
| PROCHECK (98)ψ-ϕ plot within preferred/additional allowed regions | 93.1/6.9% |
a Values in parentheses refer to the highest resolution shell.
b Rsym = Σ|I − 〈I〉|/ΣI.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|. Rfree (99) was calculated as Rwork by using 3.8% of the data selected in thin resolution shells with SFTOOLS.
d r.m.s.d. means root mean square deviation.
GTP Binding Assay
Wild-type Homo sapiens ARL6 cDNA (GenBankTM accession number NM_032146) was cloned into pRSET-6a (48). All ARL6 mutants were prepared by a modified site-directed mutagenesis method known as SLIM (49). To obtain GST-tagged protein, full-length ARL6 was cloned into pGEX-6p-1 (Amersham Biosciences).
BL21(DE3)pLysS cells were used for expression of untagged (pRSET) or GST-tagged (pGEX) wild-type and mutant ARL6. Transformed E. coli cells were cultured in 50 ml of LB broth containing 34 μg/ml chloramphenicol and 50 μg/ml ampicillin at 37 °C until A600 = 0.6. Cultures were induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside overnight at room temperature. To harvest, cells were pelleted by centrifugation at 5000 × g for 10 min, suspended in wash buffer (50 mm NaH2PO4, 2 mm dithiothreitol, 0.3% Tween 20, 4 μm ATP), and lysed by sonication. Soluble and insoluble fractions were separated by centrifugation at 20,000 × g for 10 min. GTP binding assays were done as described previously (50, 51). To assay the soluble fraction, 10 μl of lysate were spotted onto nitrocellulose membrane, which was then washed in wash buffer and allowed to incubate for 2 h at room temperature in binding buffer (50 mm NaH2PO4, 2 mm dithiothreitol, 0.3% Tween 20, 4 μm ATP, 2.5 mm MgCl2) with 2 μCi of GTP/ml. Following binding, the membrane was rinsed in wash buffer and exposed to film or a phosphor screen. To assay the pellet fraction, protein was suspended in Laemmli sample buffer and separated by SDS-PAGE. The gel was soaked in 50 mm Tris-HCl, pH 7.5, with 20% glycerol for 30 min at room temperature, and liquid was transferred onto nitrocellulose. TLC was done as described previously (50).
Mammalian Cell Culture
Eagle's minimum essential medium (Invitrogen) was used to maintain HEK-293 cells, and a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium (Invitrogen) was used for growing IMCD3 and hTERT-RPE cells. Both types of media were supplemented with 10% fetal bovine serum, and all cells were grown at 37 °C in 5% CO2. For drug treatments, nocodazole was suspended in DMSO and used at a final concentration of 0.1 μg/ml for 1 h or overnight. The myc-tagged p50 dynamitin expression construct was a kind gift from Richard Vallee (Columbia University) and was transfected into IMCD3 cells using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol, as were all other constructs used.
Immunocytochemistry and Fluorescence Microscopy
For all immunocytochemistry experiments, cells were grown on glass coverslips and rinsed with phosphate-buffered saline prior to fixation. The cells were fixed either by a 10-min incubation in −20 °C methanol, followed by a 10-min rehydration in TBS-T (20 mm Tris base, pH 7.5, 150 mm NaCl, 0.05% Tween 20), or by a 30-min fixation with 2% paraformaldehyde in PHEM (0.05 m Pipes, 0.025 m HEPES, 0.01 m EGTA, 0.01 m MgCl2, pH 7.2), followed by a 2-min extraction with 0.1% Triton X-100 in PHEM. After fixation, cells were either blocked for 1 h in TBS-T supplemented with 1% bovine serum albumin prior to primary antibody incubation or directly incubated for 60 min at room temperature with the relevant primary antibody diluted in TBS-T. Cells were then washed in TBS-T and incubated another 60 min in the appropriate secondary antibody. Nuclei were counterstained by incubating the glass coverslips in 100 nm 4′,6-diamidino-2-phenylindole for 5 min in TBS-T at room temperature. Slides were mounted in Prolong anti-fade reagent (Molecular Probes) and observed by fluorescence microscopy using standard epifluorescence, Leica LSM 410 confocal, or Leica TCS SP confocal microscope systems. Immunofluorescent digital confocal microscopy and deconvolution were performed as described previously (52).
Immunohistochemistry of mouse tissue was performed on mouse testis embedded in OCT and quick frozen prior to sectioning. Mouse kidney sections were obtained from INOVA Diagnostics, San Diego (catalogue no. 508385), and monkey brain sections were obtained from MeDiCA, Encinitas, CA (catalogue no. 0608-BR). Staining and imaging were done as described above for mammalian tissue culture cells.
Primary antibodies used for immunocytochemistry included anti-ARL6C and anti-ARL6N (Abgent), anti-γ-tubulin (Sigma), anti-acetylated tubulin (Sigma), anti-α-tubulin (Sigma), and anti-PCM1 (gift from Dr. Andreas Merdes, University of Edinburgh). Secondary antibodies used included rabbit and mouse IgG conjugated with Alexa Fluor 488 and 594 (Molecular Probes).
Statistical Analyses
Differences in cilia length were calculated using the Student's t test, whereas differences in percentage of cells with cilia were calculated using the χ2 test.
Wnt Luciferase Assays
HEK-293T cells stably expressing pTOPflash reporter (53) and hTERT-RPE cells were seeded at a density of 104 cells per well in a 24-well plate. After 24 h, cells were transfected with a total of 1 μg of DNA (Renilla luciferase control, plasmid of interest, and pTOPflash reporter plasmid where necessary) using FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. The BBS3 wild-type and mutant alleles were cloned into a pCMV and a pcDNA 6.2 green fluorescent protein backbone and tested with appropriate control vectors, respectively. The cells were stimulated with Wnt3a conditioned medium (54) 48 h post-transfection and harvested 24 h after stimulation. The luciferase activity was measured using a Dual Luciferase Reporter Assay kit (Promega) on a Tecan Genios Pro luminometer and BMG LabTech FLUOstar Omega plate reader.
RESULTS
Crystal Structure of ARL6 Displays Unique Characteristics and Suggests Abrogated Nucleotide Binding for Pathogenic Variants
To illuminate the molecular function of ARL6/BBS3 and facilitate structure-function studies, we determined the high resolution structure of this small GTPase. Using a standard sitting drop vaporization method, we obtained crystals diffracting at up to 2 Å resolution for an N-terminal truncated form of ARL6 (residues 16–186) in the GTP-bound state (ARL6-GTP) (Table 1). The crystal structure of the protein was then solved by molecular replacement, as described under “Experimental Procedures.”
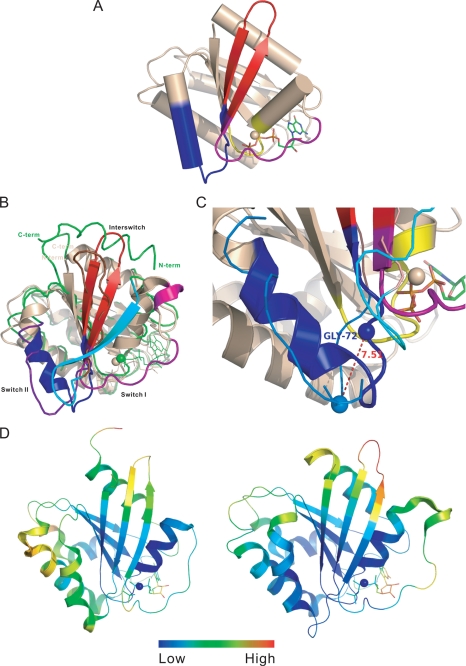
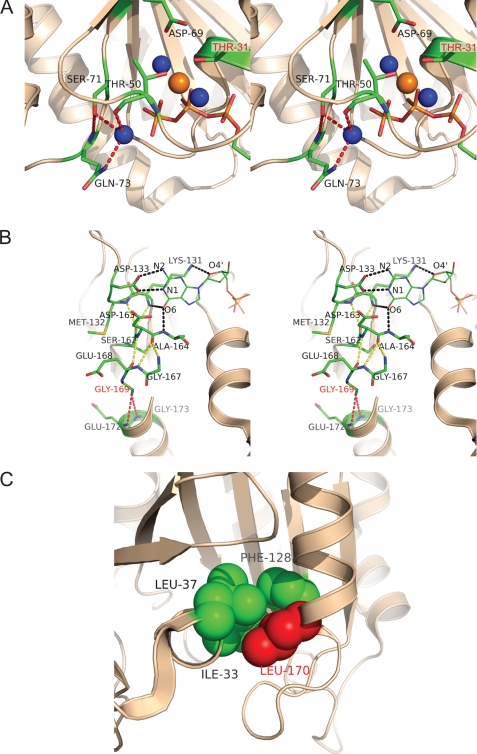
The structure of the ARL6-BBS3-GTP complex shows a typical GTPase fold with a central six-stranded β-sheet surrounded by five α-helices (Fig. 1A), but it is distinct from most small GTPases that typically hydrolyze bound GTP and are often found in the GDP-bound state. Despite the similarities of ARL6-GTP to other Ras superfamily small GTPases in the GTP or nonhydrolyzable GTP analogue-bound states, including the conformations of five conserved G-box (G1–5) sequences (55), three structural elements of ARL6-GTP, particularly in the switch I, switch II, and interswitch regions, showed dramatic conformational differences from that of ARF1 in the GDP-bound state (ARF1-GDP) (Fig. 1B). The switch I region adopted an irregular extended conformation that is consistent with being GTP-bound and did not form a β-strand to pair with β3 as in ARF1-GDP (Fig. 1B). The switch I region of ARL6 is two residues longer than other typical members of the ARF family, forming a 310-helix at residues 40–42 (Fig. 1B). The amide group of Gly-72 in the switch II region forms a hydrogen bond with the γ-phosphate group of GTP and is moved up 7.5 Å as compared to the corresponding residue Gly-70 of ARF1-GDP (Fig. 1C); this change is likely important for preferential binding of GTP. In conforming to the movement of the switch II residue Gly-72, the β-hairpin of the interswitch region (strands β2–β3) is extended and exposed to solvent (56). Interestingly, ARL6 is the only member of the Arf family small GTPases that has a serine (residue Ser-71) at the second X position of the DXXG motif of the switch II region; all other ARF-like GTPases have a glycine at the same position (57). The side chain hydroxyl group of Ser-71 forms hydrogen bonds with the amide group of Gln-73 and a water molecule proposed to attack the γ-phosphate group of GTP during hydrolysis (Fig. 2A). Consistent with the low temperature factor distribution of ARL6-GTP (Fig. 1D), these extra hydrogen bonds make the entire interaction network involving Mg2+ and GTP, especially the switch I and switch II regions, more rigid compared to other GTP-bound ARF GTPases. On the other hand, the loop connecting β2 and β3 is more flexible compared to other ARF GTPases (Fig. 1D).
FIGURE 1.
Crystal structure of the ARL6/BBS3 small GTPase. A, cylinder representation of ARL6 structure in its GTP-bound form. The structure elements that undergo fundamental changes upon GDP/GTP exchange cycle are colored purple for switch I, blue for switch II, and red for interswitch. The P-loop is shown in yellow, and the bound GTP is shown in stick representation. B, structure comparison of GTP-bound ARL6 GTPase (PDB code 2H57) with GDP-bound full-length ARF1 GTPase (PDB code 1HUR). The structure elements undergoing fundamental conformation change in GDP/GTP exchange cycle, switch I, switch II, and interswitch, are colored purple, blue, and red for ARL6 and cyan, violet blue, and chocolate red in ARF1. All other secondary structure elements of ARL6 are presented in ribbon (wheat), although for ARF1 only Cα trace is shown (green). C, major conformational change at switch II region upon change of bound nucleotide states between ARL6 (GTP-bound) and ARF1 (GDP-bound). The color scheme is the same as in A. The Cα atoms of Gly-72 of ARL6 and corresponding Gly-70 of ARF1 are shown as spheres. D, regional conformation flexibility of GTP-bound ARF1 (left, PDB code 1O3Y) compared with that of GTP-bound ARL6 (right PDB code 2H57). The ribbons are colored according to relative B-factor of the Cα atoms, where Cα atoms with highest B-factor are colored red, and Cα atoms with lowest B-factor are colored blue.
FIGURE 2.
Crystal structure of ARL6/BBS3 sheds light on the pathogenicity of variants found in BBS patients. A, stereo view of the interaction network around the active site of the ARL6 small GTPase. Water molecules are shown in blue and Mg2+ ion in orange. Polar interactions involved with attacking water molecule and Ser-71 are shown as red dashed lines. Residue Thr-31, mutation of which causes BBS, is labeled in magenta. B, stereo view of the hydrogen bonding network around the G4 and G5 boxes as well as Gly-169 of ARL6. Relevant side chains of ARL6 are shown in stick representation, and the sticks for GTP are shown with a thinner radius. Hydrogen bonds involving the GTP molecule are depicted as black dashed lines; hydrogen bonds involving Gly-169 are depicted in red, and others hydrogen bonds involving residues the G4 and G5 are in yellow. C, hydrophobic core around Leu-170 of ARL6. Side chains of Leu-37, Ile-33, Phe-128, and Leu-170 are shown as spheres.
Resolving the crystal structure of ARL6/BBS3 has not only allowed us to characterize the structure of the wild-type protein, but it has also provided insights into the pathogenicity of the single amino acid substitutions (T31M, T31R, G169A, and L170W) our group reported to cause BBS in patients (28). Several amide groups in the G1 box (GX4GKT), also known as the P-loop, form critical hydrogen bonds with the triphosphates of the GTP nucleotide. In particular, the Thr-31 side chain hydroxyl group contributes to coordination of Mg2+, β- and γ-phosphates of GTP, free water molecule, and a water molecule bridging to Asp-69, all critically important for the GTPase activity of ARL6 (Fig. 2A). It is possible that mutation of Thr-31 to arginine provides a necessary catalytic residue in cis and thus increases the GTPase activity of ARL6. Alternatively, mutation of Thr-31 to methionine may hinder the binding of Mg2+ and γ-phosphate because of the hydrophobic nature of the methionine side chain. Mutation of residue Gly-169 to an alanine was found in Bardet-Biedl syndrome 3 patients. The Gly-169 residue lies within the conserved G5 box (residues 162–165), which not only binds the GTP guanine ring directly but also forms a hydrogen-bonding network with residues of the G4 box and residues following the G5 box. These hydrogen bonds contribute to the stabilization of the bound nucleotide (Fig. 2B). Interestingly, residue Gly-169 contributes to this hydrogen bonding network, forming hydrogen bonds with Glu-172 and Gly-173 (Fig. 2B). Finally, the Leu-170 residue was found to be part of a hydrophobic core with the side chains of Ile-33 and Leu-37 (Fig. 2C). Mutation of the Leu-170 residue to a tryptophan would likely disrupt the packing of this hydrophobic core and destabilize the protein.
Therefore, our structural analyses predict that the Thr-31 mutations are likely to specifically affect nucleotide binding, while the Gly-169 and Leu-170 mutations are predicted to primarily affect protein folding, resulting in an inability to bind nucleotide and degradation by the cell machinery. Furthermore, all mutations are predicted to result in a loss-of-function.
Pathogenic ARL6 Variants Display Variable Nucleotide Binding Defects
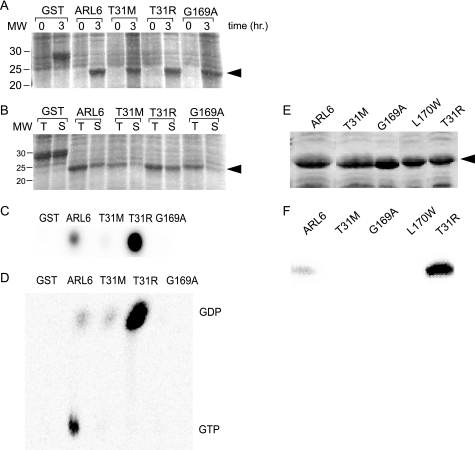
To experimentally validate the crystallography data, we performed nucleotide binding assays on ARL6 recombinant proteins. The wild-type and mutant GST-tagged ARL6 proteins were tested in dot-blot assays for binding to [α-32P]GTP, using GST alone as a negative control. As with GST alone, the GST-tagged ARL6(T31M) and ARL6(G169A) mutants displayed little or no binding compared to wild-type ARL6, whereas the ARL6(T31R) mutant bound significantly higher levels of the nucleotide (supplemental Fig. 1, A–C). Although [α-32P]GTP was used, a small amount of [α-32P]GDP is present in the source vial. Therefore, to determine the nature of the bound nucleotide, we also presented the tagged variants with [γ-32P]GTP, in which case only wild-type ARL6 displayed nucleotide binding activity above the GST-alone control (supplemental Fig. 1D). Although these results are comparable with those published recently by Kobayashi et al. (57), who used FLAG- or GST-tagged proteins, it is known that small GTPases are often sensitive to the addition of coding region at their N or C terminus and that this could interfere with their stability and/or functions. For this reason, we also carried out studies using untagged versions of wild-type and mutant ARL6 proteins (Fig. 3, A and B). Similar to the results presented above, the ARL6(T31M), ARL6(G169A), and ARL6 (L170W) mutants showed little or no binding of nucleotide present in the [α-32P]GTP mixture, as opposed to ARL6(T31R), which displayed a significantly higher affinity for the nucleotide (Fig. 3, C and F). TLC was then used to determine the nature of the labeled nucleotide bound to ARL6. The results show that although wild-type ARL6 is primarily bound to GTP, ARL6(T31R) appears strictly bound to GDP (Fig. 3D). Our combined crystallography and biochemical results therefore demonstrate that each of the presumed pathogenic ARL6 variants, in the form they would be present in patients (i.e. untagged), display abrogated nucleotide binding, with little or no binding in the case of ARL6(T31M), ARL6(G169A), and ARL6(L170W) and binding exclusively to GDP in the case of ARL6(T31R) (Fig. 3F).
FIGURE 3.
BBS3 pathogenic variants possess abrogated nucleotide binding activities. A, Coomassie-stained SDS gel showing induction of GST alone, untagged wild-type ARL6, and the T31M, T31R, and G169A ARL6 variants. B, Coomassie-stained SDS-polyacrylamide gel showing the proportion of total cell lysate (T) that remains in the soluble fraction (S) of samples from A after a 3-h induction. C, aliquot of the soluble fractions from B was spotted onto nitrocellulose and probed with [α-32P]GTP. GST, the negative control, does not bind nucleotide, whereas ARL6 binds. ARL6(T31R) binds significantly more than wild-type ARL6, and the T31M and G169A variants bind very poorly. D, TLC showing the nature of the radiolabeled nucleotide bound to soluble untagged ARL6 or variant forms of ARL6. Wild-type (untagged) ARL6 binds both GDP and GTP, but the untagged mutants (T31M and T31R) primarily bind GDP. E, Coomassie-stained SDS gel showing pellet fractions wild-type and variant forms (T31M, T31R, G169A, L170W) of the ARL6 protein, induced similarly to A in a separate experiment. F, as with E except that following SDS-PAGE the protein was transferred to nitrocellulose membrane, allowed to re-fold, and probed with [α-32P]GTP.
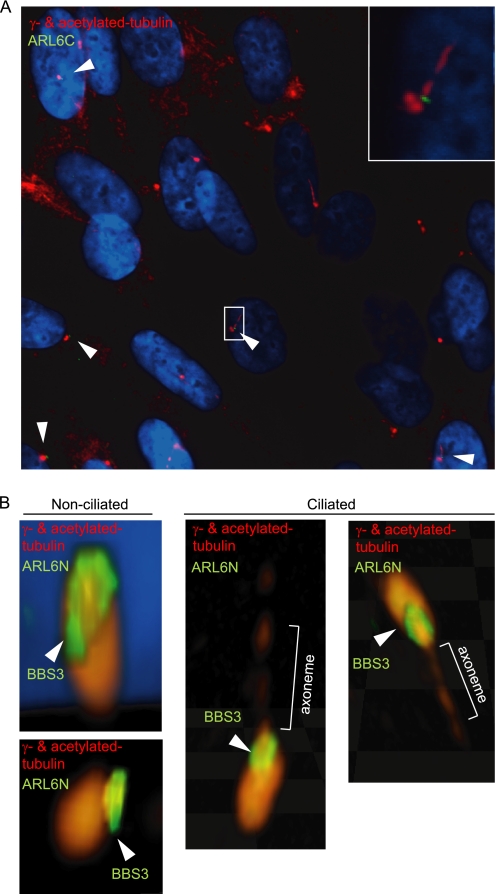
ARL6 Localizes to a Ring-like Structure at the Distal End of Basal Bodies in Ciliated Cells
Thus far, all studied mammalian BBS proteins have been found to localize either at the centrosome/basal body or in the adjoining centriolar satellites, which represent prime locations for proteins having ciliary roles. Specifically, BBS2, BBS5, and BBS13/MKS1 co-localize with the centrosome/basal body marker γ-tubulin (38, 58–60); BBS4, BBS8, and BBS14/CEP290 co-localize with PCM1 at centriolar satellites (61–63); and BBS6 is found within the pericentriolar material tube surrounding centrioles (64). To our surprise, two previous studies positioned green fluorescent protein-tagged and hemagglutinin-tagged ARL6 in the Golgi (65) or in the cytosol (66) of nonciliated cells, respectively. We therefore sought to determine the subcellular localization of endogenous ARL6 in a ciliated cell line.
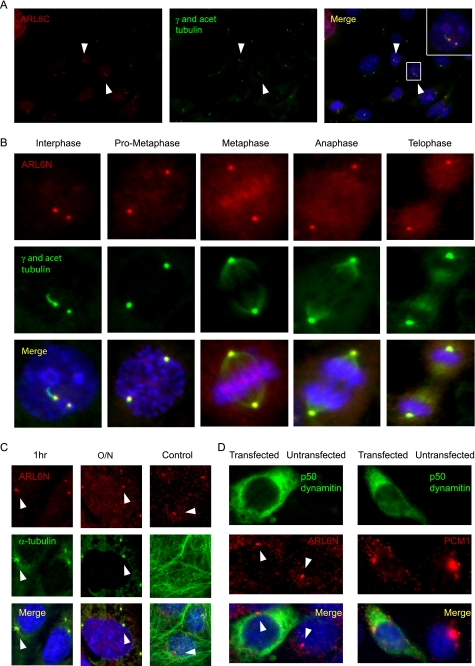
To perform our immunocytochemistry experiments, we employed polyclonal antibodies directed against N- and C-terminal epitopes of human ARL6 (named ARL6N and ARL6C, respectively); by Western blot analysis, both recognize ARL6 but not the closely related ARF6 protein, used as a control for specificity (supplemental Fig. 2). In mouse medullary collecting duct epithelial cells (IMCD3), the two anti-ARL6 antibodies specifically stain two peri-nuclear puncta that co-localize with the centrosomal marker γ-tubulin (Fig. 4, A and B). The ARL6 signal at centrosomes could be observed throughout the cell cycle, indicating a specific association with the dynamic microtubule organizing center; in interphase cells, ARL6 was associated with the basal body, which sports a cilium that is detected using an antibody against acetylated tubulin (Fig. 4B). We confirmed the centrosomal/basal body localization of ARL6 in a second ciliated cell line, namely retinal pigmented epithelial cells (hTERT-RPE) (Fig. 5). Importantly, we also observed co-localization of ARL6 with basal bodies in several ciliated mammalian tissue sections, including testis, brain, and kidney (supplemental Fig. 3, A–C). These data demonstrate that ARL6/BBS3 associates with centriolar structures/basal bodies in a variety of ciliated cells in vivo.
FIGURE 4.
ARL6 is a bona fide centrosomal/basal body-associated protein that localizes independently of the microtubule network or the dynein molecular motor. A, endogenous ARL6, observed using the ARL6C antibody (red) in ciliated IMCD3 cells, is observed at peri-nuclear puncta that co-localize with γ-tubulin (green), a centrosomal/basal body marker (yellow in the merged image). Inset, 2× magnification of area shown. DNA is stained blue with 4′,6-diamidino-2-phenylindole. Arrowheads point to ARL6 localization to centrosomes. B, endogenous ARL6 (visualized using the ARL6N antibody in IMCD3 cells; red) remains associated with the centrosome (marked using an antibody against γ-tubulin; green) throughout the cell cycle (yellow signal in merged image). Note that the interphase cell is ciliated (stained with antibody against acetylated (acet) tubulin; green). C, nocodazole treatment, but not the control treatment (DMSO), for either 1 h or overnight (O/N), is sufficient to destroy the microtubule network in IMCD3 cells assayed by α-tubulin staining (green), but it has no effect on ARL6 (red) localization. Arrowheads point to ARL6 localized to centrosomes in cells treated with nocodazole overnight. D, inhibition of the molecular motor dynein is achieved through the overexpression of green fluorescent protein-tagged p50-dynamitin (green) shown by the ability to mislocalize PCM1 (right panel), a protein known to be dependent upon dynein for its centrosomal localization (note its dispersed staining in cells expressing p50-dynamitin compared with cells not expressing the dynein inhibitor). ARL6 (left panel) is not mislocalized when p50 dynamitin is overexpressed. Arrowheads point to the proper localization of ARL6 in cells expressing p50-dynamitin.
FIGURE 5.
ARL6 localizes at the distal end of the basal body in a ring-shaped pattern. A, immunocytochemistry using the ARL6C antibody shows ARL6 localizing to the distal end of mother centrioles and basal bodies in hTERT-RPE cells. Inset, magnified image of the selected area. B, confocal microscopy images using the ARL6N antibody in hTERT-RPE cells shows that ARL6 localizes in a ring-like pattern, just distal to the mother centrioles, in nonciliated and ciliated cells.
Similar to BBS6 (64), but unlike BBS4 and its interacting protein PCM1 (63), the localization of ARL6 to the centrosome/basal body is microtubule-independent, as treatment of IMCD3 cells with the microtubule depolymerizing drug nocodazole does not affect this localization (Fig. 4C). Moreover, the localization of ARL6 is also unaffected in cells where the dynein retrograde motor is disrupted by overexpression of p50-dynamitin (Fig. 4D). Together, these results strongly support the notion that the endogenous ARL6/BBS3 protein is a bona fide centriole-associated protein that is present at the microtubule organizing center in dividing cells and at the basal body in ciliated cells, in a microtubule- and dynein-independent manner.
With the goal of precisely determining the position of ARL6 within the basal body, we carried out confocal imaging on hTERT-RPE cells stained with both anti-ARL6 antibodies. Interestingly, the ARL6 protein was found to be concentrated at one end of the microtubule-based structure. Specifically, interphase cells lacking a cilium at the time of fixation show ARL6 localizing in a ring-like pattern at the distal end of only one centriole, presumably the mother centriole; for cells that do possess a cilium, the ring of ARL6 is identically positioned, precisely at the junction between the basal body and the ciliary axoneme (Fig. 5, A and B). These results indicate that ARL6, which is not present within the BBSome complex (37), operates within a distinct and unique region of the basal body compared to all other studied BBS proteins. Its localization is consistent with it being near or at the transition fibers, which form part of the so-called ciliary gate.
Expression of Wild Type and Dominant Negative ARL6/BBS3 Variants Results in Cilia Anomalies
The unique localization of mammalian ARL6/BBS3 at the base of cilia suggested that this small GTPase possesses a role in cilium formation in mammalian cells, perhaps by way of ciliary membrane trafficking. To investigate this possibility, we took advantage of the fact that overproduction of small GTPase mutant forms is often used to probe their in vivo functions (67). Fortuitously, mutations corresponding to the Thr-31 residue, which we identified previously in the homozygous state in BBS patients (28) and leads to a GDP-locked protein (Fig. 3D), are commonly used as dominant negative forms. We therefore prepared a mammalian expression construct for ARL6(T31R), as well as another bearing a single amino acid mutation at Gln-73, predicted to yield a GTP-locked, constitutively active form of the protein (68). Although in most cases GTP-locked forms create constitutively active small GTPases, in some cases it appears that it is the cycling of nucleotide that is necessary for protein function. In those cases, the GTP-locked form acts more like a dominant negative, much like the GDP-locked form, as is the case for the GTP-locked form of ARL6 (see below). Interestingly, the GTP-locked form of ARF6 (Q67L), which also happens to be the ARF small GTPase with the highest degree of identity to ARL6, also displays this characteristic (69–71).
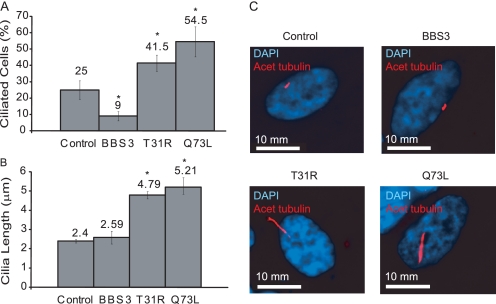
Overproduction of wild-type ARL6, or either of the variants, ARL6(T31R) or ARL6(Q73L), in hTERT-RPE cells did not result in appreciable changes to the division, microtubule cytoskeleton, or morphology of the cells compared to controls (data not shown). However, using acetylated tubulin as a marker for cilia, we observed a significantly higher proportion of cells possessing cilia in those transfected with either ARL6(T31R) or ARL6(Q73L) overexpression constructs as compared with the control cells (41.5 ± 3.5 and 54.5 ± 6.5% versus 25 ± 4%; p < 0.001 compared with control) (Fig. 6A). Remarkably, the opposite phenotype, namely a statistically significant lower proportion of cells with cilia, was detected in those transfected with the wild-type ARL6 construct (9 ± 2 versus 25 ± 4%; p < 0.001 compared with control) (Fig. 6A). Another phenotype observed was the significant difference in the length of cilia in cells expressing the mutant forms of ARL6. Whereas the average length of cilia from cells transfected with wild-type ARL6 was not significantly different from the control, the cilia of ARL6(T31R)- and ARL6(Q73L)-transfected cells were significantly longer (4.79 ± 0.19 and 5.21 ± 0.36 μm versus 2.4 ± 0.09 μm; p < 0.001) (Fig. 6B). Together, these results suggest that BBS3 acts to modulate the presence and structure of primary cilia; excess wild-type BBS3 results in fewer ciliated cells, whereas abrogation of its function leads to a larger proportion of ciliated cells with longer cilia.
FIGURE 6.
Overexpression of wild-type ARL6/BBS3 and variants results in ciliary anomalies. hTERT-RPE cells were transfected with one of three constructs, pCMV-BBS3 (overexpressing untagged and wild-type BBS3), pCMV-T31R, or pCMV-Q73L (overexpressing the untagged variants). The resulting phenotypes were quantified 2 days following transfection. A, proportion of cells possessing a cilium was determined by randomly counting >100 cells from each population. The error bars represent the standard error between duplicate experiments. B, cilium length was determined by measuring >30 random cilia from the population. The error bars represent the standard error between duplicate experiments. Asterisk represents statistically significant difference compared with control. C, representative images showing the differences in cilium length between hTERT-RPE cells that have been mock-transfected (control) or transfected to express untagged versions of wild-type ARL6, ARL6(T31R), or ARL6(Q73L) variants. DAPI, 4′,6-diamidino-2-phenylindole.
Wild-type ARL6, but Not Pathogenic Variants, Modulates Wnt Signaling
The discovery that cilia are central to various signaling pathways has greatly enhanced our understanding of the role of these organelles in development and disease. As part of its function in intracellular signaling, the cilium normally suppresses canonical, β-catenin-dependent Wnt signals (72, 73) and is required for noncanonical, β-catenin-independent Wnt signaling. Loss of several BBS proteins, including BBS2, BBS4, and BBS6, leads to increased β-catenin transcriptional activity when stimulated by canonical Wnt3a signal (73), whereas overexpression of BBS4 does not have an overt effect on β-catenin activity.5
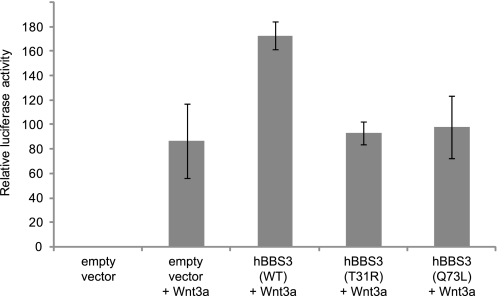
To explore the potential role of ARL6/BBS3 in Wnt signaling, we expressed ARL6/BBS3 in an hTERT-RPE1 cell line transfected with a pTOPflash reporter plasmid (Fig. 7) and a stable HEK293T Wnt reporter cell line (supplemental Fig. 4A). Interestingly, the effect of ARL6/BBS3 on β-catenin activity is dosage-dependent, because overexpression leads to an approximate 2- or 6-fold increased Wnt3a response in the hTERT-RPE1 (Fig. 7) and HEK293T (supplemental Fig. 4A) cell line compared to wild-type cells, respectively. In contrast, overexpression of ARL6(T31R) and ARL6(Q73L), now known to cause ciliary anomalies (Fig. 6), did not have an effect on β-catenin transcriptional activity (Fig. 7). Although ARL6(T31R) represents a pathogenic mutation, ARL6(Q73L) is not a mutation identified in BBS3 patients. Therefore, to further investigate whether or not Wnt signaling may be abrogated in BBS patients, we tested the response to Wnt3a ligand in additional overexpressed ARL6 pathogenic variants, namely ARL6(G169A) and ARL6(L170W). Similarly to ARL6(T31R) and ARL6(Q73L), we observed a loss of responsivity to the Wnt3a signal and β-catenin transcriptional activity compared to when wild-type ARL6 is overexpressed (supplemental Fig. 4B). Together, these results demonstrate that ARL6/BBS3 can modulate Wnt signaling in mammalian cells and that its effect may be different from that of other BBS proteins (see below).
FIGURE 7.
Overexpression of wild-type but not mutant forms of ARL6/BBS3 alters responsiveness to Wnt3a ligand. In hTERT-RPE TOPflash reporter cells, overexpression of ARL6/BBS3 leads to increased β-catenin activity in response to Wnt3a ligand but no global dysregulation of β-catenin signaling. In contrast, overexpression of ARL6(T31R) and ARL6(Q73L) has no marked effect on β-catenin transcriptional activity. Cells are consistently 70–80% confluent and ciliated as observed by staining with an anti-acetylated tubulin antibody (data not shown). Error bars represent standard deviation; only the BBS3 wild type (WT) samples are statistically different from the empty vector control (p < 0.05).
DISCUSSION
BBS3 Possesses Unique Structural Features and Nucleotide Binding Characteristics
Here, we present the crystal structure of ARL6/BBS3, the first high resolution structural information for a BBS or IFT protein. Because ARF GTPases usually have a higher affinity for GDP than GTP (for example ARL5A, PDB codes 2H16/2H17; ARL8A, PDB codes 2H18/1ZD9; ARL8B, PDB code 2AL7; and SAR1A, PDB code 2GAO), our structural data on ARL6 were unexpected, particularly as we were able to solve its structure in the GTP-bound state (74). One potential reason for this seemingly unique property of ARL6 is that its switch II region prefers a GTP-bound conformation because of the hydrogen bond between the Ser-71 side chain, which is unique among ARF/ARF-like small GTPases, and the amide group of Gln-73 (Fig. 2A). Thus, ARL6 may have higher affinity for GTP than GDP, a finding that is supported biochemically (Fig. 3 and supplemental Fig. 1). Additionally, the leucine-rich N-terminal 14 residues of ARL6, not present in our structure, are predicted to form a hydrophobic α-helix rather than an amphipathic helix, as with other ARF family small GTPases; myristoylation of the Gly-2 residue in ARL6 is also predicted to be unlikely (32). Thus, it is possible that both the nucleotide binding properties of ARL6, as well as the manner in which its membrane binding is regulated, may be different from that of other family members.
Importantly, the crystal structure data explain the effect that pathogenic mutations have on the ARL6/BBS3 protein. These mutations either affect critical residues necessary for GTP binding, i.e. Thr-31, or they affect the intramolecular hydrogen bonds in such a way as to probably cause local/global misfolding as well, i.e. Gly-169 and Leu-170. Our functional studies are consistent with this notion; the ARL6(T31M), ARL6(G169A), and ARL6(L170W) variants displayed little or no nucleotide binding, whereas the ARL6(T31R) variant was observed in a GDP-locked state that differed from its wild-type counterpart, which was mainly GTP-bound (Fig. 3). Under these conditions, the activities of the small GTPase are predicted to be impaired, a possibility that, as mentioned below, we confirmed by observing ciliary and signaling phenotypes.
ARL6/BBS3 Is a Novel Basal Body Protein That Localizes at the Entrance of the Ciliary Compartment
Our findings demonstrate that ARL6/BBS3 is found in a ring-like pattern at the distal end of the basal body and most proximal region of the ciliary axoneme (Fig. 5). Such a basal body localization for ARL6/BBS3 is consistent with previous findings for other BBS proteins (38, 58, 60–63). However, the precise position of ARL6/BBS3 at the distal end of basal bodies is unique among BBS proteins but similar to that reported for the retinitis pigmentosa 2 (RP2) homologue in C. elegans (34) and Trypanosoma brucei (75), which also form ring-shaped structures at the base of cilia. Interestingly, RP2 is known to interact with, and act as a GTPase-activator for, the small GTPase ARL3 (76), whose function in cilium formation has been documented in different organisms (33–35).
Also notable is the fact that the subcellular site of action of ARL6 is either at, or in close proximity to, the transitional fibers at the distal end of all basal bodies. The tips of these fibers, which join the ciliary membrane, are believed to be the docking sites for IFT particles (19, 22). Given that C. elegans ARL-6/BBS-3, like all other tested C. elegans BBS proteins (BBS-1, -2, -5, -7, and -8) (77, 78), associates with the IFT machinery (i.e. moves in an IFT-like fashion in sensory cilia), we speculate that ARL6/BBS3 may interact with other BBS proteins and the IFT machinery near or at this transition fiber region. The fibers also help form a ciliary gate, which restricts the entry of vesicles into the ciliary space, such that Golgi-derived vesicles bearing ciliary cargo dock and fuse in close proximity to the fibers (21, 22). Therefore, given the established role for BBS proteins in trafficking within the cell body, and IFT in cilia, ARL6/BBS3 could conceivably facilitate/modulate the transition between vesicular and intraciliary trafficking. It will be particularly challenging to dissect the specific functions of ARL6/BBS3, as one will need to take into account both pre-basal body/ciliary membrane trafficking as well as intraciliary trafficking (IFT)-associated processes. The former likely includes several small GTPases, including RAB8, RAB17, and RAB23 and, perhaps for a subset of proteins (containing VXPX motifs), ARF4 and RAB11 (37, 79, 80); the latter depends on an emerging set of players, including the RAB proteins IFT25, RAB8, and IFTA-2, as well as ARL13B (34, 36, 37, 39).
ARL6/BBS3 May Modulate Cilium Disassembly and/or Length Control
By overproducing in ciliated cells both wild-type ARL6 and two mutant forms of the ARL6 small GTPase, we uncovered a potential role for this small GTPase in cilium disassembly. Specifically, overexpression of wild-type ARL6 results in a reduction in cilium formation, a phenotype also observed upon small interfering RNA knockdown of BBS3 in a survey of all BBS genes (80); on the other hand, overexpression of ARL6(T31R) and ARL6(Q73L), which inhibit the activity of endogenous ARL6, cause an increase in cilium formation and length (Fig. 6).
From these results, we propose that ARL6 may facilitate, potentially through the trafficking of certain ciliary cargo, the resorption of cilia. This process is required for freeing the centrioles (basal body), for use as microtubule organizing centers, prior to cell cycle re-entry (81). Although such a function would be a first for BBS proteins, IFT proteins are intimately involved in ciliary resorption (17).
Interestingly, overproduction of the equivalent (presumably GDP-locked) mutant forms of ARL6(T31R) (Fig. 6B) and RAB8(T22N) (37) produces essentially opposite ciliary defects, indicating opposing functions for the two small GTPases. It is potentially relevant that in a zebrafish screen for genetic interactions between BBS genes, ARL6 was found to interact with BBS1 (82), whose gene product physically associates with RAB8 (37). On the whole, these data point to possible functional associations between the BBSome, ARL6/BBS3, and RAB8 in mediating ciliogenesis and cilium resorption. There is now a connection between the BBSome and ciliary resorption; the BBSome-associated protein BBIP10 has been shown to interact with HDAC6, whose microtubule acetylation activity affects ciliary disassembly (83, 84). Additionally, BBS proteins are also implicated in removing proteins from cilia in Chlamydomonas (100), consistent with a potential role in resorption.
Alternatively, BBS3 could participate in cilium length control, a process we know to be tightly regulated in Chlamydomonas reinhardtii. When Chlamydomonas cells are stimulated to excise their flagella, their replacements are made to pre-deflagellation lengths in under 2 h (85). Furthermore, when only one flagellum is amputated, the remaining flagellum will shorten until its counterpart reaches the same length, at which time both flagella grow out to their pre-determined length (85). At least seven proteins, encoded by four long flagella (lf) and three short flagella (shf) genes, regulate this process in C. reinhardtii (86–89). BBS3 is ideally positioned, at or near the ciliary gate, to function in ciliary length control. Specifically, it has been hypothesized that the transition fibers and machinery located within this area may tightly control the movement of components into and out of the ciliary compartment, for example IFT proteins, in a manner similar to that of the nuclear pore complex (19).
ARL6/BBS3 Modulates Wnt Signaling
Loss of BBS proteins, including BBS1, BBS4, and BBS6, leads to an augmented response to canonical Wnt signals such as Wnt3a as measured by β-catenin transcriptional activity (73). Given these results, it was unexpected that overexpression, and not knockdown, of another BBS protein, namely BBS3, led to a similar augmented response to the canonical Wnt3a ligand (Fig. 7). We propose that the increased β-catenin activity seen in wild-type BBS3-overexpressing cells is linked to their lower proportion of cilia (Fig. 6A). It has been suggested that cilia influence the balance between canonical and noncanonical Wnt signaling within the cell by favoring the noncanonical pathway (73); therefore, increased disassembly of cilia in cells overexpressing wild-type BBS3 would favor canonical Wnt signaling, as we have observed.
Although one might expect to see the opposite effect, namely decreased β-catenin activity in cells overexpressing the ARL6(T31R) and ARL6(Q73L) variants, which have also been shown here to result in ciliary phenotypes, we observed no significant effect on β-catenin transcriptional activity from overexpression of these dominant mutations (Fig. 7). Although this result is not immediately intuitive, it does fit with our hypothesized role for BBS3, which is that part of its function may be to assist in the trafficking of ciliary membrane components out of the ciliary compartment. To achieve dynamic equilibrium between both the canonical and noncanonical Wnt signaling pathways, the ciliary “switch” must function properly, and it is likely to require proper trafficking of ciliary membrane components not only into, but also out of, the ciliary compartment. If ciliary components of the Wnt pathway (e.g. inversin) (90–92), are not properly trafficked out of the cilium, the switch to noncanonical signaling will likely be impaired, and wild-type levels of β-catenin activity will be observed, as is the case with our results. This may suggest that the ciliary switch that favors noncanonical Wnt signaling may be dysfunctional when BBS3 is abrogated, a possibility that will require further investigation.
Although our results agree with current models indicating a role for cilia in Wnt signaling, it would only be pertinent to acknowledge that the requirement of cilia in restraining canonical Wnt signaling remains controversial. The controversy stems from findings that show IFT-defective mouse models and cilia-less zebrafish models exhibit wild-type expression patterns of canonical Wnt signaling components and respond normally to canonical Wnt ligand (93, 94). However, as Huang and Schier (94) acknowledge, knockdown of these IFT components results exclusively in cilia defects and leaves the basal body more or less intact. Because BBS proteins are often primarily basal body-associated and, in some cases, are involved in the trafficking of critical centrosomal components (61, 63), it could be that the involvement of BBS proteins in Wnt signaling has to do with their role at basal bodies, the foundation from which cilia are built. Therefore, we assert that if it is not cilia then basal bodies at the very least are critical for restraining canonical Wnt signaling by influencing the dynamic equilibrium between canonical and noncanonical Wnt signaling.
Interestingly, expression of Wnt3a was disrupted in the pre-migratory cardiac neural crest cells of zebrafish with morpholino-mediated knockdown of the ARL6-interacting protein (ARL6IP) (95). Furthermore, loss of ARL6IP mimicked many phenotypes observed in the knockdown of BBS proteins in Danio rerio, namely craniofacial abnormalities, defects in limb bud development, heart defects, abnormal pigmentation, and retinal degeneration (82, 95–97). The nature of this functional interaction between ARL6 and ARL6IP could therefore prove to be useful for our understanding the cellular and molecular processes that regulate cilium function, and its association with Wnt signaling.
Supplementary Material
Acknowledgments
We thank Dr. Andreas Merdes for generously providing us with the anti-PCM1 antibody and Linda Kang for technical assistance. Additionally, Y. T. and H.-W. P. thank Yang Shen and Drs. Jihong Wang and Wolfram Tempel for work on the structure determination of ARL6.
This work was supported, in whole or in part, by National Institutes of Health Grants R01HD04260 from NICHD (to N. K.) and R01DK072301 and DK075972 from NIDDK (to N. K.). This work was also supported by a Conte Center grant from the National Institute of Mental Health (to N. K.), Canadian Institutes of Health Research Grant MOP-82870 (to M. R. L.), Heart and Stroke Foundation of British Columbia and Yukon grant (to M. R. L.), Natural Sciences and Engineering Research Council of Canada grant (to J. B. R.), and in part by the Structural Genomics Consortium, a registered charity (number 1097737) that receives funds from Canadian Institutes of Health Research, Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, Ontario Innovation Trust, Ontario Ministry for Research and Innovation, Merck, Novartis Research Foundation, Swedish Agency for Innovation Systems, Swedish Foundation for Strategic Research, and the Wellcome Trust.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
The atomic coordinates and structure factors (code 2H57) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
N. Katsanis, unpublished results.
- BBS
- Bardet-Biedl syndrome
- ARF
- ADP-ribosylation factor
- IFT
- intraflagellar transport
- PDB
- Protein Data Bank
- GST
- glutathione S-transferase
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- Pipes
- 1,4-piperazinediethanesulfonic acid.
REFERENCES
- 1.Beales P. L. (2005) Curr. Opin. Genet. Dev. 15, 315–323 [DOI] [PubMed] [Google Scholar]
- 2.Blacque O. E., Leroux M. R. (2006) Cell. Mol. Life Sci. 63, 2145–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tobin J. L., Beales P. L. (2007) Pediatr. Nephrol. 22, 926–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaghloul N. A., Katsanis N. (2009) J. Clin. Invest. 119, 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006) Annu. Rev. Genomics Hum. Genet. 7, 125–148 [DOI] [PubMed] [Google Scholar]
- 6.Fliegauf M., Benzing T., Omran H. (2007) Nat. Rev. Mol. Cell Biol. 8, 880–893 [DOI] [PubMed] [Google Scholar]
- 7.Satir P., Christensen S. T. (2007) Annu. Rev. Physiol. 69, 377–400 [DOI] [PubMed] [Google Scholar]
- 8.Quinlan R. J., Tobin J. L., Beales P. L. (2008) Curr. Top. Dev. Biol. 84, 249–310 [DOI] [PubMed] [Google Scholar]
- 9.Sharma N., Berbari N. F., Yoder B. K. (2008) Curr. Top. Dev. Biol. 85, 371–427 [DOI] [PubMed] [Google Scholar]
- 10.Eggenschwiler J. T., Anderson K. V. (2007) Annu. Rev. Cell Dev. Biol. 23, 345–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes J. M., Katsanis N. (2008) Curr. Top. Dev. Biol. 85, 175–195 [DOI] [PubMed] [Google Scholar]
- 12.Pedersen L. B., Veland I. R., Schrøder J. M., Christensen S. T. (2008) Dev. Dyn. 237, 1993–2006 [DOI] [PubMed] [Google Scholar]
- 13.Marshall W. F. (2007) J. Cell. Biochem. 100, 916–922 [DOI] [PubMed] [Google Scholar]
- 14.Santos N., Reiter J. F. (2008) Dev. Dyn. 237, 1972–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blacque O. E., Cevik S., Kaplan O. I. (2008) Front. Biosci. 13, 2633–2652 [DOI] [PubMed] [Google Scholar]
- 16.Insinna C., Besharse J. C. (2008) Dev. Dyn. 237, 1982–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pedersen L. B., Rosenbaum J. L. (2008) Curr. Top. Dev. Biol. 85, 23–61 [DOI] [PubMed] [Google Scholar]
- 18.Scholey J. M. (2008) J. Cell Biol. 180, 23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deane J. A., Cole D. G., Seeley E. S., Diener D. R., Rosenbaum J. L. (2001) Curr. Biol. 11, 1586–1590 [DOI] [PubMed] [Google Scholar]
- 20.Luby-Phelps K., Fogerty J., Baker S. A., Pazour G. J., Besharse J. C. (2008) Vis. Res. 48, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deretic D., Papermaster D. S. (1991) J. Cell Biol. 113, 1281–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenbaum J. L., Witman G. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 813–825 [DOI] [PubMed] [Google Scholar]
- 23.Silverman M. A., Leroux M. R. (2009) Trends Cell Biol. 19, 306–316 [DOI] [PubMed] [Google Scholar]
- 24.Leroux M. R. (2007) Cell 129, 1041–1043 [DOI] [PubMed] [Google Scholar]
- 25.Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. (2004) Cell 117, 527–539 [DOI] [PubMed] [Google Scholar]
- 26.Jékely G., Arendt D. (2006) BioEssays 28, 191–198 [DOI] [PubMed] [Google Scholar]
- 27.Chiang A. P., Nishimura D., Searby C., Elbedour K., Carmi R., Ferguson A. L., Secrist J., Braun T., Casavant T., Stone E. M., Sheffield V. C. (2004) Am. J. Hum. Genet. 75, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan Y., Esmail M. A., Ansley S. J., Blacque O. E., Boroevich K., Ross A. J., Moore S. J., Badano J. L., May-Simera H., Compton D. S., Green J. S., Lewis R. A., van Haelst M. M., Parfrey P. S., Baillie D. L., Beales P. L., Katsanis N., Davidson W. S., Leroux M. R. (2004) Nat. Genet. 36, 989–993 [DOI] [PubMed] [Google Scholar]
- 29.Takai Y., Sasaki T., Matozaki T. (2001) Physiol. Rev. 81, 153–208 [DOI] [PubMed] [Google Scholar]
- 30.Kahn R. A., Volpicelli-Daley L., Bowzard B., Shrivastava-Ranjan P., Li Y., Zhou C., Cunningham L. (2005) Biochem. Soc. Trans. 33, 1269–1272 [DOI] [PubMed] [Google Scholar]
- 31.Zhou C., Cunningham L., Marcus A. I., Li Y., Kahn R. A. (2006) Mol. Biol. Cell 17, 2476–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillingham A. K., Munro S. (2007) Annu. Rev. Cell Dev. Biol. 23, 579–611 [DOI] [PubMed] [Google Scholar]
- 33.Cuvillier A., Redon F., Antoine J. C., Chardin P., DeVos T., Merlin G. (2000) J. Cell Sci. 113, 2065–2074 [DOI] [PubMed] [Google Scholar]
- 34.Blacque O. E., Perens E. A., Boroevich K. A., Inglis P. N., Li C., Warner A., Khattra J., Holt R. A., Ou G., Mah A. K., McKay S. J., Huang P., Swoboda P., Jones S. J., Marra M. A., Baillie D. L., Moerman D. G., Shaham S., Leroux M. R. (2005) Curr. Biol. 15, 935–941 [DOI] [PubMed] [Google Scholar]
- 35.Schrick J. J., Vogel P., Abuin A., Hampton B., Rice D. S. (2006) Am. J. Pathol. 168, 1288–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caspary T., Larkins C. E., Anderson K. V. (2007) Dev. Cell 12, 767–778 [DOI] [PubMed] [Google Scholar]
- 37.Nachury M. V., Loktev A. V., Zhang Q., Westlake C. J., Peränen J., Merdes A., Slusarski D. C., Scheller R. H., Bazan J. F., Sheffield V. C., Jackson P. K. (2007) Cell 129, 1201–1213 [DOI] [PubMed] [Google Scholar]
- 38.Li C., Inglis P. N., Leitch C. C., Efimenko E., Zaghloul N. A., Mok C. A., Davis E. E., Bialas N. J., Healey M. P., Héon E., Zhen M., Swoboda P., Katsanis N., Leroux M. R. (2008) PLoS Genet. 4, e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omori Y., Zhao C., Saras A., Mukhopadhyay S., Kim W., Furukawa T., Sengupta P., Veraksa A., Malicki J. (2008) Nat. Cell Biol. 10, 437–444 [DOI] [PubMed] [Google Scholar]
- 40.Hu J., Wittekind S. G., Barr M. M. (2007) Mol. Biol. Cell 18, 3277–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 42.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 43.Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 44.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 45.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoepfer R. (1993) Gene 124, 83–85 [DOI] [PubMed] [Google Scholar]
- 49.Chiu J., Tillett D., Dawes I. W., March P. E. (2008) J. Microbiol. Methods 73, 195–198 [DOI] [PubMed] [Google Scholar]
- 50.Ding M., Vitale N., Tsai S. C., Adamik R., Moss J., Vaughan M. (1996) J. Biol. Chem. 271, 24005–24009 [DOI] [PubMed] [Google Scholar]
- 51.Wagner P., Hengst L., Gallwitz D. (1992) Methods Enzymol. 219, 369–387 [DOI] [PubMed] [Google Scholar]
- 52.Ou Y., Rattner J. B. (2000) Cell Motil. Cytoskeleton 47, 13–24 [DOI] [PubMed] [Google Scholar]
- 53.Korinek V., Barker N., Morin P. J., van Wichen D., de Weger R., Kinzler K. W., Vogelstein B., Clevers H. (1997) Science 275, 1784–1787 [DOI] [PubMed] [Google Scholar]
- 54.Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 55.Bourne H. R., Sanders D. A., McCormick F. (1991) Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 56.Pasqualato S., Renault L., Cherfils J. (2002) EMBO Rep. 3, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi T., Hori Y., Ueda N., Kajiho H., Muraoka S., Shima F., Kataoka T., Kontani K., Katada T. (2009) Biochem. Biophys. Res. Commun. 381, 439–442 [DOI] [PubMed] [Google Scholar]
- 58.Dawe H. R., Smith U. M., Cullinane A. R., Gerrelli D., Cox P., Badano J. L., Blair-Reid S., Sriram N., Katsanis N., Attie-Bitach T., Afford S. C., Copp A. J., Kelly D. A., Gull K., Johnson C. A. (2007) Hum. Mol. Genet. 16, 173–186 [DOI] [PubMed] [Google Scholar]
- 59.Shah A. S., Farmen S. L., Moninger T. O., Businga T. R., Andrews M. P., Bugge K., Searby C. C., Nishimura D., Brogden K. A., Kline J. N., Sheffield V. C., Welsh M. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3380–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bialas N. J., Inglis P. N., Li C., Robinson J. F., Parker J. D., Healey M. P., Davis E. E., Inglis C. D., Toivonen T., Cottell D. C., Blacque O. E., Quarmby L. M., Katsanis N., Leroux M. R. (2009) J. Cell Sci. 122, 611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J., Krishnaswami S. R., Gleeson J. G. (2008) Hum. Mol. Genet. 17, 3796–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ansley S. J., Badano J. L., Blacque O. E., Hill J., Hoskins B. E., Leitch C. C., Kim J. C., Ross A. J., Eichers E. R., Teslovich T. M., Mah A. K., Johnsen R. C., Cavender J. C., Lewis R. A., Leroux M. R., Beales P. L., Katsanis N. (2003) Nature 425, 628–633 [DOI] [PubMed] [Google Scholar]
- 63.Kim J. C., Badano J. L., Sibold S., Esmail M. A., Hill J., Hoskins B. E., Leitch C. C., Venner K., Ansley S. J., Ross A. J., Leroux M. R., Katsanis N., Beales P. L. (2004) Nat. Genet. 36, 462–470 [DOI] [PubMed] [Google Scholar]
- 64.Kim J. C., Ou Y. Y., Badano J. L., Esmail M. A., Leitch C. C., Fiedrich E., Beales P. L., Archibald J. M., Katsanis N., Rattner J. B., Leroux M. R. (2005) J. Cell Sci. 118, 1007–1020 [DOI] [PubMed] [Google Scholar]
- 65.Dejgaard S. Y., Murshid A., Erman A., Kizilay O., Verbich D., Lodge R., Dejgaard K., Ly-Hartig T. B., Pepperkok R., Simpson J. C., Presley J. F. (2008) J. Cell Sci. 121, 2768–2781 [DOI] [PubMed] [Google Scholar]
- 66.Ingley E., Williams J. H., Walker C. E., Tsai S., Colley S., Sayer M. S., Tilbrook P. A., Sarna M., Beaumont J. G., Klinken S. P. (1999) FEBS Lett. 459, 69–74 [DOI] [PubMed] [Google Scholar]
- 67.Dascher C., Balch W. E. (1994) J. Biol. Chem. 269, 1437–1448 [PubMed] [Google Scholar]
- 68.Pai E. F., Krengel U., Petsko G. A., Goody R. S., Kabsch W., Wittinghofer A. (1990) EMBO J. 9, 2351–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Radhakrishna H., Donaldson J. G. (1997) J. Cell Biol. 139, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein S., Franco M., Chardin P., Luton F. (2006) J. Biol. Chem. 281, 12352–12361 [DOI] [PubMed] [Google Scholar]
- 71.Altschuler Y., Liu S., Katz L., Tang K., Hardy S., Brodsky F., Apodaca G., Mostov K. (1999) J. Cell Biol. 147, 7–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Corbit K. C., Shyer A. E., Dowdle W. E., Gaulden J., Singla V., Chen M. H., Chuang P. T., Reiter J. F. (2008) Nat. Cell Biol. 10, 70–76 [DOI] [PubMed] [Google Scholar]
- 73.Gerdes J. M., Liu Y., Zaghloul N. A., Leitch C. C., Lawson S. S., Kato M., Beachy P. A., Beales P. L., DeMartino G. N., Fisher S., Badano J. L., Katsanis N. (2007) Nat. Genet. 39, 1350–1360 [DOI] [PubMed] [Google Scholar]
- 74.Weiss O., Holden J., Rulka C., Kahn R. A. (1989) J. Biol. Chem. 264, 21066–21072 [PubMed] [Google Scholar]
- 75.Stephan A., Vaughan S., Shaw M. K., Gull K., McKean P. G. (2007) Traffic 8, 1323–1330 [DOI] [PubMed] [Google Scholar]
- 76.Veltel S., Gasper R., Eisenacher E., Wittinghofer A. (2008) Nat. Struct. Mol. Biol. 15, 373–380 [DOI] [PubMed] [Google Scholar]
- 77.Blacque O. E., Reardon M. J., Li C., McCarthy J., Mahjoub M. R., Ansley S. J., Badano J. L., Mah A. K., Beales P. L., Davidson W. S., Johnsen R. C., Audeh M., Plasterk R. H., Baillie D. L., Katsanis N., Quarmby L. M., Wicks S. R., Leroux M. R. (2004) Genes Dev. 18, 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ou G., Koga M., Blacque O. E., Murayama T., Ohshima Y., Schafer J. C., Li C., Yoder B. K., Leroux M. R., Scholey J. M. (2007) Mol. Biol. Cell 18, 1554–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yoshimura S., Egerer J., Fuchs E., Haas A. K., Barr F. A. (2007) J. Cell Biol. 178, 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mazelova J., Astuto-Gribble L., Inoue H., Tam B. M., Schonteich E., Prekeris R., Moritz O. L., Randazzo P. A., Deretic D. (2009) EMBO J. 28, 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quarmby L. M., Parker J. D. (2005) J. Cell Biol. 169, 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tayeh M. K., Yen H. J., Beck J. S., Searby C. C., Westfall T. A., Griesbach H., Sheffield V. C., Slusarski D. C. (2008) Hum. Mol. Genet. 17, 1956–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loktev A. V., Zhang Q., Beck J. S., Searby C. C., Scheetz T. E., Bazan J. F., Slusarski D. C., Sheffield V. C., Jackson P. K., Nachury M. V. (2008) Dev. Cell 15, 854–865 [DOI] [PubMed] [Google Scholar]
- 84.Pugacheva E. N., Jablonski S. A., Hartman T. R., Henske E. P., Golemis E. A. (2007) Cell 129, 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosenbaum J. L., Moulder J. E., Ringo D. L. (1969) J. Cell Biol. 41, 600–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asleson C. M., Lefebvre P. A. (1998) Genetics 148, 693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barsel S. E., Wexler D. E., Lefebvre P. A. (1988) Genetics 118, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kuchka M. R., Jarvik J. W. (1987) Genetics 115, 685–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McVittie A. (1972) J. Gen. Microbiol. 71, 525–540 [DOI] [PubMed] [Google Scholar]
- 90.Otto E. A., Schermer B., Obara T., O'Toole J. F., Hiller K. S., Mueller A. M., Ruf R. G., Hoefele J., Beekmann F., Landau D., Foreman J. W., Goodship J. A., Strachan T., Kispert A., Wolf M. T., Gagnadoux M. F., Nivet H., Antignac C., Walz G., Drummond I. A., Benzing T., Hildebrandt F. (2003) Nat. Genet. 34, 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Watanabe D., Saijoh Y., Nonaka S., Sasaki G., Ikawa Y., Yokoyama T., Hamada H. (2003) Development 130, 1725–1734 [DOI] [PubMed] [Google Scholar]
- 92.Singla V., Reiter J. F. (2006) Science 313, 629–633 [DOI] [PubMed] [Google Scholar]
- 93.Ocbina P. J., Tuson M., Anderson K. V. (2009) PLoS One 4, e6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang P., Schier A. F. (2009) Development 136, 3089–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huang H. Y., Dai E. S., Liu J. T., Tu C. T., Yang T. C., Tsai H. J. (2009) Dev. Dyn. 238, 232–240 [DOI] [PubMed] [Google Scholar]
- 96.Tobin J. L., Di Franco M., Eichers E., May-Simera H., Garcia M., Yan J., Quinlan R., Justice M. J., Hennekam R. C., Briscoe J., Tada M., Mayor R., Burns A. J., Lupski J. R., Hammond P., Beales P. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yen H. J., Tayeh M. K., Mullins R. F., Stone E. M., Sheffield V. C., Slusarski D. C. (2006) Hum. Mol. Genet. 15, 667–677 [DOI] [PubMed] [Google Scholar]
- 98.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 99.Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 100.Lechtreck K. F., Johnson E. C., Sakai T., Cochran D., Ballif B. A., Rush J., Pazour G. J., Ikebe M., Witman G. B. (2009) J. Cell Biol. 187, 1117–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.