Abstract
NUP98 is a nucleoporin that plays complex roles in the nucleocytoplasmic trafficking of macromolecules. Rearrangements of the NUP98 gene in human leukemia result in the expression of numerous fusion oncoproteins whose effect on nucleocytoplasmic trafficking is poorly understood. The present study was undertaken to determine the effects of leukemogenic NUP98 fusion proteins on CRM1-mediated nuclear export. NUP98-HOXA9, a prototypic NUP98 fusion, inhibited the nuclear export of two known CRM1 substrates: mutated cytoplasmic nucleophosmin and HIV-1 Rev. In vitro binding assays revealed that NUP98-HOXA9 binds CRM1 through the FG repeat motif in a Ran-GTP-dependent manner similar to but stronger than the interaction between CRM1 and its export substrates. Two NUP98 fusions, NUP98-HOXA9 and NUP98-DDX10, whose fusion partners are structurally and functionally unrelated, interacted with endogenous CRM1 in myeloid cells as shown by co-immunoprecipitation. These leukemogenic NUP98 fusion proteins interacted with CRM1, Ran, and the nucleoporin NUP214 in a manner fundamentally different from that of wild-type NUP98. NUP98-HOXA9 and NUP98-DDX10 formed characteristic aggregates within the nuclei of a myeloid cell line and primary human CD34+ cells and caused aberrant localization of CRM1 to these aggregates. These NUP98 fusions caused nuclear accumulation of two transcription factors, NFAT and NFκB, that are regulated by CRM1-mediated export. The nuclear entrapment of NFAT and NFκB correlated with enhanced transcription from promoters responsive to these transcription factors. Taken together, the results suggest a new mechanism by which NUP98 fusions dysregulate transcription and cause leukemia, namely, inhibition of CRM1-mediated nuclear export with aberrant nuclear retention of transcriptional regulators.
Keywords: Diseases/Cancer/Leukemia, Nucleus/Nuclear Export, Nucleus/Nuclear Pore, Nucleus/Nuclear Transport, Transcription Factors, Acute Myeloid Leukemia, CRM1, NUP98, Nucleoporin
Introduction
The nucleoporin NUP98 gene is a target of chromosomal rearrangements resulting in fusion to numerous partners with various biological functions (1, 2). NUP98 rearrangements occur in acute myeloid leukemia, myelodysplastic syndromes, chronic myelogenous leukemia, and T-cell acute lymphoblastic leukemia and are usually associated with poor prognosis. In all reported cases, an N-terminal segment of NUP98 containing FG repeat motifs and a GLEBS domain is on the N-terminal side of the fusion oncoprotein.
NUP98 fusion partners include several homeobox transcription factors. The resulting fusion oncoproteins dysregulate transcription by binding DNA through their homeodomains (3–5). Binding of the FG repeat region of NUP98 to the transcriptional co-activators cAMP-responsive element-binding protein-binding protein and p300 (CBP/p300) was correlated with transactivation by NUP98-HOXA9 (3). More recently, a leukemogenic role of histone methylation by the NSD1 domain of the NUP98-NSD1 fusion oncoprotein has been proposed (6). Transformation mechanisms involving other fusion partners without a known DNA binding or DNA modifying capability are unclear. On the other hand, accumulating studies of NUP98 fusions and their variants indicate that the N-terminal NUP98 portion shared by all oncogenic NUP98 fusions is also essential in leukemogenesis (3, 7–11). However, the mechanism by which this portion of NUP98 exerts its oncogenic effect is not well understood.
NUP98 is a component of the massive protein assemblies that constitute the nuclear pore complex (NPC)3 through which nucleocytoplasmic trafficking of macromolecules is regulated (12, 13). NUP98 has been localized to both the nuclear and cytoplasmic ends of the NPC as well as within the nucleus (14–16). Further, NUP98 is one of several nucleoporins that exhibit transcription-dependent mobility within cells (17). The functions of NUP98 are multifaceted. By forming a complex with RNA export 1 (RAE1), NUP98 is involved in RNA export (18) as well as regulation of the anaphase-promoting complex (19). In addition, it may facilitate nuclear import of some proteins (20) and may also participate in the nuclear export of others (21). Given these functions of NUP98, the question arises of whether oncogenic NUP98 fusion molecules can interfere with nucleocytoplasmic trafficking.
The present study was undertaken to examine the effect of NUP98 fusion molecules on nuclear export. It is demonstrated first that the expression of an oncogenic NUP98 fusion protein indeed inhibits CRM1 (chromosome region maintenance 1; exportin 1)-mediated nuclear export of known export substrates. Next, it is shown that the export block is linked to aberrant interactions between NUP98 fusion proteins, CRM1, and NUP214. Finally, it is shown that NUP98 fusion proteins cause nuclear accumulation of CRM1-regulated transcription factors resulting in enhanced transcription from promoters regulated by these factors. These data identify inhibition of CRM1-mediated nuclear export as a new mechanism by which NUP98 fusions may dysregulate transcription and contribute to leukemic transformation.
EXPERIMENTAL PROCEDURES
Plasmids
Enhanced green fluorescent protein (EGFP)-tagged cytoplasmic mutated nucleophosmin (NPMc) (from Dr. B. Falini, University of Perugia, Perugia, Italy) (22), GFP-tagged Rev (encoding full-length HIV-1 Rev followed by the hormone-responsive element of the rat glucocorticoid receptor and GFP; from Dr. J. Hanover, National Institutes of Health, Bethesda, MD) (23), GFP-tagged NFAT (from Dr. L. Gerace, Scripps Research Institute, La Jolla, CA) (24), and EGFP-tagged p65 subunit of NFκB (from Dr. J. Schmid, University of Vienna, Vienna, Austria) (25) were subcloned into pcDNA3.1 Hygro for in vitro transcription/translation. The coding sequence of CRM1 (from Dr. M. Fornerod, Netherlands Cancer Institute, Amsterdam, Netherlands) (26) was subcloned into pGEX-4T3 for recombinant protein production. Likewise, the C-terminal segment of NUP214 (NUP214C; amino acids 1864–2090) (from Dr. J. van Deursen, Mayo Clinic, Rochester, MN) (26) and the NUP358 sequence containing the zinc finger domain (ZFD) (NUP358ZFD; ZFD amino acids 1346–1826 plus the 33 amino acids on the N-terminal side of the ZFD and the 137 amino acids on the C-terminal side) (27) were subcloned into pGEX-6P-1. Construction of hemagglutinin (HA)-tagged NUP98-HOXA9 in pcDNA3 and MSCV-IRES-GFP was described before (9). HA-NUP98-DDX10 (28) was constructed in a similar manner using the DDX10 cDNA clone (MBA-221 from American Type Culture Collection, Manassas, VA) and subcloned into pcDNA3 and MSCV-IRES-GFP. HA-NUP98, NUP98-HOXA9ΔM (NUP98-HOXA9 without the segment for amino acids 254–444), and NUP98-HOXA9ΔNUP (NUP98-HOXA9 without the NUP98 portion) in pUHD10S (from Dr. J. van Deursen) (3) were subcloned into pcDNA3 for in vitro transcription/translation. NUP98-HOXA9ΔN (NUP98-HOXA9 without the segment for amino acids 1–253) was constructed by PCR in pGEX-4T-1 and subcloned into pcDNA3. NUP98-HOXA9ΔJ (NUP98-HOXA9 without the segment for amino acids 448–479) was constructed in pcDNA3 by excising the BglI/KasI fragment, thus replacing amino acids 448–479 with one Cys residue. GFP-tagged HA-NUP98-HOXA9 was constructed in pGEX-6P-1. All of the constructs were verified by DNA sequencing.
Localization of GFP-tagged Proteins
K562 cells were suspended at 2 × 106/100 μl in Nucleofector solution (Lonza, Cologne, Germany) and mixed with 2 μg of EGFP-NPMc, GFP-Rev, GFP-NFAT, or EGFP-NFκB(p65) and 4 μg of either empty pcDNA3 vector or vector expressing HA-NUP98-HOXA9, HA-NUP98-HOXA9 without the NUP98 portion (NUP98-HOXA9ΔNUP), or HA-NUP98-DDX10. Nucleofection was carried out according to the manufacturer's protocol, and the cells were then cultured overnight (15–18 h). Culture medium for cells nucleofected with GFP-NFAT contained 250 nm trichostatin A (Sigma-Aldrich) and 1 mm sodium butyrate to augment the expression of the transduced genes. For cells nucleofected with GFP-Rev, 1 μm dexamethasone (EMD Biosciences, Gibbstown, NJ) was added to the culture during the last 30 min to initiate nuclear import. Twenty thousand cells were collected onto a slide by Cytospin centrifugation for localization analysis. Cells that showed nuclear retention of GFP-tagged proteins in an identical area of each slide were scored using Nikon Eclipse 80i fluorescent microscope (Nikon, Melville, NY).
Recombinant Proteins
Recombinant glutathione S-transferase (GST)-CRM1 was produced from the pGEX-4T3 vector in BL21 (DE3) bacteria and purified using glutathione-Sepharose 4B beads (GE Healthcare). Recombinant NUP214C, NUP358ZFD, and GFP-NUP98-HOXA9 proteins were produced in the pGEX-6P-1 vector in BL21-CodonPlus (DE3)-RIL bacteria (Agilent Technologies, Santa Clara, CA), purified using glutathione-Sepharose 4B beads followed by digestion with Pre-Scission protease (GE Healthcare). RanGDP and RanGMPPNP were produced and purified by ion exchange chromatography as described previously (29).
Protein Binding Assays
NPMc, Rev, NFAT, NFκB, NUP98, NUP98-DDX10, NUP98-HOXA9, and its variants were produced using the TnT T7 quick coupled transcription/translation system (Promega, Madison, WI) in the presence of Tran35S-label (MP Biomedicals, Solon, OH). The amounts of translation products from different constructs were compared by SDS-PAGE and autoradiography, and the amounts used for binding assays were adjusted accordingly to contain bands of equal intensity. Binding assays were carried out essentially as described previously (30). For a binding reaction, 10 μl of beads of immobilized recombinant protein (GST control or GST-CRM1) were incubated for 1 h at 4 °C with 34-μl mixtures consisting of in vitro translated protein, transport buffer-Tween 20 (TB-T, which consists of 20 mm HEPES-KOH, pH 7.4, 110 mm potassium acetate, and 2 mm MgCl2 with 0.1% Tween 20) in the presence or absence of leptomycin B (LMB; EMD Biosciences), RanGDP, RanGMPPNP, NUP214C, or NUP358ZFD proteins. The estimated molar ratio of CRM1:NUP214C or NUP358ZFD used in binding reactions was 1:6. The supernatants and the material that remained bound to the beads after three washes in cold TB-T were separated by SDS-PAGE for autoradiography. Binding assays employing recombinant GFP-NUP98-HOXA9 was carried out in a similar manner, and bound and unbound material was analyzed by immunoblotting with anti-GFP antibody (B-2; Santa Cruz Biotechnology, Santa Cruz, CA).
Co-immunoprecipitation
K562 cells were nucleofected using 7.5 μg of DNA for 4 × 106 cells as described above with either empty pcDNA3 vector or vector expressing FLAG-tagged NUP98-HOXA9 or NUP98-DDX10 and collected after 17 h. The cells were lysed with 360 μl/4 × 106 cells in 0.8% BRIJ 97 (polyoxyethylene 10 oleyl ether) containing 20 mm HEPES-KOH, pH 7.4, 110 mm potassium acetate, 2 mm MgCl2, 5 mm EDTA, 0.1 mm GMPPNP, 2.5 mm thioglycolic acid, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture (Roche Applied Science, Indianapolis, IN). The nuclei were spun down at 800 × g for 3 min at 4 °C and then lysed in a buffer consisting of 1% BRIJ 97, 0.1% sodium deoxycholate, 0.1% SDS, 0.3 m KCl, 20 mm HEPES-KOH, pH 7.4, 110 mm potassium acetate, 2 mm MgCl2, 5 mm EDTA, 1 mm GMPPNP, 2.5 mm thioglycolic acid, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture. The lysate was centrifuged at 16,100 × g for 20 min at 4 °C, and 20 mm MgCl2 was added to the supernatant. The supernatant was incubated with anti-FLAG-agarose M2 antibody (Sigma-Aldrich) at 4 °C. The beads were washed three times with 0.4 m KCl buffer containing 0.8% BRIJ 97, 0.1% sodium deoxycholate, 0.1% SDS, 20 mm HEPES-KOH, pH 7.4, 110 mm potassium acetate, 12 mm MgCl2, 2 mm EGTA, and 2.5 mm thioglycolic acid and once in the above buffer without 0.4 m KCl. The immunoprecipitates were then heated at 100 °C for 5 min in SDS-PAGE sample buffer containing 5% 2-mercaptoethanol and subjected to SDS-PAGE followed by immunoblotting with anti-CRM1 antibody (C-20; Santa Cruz Biotechnology). The cell lysates were analyzed by immunoblotting with anti-CRM1 and anti-NUP98 (Cell Signaling Technology, Beverly, MA) antibodies.
Retroviral Transduction
The GP293 packaging cell line was transiently transfected with 4.4 μg of either control MSCV-IRES-GFP retroviral vector or vector expressing HA-tagged NUP98-DDX10 and 1.1 μg of the vesicular stomatitis virus glycoprotein plasmid (pVSV-G) using Lipofectamine and Plus reagents (Invitrogen). After 48 h, the culture supernatant, containing VSV-G pseudotyped retrovirus, was collected and used for transduction of PG13 packaging cells (CRL-10686 from American Type Culture Collection) by spinoculation in the presence of 8 μg/ml polybrene (hexadimethrine bromide; Sigma-Aldrich). PG13 cells producing MSCV-IRES-GFP/HA-NUP98-HOXA9 retrovirus were previously described (31). The PG13 culture supernatant containing GaLV-pseudotyped retrovirus was used for transduction of K562 cells and human primary CD34+ cells from mobilized peripheral blood of healthy volunteers (purchased from the Fred Hutchinson Cancer Research Center, Seattle, WA) by co-culturing for 48 h as described previously (31).
Immunofluorescence Microscopy
The cells were centrifuged onto slides by Cytospin centrifugation, fixed with 4% paraformaldehyde in Dulbecco's phosphate-buffered saline for 20 min and permeabilized with 0.1% Triton X-100 for 20 min at room temperature. Two percent normal donkey or goat serum in Dulbecco's phosphate-buffered saline with 0.1% Tween 20 was used for blocking and washing. Primary antibodies used were anti-HA (12CA5) and anti-CRM1 (C-20). The fluorescent secondary antibodies used were Alexa Fluor 647-conjugated anti-mouse IgG (Invitrogen), rhodamine-conjugated anti-goat IgG (Santa Cruz Biotechnology), and rhodamine-conjugated anti-mouse IgG (Millipore Corp., Billerica, MA). The images were captured using either an Eclipse 80i fluorescent microscope (Nikon) or an LSM510 Meta laser scanning confocal microscope (Carl Zeiss Inc., Thornwood, NY).
Luciferase Reporter Assays
K562 cells (107 cells) were transfected by electroporation using a GenePulser (Bio-Rad) with 5 μg of NFAT-pGL3 (from Dr. N. Clipstone, Loyola University, Chicago, IL) or NFκB-pTransLucent (Panomics, Fremont, CA) luciferase reporter vector and 10 μg of either empty pcDNA3 vector or vector expressing HA-tagged NUP98-HOXA9 or NUP98-DDX10. To control for efficiency of transfection, 0.125 μg of pRL-TK (Promega), which expresses Renilla luciferase, was included. Luciferase activity was measured after 48 h of culture following electroporation using the Dual-Luciferase reporter assay system (Promega), and the results were normalized to Renilla luciferase.
RESULTS
NUP98-HOXA9 Causes Nuclear Retention of CRM-1 Export Substrates
The major pathway for the export of proteins from the nucleus is mediated by the export factor CRM1 (32, 33). Two well known proteins exported through the CRM1-mediated pathway are NPMc (22) and HIV-1 Rev (23). Therefore, to determine whether the oncogenic NUP98-HOXA9 fusion protein interferes with CRM1-mediated nuclear export, its effect on the cellular localization of NPMc and Rev was examined by co-transfecting either a control vector or a vector expressing NUP98-HOXA9.
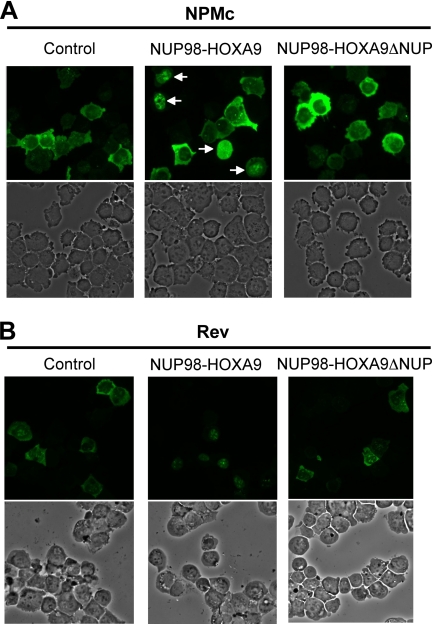
As shown in Fig. 1, expression of NUP98-HOXA9 caused significant nuclear retention of GFP-tagged NPMc (Fig. 1A) and Rev (Fig. 1B) in human myeloid K562 cells. Removal of the NUP98 portion from NUP98-HOXA9 (NUP98-HOXA9ΔNUP) abolished the nuclear retention activity. The percentage of cells that showed nuclear retention among transduced cells was scored in identical fields in each slide, and the averages obtained from three or four independent experiments are shown in Table 1. NUP98-HOXA9 was detected in virtually every cell that retained NPMc or Rev in the nucleus as shown by immunofluorescence staining of HA-NUP98-HOXA9 in transduced cells (supplemental Fig. S1).
FIGURE 1.
NUP98-HOXA9 causes nuclear retention of CRM-1 export substrates. K562 cells were nucleofected with EGFP-NPMc (A) or GFP-Rev (B) in combination with either empty pcDNA3 vector (control) or vector expressing NUP98-HOXA9 or NUP98-HOXA9 without the NUP98 portion (NUP98-HOXA9ΔNUP). The upper panels show fluorescent images, and the lower panels show corresponding phase contrast images. The arrows in the upper middle panel in A point to cells with nuclear-retained NPMc. The images were viewed using a Nikon Eclipse 80i microscope with a Nikon 40×, 0.75 numerical aperture CFI Plan Fluor DLL objective and were acquired with a Nikon Coolsnap ES camera using MetaMorph 6.3r2 software.
TABLE 1.
Nuclear retention of CRM1 export substrates by NUP98-HOXA9
The average percentages of transduced cells showing nuclear retention from three or four independent experiments are shown ± standard deviations. The p value was obtained by comparing with control using a paired two-tailed distribution t test.
| NPMc | Rev | |
|---|---|---|
| Control | 2.38 ± 2.09 | 4.18 ± 1.92 |
| NUP98-HOXA9 | 42.7 ± 7.57a | 81.7 ± 10.7b |
| NUP98HOXA9ΔNUP | 1.81 ± 1.61 | 2.87 ± 1.76 |
a p < 0.05.
b p < 0.01.
NUP98-HOXA9 Binds CRM1 through the FG Motif in a Ran-GTP-dependent Manner
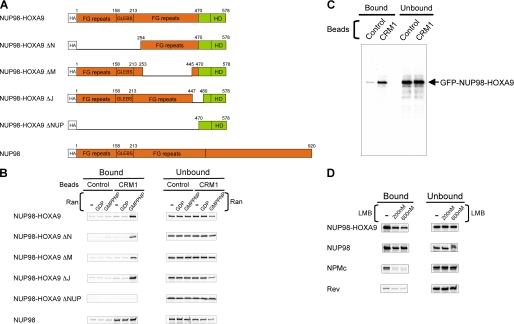
The above results indicate that NUP98-HOXA9 altered nuclear export of CRM1-dependent export substrates. This finding, as well as previous data suggesting participation of NUP98 in the nuclear export of Rev (21), raised the possibility that NUP98 fusions might interact with CRM1. To explore this possibility, NUP98, NUP98-HOXA9, and several deletion mutants were expressed by in vitro translation, and their interactions with CRM1 were studied in the presence or absence of the small GTPase Ran (Fig. 2, A and B). Ran controls loading and unloading of nuclear transport receptors with cargoes by cycling between a GTP-bound form (RanGTP) and a GDP-bound form (RanGDP). CRM1 binds export substrate (cargo) through the leucine-rich nuclear export signals (NESs) of the cargo cooperatively with RanGTP to form an export complex (32, 33). Loading of CRM1 with an export cargo requires RanGTP on the nuclear side of the NPC, whereas unloading on the cytoplasmic side requires hydrolysis to RanGDP and dissociation of the export complex. Ran loaded with the hydrolysis-resistant GTP analog GMPPNP (RanGMPPNP) was used in these binding reactions instead of RanGTP because of its better stability.
FIGURE 2.
NUP98-HOXA9 binds CRM1 through the FG motif in a Ran-GTP-dependent manner. A, schematic of NUP98-HOXA9 and its variants used in the binding assays. GLEBS, Gle2p-binding motif; HD, homeodomain. B, 35S-labeled NUP98-HOXA9 and its variants were incubated with GST (Control) or GST-CRM1 (CRM1) immobilized on glutathione-Sepharose 4B beads in the presence or absence of RanGDP or the RanGTP analog, RanGMPPNP. Approximately 30% of the total bound material and 5% of total unbound material for each reaction were analyzed as shown. Uncropped gels with their corresponding Coomassie-stained images are shown in supplemental Fig. S2. C, recombinant GFP-NUP98-HOXA9 protein was incubated with GST (Control) or GST-CRM1 (CRM1) immobilized on glutathione-Sepharose 4B beads in the presence of RanGTP. Approximately 0.67% of the total bound material and 0.13% of total unbound material for each reaction were analyzed by immunoblotting against GFP. D, 35S-labeled NUP98-HOXA9, NUP98, NPMc, and Rev were incubated with GST-CRM1 immobilized on glutathione-Sepharose 4B beads in the presence of RanGMPPNP with or without the indicated amount of LMB. Approximately 30% of the total bound material and 3% of total unbound material for each reaction were analyzed as shown. Uncropped gels with their corresponding Coomassie-stained images are shown in supplemental Fig. S3. Binding of Rev to CRM1 compared with control beads is shown in supplemental Fig. S4.
NUP98-HOXA9 bound specifically to CRM1 in the presence of RanGMPPNP but not RanGDP. This is reminiscent of binding of export cargoes to CRM1, although NUP98-HOXA9 does not contain a known leucine-rich NES (32). Removal of the NUP98 portion of NUP98-HOXA9 (NUP98-HOXA9ΔNUP) resulted in a complete loss of CRM1 binding activity. Partial but large deletions in the NUP98 portion (NUP98-HOXA9ΔN and NUP98-HOXA9ΔM) (Fig. 2A) decreased CRM1 binding (Fig. 2B), indicating that the FG repeat motifs rather than the GLEBS domain are involved in the binding. A small deletion (NUP98-HOXA9ΔJ) that encompasses the fusion point between NUP98 and HOXA9 did not affect the binding at all. Interestingly, in contrast to NUP98-HOXA9, specific binding of NUP98 to CRM1 occurred in the absence of Ran and increased only marginally by the addition of RanGMPPNP.
The in vitro translated products used in the above assays are produced in reticulocyte lysates that contain numerous mammalian proteins. To exclude the possibility that such proteins may be involved in the interaction between NUP98-HOXA9 and CRM1, recombinant GFP-tagged NUP98-HOXA9 protein was produced in bacteria, purified, and then tested for its capacity to bind to purified recombinant CRM1 in the presence of purified recombinant RanGTP. As shown in Fig. 2C, recombinant NUP98-HOXA9 specifically bound to CRM1, indicating that NUP98-HOXA9, CRM1, and RanGTP can form a trimeric complex without any other component.
Next, binding of NUP98-HOXA9 and NUP98 to CRM1 in the presence of RanGMPPNP was compared with that of the established CRM1-mediated export substrates, NPMc and Rev, (Fig. 2D). NUP98-HOXA9 and NUP98 showed more binding to CRM1 than NPMc and Rev. LMB, a potent inhibitor of CRM1-mediated nuclear export that targets the NES-binding site of CRM1, decreased the binding of NPMc and Rev to background level. In contrast, a significant fraction of both NUP98-HOXA9 and NUP98 remained bound to CRM1 in the presence of LMB. Taken together, the results shown in Fig. 2 indicate that the FG repeat region of NUP98-HOXA9 is involved in the interaction with CRM1 in a manner distinct from the standard interactions between CRM1 and export cargoes with leucine-rich NESs. In addition, a significant difference exists between NUP98 and NUP98-HOXA9 in regard to the requirement for RanGTP for CRM1 binding.
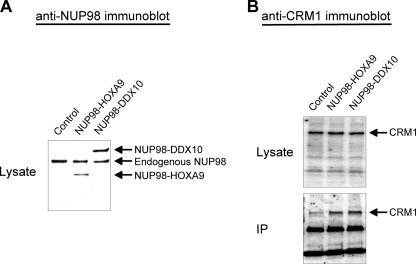
To determine whether NUP98 fusion proteins interact with endogenous CRM1 in cells, co-immunoprecipitation assays were carried out (Fig. 3). Because the FG repeat region, which is required for CRM1 binding (Fig. 2), is a part of all NUP98 fusions (1, 2), it is likely that other NUP98 fusions would also interact with CRM1. Therefore we prepared a construct expressing NUP98-DDX10, another leukemogenic fusion consisting of the FG repeat region of NUP98 fused to the putative RNA helicase DDX10 (28). FLAG-tagged NUP98-HOXA9 and NUP98-DDX10 were expressed in K562 cells (Fig. 3A). The fusion proteins were immunoprecipitated from nuclear lysates using antibody against the tag. As shown in Fig. 3B, CRM1 co-immunoprecipitated with both NUP98-HOXA9 and NUP98-DDX10, demonstrating that these fusions interact with CRM1 in vivo.
FIGURE 3.
NUP98-HOXA9 and NUP98-DDX10 co-immunoprecipitate with endogenous CRM1. A, K562 cells were nucleofected with either empty pcDNA3 vector (Control) or vector expressing FLAG-tagged NUP98-HOXA9 or NUP98-DDX10. Anti-NUP98 immunoblot of nuclear lysates shows the transduced fusion proteins as well as endogenous NUP98. B, nuclear lysates and immunoprecipitates (IP) prepared using anti-FLAG antibody from control cells or cells expressing the indicated FLAG-tagged NUP98 fusions were analyzed by anti-CRM1 immunoblotting.
NUP98 Fusion Proteins and NUP98 Interact Differently with NUP214
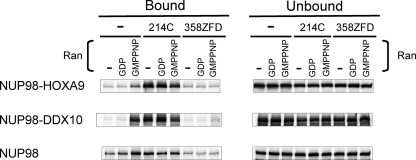
Many aspects of the mechanism of CRM1-mediated nuclear export remain unknown (33). Nucleoporins that have been reported to directly interact with CRM1 and participate in nuclear export are NUP214 and NUP358 (RanBP2), both located at the cytoplasmic side of NPC. There is strong evidence that NUP214, in a complex with NUP88, provides a terminal docking site for CRM1 export complexes (33–35). NUP358, on the other hand, may play a role in disassembling the complexes (36, 37). CRM1 binds the C-terminal FG repeat segment of NUP214 and the zinc fingers of NUP358. To determine whether NUP214 or NUP358 compete with NUP98 or NUP98 fusion proteins for CRM1 binding, binding assays were carried out in the presence or absence of CRM1-binding segments of NUP214 (NUP214C) or NUP358 (NUP358ZFD) (Fig. 4).
FIGURE 4.
NUP98 fusion proteins and NUP98 interact differently with NUP214. 35S-Labeled NUP98-HOXA9, NUP98-DDX10, and NUP98 were incubated with GST-CRM1 immobilized on glutathione-Sepharose 4B beads in the presence or absence of RanGDP or RanGMPPNP in combination with or without NUP214C or NUP358ZFD proteins as shown. Approximately 30% of the total bound material and 10% of total unbound material for each reaction were analyzed as shown. Uncropped gels with their corresponding Coomassie-stained images are shown in supplemental Fig. S5.
Both NUP98-HOXA9 and NUP98-DDX10 bound specifically to CRM1 in a RanGMPPNP-dependent manner as described above. With the addition of NUP214C, the binding of both fusion proteins to CRM1 increased and no longer required Ran. In contrast, NUP98 binding to CRM1 increased only marginally by adding the NUP214 peptide in the absence of Ran but was not affected at all in the presence of RanGDP or RanGMPPNP (Fig. 4). The result again revealed a fundamental difference between NUP98 and its oncogenic fusion molecules in their interactions with CRM1; in the presence of NUP214, binding of NUP98 fusions to CRM1 became much higher than that of NUP98. The NUP358 peptide, on the other hand, essentially abolished the interactions of NUP98 and its oncogenic fusions with CRM1 (Fig. 4).
NUP98 Fusion Proteins Co-localize with CRM1 in Nuclear Subdomains
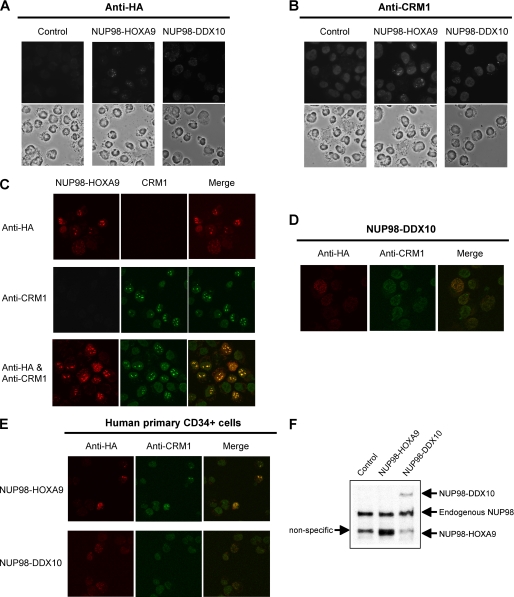
As shown above, NUP98 fusions interact aberrantly with CRM1 (Figs. 2 and 4) and cause nuclear retention of CRM1-mediated export substrates (Fig. 1). To further understand the nature of the in vivo interactions between NUP98 fusions and CRM1, K562 cells were retrovirally transduced to express NUP98-HOXA9 or NUP98-DDX10, and the locations of NUP98 fusions and endogenous CRM1 were determined by immunofluorescence staining. The NUP98 fusion proteins were localized within the nucleus and were concentrated either in relatively large sized dots for NUP98-HOXA9 or fine grains for NUP98-DDX10 (Fig. 5A). Interestingly, anti-CRM1 immunofluorescence staining revealed an intranuclear localization that followed the pattern of the corresponding NUP98 fusion (Fig. 5B). Control cells transduced with empty vector showed a homogenous nuclear stain of CRM1 without any concentration in nuclear subdomains.
FIGURE 5.
NUP98 fusion proteins co-localize with CRM1 in nuclear subdomains. A and B, K562 cells were retrovirally transduced with either empty vector (Control) or a vector expressing HA-NUP98-HOXA9 or HA-NUP98-DDX10. The cells were immunostained with anti-HA (A) or anti-CRM1 (B) antibodies in combination with rhodamine-conjugated secondary antibody. The upper panels show fluorescent images, and the lower panels show corresponding phase contrast images. The images were viewed using a Nikon Eclipse 80i microscope with a Nikon 40×, 0.75 numerical aperture CFI Plan Fluor DLL objective and were acquired with a Nikon Coolsnap ES camera using MetaMorph 6.3r2 software. C, K562 cells were retrovirally transduced to express HA-NUP98-HOXA9 and were immunostained with anti-HA antibody (top panels), anti-CRM1 antibody (middle panels), or both antibodies (bottom panels) in combination with both Alexa Fluor 647-conjugated and rhodamine-conjugated secondary antibodies. The images were acquired with a Zeiss LSM510 Meta laser scanning confocal microscope equipped with a Zeiss 63×, 1.4 numerical aperture Plan Apochromat oil objective using Zeiss LSM510 software. D, K562 cells were retrovirally transduced to express HA-NUP98-DDX10 and were immunostained with anti-HA and anti-CRM1 antibodies in combination with Alexa Fluor 647-conjugated and rhodamine-conjugated secondary antibodies. The images were acquired with a Zeiss LSM510 Meta laser scanning confocal microscope equipped with a Zeiss 63×, 1.4 numerical aperture Plan Apochromat oil objective using Zeiss LSM510 software. E, human primary CD34+ cells were retrovirally transduced to express HA-NUP98-HOXA9 or HA-NUP98-DDX10 and were immunostained with anti-HA and anti-CRM1 antibodies in combination with Alexa Fluor 647-conjugated and rhodamine-conjugated secondary antibodies. The images were acquired with a Zeiss LSM510 Meta laser scanning confocal microscope equipped with a Zeiss 63×, 1.4 numerical aperture Plan Apochromat oil objective using Zeiss LSM510 software. F, cell lysates of human primary CD34+ cells retrovirally transduced to express HA-NUP98-HOXA9 or HA-NUP98-DDX10 were analyzed by anti-HA immunoblotting.
To determine whether the similar staining patterns obtained for NUP98 fusion proteins and endogenous CRM1 are a result of co-localization, K562 cells expressing HA-tagged NUP98-HOXA9 were stained for HA, CRM1, or both and were analyzed by confocal microscopy (Fig. 5C). When cells were stained simultaneously with the two antibodies, the characteristic nuclear dots obtained by anti-HA and anti-CRM1 antibodies completely overlapped, indicating that NUP98-HOXA9 and CRM1 co-localize in the same nuclear subdomains. A similar overlap was also evident in cells expressing NUP98-DDX10 (Fig. 5D). These data demonstrate that binding of NUP98 fusions to CRM1 lead to its mislocalization within the nucleus.
To examine whether the co-localization also occurs in normal human hematopoietic cells, human primary CD34+ cells were retrovirally transduced to express either NUP98-HOXA9 or NUP98-DDX10 and stained simultaneously for the fusion proteins and CRM1. As shown in Fig. 5E, the NUP98 fusion proteins and CRM1 co-localized in these cells, exhibiting a similar punctate nuclear pattern. The expression of transduced NUP98-HOXA9 and NUP98-DDX10 proteins in the primary cells was equivalent to or less than that of endogenous NUP98, respectively (Fig. 5F).
NUP98 Fusions Enhance Transcriptional Activity by Nuclear Entrapment of Transcription Factors
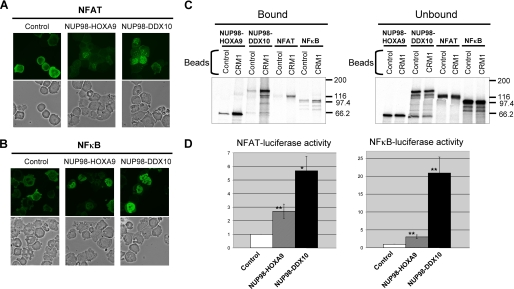
It has been reported that the NUP98 portion of oncogenic NUP98 fusions may contribute to some of their aberrant transcriptional activities (3, 8, 9, 11). Further, our results described above showed that NUP98 fusion proteins interact with CRM1 and inhibit CRM1-mediated nuclear export. Because CRM1-mediated nuclear export is known to regulate the activity of transcription factors (38), we reasoned that one of the mechanisms by which NUP98 fusions alter transcription and cause leukemia may be through aberrant nuclear accumulation of transcription factors. Therefore, the effects of NUP98 fusions on the nucleocytoplasmic trafficking of two transcription factors, NFAT and NFκB, were examined next. NFAT (24) and NFκB (39, 40) are regulated through CRM1-mediated nuclear export (32). Expression of either NUP98-HOXA9 or NUP98-DDX10 caused significant nuclear retention of GFP-tagged NFAT (Fig. 6A) and NFκB(p65) (Fig. 6B) in K562 cells. The percentage of transduced cells showing nuclear retention was scored in an identical field in each slide, and the average percentages obtained from six independent experiments are shown in Table 2. NUP98 fusion proteins were detected in virtually every cell that retained NFAT or NFκB in the nucleus as shown by immunofluorescence staining of HA-NUP98-HOXA9 and HA-NUP98-DDX10 in transduced cells (supplemental Fig. S6).
FIGURE 6.
NUP98 fusions enhance transcription by NFAT and NFκB by nuclear entrapment. A and B, K562 cells were nucleofected with GFP-NFAT (A) or EGFP-NFκB(p65) (B) in combination with either empty pcDNA3 vector (Control) or vector expressing NUP98-HOXA9 or NUP98-DDX10. The upper panels show fluorescent images, and the lower panels show corresponding phase contrast images. The images were viewed using a Nikon Eclipse 80i microscope with a Nikon 40×, 0.75 numerical aperture CFI Plan Fluor DLL objective and were acquired with a Nikon Coolsnap ES camera using MetaMorph 6.3r2 software. C, 35S-labeled NUP98-HOXA9, NUP98-DDX10, NFAT, and NFκB were incubated with GST (Control) or GST-CRM1 (CRM1) immobilized on glutathione-Sepharose 4B beads in the presence of RanGMPPNP. Approximately 30% of the total bound material and 2.1% of total unbound material for each reaction were analyzed as shown. The positions of molecular mass markers (expressed in kilodaltons) are shown. Uncropped gels with their corresponding Coomassie-stained images are shown in supplemental Fig. S6E. D, K562 cells were transfected by electroporation with either NFAT-pGL3 or NFκB-pTransLucent luciferase reporter vector in combination with either empty pcDNA3 vector (Control) or vector expressing NUP98-HOXA9 or NUP98-DDX10. The pRL-TK vector expressing Renilla luciferase was included in all samples to control for transfection efficiency, and the results were normalized to Renilla luciferase activities. The results shown are averages of three or four experiments ± standard deviations. The p value was obtained by comparing to empty pcDNA3 vector control using a paired two-tailed distribution t test. *, p < 0.05; **, p < 0.01.
TABLE 2.
Nuclear retention of transcription factors by NUP98 fusion proteins
The average percentages of transduced cells showing nuclear retention from six independent experiments are shown ± standard deviations. The p value was obtained by comparing to control using a paired two-tailed distribution t test.
| NFAT | NFκ B | |
|---|---|---|
| Control | 8.45 ± 2.82 | 4.94 ± 3.86 |
| NUP98-HOXA9 | 29.0 ± 10.2a | 46.3 ± 16.0a |
| NUP98-DDX10 | 40.7 ± 6.38a | 29.8 ± 7.92a |
a p < 0.01.
Next, binding of in vitro-translated NFAT and NFκB to CRM1 in the presence of RanGMPPNP was compared with that of NUP98-HOXA9 and NUP98-DDX10 by protein binding assays (Fig. 6C). As expected for CRM1 export substrates, NFAT and NFκB exhibited specific binding to CRM1. Their binding, however, was less than that of the NUP98 fusion proteins.
To determine whether the nuclear accumulation of transcription factors results in increased transcription from their cognate promoters, luciferase reporter assays were performed. As shown in Fig. 6D, both NFAT- and NFκB-dependent transcriptional activities were significantly augmented by NUP98-HOXA9 and NUP98-DDX10. Stimulation caused by NUP98-DDX10 was consistently higher than that by NUP98-HOXA9.
DISCUSSION
The present study demonstrates that expression of oncogenic NUP98 fusion proteins causes aberrant nuclear retention of representative CRM1 export substrates (NPMc and Rev) as well as CRM1-regulated transcription factors (NFAT and NFκB) known to play important roles in oncogenesis (40–43). NUP98 fusion proteins bound CRM1 in a manner distinct from the interactions between CRM1 and its cargoes or NUP98. Further, expression of NUP98 fusion proteins resulted in aberrant localization of CRM1 within nuclear subdomains. Therefore, perturbed nuclear export was the likely reason for nuclear accumulation of the proteins investigated. The nuclear entrapment of NFAT and NFκB caused by NUP98 fusions correlated with their enhanced transcriptional activities, supporting the hypothesis that blocked nuclear export could contribute to oncogenic transformation.
To exclude the possibility that the nuclear accumulation of NFAT and NFκB is due to an enhancement of nuclear import by NUP98 fusions, standard in vitro nuclear import assays were performed in permeabilized cells (supplemental Fig. S7). Because NFAT and NFκB are imported into the nucleus through the classical nuclear import pathway (38, 44), karyopherins (importins) α1 and β1 in conjunction with RanGDP, energy-generating system, and TRITC-BSA-NLS, a fluorescent import substrate containing classical nuclear localization signals (NLS), were used. The results, shown in supplemental Fig. S7, demonstrate that NUP98 fusions actually have a mild inhibitory effect on nuclear import. These data indicate that the increased amounts of intranuclear transcription factors observed in the presence of NUP98 fusions cannot be explained by increased nuclear import. Moreover, the augmentation of NFAT- and NFκB-dependent transcriptional activities by NUP98 fusions (Fig. 6D) cannot be explained by enhanced RNA export or stability; it has been shown that NUP98-HOXA9 does not induce luciferase activity from a control luciferase vector or from a luciferase reporter driven by HOXB6 (11), arguing against an effect on the stability or export of luciferase RNA. Furthermore, quantitation of NFAT-pGL3 and NFκB-pTransLucent luciferase transcripts in the cytoplasm and the nucleus of transduced cells (supplemental Table S1) directly confirmed that NUP98 fusion proteins did not significantly increase the fraction of these luciferase mRNAs present in the cytoplasm.
The exact mechanisms of protein translocation through the NPC are not well understood (12, 13). Much remains unknown regarding how CRM1 interacts with components of the NPC during the process of nuclear export (32, 33). The interaction of NUP214 with CRM1 and its role in CRM1-mediated export are well documented (33–35). Although there is some evidence suggesting a role for NUP98 in CRM1-mediated export (21), the mechanisms and molecular interactions underlying it are not understood. Our data show a specific interaction between NUP98 and CRM1 that is only slightly augmented by a RanGTP analog or by NUP214 and is inhibited by NUP358 (Figs. 2–4). Thus NUP98 may function in the binding and release of CRM1-nuclear export complexes at different locations in the NPC. In contrast, oncogenic NUP98 fusion proteins bound strongly to CRM1 only in the presence of the RanGTP analog. The binding of NUP98 fusion proteins to CRM1 appeared stronger than that of CRM1 export substrates and more resistant to inhibition by LMB (Figs. 2D and 6C). Interactions between leucine-rich NESs of export cargoes and CRM1 are generally weak (32), and it is interesting to note that CRM1 export substrates containing modified high affinity NESs fail to be efficiently exported (45). NUP98 fusion proteins may be trapped in the nucleus by a similar mechanism. Interestingly, binding of NUP98 fusion proteins to CRM1, but not that of NUP98, dramatically increased in the presence of the CRM1-binding domain of NUP214 (Fig. 4). The binding of NUP98 fusions also became Ran-independent in the presence of NUP214, suggesting that NUP98 fusion proteins bind CRM1 and NUP214 in a cooperative manner without Ran. The complexes in this conformation may not be able to exit the nucleus efficiently. The strong interaction of NUP98 fusion proteins with CRM1 may perturb CRM1-dependent nuclear export of normal cargoes by hindering recycling of free CRM1 for export, thus shifting the balance between import and export and leading to the nuclear accumulation of CRM1 export substrates at the steady-state level.
Upon expression of NUP98-HOXA9 and NUP98-DDX10 by retroviral transduction, the fusion proteins were found associated with characteristic dots/grains inside the nucleus of K562 and normal human CD34+ cells (Fig. 5). These aggregates were also observed previously by others in cells expressing NUP98-HOXA9 (46), NUP98-HHEX (47), and SET-CAN (NUP214) (48). In agreement with the in vitro binding assays (Figs. 2 and 4) and co-immunoprecipitation results (Fig. 3), CRM1 co-localized with the fusion oncoproteins in these nuclear dots/grains (Fig. 5). The exact nature of these nuclear subdomains is not known at present but appears to correspond to the structures termed “FG bodies” that NUP98 forms inside the nucleus of HeLa cells and that are distinct from known nuclear subdomains such as Cajal bodies, PML (promyelocytic leukemia) bodies, and Speckles (17). Transduction of NUP98 fusion oncogenes by nucleofection or retrovirus resulted in the expression of the fusion proteins at a level comparable with or less than that of endogenous NUP98 (Figs. 3A and 5F). Thus the characteristic bodies formed by NUP98 fusion proteins are unlikely to be an artifact of protein overexpression. Moreover, similar nuclear aggregates were observed in leukemic cells of an acute myeloid leukemia patient with NUP98-HOXA9 translocation (46).
Expression of NUP98 fusion proteins resulted in nuclear accumulation of NFAT and NFκB and led to the enhancement of their transcriptional activities (Fig. 6 and Table 2). NFAT contains a leucine-rich NES and is exported from the nucleus by CRM1 (24, 32, 38). The nuclear localization of NFκB is regulated by CRM1 both directly and through its interaction with IκBα (39, 40). NUP98 fusion proteins may allow an abnormally large amount of transcription factors to accumulate in nucleus by causing a shortage of CRM1 available for export. On the other hand, it is possible that NUP98 fusion proteins and CRM1 interact with trapped transcription factors cooperatively for activation. In fact, NUP98-HOXA9 can bind DNA through the HOXA9 domain and activate transcription by interacting with transcriptional co-activators (3). Furthermore, CRM1 has been recently reported to interact with the SET-NUP214 fusion protein at the promoter region of HOXA genes (49), and recent studies indicate CRM1 overexpression in some cancers (50, 51).
In summary, the present study revealed a link between the presence of NUP98 fusion proteins, perturbed nucleocytoplasmic trafficking, and activation of transcription factors whose deregulation may be important for leukemogenesis. The NUP98 fusions show strong, aberrant interactions with CRM1, Ran, and NUP214 and disrupt CRM1-mediated nuclear export. It will be of great interest to determine the exact mechanisms underlying these effects and their contribution to leukemogenesis.
Supplementary Material
Acknowledgments
We are grateful to Dr. N. Clipstone (Loyola University, Chicago, IL), Dr. J. van Deursen (Mayo Clinic, Rochester, MN), Dr. B. Falini (University of Perugia, Perugia, Italy), Dr. M. Fornerod (Netherlands Cancer Institute, Amsterdam, The Netherlands), Dr. L. Gerace (Scripps Research Institute, La Jolla, CA), Dr. J. Hanover (National Institutes of Health, Bethesda, MD), and Dr. J. Schmid (University of Vienna, Vienna, Austria) for providing reagents. We thank the Molecular Microbiology Imaging Facility at Washington University School of Medicine for the use of the confocal microscope.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL082549 and K02 HL084179 (to N. R. Y.) and T32 CA009547 (to A. M. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Table S1, and Figs. S1–S7.
- NPC
- nuclear pore complex
- CRM1
- chromosome region maintenance 1
- EGFP
- enhanced green fluorescent protein
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- LMB
- leptomycin B
- NES
- nuclear export signal
- NPMc
- mutated cytoplasmic nucleophosmin
- ZFD
- zinc finger domain
- HIV-1
- human immunodeficiency virus, type 1
- GMPPNP
- guanosine 5′-(β,γ-imido)triphosphate.
REFERENCES
- 1.Slape C., Aplan P. D. (2004) Leuk. Lymphoma 45, 1341–1350 [DOI] [PubMed] [Google Scholar]
- 2.Romana S. P., Radford-Weiss I., Ben Abdelali R., Schluth C., Petit A., Dastugue N., Talmant P., Bilhou-Nabera C., Mugneret F., Lafage-Pochitaloff M., Mozziconacci M. J., Andrieu J., Lai J. L., Terre C., Rack K., Cornillet-Lefebvre P., Luquet I., Nadal N., Nguyen-Khac F., Perot C., Van den Akker J., Fert-Ferrer S., Cabrol C., Charrin C., Tigaud I., Poirel H., Vekemans M., Bernard O. A., Berger R. (2006) Leukemia 20, 696–706 [DOI] [PubMed] [Google Scholar]
- 3.Kasper L. H., Brindle P. K., Schnabel C. A., Pritchard C. E., Cleary M. L., van Deursen J. M. (1999) Mol. Cell Biol. 19, 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pineault N., Buske C., Feuring-Buske M., Abramovich C., Rosten P., Hogge D. E., Aplan P. D., Humphries R. K. (2003) Blood 101, 4529–4538 [DOI] [PubMed] [Google Scholar]
- 5.Hirose K., Abramovich C., Argiropoulos B., Humphries R. K. (2008) Oncogene 27, 6056–6067 [DOI] [PubMed] [Google Scholar]
- 6.Wang G. G., Cai L., Pasillas M. P., Kamps M. P. (2007) Nat. Cell Biol. 9, 804–812 [DOI] [PubMed] [Google Scholar]
- 7.Kroon E., Thorsteinsdottir U., Mayotte N., Nakamura T., Sauvageau G. (2001) EMBO J. 20, 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo K. R., Sykes D. B., Pasillas M. P., Kamps M. P. (2002) Oncogene 21, 4247–4256 [DOI] [PubMed] [Google Scholar]
- 9.Ghannam G., Takeda A., Camarata T., Moore M. A., Viale A., Yaseen N. R. (2004) J. Biol. Chem. 279, 866–875 [DOI] [PubMed] [Google Scholar]
- 10.Palmqvist L., Pineault N., Wasslavik C., Humphries R. K. (2007) PLoS One 2, e768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yassin E. R., Sarma N. J., Abdul-Nabi A. M., Dombrowski J., Han Y., Takeda A., Yaseen N. R. (2009) PLoS One 4, e6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran E. J., Wente S. R. (2006) Cell 125, 1041–1053 [DOI] [PubMed] [Google Scholar]
- 13.Lim R. Y., Fahrenkrog B. (2006) Curr. Opin. Cell Biol. 18, 342–347 [DOI] [PubMed] [Google Scholar]
- 14.Powers M. A., Macaulay C., Masiarz F. R., Forbes D. J. (1995) J. Cell Biol. 128, 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontoura B. M., Dales S., Blobel G., Zhong H. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3208–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz T. U. (2005) Curr. Opin. Struct. Biol. 15, 221–226 [DOI] [PubMed] [Google Scholar]
- 17.Griffis E. R., Altan N., Lippincott-Schwartz J., Powers M. A. (2002) Mol. Biol. Cell 13, 1282–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard C. E., Fornerod M., Kasper L. H., van Deursen J. M. (1999) J. Cell Biol. 145, 237–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeganathan K. B., Malureanu L., van Deursen J. M. (2005) Nature 438, 1036–1039 [DOI] [PubMed] [Google Scholar]
- 20.Fontoura B. M., Blobel G., Yaseen N. R. (2000) J. Biol. Chem. 275, 31289–31296 [DOI] [PubMed] [Google Scholar]
- 21.Zolotukhin A. S., Felber B. K. (1999) J. Virol. 73, 120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falini B., Mecucci C., Tiacci E., Alcalay M., Rosati R., Pasqualucci L., La Starza R., Diverio D., Colombo E., Santucci A., Bigerna B., Pacini R., Pucciarini A., Liso A., Vignetti M., Fazi P., Meani N., Pettirossi V., Saglio G., Mandelli F., Lo-Coco F., Pelicci P. G., Martelli M. F. (2005) N. Engl. J. Med. 352, 254–266 [DOI] [PubMed] [Google Scholar]
- 23.Love D. C., Sweitzer T. D., Hanover J. A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kehlenbach R. H., Dickmanns A., Gerace L. (1998) J. Cell Biol. 141, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid J. A., Birbach A., Hofer-Warbinek R., Pengg M., Burner U., Furtmüller P. G., Binder B. R., de Martin R. (2000) J. Biol. Chem. 275, 17035–17042 [DOI] [PubMed] [Google Scholar]
- 26.Fornerod M., van Deursen J., van Baal S., Reynolds A., Davis D., Murti K. G., Fransen J., Grosveld G. (1997) EMBO J. 16, 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yaseen N. R., Blobel G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai Y., Hosoda F., Kobayashi H., Arai K., Hayashi Y., Kamada N., Kaneko Y., Ohki M. (1997) Blood 89, 3936–3944 [PubMed] [Google Scholar]
- 29.Bibak N., Paul R. M., Freymann D. M., Yaseen N. R. (2004) Anal. Biochem. 333, 57–64 [DOI] [PubMed] [Google Scholar]
- 30.Zhong H., Takeda A., Nazari R., Shio H., Blobel G., Yaseen N. R. (2005) J. Biol. Chem. 280, 10675–10682 [DOI] [PubMed] [Google Scholar]
- 31.Takeda A., Goolsby C., Yaseen N. R. (2006) Cancer Res. 66, 6628–6637 [DOI] [PubMed] [Google Scholar]
- 32.Kutay U., Güttinger S. (2005) Trends Cell Biol. 15, 121–124 [DOI] [PubMed] [Google Scholar]
- 33.Hutten S., Kehlenbach R. H. (2007) Trends Cell Biol. 17, 193–201 [DOI] [PubMed] [Google Scholar]
- 34.Askjaer P., Bachi A., Wilm M., Bischoff F. R., Weeks D. L., Ogniewski V., Ohno M., Niehrs C., Kjems J., Mattaj I. W., Fornerod M. (1999) Mol. Cell Biol. 19, 6276–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutten S., Kehlenbach R. H. (2006) Mol. Cell Biol. 26, 6772–6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh B. B., Patel H. H., Roepman R., Schick D., Ferreira P. A. (1999) J. Biol. Chem. 274, 37370–37378 [DOI] [PubMed] [Google Scholar]
- 37.Bernad R., van der Velde H., Fornerod M., Pickersgill H. (2004) Mol. Cell Biol. 24, 2373–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poon I. K., Jans D. A. (2005) Traffic 6, 173–186 [DOI] [PubMed] [Google Scholar]
- 39.Roth P., Xylourgidis N., Sabri N., Uv A., Fornerod M., Samakovlis C. (2003) J. Cell Biol. 163, 701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayden M. S., Ghosh S. (2008) Cell 132, 344–362 [DOI] [PubMed] [Google Scholar]
- 41.Im S. H., Rao A. (2004) Mol. Cells 18, 1–9 [PubMed] [Google Scholar]
- 42.Robbs B. K., Cruz A. L., Werneck M. B., Mognol G. P., Viola J. P. (2008) Mol. Cell Biol. 28, 7168–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baud V., Karin M. (2009) Nat. Rev. Drug Discov. 8, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torgerson T. R., Colosia A. D., Donahue J. P., Lin Y. Z., Hawiger J. (1998) J. Immunol. 161, 6084–6092 [PubMed] [Google Scholar]
- 45.Engelsma D., Bernad R., Calafat J., Fornerod M. (2004) EMBO J. 23, 3643–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore M. A., Chung K. Y., Plasilova M., Schuringa J. J., Shieh J. H., Zhou P., Morrone G. (2007) Ann. N.Y. Acad. Sci. 1106, 114–142 [DOI] [PubMed] [Google Scholar]
- 47.Jankovic D., Gorello P., Liu T., Ehret S., La Starza R., Desjobert C., Baty F., Brutsche M., Jayaraman P. S., Santoro A., Mecucci C., Schwaller J. (2008) Blood 111, 5672–5682 [DOI] [PubMed] [Google Scholar]
- 48.Saito S., Miyaji-Yamaguchi M., Nagata K. (2004) Int. J. Cancer 111, 501–507 [DOI] [PubMed] [Google Scholar]
- 49.Van Vlierberghe P., van Grotel M., Tchinda J., Lee C., Beverloo H. B., van der Spek P. J., Stubbs A., Cools J., Nagata K., Fornerod M., Buijs-Gladdines J., Horstmann M., van Wering E. R., Soulier J., Pieters R., Meijerink J. P. (2008) Blood 111, 4668–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noske A., Weichert W., Niesporek S., Röske A., Buckendahl A. C., Koch I., Sehouli J., Dietel M., Denkert C. (2008) Cancer 112, 1733–1743 [DOI] [PubMed] [Google Scholar]
- 51.van der Watt P. J., Maske C. P., Hendricks D. T., Parker M. I., Denny L., Govender D., Birrer M. J., Leaner V. D. (2009) Int. J. Cancer 124, 1829–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.