Abstract
We performed a comprehensive alanine scan of human α-defensin HNP1 and tested the ability of the resulting analogs to kill Staphylococcus aureus, inhibit anthrax lethal factor, and bind human immunodeficiency virus-1 gp120. By far, the most deleterious mutation for all of these functions was W26A. The activities lost by W26A-HNP1 were restored progressively by replacing W26 with non-coded, straight-chain aliphatic amino acids of increasing chain length. The hydrophobicity of residue 26 also correlated with the ability of the analogs to bind immobilized wild type HNP1 and to undergo further self-association. Thus, the hydrophobicity of residue 26 is not only a key determinant of the direct interactions of HNP1 with target molecules, but it also governs the ability of this peptide to form dimers and more complex quaternary structures at micromolar concentrations. Although all defensin peptides are cationic, their amphipathicity is at least as important as their positive charge in enabling them to participate in innate host defense.
Keywords: Antimicrobial Peptides, Bacterial Toxins, Defensins, HIV, Innate Immunity, Lectin, Protein-Protein Interactions, Tryptophan, X-ray Crystallography
Introduction
Antimicrobial peptides (AMPs)5 are important components of innate immunity in organisms ranging from insects to humans (1). In lower organisms, AMPs are the most effective “weapons of microbial destruction” against infectious microbes. Well known early examples include melittin from honeybee venom (2), cecropin from giant silkworm moths (3), and magainin from African clawed frogs (4). Defensins constitute a major class of antimicrobial peptides in vertebrates and are especially prominent in mammals (5–8). Defensins are 2–5-kDa, disulfide-stabilized cationic AMPs that are classified on the basis of size and disulfide topology into three structural families: α, β, and θ. Humans and other primates express multiple α- and β-defensin peptides, but θ-defensins are found only in certain nonhuman primates.
Six human α-defensins have been identified. The first three, known as human neutrophil peptides (HNPs 1–3), were originally isolated from the azurophil granules of neutrophils and are described as “natural antibiotic peptides” (9, 10). The less abundant HNP4 was found later in human neutrophils (11, 12). The final two α-defensins, HD5 and HD6, discovered through genomic approaches, are produced by Paneth cells of the small intestine (13, 14). Many human β-defensin genes exist. The few that have been characterized at the protein level are expressed predominantly in epithelial cells (15).
Despite their small size, defensins are active against a broad range of bacteria, fungi, and viruses. Although the mechanism of killing of bacteria by defensins is thought to primarily involve microbial membrane disruption (16, 17), their inhibition of infection by both enveloped and non-enveloped viruses is far more complex mechanistically (18). Defensins are also immuno-modulators that chemoattract and activate immune cells by acting on various cellular receptors and host proteins (19, 20). In addition, α-defensins can neutralize secreted bacterial toxins including anthrax lethal toxin, a binary complex of protective antigen and lethal factor (LF) (21, 22). The molecular basis for this extraordinary functional versatility is only partially understood.
Defensins adopt a three-stranded β-sheet core structure stabilized by three intramolecular disulfides. Prior mutational studies of α-defensins focused primarily on conserved elements such as disulfide bonding (23, 24), an invariant Gly residue (25), a conserved salt bridge (26–28), and the often abundant but less conserved Arg residues (29, 30). However, the loss of many of those conserved structural elements in α-defensins is often functionally inconsequential in vitro. Compelling experimental evidence links defensin cationicity to bacterial killing, an event initiated by electrostatic interactions between the positively charged peptide and the anionic and electronegative bacterial membrane. Yet cationicity alone does not explain the strain selectivity of different defensins. Furthermore, unlike human α-defensins and (perhaps) HBD3, the more positively charged β-defensins are not lectins and do not inhibit bacterial toxins and many viruses (31–36).
To gain additional insight into the molecular underpinnings of α-defensin function, we did a comprehensive Ala-scanning mutational analysis of HNP1, a prototypic human α-defensin, and examined its functional consequences. After finding the W26A mutation to be especially deleterious, we created HNP1 analogs containing a series of non-coded aliphatic amino acids of increasing hydrophobicity at position 26 and tested their activities. We found that the hydrophobicity of residue 26, which is normally imparted by Trp in HNP1, is essential for the functional versatility of this host defense peptide.
MATERIALS AND METHODS
Synthesis of Ala-scan Analogs of HNP1
Materials used in the experiments are described in the supplemental information. The amino acid sequence of HNP1 is 1ACYCRIPACIAGERRYGTCIYQGRLWAFCC30. Machine-assisted solid phase chemical synthesis of HNP1 and some of its analogs, using the 2-(1H-benzotriazolyl)-1,1,3,3-tetramethyluronium hexafluorophosphate activation/N,N-diisopropylethylamine in situ neutralization protocol developed by Kent and co-workers for Boc chemistry (37), was previously reported (38). Most Ala-substituted analogs of HNP1 were prepared essentially as described for the wild type defensin, except for R5A-HNP1, E13A-HNP1, and F28A-HNP1. Arg-5 and Glu-13 form a conserved salt bridge in HNP1, and substitution of either residue for Ala results in defensin misfolding and aggregation (26). Because the proHNP1 propeptide facilitates defensin oxidative folding (39), R5A-HNP1 and E13A-HNP1 were obtained through CNBr cleavage of their prefolded mutant prodefensins (40), which were prepared via native chemical ligation (41, 42). Without a covalently attached propeptide, F28A-HNP1 also failed to fold correctly. As was the case for R5A-HNP1 and E13A-HNP1, a 75-residue pro defensin was synthesized in which the almost C-terminal Phe was replaced by Ala. After oxidative folding, F28A-HNP1 was released by CNBr cleavage of the Met-Ala peptide bond connecting the N-terminal propeptide and the C-terminal defensin domain. For comparison, W26A-HNP1 was also prepared from a prodefensin mutant, W71A-proHNP1. Excluding the 6 Cys residues, 4 Ala residues, and 1 invariant Gly residue (25), a total of 19 highly pure and correctly folded Ala-scan analogs were generated.
Synthesis of W26X-HNP1 (X = Non-coded Amino Acid)
The synthesis of W26X-HNP1 was essentially as described for Ala-scan analogs of HNP1, except that Trp-26 was replaced by each of the four non-coded amino acids: α-aminobutyric acid (Abu, -CH2CH3), α-aminopentanoic acid (also called norvaline (Nva), -(CH2)2CH3), α-aminohexanoic acid (also called norleucine (Nle), -(CH2)3CH3), and α-aminoheptanoic acid (Ahp), -(CH2)4CH3. Correct oxidative folding of W26X-HNP1 was achieved at 0.25 mg/ml in 25% N,N-dimethylformamide containing 2 m urea, 3 mm reduced and 0.3 mm oxidized glutathione, pH 8.3, using the same procedures as previously published for wild type HNP1 (38).
Characterization of HNP1 Analogs
All defensin peptides were purified to homogeneity by preparative reverse phase HPLC, and their molecular masses were ascertained by electrospray ionization mass spectrometry. Defensin stock solutions prepared with water were quantified spectroscopically at 280 nm using molar extinction coefficients calculated according to the algorithm of Pace et al. (43). For biochemical verification of the native disulfide connectivity (Cys-1—Cys-6, Cys-2—Cys-4, Cys-3—Cys-5) in W26A-HNP1, the peptide (0.5 mg/ml) was digested by bovine trypsin and chymotrypsin (0.1 mg/ml each) at room temperature in 50 mm Tris/HCl, 20 mm CaCl2, 0.005% Triton X-100, pH 8.3. The resultant solution was analyzed on a Thermo Scientific LXQ linear ion trap mass spectrometer equipped with an Accela HPLC system using a Hypersil Gold column (1.9 μm, 2.1 × 50 mm).
NMR Spectroscopy
All NMR experiments were performed at 30 °C using a 5-mm probe on a Varian INOVA 500 spectrometer operating at a 1H resonance frequency of 499.754 MHz. HNP1 or W26A-HNP1 (∼2 mm) was dissolved in 10 mm phosphate buffer, pH 7.4, with 10% D2O. The homonuclear double quantum filtered COSY, TOCSY, and NOESY spectra were collected using standard protocol (44). Generally, 1024 complex data points were collected during the acquisition time F2, 400 complex free induction decays were collected during the evolution time F1, and a total of 32–64 transients was collected for each FID over 8000 Hz spectral width. TOCSY spectra were recorded with a mixing time of 60 ms, whereas mixing times of 100 and 250 ms were used for NOESY experiments and evaluation of spin-diffusion effects. All data were processed with program NMRPipe (Version 3.0) (45). The spin systems of all residues were identified using the Wüthrich strategy (46), aided by the CARA software (47).
Structural Studies of HNP1 Mutants
The crystal structures of the following HNP1 analogs were determined: I6A, Y16A, Y21A, Q22A, R24A, W26Abu, W26Ahp, and F28A. All crystals were grown using the hanging-drop, vapor diffusion method at room temperature. Initial screenings were performed either manually or robotically with the commercially available crystallization Sparse Matrix Screens from Hampton Research. X-ray diffraction data were collected on a rotating anode x-ray generator Rigaku-MSC Micromax 7 equipped with a Raxis-4++ image plate detector at the X-ray Crystallography Core Facility, University of Maryland at Baltimore. For I6A-HNP1, the data were remotely collected at Stanford Synchrotron Radiation Laboratory (Menlo Park, CA) using an ADSC Quantum-315R CCD detector. Data were integrated and scaled with HKL2000 (48). All structures were solved using the molecular replacement method as implemented in the program Phaser from the CCP4 suite (49). The monomer of HNP1 (PDB code 3GNY) was used as a search model. The structural refinements were performed using the program Refmac (50) coupled with a manual refitting and rebuilding with the program COOT (51). The crystallization conditions and data collection and refinement statistics are shown in supplemental Tables S1 and S2. The coordinates and structure factors have been deposited in the PDB with accession codes of 3LVX (I6A), 3LO1 (Y16A), 3LO2 (Y21A), 3H6C (Q22A), 3LO4 (R24A), 3LO6 (W26Abu), 3LO9 (W26Ahp), and 3LOE (F28A). Molecular graphics were generated using the program PyMOL (52).
Functional Assays
The inhibition of lethal factor (10 nm) by various defensins was quantified at 37 °C on a 96-well Vmax microplate reader (GE Healthcare) using an enzyme kinetic assay detailed elsewhere (35). The assay buffer was 20 mm HEPES containing 1 mm CaCl2 and 0.5% Nonidet P-40, pH 7.2. Data were presented in a percent inhibition plot versus varying defensin concentrations from which IC50 values, the concentration of defensin that reduced the enzymatic activity of LF by 50%, were derived using a nonlinear regression analysis.
Surface plasmon resonance (SPR) based LF and HIV gp120 binding studies were performed at 25 °C on a BIAcore T100 System (BIAcore, Inc., Piscataway, NY). The assay buffer was 10 mm HEPES, 150 mm NaCl, 0.05% surfactant P20, pH 7.4 (±3 mm EDTA). LF (2500 RUs) and gp120 (2830 and 3198 RUs) were immobilized on CM5 sensor chips using the amine-coupling chemistry recommended by the manufacturer. Detailed procedures for the SPR measurements were described elsewhere (35). Binding isotherms were analyzed with manufacturer-supplied software for BIAcore T100 and/or GraphPad Prism 4.0.
Virtual colony counting (54) was employed to quantify dose-dependent defensin killing of Staphylococcus aureus ATCC 29213 in 10 mm sodium phosphate, pH 7.4. Data analysis utilized a Visual Basic script to calculate the time necessary for each growth curve to reach a threshold change in optical density at 650 nm (ΔA650) of 0.02. The virtual LD50 was reported as the defensin concentration that resulted in a survival rate of 0.5.
RESULTS
Trp-26 Is a Critical Residue in HNP1
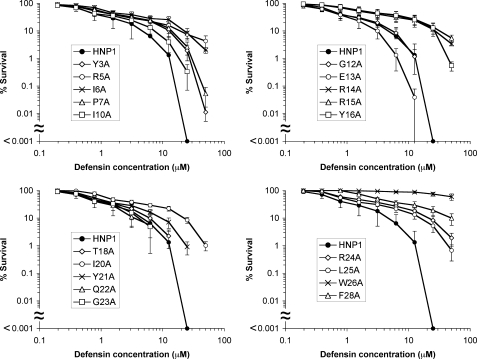
Gram-positive bacteria are much more susceptible than Gram-negative strains to α-defensin killing (54). We quantified bactericidal activity of HNP1 and its 19 Ala-scan analogs against S. aureus ATCC 29213. Shown in Fig. 1 are survival curves of S. aureus exposed to each defensin at concentrations varying from 0.195 to 50 μm. For wild type HNP1, complete killing of S. aureus, a reduction in the number of colonies by at least 6 orders of magnitude, was achieved at 25 μm. Replacement of Arg residues (Arg-5, Arg-14, Arg-15, or Arg-24) resulted in attenuated bactericidal activity, affirming the functional importance of cationicity in HNP1-mediated bacterial killing (29, 30). Conversely, substituting Ala for Glu-13, the only anionic residue of HNP1, enhanced the killing of S. aureus. Importantly, the hydrophobic residues also played critical roles in the action of HNP1 against S. aureus. Notable examples include Ile-6, Tyr-16, Ile-20, and in particular, the C-terminal hydrophobic residues Leu-25, Trp-26, and Phe-28. In fact, W26A-HNP1 barely showed any bactericidal activity at 50 μm and was the least active defensin among all 19 Ala-scan analogs of HNP1. Overall, the mutational data support the premise that the interplay between cationicity and hydrophobicity dictates S. aureus killing by α-defensins (55).
FIGURE 1.
Survival curves of S. aureus ATCC 29213 (MicroBioLogics) exposed to HNP1 and its 19 Ala-scan analogs. Strains were exposed to the peptides at concentrations varying from 0.195 to 50 μm. Except for wild type HNP1, each curve is the mean of triplicate experiments, where the error bars represent the S.D. of the measurements. The virtual colony counting assay tests a maximum of six peptides on one 96-well plate, including wild type HNP1 as a control on every plate to ensure internal consistency. The data of wild type HNP1 are presented as averages of 12 independent measurements ±S.D. Because points equivalent to zero survival cannot be plotted on a logarithmic scale, the complete killing achieved by 25 μm wild type HNP1 appears on the x axis, below the scaled portion of the results. A Student's t test was used to calculate the p values for statistical significance of wild type HNP1 versus selective defensin analogs: p = 0.00046 for R5A, p = 0.00076 for R14A, p = 0.0073 for E13A, p = 0.00068 for R15A, p = 0.00016 for R24A, p = 0.00011 for W26A, and p = 0.00063 for F28A.
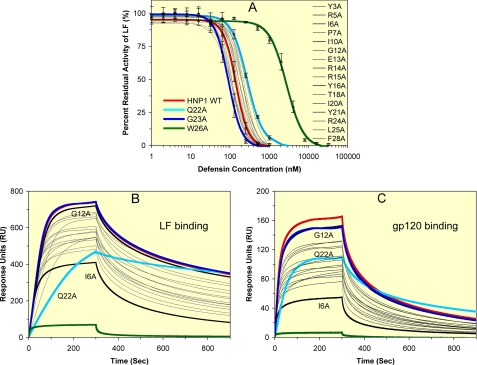
Wild type HNP1 is a potent non-competitive inhibitor of anthrax lethal factor (21). Previously, we showed that synthetic HNP1 inhibited 10 nm LF with an IC50 value of 148 nm (35). The inhibitory activity of 19 Ala-scan analogs of HNP1 against LF was quantified under identical conditions, and their IC50 values appear in Table 1. Except for the W26A mutant of HNP1, all other IC50 values fell within a 3-fold concentration range, between 92 nm for the G23A and 278 nm for the I6A mutants. Substituting alanine for Arg-5, Gly-12, Glu-13, Thr-18, Gly-23, or Leu-25 marginally improved inhibition of LF relative to the wild type peptide, and substituting alanine for Tyr-3, Ile-6, Pro-7, Arg-14, Arg-15, Tyr-16, Ile-20, Gln-22, or Phe-28 marginally decreased it. The activity of the I10A, Y21A, and R24A mutants was indistinguishable from that of wild type HNP1. As shown in Fig. 2A, all inhibition curves clustered except for that of W26A-HNP1. With W26A-HNP1 excluded, the weakest inhibitors of LF, Q22A-HNP1 and I6A-HNP1, and the strongest inhibitors, G23A-HNP1 and G12A-HNP1, displayed IC50 values within a factor of 2 of the HNP1 IC50. In sharp contrast, the W26A mutation weakened LF inhibition by almost 20 fold, yielding an IC50 of 2.8 μm for W26A-HNP1.
TABLE 1.
IC50 values of HNP1 and its 19 Ala-scan analogs for lethal factor were determined by enzyme inhibition kinetics at 37 °C in 20 mm HEPES buffer containing 1 mm CaCl2 and 0.5% Nonidet P-40, pH 7.2
The IC50 value of the wild type defensin is the mean of 24 independent measurements. The data of all other HNP1 analogs are the averages of three separate assays.
| Defensin | ICM50 | Defensin | IC50 |
|---|---|---|---|
| nm | nm | ||
| Wild type HNP1 | 148 ± 11 | Y16A | 178 ± 18 |
| Y3A | 233 ± 17 | T18A | 114 ± 12 |
| R5A | 112 ± 14 | I20A | 173 ± 10 |
| I6A | 278 ± 48 | Y21A | 143 ± 9 |
| P7A | 207 ± 19 | Q22A | 277 ± 37 |
| I10A | 153 ± 13 | G23A | 92 ± 7 |
| G12A | 109 ± 10 | R24A | 157 ± 8 |
| E13A | 111 ± 11 | L25A | 114 ± 9 |
| R14A | 177 ± 13 | W26A | 2786 ± 396 |
| R15A | 211 ± 18 | F28A | 211 ± 10 |
FIGURE 2.
Inhibition and/or binding of LF and HIV-1 gp120 by wild type HNP1 and its 19 Ala-scan analogs. A, shown is inhibition of LF activity by different concentrations of defensin. The data are the averages of three independent enzyme kinetic measurements, except for HNP1, of which the inhibition curve was obtained from 24 separate assays. For clarity, only the inhibition curves of HNP1, Q22A-HNP1, G23A-HNP1, and W26A-HNP1 are highlighted in color thick lines with error bars. WT, wild type. B and C, binding kinetics of defensins, each at 100 nm, on immobilized LF (2500 RUs) or gp120 (2830 RUs) as determined by SPR. In addition to the four color-coded defensins, HNP1 (red), Q22A-HNP1 (cyan), G23A-HNP1 (blue), and W26A-HNP1 (green), G12A-HNP1 and I6A-HNP1 are highlighted in black.
To verify the inhibitory activity data, we examined LF binding kinetics of various defensins using the surface plasmon resonance technique. Representative sensorgrams of the defensins at 100 nm are shown in Fig. 2B. Wild type HNP1, G12A-HNP1, and G23A-HNP1 showed the strongest binding to LF, whereas W26A-HNP1 was the weakest ligand of LF. The difference in RU values at 300s of association between wild type HNP1 and W26A-HNP1 was >10-fold. I6A-HNP1 and Q22A-HNP1 were the second weakest binders of LF. However, as with LF inhibition, all LF binding curves, except for that of W26A-HNP1, largely crowded together.
To investigate whether or not Trp-26 is important for HNP1 functions other than bacterial killing and LF inhibition, we tested the lectin-like properties of HNP1 analogs. The lectin-like properties of HNP1 were first demonstrated by its avid binding of glycoproteins such as HIV-1 gp120, a heavily glycosylated protein (56). Trp residues in small lectins have been shown to be important for oligosaccharide binding (57). Shown in Fig. 2C are representative binding kinetics of the defensin analogs at 100 nm on immobilized gp120. Wild type HNP1, G12A-HNP1, and G23A-HNP1 again exhibited the strongest binding to the viral protein. The W26A mutation dramatically reduced gp120-binding by 26-fold in RU at 300 s. I6A-HNP1 was the second weakest defensin in the panel, although the difference in gp120 binding between HNP1 and I6A-HNP1 was far smaller than that between HNP1 and W26A-HNP1. Taken together, the results on bacterial killing as well as inhibition and/or binding of LF and gp120 unequivocally point to Trp-26 as a critical residue in HNP1.
Structural Analysis of Ala-scan Analogs
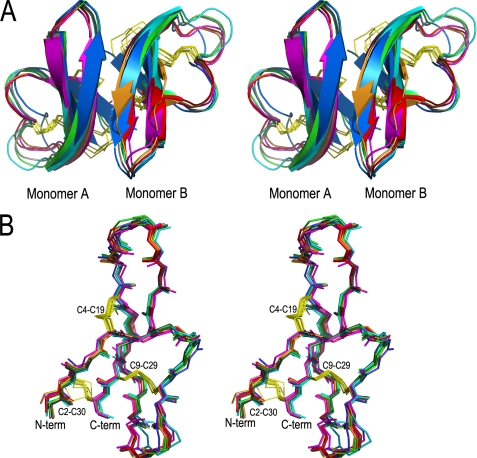
We previously determined the crystal structure of wild type HNP1 at 1.6 Å resolution (35). To ensure the correct folding of the new HNP1 analogs, we selected I6A-, Y16A-, Y21A-, Q22A-, R24A-, W26A-, and F28A-HNP1 for structural characterization by x-ray crystallography. Except for W26A-HNP1, which failed to crystallize, the structures of the remaining six HNP1 analogs were determined at resolutions ranging from 1.56 to 1.75 Å (supplemental Table S2). As shown in Fig. 3A, wild type HNP1 and its Ala-scan analogs all share the same three-stranded β-sheet fold arranged in a dimeric form, which is conserved in the α-defensin family (58). In each case, two monomers associated in a “canonical” fashion via the central (2nd) β-strands, forming a symmetric dimer stabilized by a network of intermolecular main chain-main chain H-bonds and by extensive hydrophobic interactions. Shown in Fig. 3B are superimposed monomers of wild type HNP1 and the six defensin analogs. Minimal conformational variation is observed in the backbone and three disulfides. Overall, these structural data indicate that Ala substitutions in HNP1 caused little change to its tertiary structure and had limited impact on its quaternary structure.
FIGURE 3.
Structural alignment of Cα traces of dimers and monomers of HNP1 analogs. A, shown is a stereo view of backbone-superimposed dimers of wild type HNP1 (light green), I6A-HNP1 (magenta), Y16A-HNP1 (cyan), Y21A-HNP1 (gray), Q22A-HNP1 (orange), R24A-HNP1 (red), and F28A-HNP1 (blue). B, shown is a stereo view of backbone-superimposed monomers of HNP1 and the six Ala-substituted analogs. Disulfide bonds are represented by yellow sticks.
W26A-HNP1 Is Correctly Folded
Two different synthetic approaches were used for W26A-HNP1; they are direct oxidative folding of a fully reduced W26A-HNP1 and CNBr cleavage of an oxidatively folded pro-defensin mutant, i.e. W71A-proHNP1. Both W26A-HNP1 preparations were indistinguishable on analytical reverse phase-HPLC and electrospray ionization mass spectrometry and showed identical activity in various functional assays (data not shown). To verify the correct folding of W26A-HNP1, we determined its disulfide connectivity. The mutant defensin was subjected to enzymatic digestion by trypsin and chymotrypsin followed by a liquid chromatography-MS/MS analysis. Two major fragments were generated of 830.5 and 1504.9 Da, corresponding to (CR5)(GTCIY21) (calculated monoisotopic mass 830.3 Da) and (ACY3)(IPACIAGER14)(CC30) (calculated average isotopic mass 1504.8 Da), respectively. The mass mapping of peptide fragments confirmed the existence of one native disulfide bridge between Cys-4 and Cys-19 but was insufficient for unambiguous identification of the remaining two native S-S bonds, Cys-2—Cys-30 and Cys-9—Cys-29. It did, however, exclude the possibility of a non-native connectivity, i.e. Cys-2—Cys-9, Cys-29—Cys-30. As our attempts to crystallize W26A-HNP1 were unsuccessful, we resorted to NMR spectroscopy.
Our proton assignments of wild type HNP1 were in agreement with the published results (59). Compared with the wild type, the chemical shifts of backbone protons of a majority of residues in W26A-HNP1 (Ala-1 to Cys-4, Ala-8 to Gly-17, Arg-24, and Ala-27 to Cys-30) were slightly perturbed; that is, less than 0.1 ppm for Cα protons and less than 0.2 ppm for amide protons. A contour plot of combined two-dimensional NOE and COSY spectra in the fingerprint region showed similar patterns of connectivity between wild type and the mutant HNP1. The characteristic anti-parallel β-sheet comprising residues 14–30 was clearly “visible” in the spectra of W26A-HNP1, further demonstrating its structural similarity to the wild type defensin. However, significant changes in chemical shift were observed for residues surrounding Ala-26 presumably due to the W26A mutation. Strong NOE connectivities among Trp-26, Tyr-21, and Arg-24, previously seen in wild type HNP1, were absent in W26A-HNP1, reflecting a localized conformational mobility caused by the mutation-created void in the molecule.
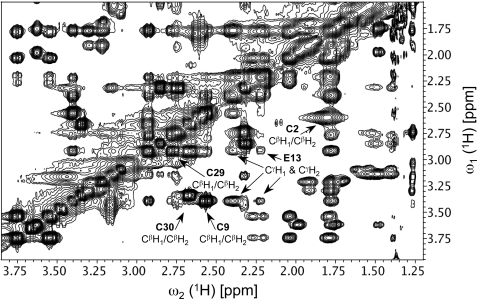
Further analysis of the proton-proton NOEs of W26A-HNP1 revealed two additional native pairs of disulfide bonds: Cys-2—Cys-30 and Cys-9—Cys-29 (Fig. 4) (ironically, we could not establish the Cys-4—Cys-19 connectivity previously identified by liquid chromatography-MS/MS due to lack of discernable signals). Three lines of evidence support the existence of the two native S-S bonds in W26A-HNP1, thereby confirming that W26A-HNP1 was correctly folded. First, both HNP1 and W26A-HNP1 had similar CβH NOE connectivity patterns for Cys-2, Cys-30, Cys-9, and Cys-29. Second, there existed strong cross-strand NOEs between the CγH of Glu-13 and the CβH of both Cys-9 and Cys-29. Third, weak cross-strand NOEs were found between the CδH or CϵH of Phe-28 and the CβH of both Cys-2 and Cys-30. It is noteworthy that direct comparison of NOEs between wild type HNP1 and W26A-HNP1 was essential to avoid ambiguous S-S bond assignments as strong CβH NOEs can exist between non-bonded Cys residues in Cys-rich peptides (60).
FIGURE 4.
Native disulfide linkages shown in a NOESY spectrum of W26A-HNP1. The Cys-9—Cys-29 linkage is observed through long-range connectivities with the side chain of Glu-13. The Cys-2—Cys-30 linkage is established through weak cross-strand NOEs between the CδH or CϵH of Phe-28 (7.05 ppm) and the CβH of both Cys-2 and Cys-30.
Functional Rescue of W26A-HNP1
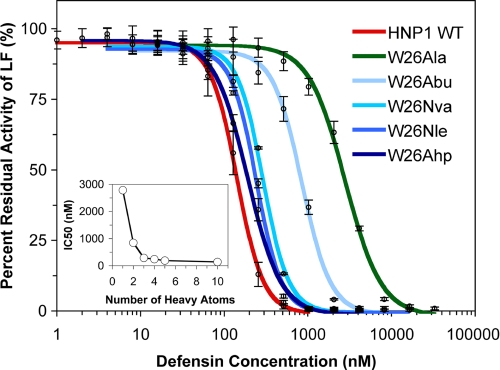
To gain additional insights into the nature of the deleterious W26A mutation, we introduced at position 26 a homologous series of unnatural amino acids with straight aliphatic side chains: Abu, Nva, Nle, and Ahp (61). Shown in Fig. 5 is dose-dependent inhibition of LF by different concentrations of W26Abu-, W26Nva-, W26Nle-, and W26Ahp-HNP1. For comparison, the data for wild type HNP1 and W26A-HNP1 are also included. The IC50 values are tabulated in Table 2. Inhibition of LF by W26X-HNP1 progressively improved as the aliphatic side chain of residue X was elongated. The most dramatic improvement in LF inhibition came from the change of Ala (-CH3) to Nva (-CH2CH2CH3), where lengthening of the side chain by each additional methylene group (-CH2) lowered IC50 by 3-fold. From Nva to Ahp, however, the improvement was modest. Nva at position 26 apparently recovered much of the lost inhibitory activity of W26A-HNP1, as the IC50 value of W26Nva-HNP1 (291 nm) was only 2-fold that of wild type HNP1 (148 nm).
FIGURE 5.
Inhibition of LF activity by different concentrations of W26X-HNP1 analogs. The data are the averages of three independent enzyme kinetic measurements. For comparison, wild type (WT) HNP1 and W26A-HNP1 are also plotted.
TABLE 2.
IC50 values of W26X-HNP1 for lethal factor were determined by enzyme inhibition kinetics at 37 °C in 20 mm HEPES buffer containing 1 mm CaCl2 and 0.5% Nonidet P-40, pH 7.2
The IC50 value of the wild type defensin is the mean of 24 independent measurements. The data of all other HNP1 analogs are averages of three separate assays.
| Defensin | IC50 | Defensin | IC50 |
|---|---|---|---|
| nm | nm | ||
| Wild type HNP1 | 148 ± 11 | W26A | 2786 ± 396 |
| W26Abu | 847 ± 89 | W26Nle | 242 ± 19 |
| W26Nva | 291 ± 26 | W26Ahp | 191 ± 27 |
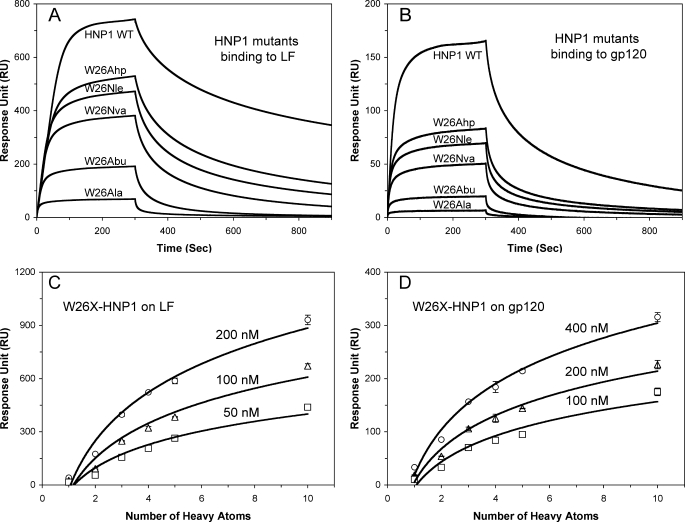
Consistent with these findings, a strong correlation existed between the hydrophobicity (or size) of the side chain of residue X and the ability of W26X-HNP1 to bind LF or HIV-1 gp120 (Figs. 6, A and B). The SPR experiments were performed at three different defensin concentrations (50, 100, 200 nm W26X-HNP1 for LF and 100, 200, 400 nm for HIV gp120). The strong correlation was illustrated by plots of RU values at 300 s of association versus the number of heavy atoms of the side chain of residue X (Figs. 6, C and D).
FIGURE 6.
Binding of LF and gp120 by W26X-HNP1 analogs. A and B, shown are binding kinetics of defensins, each at 100 nm, on immobilized LF (2500 RUs) and gp120 (2830 RUs) as determined by SPR. C and D, shown are plots of RU values at 300 s of association of W26X-HNP1 analogs at three different concentrations versus the number of heavy atoms (non-H atoms) in the side chain of residue 26 in HNP1. The data of HNP1 and W26A-HNP1 are included for comparison.
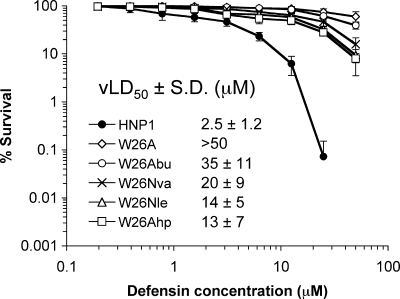
We also quantified bactericidal activity of W26X-HNP1 against S. aureus using wild type HNP1 and W26A-HNP1 as controls (Fig. 7). The relative potencies were: HNP1 (virtual LD50 = 2.5 μm) > W26Ahp-HNP1 (13 μm) ≈ W26Nle-HNP1 (14 μm) > W26Nva-HNP1 (20 μm) > W26Abu-HNP1 (35 μm) > W26A-HNP1 (>50 μm). Evidently, none of the W26X-HNP1 analogs was nearly as effective as the wild type defensin in killing the Gram-positive bacterium. Nevertheless, their bactericidal activity correlated well with the side chain size of residue X at position 26. Taken together, the LF inhibition, gp120 binding, and bacterial killing data indicate that W26A-HNP1 can be functionally rescued in large part by increasing the hydrophobicity of residue 26.
FIGURE 7.
Bactericidal activity of HNP1, W26A-HNP1, and W26X-HNP1 analogs (from 0. 195 to 50 μm) against S. aureus ATCC 29213. Each curve is the mean of three independent experiments, where the error bars represent the S.D. of the measurements. Points scored as 0 survival could not be plotted. vLD50, virtual LD50.
Structural Analysis of W26Abu- and W26Ahp-HNP1
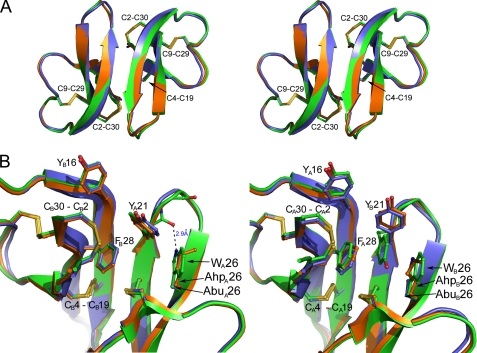
All four unnatural analogs of HNP1 crystallized, and W26Abu- and W26Ahp-HNP1 were selected for structural analysis. As shown in Fig. 8A, both analogs, although highly similar to each other, adopt dimeric structures nearly identical to wild type HNP1. Superposition of the dimers of W26Abu-HNP1 and W26Ahp-HNP1 to the wild type defensin yielded root mean square deviation values of 0.55 and 0.42 Å, respectively, between (60) equivalent Cα atoms. In wild type HNP1, Trp-26 stacks against Tyr-21, which in turn makes van der Waals contacts with Tyr-16, Phe-28, and the Cys-2—Cys-30 disulfide of the opposing monomer (Fig. 8B). Notably, Tyr-21 adopts two different conformations in wild type HNP1, 1) packing vertically against the aromatic rings of Phe-28 and Trp-26 in monomer A and 2) sandwiching horizontally between them in monomer B. Vertical packing of aromatic side chains commonly occurs in proteins and is energetically more favorable than parallel stacking. In HNP1, the vertical packing mode of Tyr-21 is further stabilized by an H-bond between Trp-26 Nϵ1 and Tyr-21 O (Fig. 8B). As monomer B of HNP1 is involved in crystal contacts with symmetry-related molecules, the parallel conformation of Tyr-21 may reflect the intrinsic properties of crystal packing.
FIGURE 8.
Crystal structures of W26Abu-HNP1 and W26Ahp-HNP1. A, shown is a stereo view of superimposed backbone structures of HNP1 (green), W26Abu-HNP1 (slate), and W26Ahp-HNP1 (orange) dimers. B, shown is a close-up view of the dimer interfaces in monomer A (left panel) and monomer B (right panel). The residues and disulfide bonds at the dimer interface are highlighted as sticks. H-bonds that stabilize the Trp-26 conformation in wild type HNP1 are shown as black dashes.
Mutation of Trp-26 to Abu and Ahp caused little change to defensin tertiary and quaternary structures. However, the loss of the bulky tryptophan side chain at position 26 did “loosen up” Tyr-21, as evidenced by its high B-factor values and poorly defined electron density (supplemental Fig. S1), particularly in monomer A of W26Abu-HNP1 structure. The increased local structural mobility seen in W26Abu-HNP1, supported by the NMR study of W26A-HNP1, largely subsided upon introduction of the significantly longer Ahp at position 26, resulting in a lower B-factor and root mean square deviation values (0.42 Å for W26Ahp-HNP1 versus 0.55 Å for W26Abu-HNP1 in relation to the wild type defensin). Interestingly, the Y16A, Y21A, and F28A mutations did not induce significant increase in mobility of neighboring aromatic residues such as Trp-26; Tyr-21 exists only in the “vertical” conformation in both W26Abu- and W26Ahp-HNP1 structures. Overall, the structural data suggest that Trp-26 plays a critical role in stabilizing the hydrophobic interface of HNP1 dimer.
Trp-26 Mediates HNP1 Self-association
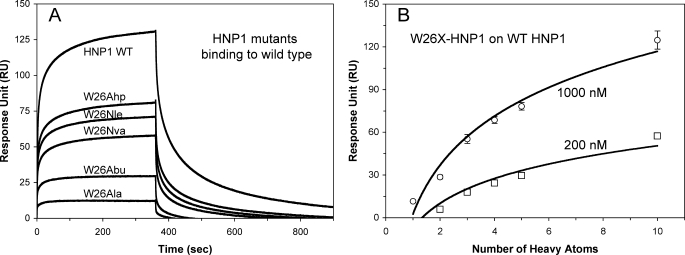
We have previously shown that α-defensins HNP1 and HD5 self-associate at high nm to low μm concentrations, particularly in the presence of target proteins (34, 62). To further examine the structural and functional role of Trp-26, we compared binding kinetics of HNP1, W26A-HNP1, and W26X-HNP1 on immobilized HNP1. Representative sensorgrams of the defensins at 1 μm are shown in Fig. 9A. Wild type HNP1 exhibited the strongest self-association on its surface, which was substantially diminished by the W26A mutation. At 300 s, the 10.8-fold difference in RU between HNP1 and W26A-HNP1 was comparable with their inhibition of LF (19.6-fold difference in their IC50), their binding of LF (a 22.8-fold difference in RU bound at 200 nm), and their binding of gp120 binding (a 12.1-fold difference in RU bound at 200 nm). Importantly, W26X-HNP1 binding to immobilized HNP1 strongly correlated with the side chain size of residue X (Fig. 9B). These data indicate that the hydrophobicity of residue 26 in HNP1 dictates its self-association at the micromolar concentrations likely to exist when it acts in vivo. We attribute our ability to crystallize W26Abu-HNP1 (and HNP1 analogs other than W26A-HNP1) as structurally conserved dimers to carrying out crystallization at millimolar concentrations that were several orders of magnitude higher than biologically active concentrations of defensins.
FIGURE 9.
Self-association of HNP1, W26A-HNP1, and W26X-HNP1 analogs on wild type HNP1 surface. A, shown are binding kinetics of the defensins at 1 μm each on 285 RUs of immobilized HNP1. B, shown are plots of RU values at 300 s of association of the defensins at 200 nm and 1 μm versus the number of heavy atoms (non-H atoms) in the side chain of residue 26 in HNP1. The RU values are the average readings from three separate experiments performed at room temperature. WT, wild type.
DISCUSSION
Defensins are structurally conserved yet functionally diverse. To gain insights into the molecular determinants of defensin function, we prepared 19 Ala-substituted analogs of HNP1, sparing only the 6 cysteine residues, the 4 alanines, and the 1 invariant glycine that exists in the wild type peptide. When we characterized their ability to kill bacteria, inhibit LF, and bind HIV-1 gp120, Trp-26 emerged as the most critical residue in HNP1. Mutational analysis at position 26 using the non-coded amino acids Abu, Nva, Nle, and Ahp further established hydrophobicity as an important molecular signature that imparted functional versatility to the α-defensin. This observation, although seemingly surprising and counter-intuitive at first, is strongly supported by our experimental findings and published reports.
Some Structurally Conserved Elements, although Essential for Defensin Biosynthesis, Do Not Contribute to HNP1 Function
Earlier structure-activity studies of α-defensins, including our own, typically focused on their conserved structural elements (23–28), including an invariant Gly-17, a salt bridge between Arg-5 and Glu-13 (using HNP1 numbering), and disulfide bonding. Gly-17, part of an atypical β-bulge structure, is critical for α-defensin folding, whereas the salt bridge stabilizes α-defensins to prevent their in vivo degradation by proteases. As was demonstrated previously and again confirmed by the current study, loss of the Arg-5–Glu-13 salt bridge generally has little impact on defensin function in vitro. By contrast, the loss of disulfide bonding causes collapse of defensin tertiary structure and indiscriminately inactivates α-defensins in several biological assays such as S. aureus killing, LF inhibition, and gp120 binding (35), except for killing certain Gram-negative bacteria such as Escherichia coli. Disulfide bonding is important for defensin biosynthesis and in vivo stability (23). Despite its vital role in maintaining defensin structure, disulfide bonding itself is rarely targeted for mutational analysis as it generally confers little functional specificity.
Hydrophobicity Rather Than Cationicity Dictates HNP1 Strain Selectivity in Bacterial Killing
Many earlier mutational studies also aimed to probe abundant but less conserved Arg residues in α-defensins (29, 30). Although the importance of cationicity is self-evident in defensin-mediated bacterial killing, cationicity alone does not explain defensin strain selectivity. Human β-defensins (HBDs) are longer, more cationic, more polar, and less hydrophobic than human α-defensins (64). Whereas α-defensins are generally more active against Gram-positive bacteria (54), β-defensins tend to be more effective against Gram-negatives (65, 66). The most prominent exception to this “rule” is HBD3, which carries an unusually high net positive charge (+11) and is effective against both Gram-positive and Gram-negative bacteria (35). The more typical HBD2 has a net charge of +6 and kills only Gram-negative strains (67). Because HNP1 (net charge, +3) was more effective than HBD3 against S. aureus, cationicity alone cannot account for this strain selectivity.
A hydrophobic cluster (25LWAF28) immediately precedes the last two Cys residues of HNP1. Trp-26 and Phe-28 were the two most critical residues for HNP1 to kill S. aureus, whereas Leu-25 was as important as any Arg residues in the sequence (Fig. 1). The identical residues exist in HNP2 and HNP3, and similarly placed hydrophobic clusters exist in HNP4 and HD5. Sequences of the known α-defensins from human, mouse, rat, rhesus macaque, rabbit, chimpanzee, and guinea pig indicate that the most common residues at position 26 (using HNP1 numbering) are Trp, Tyr, and Phe. Human α-defensin 6 (HD6), which has the weakest direct microbicidal activity of any α-defensin (34), contains 25NHRF28. By contrast, amino acids in the same region of β-defensins are largely polar and/or cationic (33). Thus, we hypothesize that hydrophobicity and/or amphiphilicity play key roles in the effectiveness of α-defensins such as HNP1 and HD5 against Gram-positive bacteria (34). Consistent with this hypothesis are recent findings on the 79-residue “big defensin” derived from hemocytes of the Japanese horseshoe crab (68, 69). Big defensin has two domains. In its globular N-terminal domain, 70% of the residues are nonpolar. In contrast, its C-terminal, β-defensin domain is more cationic and polar. The N-terminal domain strictly confers activity against Gram-positive bacteria, whereas the C-terminal domain is solely responsible for killing Gram-negative bacteria.
Trp Is a Common “Functional Hot-spot” Residue in Proteins
Wells and Clackson (70) first proposed the functional hot spot concept of binding energy in proteins, characterized by a small set of contact residues at a large protein-protein interface that make a dominant contribution to the binding free energy. Using Ala-scan mutagenesis, Clackson and Wells (70) demonstrated that two Trp residues from the human growth hormone receptor dominated the hormone-receptor interaction despite the fact that more than 30 side chains from each protein made contact in the complex structure. Many subsequent studies suggest that the presence of a small set of functional hot spot residues, particularly bulky hydrophobic amino acids, is a general characteristic of most protein-protein interfaces (71). The tumor suppressor protein p53 and its high affinity negative regulator MDM2 is another intensively studied protein-protein interacting system where more than half of the total binding free energy is contributed by Trp-23 of p53 buried inside a hydrophobic cavity of MDM2 (71–73). Not surprisingly, replacing the only Trp residue in HNP1 was detrimental to multiple defensin functions.
How Does Trp-26 Contribute to Defensin Function?
Trp-26 may contribute to defensin function at multiple levels. First, Trp can directly interact with target molecules of a hydrophobic nature, such as bacterial membrane and cell wall components, where its bulky hydrophobic side chain provides high binding energy. This should be relevant to bacterial killing as membrane permeabilization necessitates spatial segregation of cationic residues from hydrophobic moieties for optimal interactions with phospholipids. Second, Trp-26 structurally stabilizes HNP1 dimers, which, in turn, may form multimeric pores in the microbial membrane (74), causing leakage of intracellular contents and cell lysis (16, 17). Defensin dimerization could conceivably enhance HNP1 interactions with other target molecules as well due to the effect of multivalency. Third, Trp-26 mediates HNP1 self-association, a distinct molecular event shown to be important for multivalent binding of defensins to a variety of proteins such as bacterial toxins, cell surface receptors, viral components, etc. (34, 62). It has been suggested that the ability of HNP1 to self-associate on target surfaces may afford sufficient molecular complexity at the quaternary structural level, thereby contributing to enhanced structural diversity and functional versatility (35).
A Caveat
Despite being conserved structurally, α-defensins are highly variable in amino acid sequence and composition. For example, mouse α-defensins, also known as cryptdins, are significantly more cationic than HNP1; many studies have shown that cationicity plays a pivotal role in the action of cryptdins against bacteria (30, 75, 76). In fact, functional inhibition of cryptdins by their heavily polar and anionic propeptide, is achieved mainly through charge neutralization (75), whereas intramolecular or intermolecular inhibition of HNP1 activity by its heavily nonpolar propeptide is mediated predominantly by hydrophobic force (77). For these reasons, the tenet that Trp-26 and hydrophobicity dictate HNP1 function should not be expected to apply to all α-defensins.
It is noteworthy that Phe-28 is also an important residue for HNP1 dimerization as indicated by structural analysis. SPR studies showed that among all Ala-scan analogs tested, F28A-HNP1 had the second worse tendency to self-associate on the HNP1 surface (data not shown), coinciding with its second weakest bactericidal activity against S. aureus (Fig. 1). In contrast, the F28A mutation did not debilitate HNP1 to inhibit LF and bind gp120. These contrasting results suggest that S. aureus killing may be more sensitive than LF inhibition and gp120 binding to the structural integrity and/or stability of defensin dimers. Similarly, although Nva26 largely rescued W26A-HNP1 with respect to LF inhibition and gp120 binding, W26Abu-HNP1 remained extremely weak against the bacterium, indicative of less tolerance of S. aureus killing toward “molecular defects” in defensins at the quaternary structure level. Further investigation is warranted to better understand how defensins function as dimers and/or oligomers.
Finally, Ala-scanning mutagenesis, although a powerful tool for identifying binding epitopes and functional residues in proteins, has some limitations. It can generate aberrant mutational data at positions where the side chains interact, a major known cause of non-additivity of mutational effects in proteins (78, 79). To accurately dissect functional contributions of interacting residues in a protein, double mutations need to be introduced (53, 63). As Trp-26 makes contacts with several neighboring residues, it remains interesting to examine the activity of some double mutants of HNP1 in future studies.
In conclusion, we identified Trp-26 as a crucial molecular determinant of HNP1 that enhances the ability of this peptide to kill S. aureus, inhibit anthrax lethal factor, and bind HIV-1 gp120. Identification of this functional hot spot in HNP1 underscores the contribution of this hitherto largely unappreciated hydrophobic residue in the actions of this α-defensin against a variety of molecular, bacterial, and viral targets. The overall findings provide important insights into the mechanistic complexity of defensin-mediated host defense at the molecular level. Finally, although calling HNP1 a “cationic antimicrobial peptide” has a historical basis, this classification provides little insight into (and may actually obscure) the molecular mechanisms that underlie its many and diverse activities.
Supplementary Material
Acknowledgments
We thank the X-ray Crystallography Core Facility of the University of Maryland at Baltimore for providing crystallographic equipment and resources. We also thank Prof. Edvin Pozharski of the University of Maryland School of Pharmacy for help with synchrotron data collection.
This work was supported, in whole or in part, by National Institutes of Health Grants AI072732 and AI061482 (to W. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Materials and Methods, Tables 1 and 2, and Fig. 1.
- AMP
- antimicrobial peptides
- HNP
- human neutrophil peptide
- HIV
- human immunodeficiency virus
- LF
- lethal factor
- Abu
- aminobutyric acid
- Nva
- norvaline, α-aminopentanoic acid
- Nle
- norleucine, α-aminohexanoic acid
- Ahp
- α-aminoheptanoic acid
- HPLC
- high performance liquid chromatography
- TOCSY
- two-dimensional total correlation spectroscopy
- NOE
- nuclear Overhauser effect
- NOESY
- two-dimensional NOE spectroscopy
- SPR
- surface plasmon resonance
- HBD
- human β-defensins
- MS
- mass spectrometry
- RU
- response units.
REFERENCES
- 1.Zasloff M. (2002) Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 2.Habermann E. (1972) Science 177, 314–322 [DOI] [PubMed] [Google Scholar]
- 3.Steiner H., Hultmark D., Engström A., Bennich H., Boman H. G. (1981) Nature 292, 246–248 [PubMed] [Google Scholar]
- 4.Zasloff M. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5449–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selsted M. E., Ouellette A. J. (2005) Nat. Immunol. 6, 551–557 [DOI] [PubMed] [Google Scholar]
- 6.Ganz T. (2003) Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 7.Lehrer R. I. (2004) Nat. Rev. Microbiol. 2, 727–738 [DOI] [PubMed] [Google Scholar]
- 8.Bevins C. L. (2006) Biochem. Soc. Trans. 34, 263–266 [DOI] [PubMed] [Google Scholar]
- 9.Ganz T., Selsted M. E., Szklarek D., Harwig S. S., Daher K., Bainton D. F., Lehrer R. I. (1985) J. Clin. Invest. 76, 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selsted M. E., Harwig S. S., Ganz T., Schilling J. W., Lehrer R. I. (1985) J. Clin. Invest. 76, 1436–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabay J. E., Scott R. W., Campanelli D., Griffith J., Wilde C., Marra M. N., Seeger M., Nathan C. F. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 5610–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilde C. G., Griffith J. E., Marra M. N., Snable J. L., Scott R. W. (1989) J. Biol. Chem. 264, 11200–11203 [PubMed] [Google Scholar]
- 13.Jones D. E., Bevins C. L. (1992) J. Biol. Chem. 267, 23216–23225 [PubMed] [Google Scholar]
- 14.Jones D. E., Bevins C. L. (1993) FEBS Lett. 315, 187–192 [DOI] [PubMed] [Google Scholar]
- 15.Semple C. A., Taylor K., Eastwood H., Barran P. E., Dorin J. R. (2006) Biochem. Soc. Trans. 34, 257–262 [DOI] [PubMed] [Google Scholar]
- 16.Lehrer R. I., Barton A., Daher K. A., Harwig S. S., Ganz T., Selsted M. E. (1989) J. Clin. Invest. 84, 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagan B. L., Selsted M. E., Ganz T., Lehrer R. I. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 210–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klotman M. E., Chang T. L. (2006) Nat. Rev. Immunol. 6, 447–456 [DOI] [PubMed] [Google Scholar]
- 19.Yang D., Biragyn A., Hoover D. M., Lubkowski J., Oppenheim J. J. (2004) Annu. Rev. Immunol. 22, 181–215 [DOI] [PubMed] [Google Scholar]
- 20.Rehaume L. M., Hancock R. E. (2008) Crit. Rev. Immunol. 28, 185–200 [DOI] [PubMed] [Google Scholar]
- 21.Kim C., Gajendran N., Mittrücker H. W., Weiwad M., Song Y. H., Hurwitz R., Wilmanns M., Fischer G., Kaufmann S. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4830–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer-Scholl A., Hurwitz R., Brinkmann V., Schmid M., Jungblut P., Weinrauch Y., Zychlinsky A. (2005) PLoS Pathog. 1, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maemoto A., Qu X., Rosengren K. J., Tanabe H., Henschen-Edman A., Craik D. J., Ouellette A. J. (2004) J. Biol. Chem. 279, 44188–44196 [DOI] [PubMed] [Google Scholar]
- 24.de Leeuw E., Burks S. R., Li X., Kao J. P., Lu W. (2007) FEBS Lett. 581, 515–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie C., Prahl A., Ericksen B., Wu Z., Zeng P., Li X., Lu W. Y., Lubkowski J., Lu W. (2005) J. Biol. Chem. 280, 32921–32929 [DOI] [PubMed] [Google Scholar]
- 26.Wu Z., Li X., de Leeuw E., Ericksen B., Lu W. (2005) J. Biol. Chem. 280, 43039–43047 [DOI] [PubMed] [Google Scholar]
- 27.Rosengren K. J., Daly N. L., Fornander L. M., Jönsson L. M., Shirafuji Y., Qu X., Vogel H. J., Ouellette A. J., Craik D. J. (2006) J. Biol. Chem. 281, 28068–28078 [DOI] [PubMed] [Google Scholar]
- 28.Rajabi M., de Leeuw E., Pazgier M., Li J., Lubkowski J., Lu W. (2008) J. Biol. Chem. 283, 21509–21518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zou G., de Leeuw E., Li C., Pazgier M., Li C., Zeng P., Lu W. Y., Lubkowski J., Lu W. (2007) J. Biol. Chem. 282, 19653–19665 [DOI] [PubMed] [Google Scholar]
- 30.Tanabe H., Qu X., Weeks C. S., Cummings J. E., Kolusheva S., Walsh K. B., Jelinek R., Vanderlick T. K., Selsted M. E., Ouellette A. J. (2004) J. Biol. Chem. 279, 11976–11983 [DOI] [PubMed] [Google Scholar]
- 31.Hazrati E., Galen B., Lu W., Wang W., Ouyang Y., Keller M. J., Lehrer R. I., Herold B. C. (2006) J. Immunol. 177, 8658–8666 [DOI] [PubMed] [Google Scholar]
- 32.Buck C. B., Day P. M., Thompson C. D., Lubkowski J., Lu W., Lowy D. R., Schiller J. T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1516–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giesemann T., Guttenberg G., Aktories K. (2008) Gastroenterology 134, 2049–2058 [DOI] [PubMed] [Google Scholar]
- 34.Lehrer R. I., Jung G., Ruchala P., Wang W., Micewicz E. D., Waring A. J., Gillespie E. J., Bradley K. A., Ratner A. J., Rest R. F., Lu W. (2009) Infect. Immun. 77, 4028–4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei G., de Leeuw E., Pazgier M., Yuan W., Zou G., Wang J., Ericksen B., Lu W. Y., Lehrer R. I., Lu W. (2009) J. Biol. Chem. 284, 29180–29192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith J. G., Nemerow G. R. (2008) Cell Host Microbe 3, 11–19 [DOI] [PubMed] [Google Scholar]
- 37.Schnölzer M., Alewood P., Jones A., Alewood D., Kent S. B. (1992) Int. J. Pept. Protein Res. 40, 180–193 [DOI] [PubMed] [Google Scholar]
- 38.Wu Z., Powell R., Lu W. (2003) J. Am. Chem. Soc. 125, 2402–2403 [DOI] [PubMed] [Google Scholar]
- 39.Wu Z., Li X., Ericksen B., de Leeuw E., Zou G., Zeng P., Xie C., Li C., Lubkowski J., Lu W. Y., Lu W. (2007) J. Mol. Biol. 368, 537–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Z., Prahl A., Powell R., Ericksen B., Lubkowski J., Lu W. (2003) J. Pept. Res. 62, 53–62 [DOI] [PubMed] [Google Scholar]
- 41.Dawson P. E., Kent S. B. (2000) Annu. Rev. Biochem. 69, 923–960 [DOI] [PubMed] [Google Scholar]
- 42.Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. (1994) Science 266, 776–779 [DOI] [PubMed] [Google Scholar]
- 43.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braunschweiler L., Ernst R. R. (1983) J. Magn. Reson. 53, 521–528 [Google Scholar]
- 45.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 46.Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 47.Keller R. (2004) The Computer Aided Resonance Assignment Tutorial, 1st Ed., CANTINA Verlag, Switzerland [Google Scholar]
- 48.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 49.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 50.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 52.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 53.Qasim M. A., Lu W., Lu S. M., Ranjbar M., Yi Z., Chiang Y. W., Ryan K., Anderson S., Zhang W., Qasim S., Laskowski M., Jr. (2003) Biochemistry 42, 6460–6466 [DOI] [PubMed] [Google Scholar]
- 54.Ericksen B., Wu Z., Lu W., Lehrer R. I. (2005) Antimicrob. Agents Chemother. 49, 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujii G., Selsted M. E., Eisenberg D. (1993) Protein Sci 2, 1301–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., Cole A. M., Hong T., Waring A. J., Lehrer R. I. (2003) J. Immunol. 170, 4708–4716 [DOI] [PubMed] [Google Scholar]
- 57.Siebert H. C., Lu S. Y., Wechselberger R., Born K., Eckert T., Liang S., von der Lieth C. W., Jiménez-Barbero J., Schauer R., Vliegenthart J. F., Lütteke T., Kozár T. (2009) Carbohydr. Res. 344, 1515–1525 [DOI] [PubMed] [Google Scholar]
- 58.Szyk A., Wu Z., Tucker K., Yang D., Lu W., Lubkowski J. (2006) Protein Sci. 15, 2749–2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang X. L., Selsted M. E., Pardi A. (1992) Biochemistry 31, 11348–11356 [DOI] [PubMed] [Google Scholar]
- 60.Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. (2003) J. Biol. Chem. 278, 8606–8616 [DOI] [PubMed] [Google Scholar]
- 61.Lu W., Apostol I., Qasim M. A., Warne N., Wynn R., Zhang W. L., Anderson S., Chiang Y. W., Ogin E., Rothberg I., Ryan K., Laskowski M., Jr. (1997) J. Mol. Biol. 266, 441–461 [DOI] [PubMed] [Google Scholar]
- 62.Lehrer R. I., Jung G., Ruchala P., Andre S., Gabius H. J., Lu W. (2009) J. Immunol. 183, 480–490 [DOI] [PubMed] [Google Scholar]
- 63.Lu S. M., Lu W., Qasim M. A., Anderson S., Apostol I., Ardelt W., Bigler T., Chiang Y. W., Cook J., James M. N., Kato I., Kelly C., Kohr W., Komiyama T., Lin T. Y., Ogawa M., Otlewski J., Park S. J., Qasim S., Ranjbar M., Tashiro M., Warne N., Whatley H., Wieczorek A., Wieczorek M., Wilusz T., Wynn R., Zhang W., Laskowski M., Jr. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1410–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pazgier M., Hoover D. M., Yang D., Lu W., Lubkowski J. (2006) Cell. Mol. Life Sci. 63, 1294–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harder J., Bartels J., Christophers E., Schröder J. M. (1997) Nature 387, 861. [DOI] [PubMed] [Google Scholar]
- 66.Harder J., Bartels J., Christophers E., Schroder J. M. (2001) J. Biol. Chem. 276, 5707–5713 [DOI] [PubMed] [Google Scholar]
- 67.Schibli D. J., Hunter H. N., Aseyev V., Starner T. D., Wiencek J. M., McCray P. B., Jr., Tack B. F., Vogel H. J. (2002) J. Biol. Chem. 277, 8279–8289 [DOI] [PubMed] [Google Scholar]
- 68.Kouno T., Fujitani N., Mizuguchi M., Osaki T., Nishimura S., Kawabata S., Aizawa T., Demura M., Nitta K., Kawano K. (2008) Biochemistry 47, 10611–10619 [DOI] [PubMed] [Google Scholar]
- 69.Saito T., Kawabata S., Shigenaga T., Takayenoki Y., Cho J., Nakajima H., Hirata M., Iwanaga S. (1995) J Biochem. 117, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 70.Clackson T., Wells J. A. (1995) Science 267, 383–386 [DOI] [PubMed] [Google Scholar]
- 71.Kortemme T., Baker D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 73.Massova I., Kollman P. A. (1999) J. Am. Chem. Soc. 121, 8133–8143 [Google Scholar]
- 74.Hill C. P., Yee J., Selsted M. E., Eisenberg D. (1991) Science 251, 1481–1485 [DOI] [PubMed] [Google Scholar]
- 75.Figueredo S. M., Weeks C. S., Young S. K., Ouellette A. J. (2009) J. Biol. Chem. 284, 6826–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Llenado R. A., Weeks C. S., Cocco M. J., Ouellette A. J. (2009) Infect. Immun. 77, 5035–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zou G., de Leeuw E., Lubkowski J., Lu W. (2008) J. Mol. Biol. 381, 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dill K. A. (1997) J. Biol. Chem. 272, 701–704 [DOI] [PubMed] [Google Scholar]
- 79.Wells J. A. (1990) Biochemistry 29, 8509–8517 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.