Abstract
Effectors of the type III secretion systems (T3SS) are key elements in the interaction between many Gram-negative pathogens and their hosts. SlrP is an effector that is translocated into the eukaryotic host cell through the two virulence-associated T3SS of Salmonella enterica. We found previously that this effector is an E3 ubiquitin ligase for mammalian thioredoxin. Here, we identified ERdj3, an endoplasmic reticulum lumenal chaperone of the Hsp40/DnaJ family, as a new target for SlrP. Experiments with truncated forms of ERdj3 showed that domain II was essential for the interaction with SlrP. Confocal microscopy and subcellular fractionation demonstrated that, in transfected HeLa cells, SlrP was partially located in the endoplasmic reticulum. The presence of SlrP interfered with the binding of ERdj3 to a denatured substrate. Taken together, these data suggest that the role of SlrP in the interaction between Salmonella and the host cell is exerted through the modulation of the function of two independent targets: thioredoxin in the cytosol, and ERdj3 in the endoplasmic reticulum.
Keywords: Chaperone Chaperonin, Endoplasmic Reticulum (ER), Epithelial Cell, Protein Translocation, Protein-Protein Interactions, Salmonella, Type III Secretion
Introduction
Salmonella is a genus of Gram-negative bacterial pathogens that belongs to the family Enterobacteriaceae. The genus includes thousands of serovars that can infect a wide variety of animals and produce different diseases, from gastroenteritis to typhoid fever, depending on the serovar and the host. Many Gram-negative pathogens rely on type III secretion systems (T3SS)3 for the delivery into the host cells of effector proteins that direct the different stages of the infection at the cellular level. Salmonella enterica possesses two distinct T3SS essential for virulence, encoded by genes located in the Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2), respectively. More than thirty T3SS effectors are known in S. enterica and some of them have been shown to manipulate key host functions, including signal transduction, membrane traffic and pro-inflammatory immune responses (1, 2). However, the functions and cellular targets for many effectors are still unknown or incompletely understood.
SlrP (for Salmonella leucine-rich repeat protein) was identified by signature-tagged mutagenesis as a S. enterica serovar Typhimurium host range factor (3). This is one of the few effectors that can be secreted by the two virulence-related T3SS that are present in S. enterica (4). The gene slrP is located outside SPI-1 and SPI-2, in a 2.9-kb DNA region with features of horizontal acquisition. An slrP-null mutant has no difference in virulence with the wild-type strain when infecting calves but it is 6-fold attenuated for mouse virulence after oral infection (3). The predicted protein, SlrP, has a complete sequence of 765 amino acid residues that can be divided into several domains (5). The N-terminal 191 (and probably less) amino acid residues are sufficient for secretion and translocation into the eukaryotic host cells and show similarity to the N termini of Salmonella effectors SspH1, SspH2, SseI, SseJ, SifA, and SifB (4). The central domain (amino acids 176–564) contains several copies of a leucine-rich repeat signature, a protein motif frequently involved in protein-protein interactions (6, 7). The leucine-rich repeat domain is also found in other T3SS effectors including SspH1 and SspH2 from S. enterica serovar Typhimurium, YopM from Yersinia species and the IpaH family from Shigella flexneri (5). Finally, the C-terminal domain is also conserved in the effectors SspH1, SspH2, and the IpaH family. Recent studies uncovered E3 ubiquitin-ligase activities for effectors possessing this domain, including IpaH9.8, SspH1, and SspH2 (8, 9). Structural analysis showed that these effectors define a new class of ubiquitin ligases (8, 10, 11). In a previous work, we demonstrated that SlrP was able to interact with mammalian thioredoxin-1 (Trx). We also showed that SlrP is an E3 ubiquitin ligase that can use Trx as a substrate (12).
Below we show that ERdj3, a member of the Hsp40/DnaJ family of chaperones that is located in the endoplasmic reticulum in mammalian cells, is a second cellular target for SlrP. Our experiments suggest that this Salmonella effector can interfere with the function of ERdj3 by competing with the binding of unfolded client proteins.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Yeast Strains, and Plasmids
Microbial strains and plasmids used in this study are listed and described in Table 1. Plasmid pIZ1729 is a derivative of pCS2 coding for ERdj3 with a C-terminal 3×HA tag. To construct this plasmid, 3×HA was amplified from plasmid pCS2+HA3 using primers 3HA5′ and 3HA3′, and ERdj3 cDNA was amplified from plasmid pIZ1706 using primers dnajbam5′ and dnajxho3′. The first amplicon was digested with XhoI and XbaI and the second with BamHI and XhoI and both products were ligated together with vector pCS2+HA3 digested with BamHI and XbaI. All constructs were confirmed by sequencing.
TABLE 1.
Bacterial strains, yeast strains, and plasmids used in this study
| Strain/plasmid | Relevant characteristics | Source/Ref. |
|---|---|---|
| E. coli | ||
| DH5α | supE44 Δ lacU169 (Ø80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | (40) |
| BL21 (DE3) | F−ompT gal dcm lon hsdSB (rB− mB−; E. coli B strain), with DE3, a λ prophage carrying the T7 RNA pol gene | Stratagene |
| S. enterica | ||
| 14028 | Wild type | ATCC |
| SV5193 | 14028 slrP::3xFLAG, Kmr | (12) |
| S. cerevisiae | ||
| AH109 | MATa, trp1–901, leu2–3, 112, ura3–52, his3–200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2 URA3:: MEL1UAS-MEL1TATA-LacZ MEL1 | A. Holtz, unpublished |
| Y187 | MATα, ura3–52, his3–200, ade2–101, trp1–901, leu2–3, 112, gal4Δ, gal80Δ, met–, URA3:: GAL1UAS-GAL1TATA-LacZ MEL1 | (41) |
| Plasmids | ||
| pcDNA3 | Transient or stable transfection vector, Ampr | Invitrogen |
| pCS2+HA3 | Transient transfection vector, Ampr | F. Romero |
| pGADT7 | GAL4(768–881) AD, LEU2, Ampr | Clontech |
| pGBT10 | GAL4(1–147) DB, TRP1, Ampr | Clontech |
| pGEX-4T-1 | GST fusion vector, Ampr | Pharmacia |
| pGEX-4T-3 | GST fusion vector, Ampr | Pharmacia |
| pIZ1623 | pGEX-4T-3-SlrP | (12) |
| pIZ1627 | pGBT10-SlrP | (12) |
| pIZ1706 | pGADT7-ERdj3, clone obtained in the two-hybrid screen | This study |
| pIZ1708 | pGEX-4T-1-ERdj3 | This study |
| pIZ1720 | pCS2-SlrP-3xFLAG | (12) |
| pIZ1725 | pcDNA3-SlrP-3xFLAG | (12) |
| pIZ1729 | pCS2-ERdj3–3xHA | This study |
| pIZ1780 | pcDNA3-SlrP(C546A)-3xFLAG | (12) |
| pIZ1791 | pGADT7-ERdj3(23–358) | This study |
| pIZ1792 | pGADT7-ERdj3(88–358) | This study |
| pIZ1793 | pGADT7-ERdj3(129–358) | This study |
| pIZ1794 | pGADT7-ERdj3(160–358) | This study |
| pIZ1795 | pGADT7-ERdj3(201–358) | This study |
| pIZ1796 | pGADT7-ERdj3(251–358) | This study |
| pIZ1797 | pGEX-4T-1-ERdj3(23–358) | This study |
| pIZ1798 | pGEX-4T-1-ERdj3(88–358) | This study |
| pIZ1799 | pGEX-4T-1-ERdj3(129–358) | This study |
| pIZ1800 | pGEX-4T-1-ERdj3(160–358) | This study |
| pIZ1801 | pGEX-4T-1-ERdj3(201–358) | This study |
| pIZ1802 | pGEX-4T-1-ERdj3(251–358) | This study |
| pIZ1804 | pGBT10-SlrP(C546A) | This study |
| pQE10-BiP | L. Hendershot | |
DNA Amplification with the Polymerase Chain Reaction and Oligonucleotides
Amplification reactions were carried out in a Perkin Elmer Gene-Amp PCR System 2400 (PerkinElmer Life Sciences). The final volume of reactions was 50 μl, and the final concentration of MgCl2 was 1.5 mm. Reagents were used at the following concentrations: dNTPs, 200 μm; primers, 1 μm; and Taq polymerase (KAPA HiFi DNA polymerase, Kapa Biosystems), 1 unit per reaction. The thermal program included the following steps: (i) initial denaturation, 2 min at 94 °C; (ii) 25 cycles of denaturation (94 °C, 30 s), annealing (55 °C, 30 s), and extension (72 °C, 1 to 3 min); and (iii) final incubation at 72 °C for 7 min, to complete extension. Primers are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide/use | Sequence 5′-3′ (restriction sites are underlined) |
|---|---|
| Construction of pIZ1708 | |
| dnaj5′ | ATGCGAATTCATGGCTCCGCAGAACCTGAG |
| dnaj3′ | TGACCTCGAGGTCCAATTTTATTCACTCTC |
| Construction of pIZ1729 | |
| 3HA5′ | ATCGCTCGAGTACCCATACGATGTTCC |
| 3HA3′ | TGCATCTAGATCAGCCAGCGTAGTCTGGTAC |
| Dnajbam5′ | CTGAGGATCCACCATGGCTCCGCAGAACCTGAG |
| Dnajxho3′ | TGACCTCGAGATATCCTTGCAGTCCATTGTATAC |
| Construction of pIZ1791 and pIZ1797 | |
| dnaj23eco5′ | CATGGAATTCGGACGAGATTTCTATAAGATC |
| dnaj3′ | as above |
| Construction of pIZ1792 and pIZ1798 | |
| dnaj88eco5′ | CATGGAATTCACTTATGGTGAAGAAGGATTAAAAG |
| dnaj3′ | as above |
| Construction of pIZ1793 and pIZ1799 | |
| dnaj129eco5′ | CATGGAATTCCCAAGAGGAAGTGATATTATTG |
| dnaj3′ | as above |
| Construction of pIZ1794 and pIZ1800 | |
| dnaj160eco5′ | GATCGAATTCGCAAGGCAGGCTCCTGGCAAAC |
| dnaj3′ | as above |
| Construction of pIZ1795 and pIZ1801 | |
| dnaj201eco5′ | CATGGAATTCCTAGTGAATGAAGAACGAAC |
| dnaj3′ | as above |
| Construction of pIZ1796 and pIZ1802 | |
| dnaj251eco5′ | CATGGAATTCTTTGAAAGGAGAGGAGATGATTTG |
| dnaj3′ | as above |
| Sequencing of pGADT7 derivatives | |
| T7 | TAATACGACTCACTATAGGG |
| Sequencing of pGEX derivatives | |
| pgex5′ | GGGCTGGCAAGCCACGTTTGGTG |
| pgex3′ | CCGGGAGCTGCATGTGTCAGAGG |
DNA Sequencing and Sequence Analysis
cDNA from the positive clone obtained in the two-hybrid screen, and PCR constructs were sequenced with an automated DNA sequencer (Stab Vida, Oeiras, Portugal). Sequence analysis was performed with molecular biology algorithms from the National Center for Biotechnology Information (NCBI) and the European Bioinformatics Institute (EBI).
Bacterial Culture
The standard culture medium for S. enterica and Escherichia coli was LB broth. Solid LB contained agar 1.5% final concentration. Antibiotics were used at the following concentrations: kanamycin (Km), 50 μg/ml; ampicillin (Amp), 100 μg/ml.
Yeast Two-hybrid Methods
Plasmids were introduced into Saccharomyces cerevisiae strains using the lithium acetate procedure, as previously described (13). A human HeLa Matchmaker cDNA library (Clontech), prepared using the vector pGADT7 and pretransformed in S. cerevisiae strain Y187, was screened using yeast mating, according to the manufacturer instructions. Briefly, S. cerevisiae strain AH109 transformed with pIZ1627 was grown at 30 °C overnight with shaking in yeast drop-out medium lacking tryptophan. The culture was concentrated, mixed with 1 ml of the library in 45 ml of 2×YPDA, containing 2% yeast extract, 4% peptone, 4% glucose, and 0.006% adenine hemisulfate, and incubated at 30 °C overnight with gentle swirling. The mating mixture was plated in SD medium lacking leucine, tryptophan and histidine (Clontech). Plates were incubated at 30 °C for 8 days and then colonies were patched on the same medium and replica-plated on medium lacking tryptophan, leucine, histidine and adenine and in medium lacking leucine and tryptophan and supplemented with X-α-Gal, to check for the expression of the ADE2 and the MEL1 reporters, respectively. Positive clones were rescued, tested for specificity using empty pGBT10, and sequenced with primer T7.
Cell Culture, Lysis, and Transfection
HeLa cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. 2 mm l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin were included in the culture medium. All cells were maintained in a 5% CO2 humidified atmosphere at 37 °C. For cell lysis, 2 × 107 to 108 cells per ml were incubated at 4 °C in Nonidet P-40 buffer (10 mm Tris-HCl pH 7.4, 150 mm NaCl, 10% glycerol, 1% Nonidet P-40, 1% aprotinin, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, and 1 μg/ml leupeptin) for 20 min. The extract was centrifuged at 20,000 × g for 20 min, and the supernatant was stored at −80 °C. For transient transfection assays, 2–5 × 106 HeLa cells/assay were resuspended in 200 μl of 15 mm Hepes-buffered serum-containing medium, mixed with 50 μl of 210 mm NaCl containing 5–10 μg of plasmid DNA and electroporated using a BTX Electrocell Manipulator 600 set at 240 V, 720 Ω, 950 μF. Cells were processed 24 h after electroporation.
Antibodies and Chemicals
Immobilized thyroglobulin conjugated to 4% cross-linked agarose beads, mouse monoclonal anti-FLAG M2, anti-FLAG M2 Cy3 conjugate, and rabbit polyclonal anti-ERdj3 antibodies were from Sigma. Protein A/G plus-agarose, protein A-agarose, and anti-Trx rabbit polyclonal antibody were from Santa Cruz Biotechnology. Anti-HA-peroxidase rat monoclonal antibody (clone 3F10) was from Roche Diagnostics GmbH. Alexa Fluor® 488-labeled antibody HA.11 was from Covance. Peroxidase-linked anti-rabbit IgG was from GE Healthcare and peroxidase-linked anti-mouse IgG was from Bio-Rad.
GST and His6 Fusion Proteins, Electrophoresis, and Immunoblot
Expression of the GST fusion proteins was induced by the addition of 1 mm isopropyl-β-d-thiogalactoside to E. coli BL21 (DE3) containing pGEX-4T-1, pGEX-4T-3, or their derivatives and the fusion proteins were isolated from bacterial lysates by affinity chromatography with glutathione-agarose beads (Sigma). 6His-BiP was produced after addition of 1 mm isopropyl-β-d-thiogalactoside to E. coli M15/pREP4 containing plasmid pQE10-BiP (a generous gift of Dr. L. Hendershot, University of Tennessee, Memphis), purified on Ni-NTA agarose beads (Qiagen) and eluted with 200 mm imidazole in binding buffer (100 mm KCl, 20 mm Tris, pH 7.0, 5 mm MgCl2). For lysis, bacteria were sonicated in Nonidet P-40 buffer. For some binding experiments, lysates from S. enterica strain SV5193 were incubated for 2 h with fusion protein bound to glutathione-coupled agarose beads. The precipitates were washed six times in Nonidet P-40 buffer followed by SDS-PAGE (polyacrylamide gel electrophoresis). The gel was blotted onto a nitrocellulose membrane (Amersham Biosciences) and probed with primary antibodies. Goat anti-mouse horseradish peroxidase-conjugated antibodies (Bio-Rad) were used as secondary antibodies. Detection was via chemiluminescence procedures (Pierce).
Coimmunoprecipitation Experiments
For each experiment, a lysate from 5 × 106 HeLa cells cotransfected with plasmids pIZ1720 and pIZ1729, expressing SlrP-3×FLAG and ERdj3–3xHA, respectively, and a lysate from 5 × 106 HeLa cells transfected with plasmid pIZ1729 only, were incubated with 4 μl of normal mouse IgG (Santa Cruz Biotechnology) for 1 h at 4 °C, and with 20 μl of protein A/G plus-agarose beads (Santa Cruz Biotechnology) for 2 h at 4 °C. After centrifugation the supernatants were incubated with 4 μl of monoclonal anti-FLAG antibodies for 2 h at 4 °C, then overnight with 40 μl of protein A/G plus-agarose beads, and finally centrifuged. The beads were washed three times in Nonidet P-40 buffer containing 1% sodium deoxycholate and 300 mm NaCl, twice in Nonidet P-40 buffer containing 1% sodium deoxycholate and 150 mm NaCl, and once in Nonidet P-40 buffer containing 1% sodium deoxycholate and 10 mm NaCl. Proteins were eluted and dissolved into Laemmli sample buffer (50 mm Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 0.0005% bromphenol blue) containing 5% β-mercaptoethanol, incubated at 95 °C for 5 min and subjected to SDS-PAGE. Proteins were transferred to a nitrocellulose filter and probed with anti-FLAG (Sigma) and anti-HA-peroxidase (clone 3F10, Roche).
Immunofluorescence and Confocal Microscopy
Direct immunofluorescence was performed with cells cultured on permanox Lab-Tek® chamber slides (Nunc). Cells were fixed for 6 min in ice-cold methanol and rinsed thoroughly with phosphate-buffered saline (PBS) and PBS containing 0.1% Tween 20. Cells were incubated for 30 min at room temperature with 3% bovine serum albumin (BSA) in PBS-Tween and 1 h in antibody diluted in PBS-Tween-BSA. Antibodies were anti-FLAG M2 monoclonal antibody-Cy3 conjugate (Sigma) and anti-HA Alexa Fluor® 488 labeled antibody (Covance) diluted 1:1000. After immunostaining, cells were rinsed with PBS and mounted for microscopy using ProLong Gold (Invitrogen). All images were acquired using a Leica TCS SP2 confocal microscope.
Microsomal Preparation
HeLa cells (3 × 108) were detached with trypsin, washed with PBS, and centrifuged to calculate the packed cell volume (PCV). Cells were suspended in a volume of hypotonic extraction buffer (10 mm HEPES, pH 7.8, 25 mm potassium chloride, and 1 mm EGTA) equivalent to 3 times the PCV and incubate for 20 min at 4 °C to allow the cells to swell. The cells were centrifuged at 600 × g for 5 min, the supernatant was removed by aspiration, and the new PCV was measured. The cells were suspended in a volume of isotonic extraction buffer (10 mm HEPES, pH 7.8, 250 mm sucrose, 25 mm potassium chloride, and 1 mm EGTA) equivalent to 2 times the new PCV and broken in a 7 ml homogenizer (Kontes) with 10 strokes. The homogenate was centrifuged at 1000 × g for 10 min at 4 °C and the supernatant was transferred to another centrifuge tube. After a centrifugation at 12,000 × g for 15 min at 4 °C, the supernatant fraction, which is the postmitochondrial fraction, was centrifuged for 60 min at 100,000 × g at 4 °C. The supernatant is the cytosol fraction. The pellet from this last centrifugation, which is the microsomal fraction, was suspended in isotonic extraction buffer.
ERdj3-BiP Binding Assays
Binding assays were performed as previously described (14) with appropriate modifications. Briefly, GST-ERdj3–1 (with amino acids 23–765 of ERdj3), GST-ERdj3–3 (with amino acids 129–765 of ERdj3) and GST-SlrP were purified on glutathione-agarose beads as described above. 50 μg of immobilized GST-ERdj3–1 or GST-ERdj3–3 were incubated with 50 μg of soluble His6-BiP, in the presence or absence of 1 mm ATP and 50 μg of GST-SlrP, in a total volume of 500 μl of binding buffer. After incubating at 4 °C, for 2 h, in a rotating platform, the beads were washed five times with 1 ml of binding buffer. Bound proteins were resolved by SDS-PAGE and detected by Coomassie Blue staining.
ATPase Assays
In vitro assays were performed as previously described (14) with some modifications to adapt the protocol to a non-radioactive procedure, as follows. GST-SlrP, GST-ERdj3–1, GST, and His6-BiP were purified as described above and dialyzed overnight into ATPase assay buffer (50 mm HEPES, pH 6.8, 50 mm NaCl, 2 mm MgCl2). Assays were performed in a final volume of 100 μl consisting of ATPase buffer, with 20 mm KCl, and 20 μm ATP. Reactions contained 1 μg of 6His-BiP and different combinations of GST, GST-ERdj3, and GST-SlrP (2.5 μg each). Comparison was made with a reaction containing neither His6-BiP nor a GST-containing protein. After incubating at 25 °C for 75 min, the concentration of phosphate released in the reactions was measured using the Malachite Green Phosphate Assay Kit (Cayman Chemical) according to the manufacturer's instructions.
Thyroglobulin Binding Assay
A method previously described (15) was used with some modifications. Immobilized thyroglobulin was denatured (dTg) in 6 m urea, 1 m β-mercaptoethanol, for 40 min at room temperature with rocking. Beads were washed and equilibrated in Nonidet P-40 buffer. Aliquots of dTg were incubated with total HeLa cell lysates for 3 h at 4 °C with rocking. After five washes with 1 ml of Nonidet P-40 buffer, proteins bound to beads were analyzed by SDS-PAGE (10%) and immunoblot.
RESULTS
Salmonella SlrP Interacts with Human ERdj3 in the Yeast Two-hybrid System
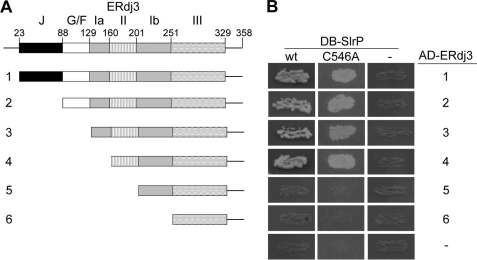
In an attempt to identify mammalian partners for Salmonella T3SS effector SlrP, a screen of a HeLa cDNA library was performed using SlrP as bait in the yeast two-hybrid system (12). The cDNA insert from one of the clones obtained in medium lacking histidine, used to select for the interactions, contained the entire coding sequence for human ERdj3 (Fig. 1A). Colonies from this clone were also able to grow in the absence of adenine and were blue in the presence of X-α-Gal, indicating that they expressed three different reporter genes used in this system: HIS3, ADE2, and MEL1. The specificity of the SlrP/ERdj3 interaction was confirmed by the lack of reporter gene activation when the empty vector (pGBT10) was used instead of the SlrP plasmid (Fig. 1B).
FIGURE 1.
Interaction of Salmonella SlrP with human ERdj3 in the yeast two-hybrid system. A, diagram of the domains of full-length and deleted versions (1–6) of ERdj3. The domains of ERdj3 are indicated as: J, J domain; G/F, glycine-, and phenylalanine-rich domain; Ia, domain Ia; II, domain II; Ib, domain Ib; III, domain III. B, diploids were obtained by conjugation between strain AH109, containing derivatives of pGBT10 (DB-SlrP), and strain Y187, containing derivatives of pGADT7 (AD-ERdj3), as indicated. The interaction between the two hybrid proteins is shown by the growth in the absence of histidine and adenine. DB, fusion with the DNA-binding domain of Gal4; AD, fusion with the activation domain of Gal4; wt, wild-type SlrP; C546A, C546A mutant of SlrP; 1 to 6 correspond to ERdj3 subclones represented in A; −, empty vectors.
Domain II of ERdj3 Is Essential for Association with SlrP
ERdj3 is a molecular chaperone of the Hsp40/DnaJ family. It contains an N-terminal signal peptide, necessary for its localization in the endoplasmic reticulum. The signal peptide is absent in the mature form of the protein (14). ERdj3 also possesses a J domain that binds to BiP, the mammalian endoplasmic reticulum Hsp70/DnaK chaperone, a Gly/Phe-rich domain, and a C-terminal substrate binding fragment. The C-terminal fragment contains three domains called I, II, and III. Domain I is interrupted by the Cys-rich domain II, which is a hallmark of type I DnaJ proteins (Fig. 1A). On the basis of the previously defined domain boundaries (16), we made sequential deletion mutants, from ERdj3–1 to ERdj3–6. Construction 1 corresponds to the mature form of the protein, without signal peptide. Construction 6 corresponds to domain III plus the 30 C-terminal amino acids (see Fig. 1A for additional details). Each of the ERdj3 constructs was tested in the two-hybrid system in yeasts for interaction with SlrP. Empty vectors were used as controls (Fig. 1B, third column and seventh row). Growth in the absence of histidine and adenine showed that all constructs, except 5 and 6, were able to interact with SlrP in the two-hybrid system (Fig. 1B, first column). Since mutants 5 and 6 lack domain II, these results indicate that this domain of ERdj3 is essential for the association with SlrP. In addition, a non-catalytic mutant form of SlrP in which Cys-546 was changed to Ala exhibited the same pattern of interaction as the wild type (Fig. 1B, second column; see “Discussion”).
Interaction of SlrP and ERdj3 Is Confirmed by Pull-down Experiments
To confirm the results obtained with the two-hybrid approach, the prokaryotic GST expression system was used to produce recombinant ERdj3 as GST fusion protein. GST-ERdj3 purified and immobilized on glutathione-agarose beads was incubated with a lysate of strain SV5193, an S. enterica serovar Typhimurium strain that codes for a 3×FLAG-tagged form of SlrP (12). After washing, the complexes were analyzed by immunoblot with anti-FLAG monoclonal antibodies. As seen in Fig. 2A, SlrP-3×FLAG interacted with GST-ERdj3 but not with GST, showing that the interaction was specific. A similar analysis was carried out with ERdj3 deletion derivatives 1 to 6 expressed in the form of GST fusion proteins. Constructs 1–4, but not 5 and 6, were able to interact with SlrP-3×FLAG (Fig. 2B, upper panel). The amount and size of the chimeric proteins used in the binding experiments was assessed by reversible staining of the nitrocellulose filter with Ponceau S red (Fig. 2B, lower panel). Taken together, these findings confirm that ERdj3 associates with SlrP and that domain II of ERdj3 is essential for this interaction.
FIGURE 2.
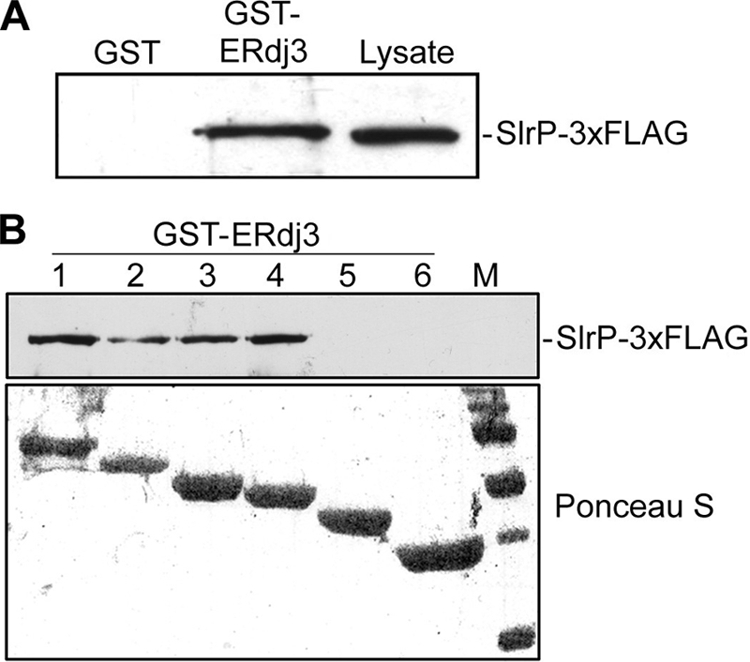
Interaction of ERdj3 and SlrP in vitro. A, in vitro binding of SlrP-3×FLAG to GST-ERdj3. Expression of GST and GST-ERdj3 proteins was induced with isopropyl-1-thio-β-d-galactopyranoside, and proteins were isolated from bacterial lysates by affinity chromatography with glutathione-agarose beads. 4 μg of each GST protein were incubated with lysates from 109 cfu of S. enterica serovar Typhimurium strain SV5193 (14028 slrP::3×FLAG) prepared in Nonidet P-40 lysis buffer. After washing, proteins eluted in sample buffer were resolved in 12% SDS-PAGE, blotted on nitrocellulose filters, and developed with monoclonal anti-FLAG antibody. Lysate from 108 cfu of the same strain is also included for reference. B, domain II of ERdj3 is essential for interaction with SlrP. Deleted derivatives of ERdj3 1–6 (see Fig. 1A) were expressed in fusion with GST and used for binding experiments with lysates from 109 cfu of Salmonella strain SV5193 as in A. M, molecular mass markers: visible bands correspond to 150, 100, 75, 50, 37, and 25 kDa.
SlrP and ERdj3 Interact in HeLa Cells
To investigate if this interaction could exist in vivo, in the host cell, coimmunoprecipitation experiments were carried out. HeLa cells were co-transfected with a plasmid coding for SlrP-3×FLAG and another plasmid coding for ERdj3–3×HA. Lysates were prepared, anti-FLAG antibodies were used for immunoprecipitation, and the presence of ERdj3 in the immunoprecipitation complexes was investigated by immunoblot with anti-HA antibodies (Fig. 3). ERdj3–3×HA was clearly detected in the anti-FLAG immunoprecipitates. As a control of specificity, the same experiment was performed with a lysate from HeLa cells transfected with the plasmid coding for ERdj3–3×HA only. In this case the tagged ERdj3 protein was not detectable in the immunoprecipitates. These findings suggest that the complex SlrP/ERdj3 may exist in host cells, confirming and extending the results obtained with the two-hybrid system and GST fusion proteins.
FIGURE 3.
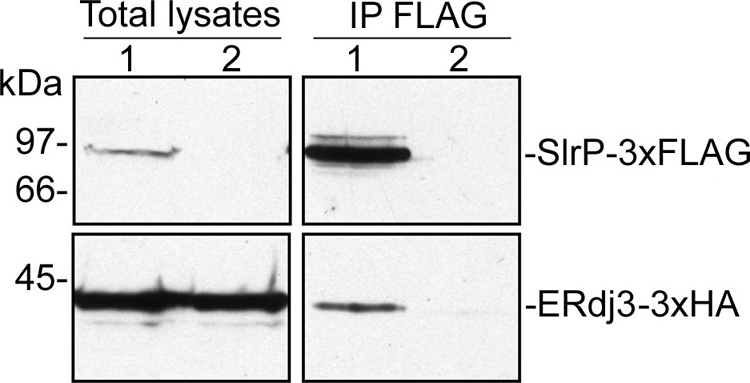
Coimmunoprecipitation of SlrP and human ERdj3. HeLa cells were transiently co-transfected with 4 μg of two derivatives of plasmid pCS2, one expressing SlrP-3×FLAG, and the other ERdj3–3×HA (lane 1), or transfected with the plasmid expressing ERdj3–3×HA only (lane 2). Nonidet P-40 lysates from 5 × 106 transfected cells were subjected to immunoprecipitation (IP, right panels) with anti-FLAG monoclonal antibodies and, after the stringent washings described under “Experimental Procedures,” resolved by 10% SDS-PAGE, transferred to nitrocellulose membranes, and developed with monoclonal anti-FLAG for the upper part of the membrane, and monoclonal anti-HA for the lower part. Lysates from 5 × 105 transfected cells are shown in the left panels.
SlrP Is Partially Located in the Endoplasmic Reticulum
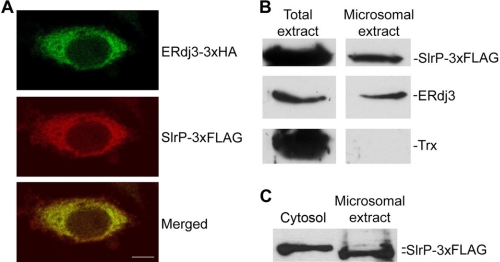
SlrP has been described as evenly distributed in the cytoplasm when expressed in mammalian cells (17). On the other hand, ERdj3, a member of the Hsp40 family of cochaperones, is exclusively located in the endoplasmic reticulum (18). The interaction described above could only take place in vivo if SlrP is, at least partially, located in the endoplasmic reticulum. This reasoning prompted us to investigate the subcellular localization of SlrP in HeLa cells. First, the localization of SlrP and ERdj3 was studied in cells transfected with the plasmids coding for SlrP-3×FLAG and ERdj3–3×HA using immunofluorescence and confocal microscopy. As seen in Fig. 4A, although SlrP is broadly distributed in the cytoplasm, colocalization with ERdj3 proteins is observed in the endoplasmic reticulum. Next, the localization of SlrP-3×FLAG was examined by subcellular fractionation. Microsome fraction was prepared from HeLa cells stably transfected with the plasmid coding for SlrP-3×FLAG, and the presence of SlrP-3×FLAG and ERdj3 in this fraction was assessed by immunoblot with anti-FLAG and anti-ERdj3 antibodies, respectively. Comparison with total extract reveals that ERdj3 is essentially located in microsomes, as expected, and that a significant part of the SlrP protein is in the same fraction (Fig. 4B). The absence of thioredoxin, a cytosolic, and nuclear protein, provided the control for the purity of the microsome fraction. Localization in the endoplasmic reticulum usually requires an N-terminal signal peptide that is cleaved upon translocation into this cellular compartment (19). The presence of such a signal in SlrP would produce two different molecular weight forms, one in the cytosol and the other in the endoplasmic reticulum. To assess this possibility we compared cytosolic and microsomal fractions of HeLa cells stably expressing SlrP-3×FLAG by Western blot with anti-FLAG antibodies (Fig. 4C). Proteins were separated in a 10% polyacrylamide gel instead of the 15% gel used in Fig. 4B to get a better resolution of the SlrP forms. Fig. 4C clearly shows a higher molecular weight form of SlrP in the cytosolic fraction and a lower molecular weight form in the microsomal fraction, which is in agreement with our hypothesis.
FIGURE 4.
Colocalization of SlrP and ERdj3 in HeLa cells. A, microscopic analysis. HeLa cells transiently transfected with plasmids expressing SlrP-3×FLAG and ERdj3–3×HA were observed in a laser-scanning confocal microscope after permeabilization and staining with Alexa Fluor® 488 labeled anti-HA (green, upper panel) and Cy3-conjugated anti-FLAG (red, center panel). Superposition of the two labelings is shown in the lower panel. Yellowish staining indicates colocalization of SlrP and ERdj3 in the endoplasmic reticulum. Scale bar, 10 μm. B, detection of SlrP in the endoplasmic reticulum of HeLa-transfected cells. Total lysate (left panels) and microsomal fraction (right panels) from 107 HeLa cells stably expressing SlrP-3×FLAG were submitted to 15% SDS-PAGE, transferred to nitrocellulose membrane, and then probed with anti-FLAG monoclonal antibody to detect SlrP. The filter was also incubated with anti-ERdj3 and anti-Trx polyclonal antibodies to detect endogenous ERdj3, and endogenous Trx (a cytosolic and partially nuclear protein), respectively, as a control of the purity of the microsomal fraction. C, comparison of the migration of SlrP present in cytosolic and microsomal fractions of transfected HeLa cells. A cytosolic fraction (106 cells) and a microsomal fraction (107 cells) obtained from HeLa cells stably expressing SlrP-3×FLAG were submitted to 10% SDS-PAGE, transferred to nitrocellulose membrane and then probed with anti-FLAG monoclonal antibody to detect SlrP.
SlrP Interferes with the Function of ERdj3 in a Ubiquitination-independent Manner
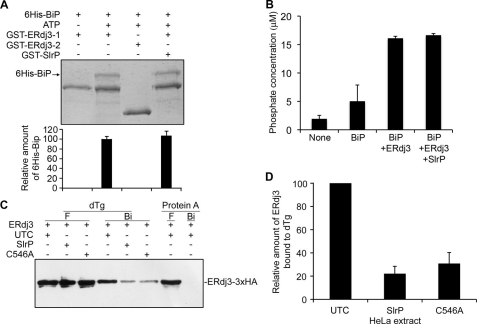
ERdj3 interacts with BiP and stimulates its ATPase activity (14). In addition ERdj3 binds directly to unfolded substrates (16). We hypothesized that the interaction between SlrP and ERdj3 could in some way alter the function of the host protein. To test our hypothesis, first we examined the binding of ERdj3 to BiP using GST-ERdj3 and His6-BiP, as previously described (14). 6His-BiP was able to bind to GST-ERdj3–1 (the mature form of the protein), but not to GST-ERdj3–3 (which lacks the J domain), in an ATP-dependent manner, as expected, but the presence of GST-SlrP in the binding experiments did not alter the results (Fig. 5A).
FIGURE 5.
Effect of SlrP on ERdj3 function. A, binding of ERdj3 to BiP. 50 μg of GST-ERdj3–1 (amino acids 23–358) or GST-ERdj3–3 (amino acids 129–358) immobilized in glutathione-agarose beads were incubated with 50 μg of His6-BiP in the presence or absence of 1 mm ATP and 50 μg of GST-SlrP as indicated. After incubating at 4 °C for 2 h, the beads were washed several times with binding buffer, and bound proteins were released by boiling for 5 min in sample buffer and subjected to 10% SDS-PAGE. Proteins were detected by Coomassie Blue staining. One stained gel is shown together with a graphical representation (below) of means ± S.D., from two independent experiments, of the quantification of bands of His6-Bip. B, stimulation of BiP ATPase activity by ERdj3. ATPase assays were performed with either no additions (None), 1 μg of His6-BiP plus 2.5 μg of GST (BiP), 1 μg of His6-BiP plus 2.5 μg of GST-ERdj3–1 plus 2.5 μg of GST (BiP+ERdj3), or 1 μg of His6-BiP plus 2.5 μg of GST-ERdj3–1 plus 2.5 μg GST-SlrP (BiP+ERdj3+SlrP). The amount of released phosphate was determined after incubation at 25 °C for 75 min with a non-radioactive procedure (see “Experimental Procedures”). Values are the mean of three separate experiments with an error bar representing S.D. C, binding of ERdj3 to dTg. Lysates from HeLa cells transiently expressing ERdj3–3×HA were mixed with lysates from untransfected cells (UTC) or with lysates transfected with SlrP or SlrPCys546Ala (C546A), as indicated, and then incubated with dTg or native protein A immobilized on agarose beads. After several washes with Nonidet P-40 buffer, proteins bound to the beads (Bi) were released by boiling in sample buffer, separated by using 10% SDS-PAGE and immunoblotted with anti-HA monoclonal antibodies. Part of the initial flow-through (F) obtained after the incubations was also included. D, image quantification of the relative amount of ERdj3 bound to immobilized dTg. The results are the mean ± S.D. of three independent experiments. The highest level was set to 100 for each experiment.
Next, we studied the effect of SlrP on the ability of ERdj3 to stimulate the ATPase activity of BiP. In vitro reactions were performed with 6His-BiP in the absence or presence of GST-ERdj3–1 and GST-SlrP, and the concentration of released phosphate was measured as described under “Experimental Procedures.” As seen in Fig. 5B some phosphate was detected in the absence of BiP, probably because of spontaneous release in the ATP preparation. The presence of BiP increased the concentration of released phosphate and ERdj3 was able to stimulate the ATPase activity of BiP to the same level irrespective of the presence of SlrP.
Finally, we assessed the binding of ERdj3 to immobilized, denatured thyroglobulin (dTg), a known substrate for this chaperone (15), in the presence or absence of SlrP. Fig. 5C shows that, when a total extract of HeLa cells transfected with ERdj3–3×HA was incubated with immobilized dTg, binding of a fraction of ERdj3–3×HA was detected, as expected. There was no significant binding to native protein A immobilized on agarose, which was used as a negative control. Preincubation with an extract from HeLa cells expressing SlrP-3×FLAG significantly reduced the binding of ERdj3 to dTg (Fig. 5D). Similar results were obtained when the experiment was repeated using an extract of HeLa cells expressing a mutant form of SlrP in which Ala substituted for Cys-546, a residue necessary for full ubiquitin ligase activity (12). No effect was observed when preincubation was carried out with an extract of non-transfected HeLa cells, used as control. The results presented in this section suggest that the interaction of SlrP with ERdj3 could have a functional significance decreasing the ability of this chaperone to bind denatured substrates, and that this effect is independent of the E3 ubiquitin ligase activity of SlrP.
DISCUSSION
Identification of eukaryotic targets for individual T3SS effectors is an essential step in the understanding of the function of these virulence factors. In a previous work, we identified human Trx as a partner for SlrP (12). Now, we have identified a second cellular target, ERdj3. The interaction of SlrP with this protein has been confirmed through three independent approaches: in vivo interaction in yeast with the two-hybrid system (Fig. 1), pull-down experiments with GST fusion proteins produced in bacteria (Fig. 2), and coimmunoprecipitation from transfected human epithelial HeLa cells (Fig. 3).
ERdj3/HEDJ is a protein of the Hsp40/DnaJ family localized in the endoplasmic reticulum (14). ERdj3 is an important chaperone/co-chaperone that associates with a number of unfolded proteins that are substrates of the Hsp70/DnaK family member GRP78/BiP (20–23). In addition, ERdj3 is able to bind client proteins directly and independently of BiP (16).
The interaction of SlrP with an endoplasmic reticulum protein prompted us to investigate its subcellular localization. A previous report, using conventional immunofluorescence microscopy, showed that HA-tagged SlrP was present in the cytoplasm and excluded from the nucleus of transiently transfected Raw-TT10 and CHO-K1 cells (17). Our confocal microscopy experiments confirm that SlrP is evenly distributed in the cytoplasm of HeLa cells but also show a significant overlap with ERdj3 (Fig. 4A). The partial localization of SlrP in the endoplasmic reticulum was confirmed by analysis of a microsomal fraction obtained from transfected HeLa cells. Quantitative comparison of the bands in Fig. 4B indicates that at least 10% of SlrP is in the microsomal fraction. When trying to identify a signal sequence in SlrP that could direct translocation into the endoplasmic reticulum with the program Sig-Pred (J. R. Bradford, University of Leeds) a putative signal (ISAFFSSEHQVEA) was detected. However when other programs, including SignalP (24), were used, no signal was predicted. The presence of an imperfect signal sequence could explain the partial localization of SlrP in the endoplasmic reticulum shown by our experiments. In fact, different signal sequences have different efficiencies in initiating translocation (25, 26), which could explain previous examples of proteins residing in multiple compartments (27). In the case of SlrP, this would allow interaction with two different cellular targets, Trx and ERdj3, which are located in different cellular compartments, in a non-competitive way. The results in Fig. 4C, showing different molecular weights for the fractions of SlrP present in the endoplasmic reticulum and in the cytosol, suggest that there is in fact a signal peptide in SlrP that is cleaved during translocation into the endoplasmic reticulum.
Members of the DnaJ family have been classified as type I, II, or III. Type I proteins contain three regions in common: a J domain, a Gly/Phe-rich domain, and a Cys-rich domain of four CXXCXG. Type II proteins have the J and Gly/Phe domains, whereas type III proteins have only the J domain in common (28, 29). Although ERdj3 contains the typical J and Gly/Phe domains, it has an atypical Cys-rich domain with one CXC and one CXXC motifs, whose presence and redox state contribute to substrate interactions (15). ERdj3 is similar to the yeast DnaJ protein Ydj1(30, 31). Structural studies on this protein established that its C terminus can be divided into three domains, called domain I, II, and III that are also found in ERdj3. Domain I and domain II of ERdj3 are essential for substrate binding (16). Our experiments with truncated forms of ERdj3 (Figs. 1 and 2) indicated that domain II, but not domain I, of ERdj3, is essential for the interaction with SlrP. These results suggest that SlrP is not just acting as a denatured substrate and that the ERdj3/SlrP interaction is specific. The absence of domains J and Gly/Phe-rich, did not affect the interaction. In agreement with these results, interaction of ERdj3 with BiP and stimulation of its ATPase activity, which are mediated by the J domain, were not affected by the presence of SlrP (Fig. 5, A and B). However, SlrP significantly reduced interaction of ERdj3 with a denatured substrate (thyroglobulin) (Fig. 5, C and D). This latter result means that the interaction of this Salmonella effector with ERdj3 can have important functional consequences: since domain II is essential both for the interaction with SlrP and with denatured client proteins, competition for this domain in ERdj3 could lead to accumulation of unfolded proteins in the endoplasmic reticulum. This accumulation could initiate activation of the unfolded protein response. Finally, if the protein folding defect is not resolved, chronic activation of the unfolded protein responses can eventually induce apoptosis (reviewed in Ref. 32). Salmonella infections of cultured cells usually lead, in the short or the long term, to apoptosis or other forms of cell death (33–35). In addition we have recently shown that expression of SlrP in HeLa cells makes them more prone to death (12). It is tempting to speculate that the interaction of SlrP with ERdj3, by competing with substrates and promoting accumulation of unfolded proteins in the endoplasmic reticulum, could contribute to cell death as observed in HeLa cells transfected with a plasmid expressing SlrP or infected with Salmonella.
It has been previously shown that S. enterica serovar Typhimurium inhibits antigen presentation by dendritic cells and antigen-dependent T-cell proliferation, and that the SPI2-encoded T3SS is involved in this inhibition (36, 37). Interestingly, SlrP is one of the effectors of this secretion system that are involved in the interference with antigen presentation (38). The interaction of SlrP with ERdj3 and their competition for unfolded substrates could also have a connection with these findings, because endoplasmic reticulum stress impairs major histocompatibility complex class I-peptide presentation (39).
SlrP is an E3 ubiquitin ligase that can use Trx as a substrate (12). The interaction demonstrated in this study suggests that ERdj3 might be another substrate for SlrP-dependent ubiquitination. In fact, ubiquitinated forms of ERdj3 are detected after in vitro reactions using SlrP as an E3 ubiquitin ligase (data not shown). However, in vivo ubiquitination of ERdj3 is unlikely because this is an endoplasmic reticulum lumenal protein and ubiquitination has not been described in this cell compartment. On these grounds, we favor the idea that the role of SlrP in the endoplasmic reticulum is unrelated to its ligase activity. This view is reinforced by the results obtained with the ubiquitin ligase inactive mutant of SlrP in which Cys-546 has been changed to Ala: this mutant is as able as the wild-type SlrP to interact with ERdj3 (Fig. 1) and to interfere with denatured substrate interaction (Fig. 5). In addition, this mutant is also able to increase spontaneous cell death in transfected HeLa cells on reaching confluence, although not at the same level as the wild type (12). A role unrelated to its ubiquitin ligase activity has also been suggested for the C-terminal domain of SspH2, a T3SS Salmonella effector that has similarities with SlrP (8).
Taken together, the data shown above and our previous results studying the interaction of SlrP with Trx (12), our present working model suggests that, after being translocated into the host cell through both the SPI-1- and the SPI-2-dependent T3SS of Salmonella, SlrP promotes death of the cell by two complementary mechanisms: (i) interaction with Trx in the cytosol leads to ubiquitination of this target and causes a decrease in its redox activity; (ii) interaction with ERdj3 in the endoplasmic reticulum interferes, in a ubiquitin ligase independent way, with the function of this chaperone, causing the accumulation of unfolded proteins in this cell compartment. Additionally, this latter effect could explain the SlrP-dependent inhibition of antigen presentation by dendritic cells.
In summary, the present study demonstrate the existence of a second cell host target, ERdj3, for the Salmonella T3SS effector SlrP, and support a model in which SlrP has a role in the endoplasmic reticulum that could have important physiological consequences for the infected cells. The identification of the targets of T3SS effectors and the elucidation of the molecular details of the interactions, as well as of their functional results, are important contributions to the understanding of the mechanisms that these fascinating molecules use to manipulate the host eukaryotic cells behavior.
Acknowledgments
We thank F. Romero (Universidad de Sevilla, Spain) and L. Hendershot (St. Jude Children's Research Hospital, Memphis, TN) for kindly providing some plasmids, F. Romero and J. Casadesús (Universidad de Sevilla, Spain) for critical reading of the manuscript, and V. Goder (Universidad de Sevilla, Spain) for helpful discussions.
This work was supported by Grants SAF2007-60738 from the Spanish Ministry of Science and Innovation and the European Regional Development Fund and P08-CVI-03487 from the Consejería de Innovación, Ciencia y Empresa, Junta de Andalucía, Spain.
- T3SS
- type three secretion system
- SPI
- Salmonella pathogenicity island
- Trx
- thioredoxin
- Km
- kanamycin
- Amp
- ampicillin
- PBS
- phosphate-buffered saline
- PCV
- packed cell volume
- DB
- DNA binding domain
- AD
- activation domain
- NP40
- Nonidet P40
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- SlrP
- Salmonella leucine-rich repeat protein
- cfu
- colony-forming unit.
REFERENCES
- 1.Haraga A., Ohlson M. B., Miller S. I. (2008) Nat. Rev. Microbiol. 6, 53–66 [DOI] [PubMed] [Google Scholar]
- 2.McGhie E. J., Brawn L. C., Hume P. J., Humphreys D., Koronakis V. (2009) Curr. Opin. Microbiol. 12, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsolis R. M., Townsend S. M., Miao E. A., Miller S. I., Ficht T. A., Adams L. G., Bäumler A. J. (1999) Infect. Immun 67, 6385–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao E. A., Miller S. I. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7539–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miao E. A., Scherer C. A., Tsolis R. M., Kingsley R. A., Adams L. G., Bäumler A. J., Miller S. I. (1999) Mol. Microbiol. 34, 850–864 [DOI] [PubMed] [Google Scholar]
- 6.Bella J., Hindle K. L., McEwan P. A., Lovell S. C. (2008) Cell Mol. Life Sci. 65, 2307–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobe B., Kajava A. V. (2001) Curr. Opin. Struct. Biol. 11, 725–732 [DOI] [PubMed] [Google Scholar]
- 8.Quezada C. M., Hicks S. W., Galán J. E., Stebbins C. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rohde J. R., Breitkreutz A., Chenal A., Sansonetti P. J., Parsot C. (2007) Cell Host Microbe 1, 77–83 [DOI] [PubMed] [Google Scholar]
- 10.Singer A. U., Rohde J. R., Lam R., Skarina T., Kagan O., Dileo R., Chirgadze N. Y., Cuff M. E., Joachimiak A., Tyers M., Sansonetti P. J., Parsot C., Savchenko A. (2008) Nat. Struct. Mol. Biol. 15, 1293–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu Y., Li H., Hu L., Wang J., Zhou Y., Pang Z., Liu L., Shao F. (2008) Nat. Struct. Mol. Biol. 15, 1302–1308 [DOI] [PubMed] [Google Scholar]
- 12.Bernal-Bayard J., Ramos-Morales F. (2009) J. Biol. Chem. 284, 27587–27595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherman F., Fink G. R., Hicks J. B. (1986) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 14.Yu M., Haslam R. H., Haslam D. B. (2000) J. Biol. Chem. 275, 24984–24992 [DOI] [PubMed] [Google Scholar]
- 15.Marcus N. Y., Marcus R. A., Schmidt B. Z., Haslam D. B. (2007) Arch. Biochem. Biophys. 468, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y., Zhuang M., Hendershot L. M. (2009) Biochemistry 48, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haraga A., Miller S. I. (2003) Infect Immun 71, 4052–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakanishi K., Kamiguchi K., Torigoe T., Nabeta C., Hirohashi Y., Asanuma H., Tobioka H., Koge N., Harada O., Tamura Y., Nagano H., Yano S., Chiba S., Matsumoto H., Sato N. (2004) Cell Stress Chaperones 9, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martoglio B., Dobberstein B. (1998) Trends Cell Biol. 8, 410–415 [DOI] [PubMed] [Google Scholar]
- 20.Bies C., Guth S., Janoschek K., Nastainczyk W., Volkmer J., Zimmermann R. (1999) Biol. Chem. 380, 1175–1182 [DOI] [PubMed] [Google Scholar]
- 21.Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. (2002) Mol. Biol. Cell 13, 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen Y., Hendershot L. M. (2005) Mol. Biol. Cell 16, 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu M., Haslam D. B. (2005) Infect. Immun. 73, 2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. (2004) J. Mol. Biol. 340, 783–795 [DOI] [PubMed] [Google Scholar]
- 25.Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. (2006) Cell 127, 999–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S. J., Mitra D., Salerno J. R., Hegde R. S. (2002) Dev. Cell 2, 207–217 [DOI] [PubMed] [Google Scholar]
- 27.Johnson S., Michalak M., Opas M., Eggleton P. (2001) Trends Cell Biol. 11, 122–129 [DOI] [PubMed] [Google Scholar]
- 28.Cheetham M. E., Caplan A. J. (1998) Cell Stress Chaperones 3, 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan C. Y., Lee S., Cyr D. M. (2003) Cell Stress Chaperones 8, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Qian X., Sha B. (2003) Structure 11, 1475–1483 [DOI] [PubMed] [Google Scholar]
- 31.Li J., Sha B. (2005) Biochem. J. 386, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malhotra J. D., Kaufman R. J. (2007) Semin Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Collier-Hyams L. S., Zeng H., Sun J., Tomlinson A. D., Bao Z. Q., Chen H., Madara J. L., Orth K., Neish A. S. (2002) J. Immunol. 169, 2846–2850 [DOI] [PubMed] [Google Scholar]
- 34.Fink S. L., Cookson B. T. (2007) Cell Microbiol. 9, 2562–2570 [DOI] [PubMed] [Google Scholar]
- 35.Kim J. M., Eckmann L., Savidge T. C., Lowe D. C., Witthöft T., Kagnoff M. F. (1998) J. Clin. Invest. 102, 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheminay C., Möhlenbrink A., Hensel M. (2005) J. Immunol. 174, 2892–2899 [DOI] [PubMed] [Google Scholar]
- 37.Tobar J. A., Carreño L. J., Bueno S. M., González P. A., Mora J. E., Quezada S. A., Kalergis A. M. (2006) Infect. Immun. 74, 6438–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Halici S., Zenk S. F., Jantsch J., Hensel M. (2008) Infect. Immun. 76, 4924–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granados D. P., Tanguay P. L., Hardy M. P., Caron E., de Verteuil D., Meloche S., Perreault C. (2009) BMC Immunol. 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanahan D. (1983) J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 41.Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]