Abstract
ATP-binding cassette transporter A1 (ABCA1)-mediated lipid efflux to apolipoprotein A1 (apoA-I) initiates the biogenesis of high density lipoprotein. Here we show that the Rho guanine nucleotide exchange factors PDZ-RhoGEF and LARG bind to the C terminus of ABCA1 by a PDZ-PDZ interaction and prevent ABCA1 protein degradation by activating RhoA. ABCA1 is a protein with a short half-life, and apoA-I stabilizes ABCA1 protein; however, depletion of PDZ-RhoGEF/LARG by RNA interference suppressed the apoA-I stabilization of ABCA1 protein in human primary fibroblasts. Exogenous PDZ-RhoGEF expression activated RhoA and increased ABCA1 protein levels and cholesterol efflux activity. Likewise, forced expression of a constitutively active RhoA mutant significantly increased ABCA1 protein levels, whereas a dominant negative RhoA mutant decreased them. The constitutively active RhoA retarded ABCA1 degradation, thus accounting for its ability to increase ABCA1 protein. Moreover, stimulation with apoA-I transiently activated RhoA, and the pharmacological inhibition of RhoA or the dominant negative RhoA blocked the ability of apoA-I to stabilize ABCA1. Finally, depletion of RhoA or RhoGEFs/RhoA reduces the cholesterol efflux when transcriptional regulation via PPARγ is eliminated. Taken together, our results have identified a novel physical and functional interaction between ABCA1 and PDZ-RhoGEF/LARG, which activates RhoA, resulting in ABCA1 stabilization and cholesterol efflux activity.
Keywords: Diseases/Atherosclerosis, Lipid/Cholesterol, Lipoprotein/HDL, Oncogene/Rho, Protein/Protein-Protein Interactions, ABC Transporter
Introduction
Homeostasis of cellular cholesterol is critical for human physiology, and disturbance of the regulatory processes that maintain cholesterol homeostasis can cause cardiovascular disease (1, 2). In particular, high density lipoprotein (HDL)2 cholesterol levels are inversely correlated with coronary heart disease risk in humans and in animal models, and HDL elevation decreases formation and progression of foam cell lesions (3, 4). ATP-binding cassette transporter A1 (ABCA1) mediates the active transfer of excess cholesterol from cells to extracellular apolipoproteins, primarily apolipoprotein A-I (apoA-I), to form nascent HDL particles. The physiological importance of this cholesterol efflux is clearly demonstrated in patients with Tangier disease, a rare genetic condition caused by loss-of-function mutations in ABCA1 that is characterized by a near absence of HDL and a massive deposition of cholesterol esters in peripheral tissues (5–7).
The activity of ABCA1 is regulated both at the transcriptional level and at the post-translational level. We and others have shown that a complex network of protein-protein interactions mediates the post-translational regulation of ABCA1 (8–13). In particular, ABCA1 contains a PDZ protein-binding motif located in the terminal four residues of the cytoplasmic tail of the transporter, and we have previously identified that the PDZ protein β1-syntrophin bound ABCA1 through this motif (8). The ABCA1/β1-syntrophin interaction increased cholesterol efflux by increasing the cell surface expression of ABCA1 and protecting it from degradation. In addition, we also identified that a subunit of the serine palmitoyltransferase enzyme, SPTLC1 (serine palmitoyltransferase long chain base subunit 1), was associated with ABCA1, and this interaction reduced the ABCA1 activity by trapping ABCA1 in the endoplasmic reticulum (9).
Here we show that PDZ-RhoGEF (Rho guanine nucleotide exchange factor 11) and leukemia-associated RhoGEF (LARG, Rho guanine nucleotide exchange factor 12) bind ABCA1 and regulate ABCA1 protein levels and cholesterol efflux activity. The RhoGEFs are known to control the activation of Rho GTPases by stimulating the exchange of GDP for GTP (14, 15). Members of the RhoGEF family have a Dbl homology (DH) domain and a pleckstrin homology (PH) domain that are responsible for the guanine nucleotide exchange factor activity. PDZ-RhoGEF and LARG, together with p115-RhoGEF, constitute a unique subfamily of RhoGEFs characterized by the presence of a regulator of G protein signaling domain, and their guanine nucleotide exchange factor activity is specific for RhoA compared with Rac1 and Cdc42 (16–22). PDZ-RhoGEF and LARG, but not p115-RhoGEF, both have an N-terminal PDZ domain, which allows them to associate with transmembrane proteins or receptors including Plexin-B1 (23–27), insulin-like growth factor (IGF-1) receptor (28), lysophophatidic acid receptor (29), and CD44 (30). Through these interactions PDZ-RhoGEF and LARG couple extracellular signaling processes to the activation of RhoA. Thus, the PDZ-RhoGEF and LARG interaction to ABCA1 implies that 1) ABCA1 may act not only as a lipid transporter but may also act as a receptor for RhoA signaling and 2) RhoA may regulate the activity and expression of ABCA1 protein.
In this report, we have examined the functional significance of the interaction of PDZ-RhoGEF/LARG to ABCA1 on the activity of cholesterol efflux and ABCA1 regulation. Our results suggest a mechanism by which the ABCA1/PDZ-RhoGEF complex couples the binding of apoA-I to ABCA1 to the activation of RhoA and the subsequent inhibition of ABCA1 protein turnover.
EXPERIMENTAL PROCEDURES
Materials
The following reagents were purchased from the indicated suppliers: mouse monoclonal anti-Myc antibody (4A6) (Upstate); rabbit anti-HA (Clontech); Alexa Fluor 555 goat anti-mouse IgG, Lipofectamine 2000 (Invitrogen); M2 anti-FLAG mouse monoclonal agarose-conjugated antibody, mouse anti-β-actin antibody, protease inhibitor mixture, and cycloheximide (Sigma); mouse anti-GTRAP48 (rat PDZ-RhoGEF) (BD Biosciences); mouse monoclonal anti-ABCA1 (Abcam); rabbit polyclonal anti-ABCA1 antibody (Affinity Bioreagents); mouse monoclonal anti-RhoA, cell-permeable C3-transferase, and Rho activation assay biochemistry kit (Cytoskeleton Inc., Denver, CO); anti-calnexin (StressGen Biotechnologies); SMARTpool siRNA duplexes (Dharmacon); apoA-I (Intracell); radionucleotides (PerkinElmer Life Sciences); human dermal fibroblast Nucleofector kit (Amaxa); and TrueBlot anti-rabbit Ig IP beads (eBioscience).
DNA Constructs
The FLAG-tagged ABCA1 and β1-syntrophin constructs have been previously described (31, 32). The cDNA of full-length PDZ-RhoGEF (KIAA0380) was kindly provided by Takahiro Nagase (Kazusa DNA Research Institute). To create the ΔDH/PH mutant of PDZ-RhoGEF cDNA, full-length PDZ-RhoGEF cDNA was digested with EcoNI (there are two cutting sites at either ends of the sequence coding the tandem of DH and PH domain), and the backbone vector was self-ligated. The cDNA coding for human RhoA was amplified by reverse transcription-PCR from total RNA isolated from HeLa cell line. The T19N and Q63L mutants of RhoA were generated by site-directed PCR mutagenesis. The cDNAs were subcloned into pCMV-Tag3 (Stratagene) or pCMV-HA (Clontech) to produce Myc- or HA-tagged versions, respectively. Enhanced green fluorescent protein (EGFP) or red fluorescence protein (RFP) was fused at the N terminus of ABCA1 or PDZ-RhoGEF in pcDNA3.1 backbone. The correct DNA sequences of all constructs were confirmed by DNA sequencing.
Cell Culture
HEK293 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The cDNA transfections were performed using Lipofectamine 2000 reagent. THP-1 cells were maintained RPMI medium containing 10% fetal bovine serum. Differentiation of THP-1 cells was induced by phorbol 12-myristate 13-acetate (PMA) for 72 h. The differentiated cells were cultured in RPMI medium containing 0.2% bovine serum albumin for 24 h and then used for the experiments (33). Primary human dermal fibroblasts (NHDF-Neo, CC-2509) were purchased from Lonza (Walkersville, MD). The cells were maintained in FGM-2 medium with provided supplement (Lonza).
Overlay Assays
The interaction of the ABCA1 or ABCA7 C terminus with the PDZ domains of PDZ-RhoGEF or LARG was analyzed using a biotinylated peptide representing the final 20 amino acids of the ABCA1 (Bio-VDVAVLTSFLQDEKVKESYV) or ABCA7 (Bio-QHPKRVSRFLEDPSSVETVI). The PDZ array membrane (Panomics TransSignalTM PDZ Domain Array IV) that includes affinity purified PDZ domains of PDZ-RhoGEF (GEF11) and LARG (GEF12) spotted in duplicate (total 34 PDZ domains from 24 proteins) were blocked overnight in blocking buffer at 4 °C with gentle agitation. 15 μl of the peptides (50 μm) were mixed with 15 μl of NeutrAvidin-horseradish peroxidase (1 mg/ml; Pierce) for 30 min at 4 °C and diluted into 5 ml of 1× blocking buffer and incubated with the arrays for 2 h at room temperature (final peptide concentration, 150 nm). After washing, the binding of the peptides was detected with enhanced chemiluminescence and quantitated using an Alpha Innotech FluorChem 8800 imager. The binding of full-length PDZ-RhoGEF to the ABCA1 C terminus was determined using the overlay technique as previously described (8). In brief, equal amounts of purified His-tagged bacterial polypeptides representing the last 185 amino acids of ABCA1, with or without the PDZ motif (wild type, Δ4), were separated by SDS-PAGE and transferred to nitrocellulose. Blocked membranes (5% dried milk proteins, 1% bovine serum albumin, 1× phosphate-buffered saline, overnight at 40 °C) were incubated with 200 mg of a total cellular lysates diluted in 20 ml of blocking buffer. The lysates were obtained from 293-EBNA-T cells transfected with cDNA of HA-tagged versions of full-length PDZ-RhoGEF for 2 h at room temperature. After washing (1× phosphate-buffered saline, 0.1% Tween 20), binding of the PDZ-RhoGEF was detected using an anti-HA monoclonal antibody. The gels run in parallel and Coomassie-stained were used to demonstrate equal loading of the wild type and PDZ motif-deleted His-tagged polypeptides.
Immunoprecipitations
cDNA-transfected 293 cells or primary human fibroblasts were lysed in immunoprecipitation buffer (1% Triton X-100, 50 mm HEPES, pH 7.0, 140 mm NaCl, 3 mm MgCl2, 10% glycerol, and 1% protease inhibitor mixture). The lysates were clarified by centrifugation, and ABCA1 interacting proteins complexes were co-precipitated with anti-FLAG M2 monoclonal antibody agarose. For the analysis of protein interactions in primary human fibroblasts, clarified lysates were incubated with rabbit anti-ABCA1 antibody (Affinity Bioreagents). Immunocomplexes were captured with TrueBlot anti-rabbit Ig beads and then analyzed by SDS-PAGE and immunoblotting using the indicated antibodies.
Immunofluorescence
Tet-on 293 cells that inducibly expressing EGFP-tagged ABCA1 were generated (Flp-InTM T-RExTM; Invitrogen). RFP-tagged PDZ-RhoGEF was transiently transfected by LF2000 in cells where EGFP-ABCA1 expression was induced by doxycycline. The cells were grown on coverslips and visualized using confocal microscopy (LSM5 510 META; Zeiss).
Cholesterol Efflux Assays
Cholesterol efflux was measured as previously described (34). In brief, 293 cells were seeded into 24-well poly-d-lysine-coated tissue culture plates and were transfected in triplicate with empty vector or the indicated cDNAs using LF2000. Primary human fibroblasts were transfected with a siRNA pool of four duplex (Dharmacon). The siRNA transfection was performed by nucleofection using a human dermal fibroblast Nucleofector kit (Amaxa Biosystems, Cologne, Germany) according to the manufacturer's protocols. After 24 h of transfection, the cells were incubated in culture medium containing 0.5 μCi/ml [3H]cholesterol for 24 h. The cells were equilibrated in containing 2 mg/ml fatty acid-free bovine serum albumin and then incubated with or without 10 μg/ml apoA-I for 20 h (293) or 6 h (primary human fibroblasts). The medium was collected and cleared of cellular debris by an 800 × g spin for 10 min, the cell layers were dissolved in 0.1 n NaOH, and the percentage of cholesterol efflux was calculated by scintillation counting.
Determination of Activated RhoA
RhoA activation was determined using a Rho activation assay biochemistry kit (Cytoskelton) according to the provided protocol. Briefly, the cells were incubated in serum-free medium for 3 h and then lysed, GTP-bound RhoA was precipitated with Rhotekin-RBD beads, and immunoprecipitated RhoA was detected by immunoblotting using anti-HA antibody or anti-RhoA antibody. For the experiments of the stimulation with apoA-I, 10 μg/ml of apoA-I was added to cells for indicated times after the 3-h serum-free incubation period.
ABCA1 Degradation
293 cells were transfected with ABCA1 and either cDNAs for wild type RhoA, RhoA-CA, RhoA-DN, or empty vector. At 24 h after transfection, cycloheximide (100 μg/ml) was added to block protein synthesis. The amount of ABCA1 in cell lysate was measured by immunoblotting using anti-ABCA1 antibody after the indicated times. The detected signal was directly quantitated on a LAS-3000 imager (Fujifilm).
ABCA1 Stabilization by ApoA-I
PMA-differentiated THP-1 cells were treated with cell-permeable C3-transferase (2 μg/ml) for 4 h and then incubated with apoA-I (100 μg/ml) for 1 h. The siRNA-transfected primary human fibroblasts were incubated with 10 μg/ml apoA-I for 1 h. The cDNA-transfected 293 cells were incubated with cycloheximide in the presence or absence of apoA-I for 4 h. The expression of proteins was detected by indicated antibodies and quantitated on a LAS-3000 imager.
RNA Extraction and Quantitative Real-time PCR
Total RNA was isolated using the RNeasy Mini kit according to the provided protocol (Qiagen). Quantitative real-time RT-PCR was performed in an ABI PRISM 7000 sequence detection system using the one-step RT-PCR Master Mix reagent kit (Applied Biosystems). The ABCA1 primers and probe used were: forward primer, 5′-AGGTTTGGAGATGGTTATACAATAGTTG-3′; reverse primer, 5′-CTTTTAGGACACTTCCCGGAAA-3′; and probe, 5′-FAM-ACGAATAGCAGGCTCCAACCCTGACC-TAMRA-3′. The data were normalized for 18 S rRNA levels and were presented as fold change compared with normalized ABCA1 message levels in the control cells.
Statistical Analysis
Data from cholesterol efflux assays were found to have equal variance and were further compared by a two-tailed Student's t test. Statistical significance were defined by a p of <0.05.
RESULTS
ABCA1 Interacts with PDZ-RhoGEF and LARG
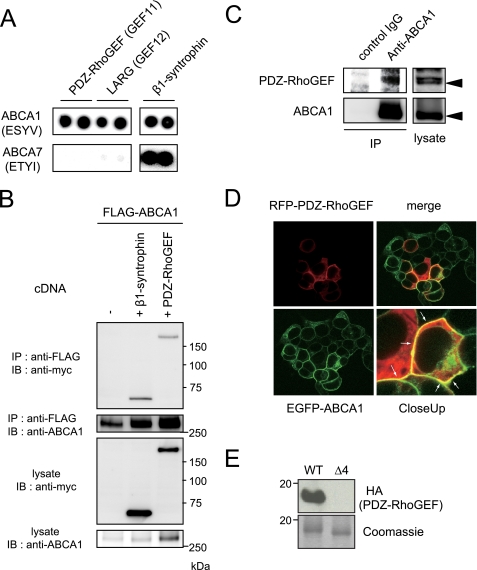
ABCA1 has a highly conserved 46-amino acid C-terminal domain, which resides in the cytoplasmic space and is essential for ABCA1 to bind and transfer lipid to its efflux acceptor apoA-I (31). The final four residues of this domain conform to a type I PDZ protein interaction motif, and we have used mass spectrometry and PDZ protein arrays to screen for proteins that interact with this domain (8). Along with syntrophins these screens indicate PDZ-RhoGEF and LARG may also bind ABCA1 through this motif (8). To further investigate the specificity of the PDZ-RhoGEF and LARG interactions, we used 20-mer biotinylated peptides representing the C terminus of ABCA7, which is a close homologue of ABCA1 (35–38), to probe membranes that had been spotted in duplicate with recombinant proteins of PDZ domains from PDZ-RhoGEF, LARG, and β1-syntrophin expressed in Escherichia coli. The ABCA1 peptide strongly interacted with all the three PDZ domains, whereas the ABCA7 peptide bound PDZ domain from β1-syntrophin but not those from PDZ-RhoGEF and LARG (Fig. 1A), indicating the PDZ domains of PDZ-RhoGEF and LARG specifically bound the ABCA1 C terminus. To investigate whether ABCA1 physically interacts with PDZ-RhoGEF or LARG in a cellular context, we performed immunoprecipitations in cDNA-transfected HEK293 cells. The PDZ-RhoGEF co-precipitated ABCA1, as did β1-syntrophin (Fig. 1B, top panel). A similar interaction was also observed between LARG and ABCA1 (data not shown). Significantly, in these assays PDZ-RhoGEF showed a greater potency than β1-syntrophin to enhance the ABCA1 protein levels (Fig. 1B, bottom panel). We then tested whether endogenous ABCA1 interacts with endogenous PDZ-RhoGEF in primary human fibroblasts, which have been extensively used in studies of ABCA1 function in patients with Tangier disease (39, 40). As shown in Fig. 1C, PDZ-RhoGEF in the primary fibroblasts co-precipitated ABCA1, indicating that ABCA1 and PDZ-RhoGEF form a complex in primary cells at endogenous protein expression levels. Next we observed the subcellular localization of ABCA1 and PDZ-RhoGEF by confocal microscopy. In cells expressing EGFP-tagged ABCA1 and RFP-tagged PDZ-RhoGEF, the proteins prominently co-localized at the cell surface (Fig. 1D, white arrows). To examine the role of the PDZ-binding motif in the interaction between the ABCA1 C terminus and PDZ-RhoGEF, we performed an overlay assay using purified His-tagged bacterial polypeptides representing the last 185 amino acids of the wild type ABCA1 C terminus or a truncated mutant lacking the PDZ-binding motif (Δ4). After SDS-PAGE and transferring these peptides to nitrocellulose, the resulting membranes were incubated with a lysate from 293-EBNA-T cells expressing the full-length HA-tagged PDZ-RhoGEF. PDZ-RhoGEF bound the wild type ABCA1 polypeptide but not the polypeptide lacking the PDZ-binding motif as determined by immunoblotting with anti-HA antibody (Fig. 1E, upper panel). Gels run in parallel and stained for total protein demonstrated the failure of PDZ-RhoGEF to bind the ABCA1-Δ4 polypeptide was not due to a loading artifact (Fig. 1E, lower panel). In aggregate these experiments showed that the ABCA1/PDZ-RhoGEF complex can be detected in primary cells at physiological expression levels and that the PDZ domain of PDZ-RhoGEF directly interacts with the ABCA1 C-terminal PDZ-binding motif.
FIGURE 1.
ABCA1 and PDZ-RhoGEF interact in a cellular context in a manner that requires the ABCA1 C-terminal PDZ-binding motif. A, biotinylated peptides representing the ABCA1 or ABCA7 C termini were used to probe against PDZ domains of PDZ-RhoGEF (GEF11), LARG (GEF12), or β1-syntrophin spotted in duplicate. Binding was detected with avidin-horseradish peroxidase. B, co-precipitation of ABCA1 and PDZ-RhoGEF. 293 cells were co-transfected with FLAG-tagged ABCA1 and either vector, Myc-tagged β1-syntrophin, or PDZ-RhoGEF. The cells were lysed and ABCA1 was immunoprecipitated with anti-FLAG antibody. Shown are Western blots of lysates or immunoprecipitated samples stained with the indicated antibodies. C, the endogenous PDZ-RhoGEF interacts with the endogenous ABCA1 in primary human fibroblasts. Cellular ABCA1 expression was stimulated by LXR and RXR ligands. ABCA1 was immunoprecipitated with anti-ABCA1 antibody. Shown are Western blots of lysates or immunoprecipitated samples stained with the indicated antibodies. D, co-localization of EGFP-tagged ABCA1 and RFP-tagged PDZ-RhoGEF in 293 cells. E, purified His-tagged polypeptides representing the wild-type ABCA1 C terminus with (WT) and without the PDZ motif (Δ4) were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was incubated with lysates expressing HA-tagged PDZ-RhoGEF, and binding was detected using an anti-HA antibody. Coomassie-stained gels run in parallel show equal loading of the ABCA1 peptides. IB, immunoblotting; IP, immunoprecipitation.
PDZ-RhoGEF/LARG Increases ABCA1 Protein Levels
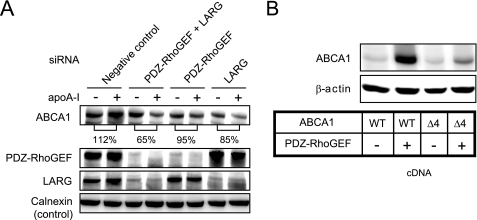
The binding of apoA-I to ABCA1 increases the ABCA1 protein levels by protecting it from calpain-mediated degradation (33, 41). To study the role of PDZ-RhoGEF/LARG in the apoA-I-mediated ABCA1 stabilization, we experimentally down-regulated PDZ-RhoGEF and/or LARG in primary human fibroblasts. We found that the knockdown of these proteins disrupted the ability of apoA-I to increase ABCA1 protein expression (Fig. 2A). Conversely, co-transfection of PDZ-RhoGEF with ABCA1 in 293 cells significantly increased the ABCA1 protein level (Fig. 2B), whereas the ability of PDZ-RhoGEF to enhance ABCA1 protein level was greatly attenuated in cells expressing the Δ4 mutant of ABCA1 (Fig. 2B). ABCA1 protein levels correlate well with cholesterol efflux activity (supplemental Fig. S1A). These results indicate that the binding of PDZ-RhoGEF/LARG to ABCA1 results in increased ABCA1 protein levels.
FIGURE 2.
PDZ-RhoGEF/LARG is associated with the increase of ABCA1 protein levels. A, knockdown of RhoGEF expression by siRNA suppressed the apoA-I-mediated ABCA1 stabilization in primary human fibroblast. The cells were transfected with either a nontargeting pool of siRNA duplexes (negative control), siRNA duplex targeting PDZ-RhoGEF, or LARG, or a mixture of siRNA duplexes targeting both PDZ-RhoGEF and LARG. After 48 h transfection, the cells were incubated with apoA-I for 1 h. The level of ABCA1 protein was quantified using a LAS-3000 imager. B, PDZ-RhoGEF expression increased ABCA1 protein level in cells expressing wild-type (WT) ABCA1 but in cells expressing the ABCA1-Δ4 mutant, which disrupts the ABCA1 PDZ-binding motif. 293 cells were transfected with wild-type or Δ4 mutant of ABCA1 cDNA either alone or together with PDZ-RhoGEF cDNA. The results are representatives of two or more experiments.
RhoA Activation Is Required for PDZ-RhoGEF-mediated Up-regulation of ABCA1 Protein Levels
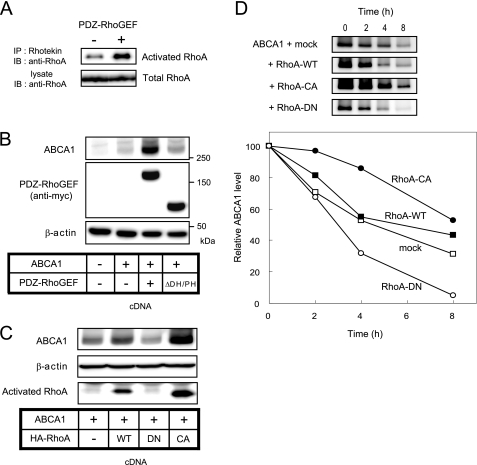
PDZ-RhoGEF facilitates the GDP/GTP exchange to activate RhoA (14, 15). Therefore, we tested whether RhoA was activated in PDZ-RhoGEF-transfected cells. As shown in Fig. 3A, PDZ-RhoGEF expression increased the amount of activated RhoA without changing total RhoA levels. Next, we used a PDZ-RhoGEF mutant lacking the DH and PH domain (ΔDH/PH), which does not have GEF activity. The ability of PDZ-RhoGEF to enhance ABCA1 expression was greatly attenuated in cells expressing the PDZ-RhoGEF mutant (Fig. 3B, ΔDH/PH). The efflux activity was reduced as well (supplemental Fig. S1B, ΔDH/PH). This suggests that the enhancement of the ABCA1 protein level by PDZ-RhoGEF is through a mechanism that activates RhoA. To further investigate whether RhoA activation contributed to the up-regulation of ABCA1, the ABCA1 protein levels were assessed in cells co-expressing ABCA1 and either wild-type RhoA, constitutively active RhoA, dominant negative RhoA, or empty vector. Wild-type RhoA, and more prominently the constitutively active RhoA (HA-RhoA-Q63L, CA), increased the ABCA1 protein level, whereas the dominant negative RhoA (HA-RhoA-T19N, DN) decreased the level of ABCA1 (Fig. 3C). These data indicate that RhoA activation is required for the up-regulation of ABCA1 protein levels mediated by PDZ-RhoGEF.
FIGURE 3.
RhoA activation by PDZ-RhoGEF increase the ABCA1 protein levels by blocking its degradation. A, the expression of PDZ-RhoGEF increases the amount of activated RhoA. The transfected cells were lysed, and activated RhoA was precipitated with Rhotekin-RBD beads. Precipitates or lysates were immunoblotted with anti-RhoA to detect activated and total RhoA, respectively. B, the ability of PDZ-RhoGEF to enhance ABCA1 expression was attenuated in cells expressing PDZ-RhoGEF ΔDH/PH mutant. 293 cells were transfected with vector alone, ABCA1 cDNA alone, or ABCA1 cDNA together with PDZ-RhoGEF cDNA or PDZ-RhoGEF cDNA mutant lacking the DH and PH domain (ΔDH/PH). C, RhoA directly regulates ABCA1 protein expression. The cells were co-transfected with ABCA1 and either HA-tagged wild-type RhoA (WT), a dominant negative form of RhoA (DN), a constitutively active form of RhoA (CA), or with empty vector. The results in A–C are representative of two or more experiments. D, activated RhoA blocks, and inactivated RhoA enhances the ABCA1 degradation. The cells were co-transfected with ABCA1 and wild-type RhoA, RhoA-CA, RhoA-DN, or empty vector. Cycloheximide was added to block protein synthesis 24 h after transfection. The cells were lysed after the indicated times, and the lysates were immunoblotted with anti-ABCA1 antibody. Shown are representative images, and graphed are the average values expressed as percentage of increase of the amount of ABCA1 quantified by LAS-3000 imager in two (mock and RhoA-DN) or three (wild type RhoA and RhoA-CA) separate experiments. IB, immunoblotting; IP, immunoprecipitation.
To investigate whether the activated RhoA increases ABCA1 protein levels by reducing its degradation, ABCA1 protein turnover was monitored. After the addition of cycloheximide to block protein synthesis, ABCA1 protein levels were quantified by immunoblots (Fig. 3D). In cells expressing empty vector, ABCA1 protein was degraded with a short half-life (∼4–5 h), indicating that ABCA1 protein turns over rapidly in 293 cells. In cells expressing wild-type RhoA, ABCA1 decreased by 45% after 4 h of cycloheximide treatment, and that in RhoA-CA cells decreased by 14%, showing that the turnover of ABCA1 was markedly delayed in cells expressing RhoA-CA. On the other hand, ABCA1 level in cells expressing RhoA-DN decreased by 95% at 8 h, whereas more than 40% of ABCA1 still remained in cells expressing wild-type RhoA, thus indicating that ABCA1 turnover was facilitated in cells expressing RhoA-DN. This result indicates that activation of RhoA prevents, and inactivation of RhoA enhances, the degradation of the ABCA1 protein.
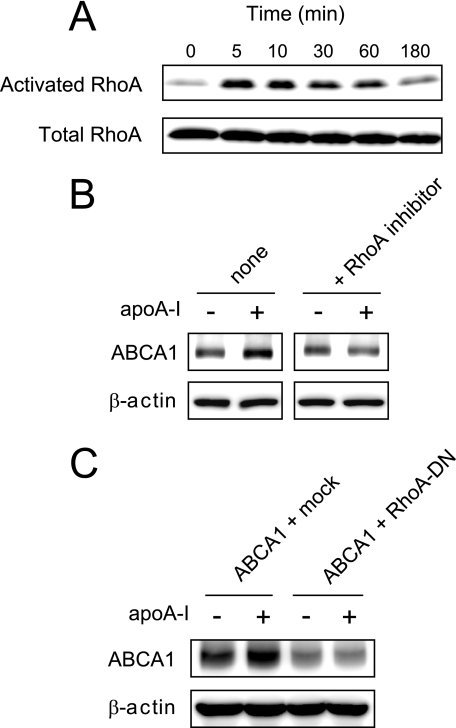
We further investigated the role of RhoA in the apoA-I-mediated ABCA1 up-regulation. apoA-I caused a rapid (peak at 5 min) and transient activation of RhoA in PMA-differentiated THP-1 macrophages (Fig. 4A). In THP-1 cells, the addition of apoA-I led to a stabilization of ABCA1 protein as previously reported (33, 41), and this effect was impaired by pretreatment with exoenzyme C3 transferase, a RhoA selective inhibitor (Fig. 4B). In 293 cells transfected with ABCA1, apoA-I increased the ABCA1 expression, whereas the co-expression of RhoA-DN abolished the ability of apoA-I to increase ABCA1 (Fig. 4C). Thus, apoA-I activates RhoA, and the RhoA activation is required for the apoA-I-mediated stabilization of ABCA1. Collectively, these results indicate that the apoA-I binding and stabilization of ABCA1 is associated with the activation of RhoA, which depends upon the interaction between PDZ-RhoGEF/LARG and ABCA1 C terminus.
FIGURE 4.
ApoA-I-mediated activation of RhoA is critical for ABCA1 protein stabilization. A, apoA-I transiently activates RhoA. PMA-differentiated THP-1 cells were incubated with apoA-I for indicated times. Shown are immunoblots of lysates (total RhoA) or immunoprecipitated samples (activated RhoA) stained with anti-RhoA antibody. B, RhoA selective inhibitor, exoenzyme C3 transferase, abolishes the apoA-I-mediated stabilization of ABCA1. PMA-differentiated THP-1 cells were incubated with apoA-I for 1 h. Prior to adding apoA-I, the cells were pretreated with exoenzyme C3 transferase for 4 h or not. C, the ability of apoA-I to stabilize ABCA1 protein is lost in cells expressing RhoA-DN. The cells were co-transfected with ABCA1 and either empty vector or RhoA-DN. The cells were incubated with cycloheximide in the presence or absence of apoA-I for 4 h. The results are representative of two or more experiments.
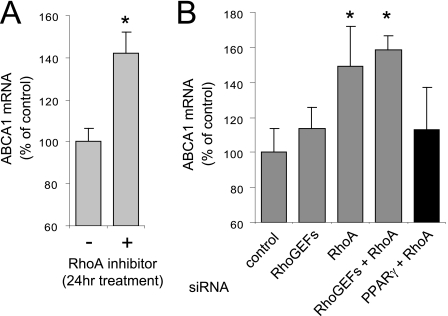
Prolonged Inhibition of RhoA Induces ABCA1 mRNA Expression
In contrast to our results that the transitory activation of RhoA by apoA-I positively regulates ABCA1 protein expression, RhoA inactivation by inhibitors or siRNA results in an increased expression of ABCA1 mRNA by a mechanism that depends on the activity of the nuclear hormone receptor called peroxisome proliferator-activated receptor γ (PPARγ), which is considered to be another mechanism to regulate ABCA1 expression by the activity of RhoA (42, 43). Indeed, we confirmed that ABCA1 mRNA was significantly increased in PMA-differentiated macrophages in which RhoA was inhibited by exoenzyme C3 transferase for 24 h (Fig. 5A). We also examined whether the ABCA1 mRNA expression was increased in the cells treated with siRNAs against RhoGEFs (PDZ-RhoGEF + LARG) and/or RhoA. RhoGEFs siRNA transfection slightly increased the ABCA1 mRNA level, and RhoA or RhoGEF/RhoA (PDZ-RhoGEF + LARG + RhoA) siRNA transfection significantly increased it (Fig. 5B, gray bars). Consistent with the increase of ABCA1 mRNA induced by RhoA inhibition being dependent PPARγ activity, the stimulation of ABCA1 mRNA by silencing of RhoA was attenuated when the expression of PPARγ was simultaneously suppressed (Fig. 5B, black bar). These results indicate that RhoA activation leads to ABCA1 protein stabilization; however, prolonged RhoA inactivation causes the up-regulation of the ABCA1 mRNA levels via PPARγ, indicating two opposing mechanisms that modulate ABCA1 expression by RhoA.
FIGURE 5.
Inhibition of RhoA signaling causes an increase in ABCA1 mRNA. A, RhoA inhibition by exoenzyme C3 transferase increased the ABCA1 mRNA in PMA-differentiated THP-1 macrophages. The cells were treated with exoenzyme C3 transferase for 24 h. mRNA was extracted and measured by the quantitative reverse transcription-PCR. Ratios of the ABCA1 to 18 S transcripts are expressed as percentages of the control treated cells. B, primary human fibroblasts were transfected with either a nontargeting pool of siRNA duplexes (control) or with siRNA duplexes targeting RhoGEFs (PDZ-RhoGEF + LARG), RhoA, RhoGEFs/RhoA (PDZ-RhoGEF + LARG + RhoA), or RhoA/PPARγ. RhoA or RhoGEF/RhoA silencing significantly increase the ABCA1 mRNA (gray bars). The simultaneous knockdown of PPARγ with RhoA attenuated the enhancement of ABCA1 mRNA (black bar). The error bars denote standard deviations of triplicate samples. *, p < 0.05.
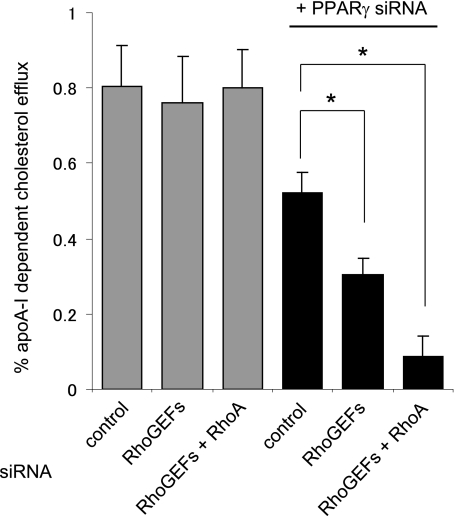
RhoGEF or RhoGEF/RhoA Silencing Decreases the Efflux Activity in PPARγ-depleted Cells
Finally, we examined cholesterol efflux activity in the cells treated with siRNA against RhoGEFs and RhoA. Because the prolonged inactivation by silencing RhoA results in up-regulation of ABCA1 mRNA via PPARγ (Fig. 5B), we measured the cholesterol efflux in the cells in which PPARγ was co-silenced to block the ABCA1 mRNA up-regulation to deconvolute these two opposing effects on ABCA1 expression. Consistent with the down-regulation of ABCA1 protein level (Fig. 2A), silencing of RhoGEFs significantly, and co-silencing of RhoGEFs/RhoA more markedly, reduced the cholesterol efflux mediated by ABCA1 (Fig. 6, black bars). The reduction of cholesterol efflux activity by silencing RhoGEFs/RhoA was not observed when PPARγ expression remained intact (Fig. 6, gray bars). This indicates that in the absence of PPARγ activity, the loss of RhoA activation and the subsequent stabilization of the ABCA1 protein level significantly reduces ABCA1 efflux activity to apoA-I.
FIGURE 6.
RhoA or RhoGEF/RhoA silencing decreases the cholesterol efflux when PPARγ activity is eliminated. In RhoGEF- or RhoGEF/RhoA-depleted primary human fibroblasts, cholesterol efflux was not altered because of mRNA compensation of ABCA1 by RhoA inhibition (gray bars). When PPARγ was simultaneously knocked down to extinguish the mRNA compensation, RhoGEF or RhoGEF/RhoA silencing causes a significant reduction of efflux activity (black bars). The error bars denote standard deviations of triplicate samples. *, p < 0.05.
DISCUSSION
Accumulating evidence suggests that protein-protein interactions in a network play significant roles in regulating ABCA1 activity (8–13). Here we show that PDZ-RhoGEF physically and functionally interacts with ABCA1. The C terminus of ABCA1 contains a strongly conserved type I PDZ-binding motif, and this motif was found to be required for binding to PDZ-RhoGEF. PDZ-RhoGEF or LARG has been reported to bind transmembrane proteins and receptors including plexin-B1, a receptor for Semaphorin 4D. Semaphorin 4D binding to plexin-B1 stimulates RhoA activation, thus mediating repulsive signals in growth cone guidance of neurons (25). LARG and likely PDZ-RhoGEF associate with the IGF-1 receptor, and IGF-1 activates RhoA and downstream effectors via the LARG/IGF-1 receptor complex and thereby regulates cytoskeletal rearrangements (28). To our knowledge, our results are the first to demonstrate a RhoGEF interaction with an ABC transporter, which is of particular interest in terms of the signaling mechanism by which ABCA1 activates RhoA and which led to the stabilization of ABCA1 protein. In this regard, we found that PDZ-RhoGEF activates RhoA, and this is required for the ability of apoA-I to block ABCA1 protein degradation. Together with the previous observations that the apoA-I stimulation prolongs ABCA1 protein half-life by inhibiting a calpain-mediated degradation pathway (33, 41), our findings indicate that the stabilization of ABCA1 protein mediated by apoA-I depends upon the RhoA activation and the ABCA1/RhoGEF complex.
We previously reported that β1-syntrophin binds ABCA1 and enhances cholesterol efflux by increasing the expression of ABCA1 by protecting it from degradation (8). Syntrophins associate with utrophin, a large scaffolding molecule that links to actin cytoskeleton; thus cell surface ABCA1 is likely stabilized by being anchored to the actin cytoskeleton. We demonstrated here that PDZ-RhoGEF or LARG, which binds to ABCA1, mediates RhoA activation and increases the ABCA1 protein level. These results indicate that ABCA1 is stabilized by multiple mechanisms depending upon its interacting partners. The multiple mechanisms for stabilization could be critically important for proteins like ABCA1, which turns over with a short half-life. Indeed, we demonstrated that the knockdown of PDZ-RhoGEF, LARG (Fig. 6, black bars), or β1- or β2-syntrophin (8) in primary human fibroblasts significantly decreased the cholesterol efflux, indicating the importance of these factors for ABCA1 regulation under endogenous expression levels.
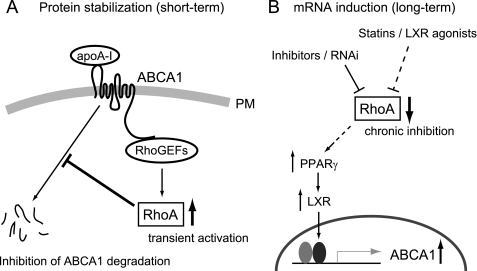
Our data that ABCA1 interacts with PDZ-RhoGEF and that apoA-I induces the RhoA activation suggests ABCA1 is not only functioning as a cholesterol transporter but also as a receptor for apolipoproteins to initiate the RhoA signaling (Fig. 7A). This model is reasonable because apoA-I can directly bind to ABCA1 (32, 44), and apoA-I stimulates signaling of Cdc42, another Rho GTPase, in normal fibroblasts but not in Tangier disease fibroblasts lacking functional ABCA1 (45). Likewise, the cAMP-dependent protein kinase, protein kinase C, and JAK2 kinases are also activated by apoA-I in an ABCA1-dependent manner (46–48). Thus, ABCA1 may act as an apoA-I receptor to transmit signals to the cytoplasm by associating with RhoGEFs, which further modulates the cholesterol efflux process in a feed forward manner by inhibiting the degradation of ABCA1. This mechanism may suggest a model in which the rapid degradation of ABCA1 protein is blocked by the transitory and proximal activation of RhoA during the efflux process when ABCA1 transfers lipid to the bound apoA-I.
FIGURE 7.
Model for the post-translational (A) and transcriptional (B) regulation of ABCA1 by RhoA. A, transient activation of RhoA by apoA-I through the binding to the ABCA1/RhoGEF protein complex blocks ABCA1 protein degradation. B, chronic inhibition of RhoA induces ABCA1 transcription through PPARγ/LXR pathway.
This ABCA1 stabilization is likely to be mediated by associating the transporter with the cytoskeletal network because it has reported that the RhoGTPases regulate cytoskeletal dynamics in response to apoA-I binding (45, 49) and agents that disrupt the cytoskeleton (e.g. cytochalasin D) also affect cholesterol efflux (50). RhoA was transiently activated by apoA-I with a peak at 5 min and then declined to the basal level (Fig. 4A), indicating that RhoA is rapidly deactivated, presumably by the action of RhoGAP proteins (15). Although further work will be needed to validate such a model, these studies may be useful in delineating the potential link between the cytoskeletal dynamics and lipid homeostasis in cells.
Although RhoGEFs stabilize ABCA1 protein in the short term (Fig. 7A), the mechanism by which the RhoGEFs and RhoA influence ABCA1 function is complex because our work and previous reports indicate that chronic inhibition of RhoA can also induce ABCA1 expression at the transcriptional level by a mechanism that depends upon PPARγ (Fig. 7B). This transcriptional effect needs a longer time compared with the ABCA1 stabilization and is indirect in that it occurs through multiple signaling processes including PPARγ and LXR (42, 43). This long term response may be used by cells to dampen inflammatory signaling pathways because ABCA1 can reduce inflammatory cytokine expression by modifying cell surface lipid raft domains independent of apoA-I (51–54) and LXR agonist treatment of mice causes an anti-inflammatory response that is associated with the inactivation of RhoA (55). Hence, in the pathophysiologic conditions such as the chronic inflammatory environment of the atherosclerotic plaque, RhoA could be inactivated for a longer time frame, leading to the transcriptional induction of ABCA1.
In summary, our results show that PDZ-RhoGEF binds to ABCA1 and positively regulates cholesterol efflux via RhoA activation and stabilization of ABCA1 protein. The binding of apoA-I to the ABCA1/RhoGEF complex stimulates RhoA activation, suggesting that ABCA1 may be playing a receptor function in this RhoA signaling pathway. Given the broad distribution of ABCA1, PDZ-RhoGEF, and RhoA among various tissues, this suggests that a wide range of cells may use this pathway to regulate RhoA signaling and ABCA1 efflux activity.
Supplementary Material
Acknowledgments
We greatly appreciate the technical assistance of Dr. Sang-Kee Jung, Dr. Kaori Iwasaki, and Dr. Hong-Yan Cui.
This work was supported by National Institutes of Health Grant HL-074136 (to M. L. F.). This work was also supported by grants-in-aid for scientific research from the Japan Society for the Promotion of Science, by a research fund from Japan Health Sciences Foundation, by a Takeda Science Foundation grant (to K. O.), and by American Heart Association Grant 09GRNT2260352 (to M. L. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- HDL
- high density lipoprotein
- ABCA1
- ATP-binding cassette transporter subfamily A member 1
- apoA-I
- apolipoprotein A-I
- PMA
- phorbol 12-myristate 13-acetate
- DH
- Dbl homology
- PH
- pleckstrin homology
- IGF
- insulin-like growth factor
- HA
- hemagglutinin
- EGFP
- enhanced green fluorescent protein
- RFP
- red fluorescence protein
- siRNA
- small interfering RNA
- CA
- constitutively active
- DN
- dominant negative
- PPAR
- peroxisome proliferator-activated receptor.
REFERENCES
- 1.Oram J. F., Vaughan A. M. (2006) Circ. Res. 99, 1031–1043 [DOI] [PubMed] [Google Scholar]
- 2.Maxfield F. R., Tabas I. (2005) Nature 438, 612–621 [DOI] [PubMed] [Google Scholar]
- 3.Miller N. E., Thelle D. S., Forde O. H., Mjos O. D. (1977) Lancet 1, 965–968 [DOI] [PubMed] [Google Scholar]
- 4.Nissen S. E., Tsunoda T., Tuzcu E. M., Schoenhagen P., Cooper C. J., Yasin M., Eaton G. M., Lauer M. A., Sheldon W. S., Grines C. L., Halpern S., Crowe T., Blankenship J. C., Kerensky R. (2003) J. Am. Med. Assoc. 290, 2292–2300 [DOI] [PubMed] [Google Scholar]
- 5.Brooks-Wilson A., Marcil M., Clee S. M., Zhang L. H., Roomp K., van Dam M., Yu L., Brewer C., Collins J. A., Molhuizen H. O., Loubser O., Ouelette B. F., Fichter K., Ashbourne-Excoffon K. J., Sensen C. W., Scherer S., Mott S., Denis M., Martindale D., Frohlich J., Morgan K., Koop B., Pimstone S., Kastelein J. J., Genest J., Jr., Hayden M. R. (1999) Nat. Genet. 22, 336–345 [DOI] [PubMed] [Google Scholar]
- 6.Bodzioch M., Orsó E., Klucken J., Langmann T., Böttcher A., Diederich W., Drobnik W., Barlage S., Büchler C., Porsch-Ozcürümez M., Kaminski W. E., Hahmann H. W., Oette K., Rothe G., Aslanidis C., Lackner K. J., Schmitz G. (1999) Nat. Genet. 22, 347–351 [DOI] [PubMed] [Google Scholar]
- 7.Rust S., Rosier M., Funke H., Real J., Amoura Z., Piette J. C., Deleuze J. F., Brewer H. B., Duverger N., Denèfle P., Assmann G. (1999) Nat. Genet. 22, 352–355 [DOI] [PubMed] [Google Scholar]
- 8.Okuhira K., Fitzgerald M. L., Sarracino D. A., Manning J. J., Bell S. A., Goss J. L., Freeman M. W. (2005) J. Biol. Chem. 280, 39653–39664 [DOI] [PubMed] [Google Scholar]
- 9.Tamehiro N., Zhou S., Okuhira K., Benita Y., Brown C. E., Zhuang D. Z., Latz E., Hornemann T., von Eckardstein A., Xavier R. J., Freeman M. W., Fitzgerald M. L. (2008) Biochemistry 47, 6138–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munehira Y., Ohnishi T., Kawamoto S., Furuya A., Shitara K., Imamura M., Yokota T., Takeda S., Amachi T., Matsuo M., Kioka N., Ueda K. (2004) J. Biol. Chem. 279, 15091–15095 [DOI] [PubMed] [Google Scholar]
- 11.Hozoji M., Munehira Y., Ikeda Y., Makishima M., Matsuo M., Kioka N., Ueda K. (2008) J. Biol. Chem. 283, 30057–30063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buechler C., Bared S. M., Aslanidis C., Ritter M., Drobnik W., Schmitz G. (2002) J. Biol. Chem. 277, 41307–41310 [DOI] [PubMed] [Google Scholar]
- 13.Bared S. M., Buechler C., Boettcher A., Dayoub R., Sigruener A., Grandl M., Rudolph C., Dada A., Schmitz G. (2004) Mol. Biol. Cell 15, 5399–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 15.Siderovski D. P., Willard F. S. (2005) Int. J. Biol. Sci. 1, 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hart M. J., Jiang X., Kozasa T., Roscoe W., Singer W. D., Gilman A. G., Sternweis P. C., Bollag G. (1998) Science 280, 2112–2114 [DOI] [PubMed] [Google Scholar]
- 17.Kozasa T., Jiang X., Hart M. J., Sternweis P. M., Singer W. D., Gilman A. G., Bollag G., Sternweis P. C. (1998) Science 280, 2109–2111 [DOI] [PubMed] [Google Scholar]
- 18.Fukuhara S., Murga C., Zohar M., Igishi T., Gutkind J. S. (1999) J. Biol. Chem. 274, 5868–5879 [DOI] [PubMed] [Google Scholar]
- 19.Rümenapp U., Blomquist A., Schwörer G., Schablowski H., Psoma A., Jakobs K. H. (1999) FEBS Lett. 459, 313–318 [DOI] [PubMed] [Google Scholar]
- 20.Fukuhara S., Chikumi H., Gutkind J. S. (2000) FEBS Lett. 485, 183–188 [DOI] [PubMed] [Google Scholar]
- 21.Togashi H., Nagata K., Takagishi M., Saitoh N., Inagaki M. (2000) J. Biol. Chem. 275, 29570–29578 [DOI] [PubMed] [Google Scholar]
- 22.Reuther G. W., Lambert Q. T., Booden M. A., Wennerberg K., Becknell B., Marcucci G., Sondek J., Caligiuri M. A., Der C. J. (2001) J. Biol. Chem. 276, 27145–27151 [DOI] [PubMed] [Google Scholar]
- 23.Aurandt J., Vikis H. G., Gutkind J. S., Ahn N., Guan K. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perrot V., Vazquez-Prado J., Gutkind J. S. (2002) J. Biol. Chem. 277, 43115–43120 [DOI] [PubMed] [Google Scholar]
- 25.Swiercz J. M., Kuner R., Behrens J., Offermanns S. (2002) Neuron 35, 51–63 [DOI] [PubMed] [Google Scholar]
- 26.Hirotani M., Ohoka Y., Yamamoto T., Nirasawa H., Furuyama T., Kogo M., Matsuya T., Inagaki S. (2002) Biochem. Biophys. Res. Commun. 297, 32–37 [DOI] [PubMed] [Google Scholar]
- 27.Driessens M. H., Olivo C., Nagata K., Inagaki M., Collard J. G. (2002) FEBS Lett. 529, 168–172 [DOI] [PubMed] [Google Scholar]
- 28.Taya S., Inagaki N., Sengiku H., Makino H., Iwamatsu A., Urakawa I., Nagao K., Kataoka S., Kaibuchi K. (2001) J. Cell Biol. 155, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamada T., Ohoka Y., Kogo M., Inagaki S. (2005) J. Biol. Chem. 280, 19358–19363 [DOI] [PubMed] [Google Scholar]
- 30.Bourguignon L. Y., Gilad E., Brightman A., Diedrich F., Singleton P. (2006) J. Biol. Chem. 281, 14026–14040 [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald M. L., Okuhira K., Short G. F., 3rd, Manning J. J., Bell S. A., Freeman M. W. (2004) J. Biol. Chem. 279, 48477–48485 [DOI] [PubMed] [Google Scholar]
- 32.Fitzgerald M. L., Morris A. L., Rhee J. S., Andersson L. P., Mendez A. J., Freeman M. W. (2002) J. Biol. Chem. 277, 33178–33187 [DOI] [PubMed] [Google Scholar]
- 33.Arakawa R., Yokoyama S. (2002) J. Biol. Chem. 277, 22426–22429 [DOI] [PubMed] [Google Scholar]
- 34.Fitzgerald M. L., Mendez A. J., Moore K. J., Andersson L. P., Panjeton H. A., Freeman M. W. (2001) J. Biol. Chem. 276, 15137–15145 [DOI] [PubMed] [Google Scholar]
- 35.Abe-Dohmae S., Ikeda Y., Matsuo M., Hayashi M., Okuhira K., Ueda K., Yokoyama S. (2004) J. Biol. Chem. 279, 604–611 [DOI] [PubMed] [Google Scholar]
- 36.Kim W. S., Fitzgerald M. L., Kang K., Okuhira K., Bell S. A., Manning J. J., Koehn S. L., Lu N., Moore K. J., Freeman M. W. (2005) J. Biol. Chem. 280, 3989–3995 [DOI] [PubMed] [Google Scholar]
- 37.Jehle A. W., Gardai S. J., Li S., Linsel-Nitschke P., Morimoto K., Janssen W. J., Vandivier R. W., Wang N., Greenberg S., Dale B. M., Qin C., Henson P. M., Tall A. R. (2006) J. Cell Biol. 174, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwamoto N., Abe-Dohmae S., Sato R., Yokoyama S. (2006) J. Lipid Res. 47, 1915–1927 [DOI] [PubMed] [Google Scholar]
- 39.Rogler G., Trümbach B., Klima B., Lackner K. J., Schmitz G. (1995) Arterioscler. Thromb. Vasc. Biol. 15, 683–690 [DOI] [PubMed] [Google Scholar]
- 40.Remaley A. T., Schumacher U. K., Stonik J. A., Farsi B. D., Nazih H., Brewer H. B., Jr. (1997) Arterioscler. Thromb. Vasc. Biol. 17, 1813–1821 [DOI] [PubMed] [Google Scholar]
- 41.Wang N., Chen W., Linsel-Nitschke P., Martinez L. O., Agerholm-Larsen B., Silver D. L., Tall A. R. (2003) J. Clin. Invest. 111, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Argmann C. A., Edwards J. Y., Sawyez C. G., O'Neil C. H., Hegele R. A., Pickering J. G., Huff M. W. (2005) J. Biol. Chem. 280, 22212–22221 [DOI] [PubMed] [Google Scholar]
- 43.Yano M., Matsumura T., Senokuchi T., Ishii N., Murata Y., Taketa K., Motoshima H., Taguchi T., Sonoda K., Kukidome D., Takuwa Y., Kawada T., Brownlee M., Nishikawa T., Araki E. (2007) Circ. Res. 100, 1442–1451 [DOI] [PubMed] [Google Scholar]
- 44.Wang N., Silver D. L., Thiele C., Tall A. R. (2001) J. Biol. Chem. 276, 23742–23747 [DOI] [PubMed] [Google Scholar]
- 45.Nofer J. R., Remaley A. T., Feuerborn R., Wolinnska I., Engel T., von Eckardstein A., Assmann G. (2006) J. Lipid Res. 47, 794–803 [DOI] [PubMed] [Google Scholar]
- 46.Yamauchi Y., Hayashi M., Abe-Dohmae S., Yokoyama S. (2003) J. Biol. Chem. 278, 47890–47897 [DOI] [PubMed] [Google Scholar]
- 47.Tang C., Vaughan A. M., Oram J. F. (2004) J. Biol. Chem. 279, 7622–7628 [DOI] [PubMed] [Google Scholar]
- 48.Haidar B., Denis M., Marcil M., Krimbou L., Genest J., Jr. (2004) J. Biol. Chem. 279, 9963–9969 [DOI] [PubMed] [Google Scholar]
- 49.Nofer J. R., Feuerborn R., Levkau B., Sokoll A., Seedorf U., Assmann G. (2003) J. Biol. Chem. 278, 53055–53062 [DOI] [PubMed] [Google Scholar]
- 50.Walter M., Forsyth N. R., Wright W. E., Shay J. W., Roth M. G. (2004) J. Biol. Chem. 279, 20866–20873 [DOI] [PubMed] [Google Scholar]
- 51.Yvan-Charvet L., Ranalletta M., Wang N., Han S., Terasaka N., Li R., Welch C., Tall A. R. (2007) J. Clin. Invest. 117, 3900–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Francone O. L., Royer L., Boucher G., Haghpassand M., Freeman A., Brees D., Aiello R. J. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 1198–1205 [DOI] [PubMed] [Google Scholar]
- 53.Koseki M., Hirano K., Masuda D., Ikegami C., Tanaka M., Ota A., Sandoval J. C., Nakagawa-Toyama Y., Sato S. B., Kobayashi T., Shimada Y., Ohno-Iwashita Y., Matsuura F., Shimomura I., Yamashita S. (2007) J. Lipid Res. 48, 299–306 [DOI] [PubMed] [Google Scholar]
- 54.Zhu X., Lee J. Y., Timmins J. M., Brown J. M., Boudyguina E., Mulya A., Gebre A. K., Willingham M. C., Hiltbold E. M., Mishra N., Maeda N., Parks J. S. (2008) J. Biol. Chem. 283, 22930–22941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smoak K., Madenspacher J., Jeyaseelan S., Williams B., Dixon D., Poch K. R., Nick J. A., Worthen G. S., Fessler M. B. (2008) J. Immunol. 180, 3305–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.