Abstract
We have reconstituted human mitochondrial transcription in vitro on DNA oligonucleotide templates representing the light strand and heavy strand-1 promoters using protein components (RNA polymerase and transcription factors A and B2) isolated from Escherichia coli. We show that 1 eq of each transcription factor and polymerase relative to the promoter is required to assemble a functional initiation complex. The light strand promoter is at least 2-fold more efficient than the heavy strand-1 promoter, but this difference cannot be explained solely by the differences in the interaction of the transcription machinery with the different promoters. In both cases, the rate-limiting step for production of the first phosphodiester bond is open complex formation. Open complex formation requires both transcription factors; however, steps immediately thereafter only require transcription factor B2. The concentration of nucleotide required for production of the first dinucleotide product is substantially higher than that required for subsequent cycles of nucleotide addition. In vitro, promoter-specific differences in post-initiation control of transcription exist, as well as a second rate-limiting step that controls conversion of the transcription initiation complex into a transcription elongation complex. Rate-limiting steps of the biochemical pathways are often those that are targeted for regulation. Like the more complex multisubunit transcription systems, multiple steps may exist for control of transcription in human mitochondria. The tools and mechanistic framework presented here will facilitate not only the discovery of mechanisms regulating human mitochondrial transcription but also interrogation of the structure, function, and mechanism of the complexes that are regulated during human mitochondrial transcription.
Keywords: Mitochondrial DNA, RNA Polymerase, Transcription Initiation Factors, Transcription Promoter, Transcription Regulation, h-mtRNAP (POLRMT), h-mtTFA, h-mtTFB2, Human Mitochondrial Transcription, Open Complex Formation
Introduction
Expression and maintenance of the mitochondrial genome (mtDNA) are essential for mitochondria to produce energy, function in other aspects of intermediary metabolism, contribute to pathogen sensing, and integrate into signal-transduction pathways, especially those regulated by circadian rhythm and nutritional status (1, 2). Mitochondrial dysfunction caused by defects in mtDNA replication and translation attributable to mutations in mitochondrial and nuclear genes contributes to numerous diseases, including muscular dystrophies, cardiac diseases, neurodegenerative disorders, diabetes, and certain cancers (3–8). A link between the cis- and trans-acting factors required for mitochondrial transcription and disease has yet to be established, likely because of limited information on the mechanism and regulation of mitochondrial transcription (1, 4, 9–11).
Human mtDNA is a 16,569-bp, double-stranded, circular DNA that encodes 13 proteins essential for oxidative phosphorylation, as well as the 2 ribosomal RNAs and 22 transfer RNAs needed for production of these proteins in mitochondria (11, 12). The control (or D-loop) region of mtDNA contains cis-acting sequences required for genome replication and transcription. Three promoters exist in the control region as follows: the light strand promoter (LSP)2 and heavy strand promoters (HSP) 1 and 2 (11, 12). These promoters give rise to polycistronic RNAs that are further processed for proper gene expression (11). HSP1 is located upstream of HSP2 and directs the synthesis of the two ribosomal RNAs. Transcription from HSP1 is terminated at the end of the 16 S rRNA gene by the action of a terminator whose function requires binding by the mitochondrial termination factor (13). Premature termination of transcription initiated from LSP is caused by a conserved, GC-rich sequence referred to as conserved sequence box II (CSBII), and this product may be the primer for replication of the leading strand of mtDNA (14).
The human mitochondrial transcription machinery consists of the mitochondrial RNA polymerase (h-mtRNAP or POLRMT), which is related to the single-subunit RNA polymerase of bacteriophage T7, mitochondrial transcription factor A (h-mtTFA also known as TFAM), and mitochondrial transcription factor B1 or B2 (h-mtTFB1, h-mtTFB2). The h-mtRNAP is a 130-kDa protein with a carboxyl-terminal core polymerase domain that exhibits homology to T7 RNAP and an amino-terminal domain that is unique to T7-RNAP-like enzymes that require transcription factors for function (11). The amino-terminal domain contains pentatricopeptide repeats, which are known to bind RNA, that have been proposed to couple transcription to translation (4, 11, 15). The h-mtTFA is a member of the high mobility group family of proteins (16), which bind, bend, and wrap DNA (17). Binding of h-mtTFA to promoters LSP and HSP1 upstream of the transcription start site (TSS) has been demonstrated by DNase I footprinting (18). In addition to transcription initiation, h-mtTFA serves a role in DNA packaging and maintenance of mtDNA (19, 20). h-mtTFB1 and h-mtTFB2 are two homologous proteins, with sequence homology to ribosomal RNA dimethyltransferases (21, 22). h-mtTFB2 has been reported to have substantially higher, transcription-stimulating activity in vitro than h-mtTFB1 (21). However, studies in vivo only support a role for mtTFB2 in transcription, with mtTFB1 contributing to translation (23–26). Recently, it was shown that the amino-terminal region of h-mtTFB2 interacts with the TSS, stabilizing the melted promoter and the initiating nucleotide (27).
Human mitochondrial transcription has been studied for quite some time on plasmid-based templates by using cell-free extracts and proteins purified from eukaryotic expression systems (21, 28, 29). All of these studies have used production of run-off transcription products to report on events occurring during transcription initiation, and most of these studies have focused on one or two promoters in isolation. Very little is known about the structure-function relationships of the protein components of human mitochondrial transcription (27, 29, 30), likely a reflection of the low yields and intense labor associated with production of recombinant proteins in eukaryotic systems.
Here we show that post-translational modifications of the human mitochondrial transcription machinery are not required for specific and efficient transcription initiation in vitro as the proteins produced in Escherichia coli appear to be completely active. LSP- and HSP1-based DNA oligonucleotide templates can be used to monitor transcription initiation and/or transcription elongation. Using this system, we show for the first time that the transcription factors and polymerase are required in a 1:1:1 stoichiometry relative to template for assembly of a functional transcription complex. Formation of the first dinucleotide is limited by open complex formation and requires high concentrations of ATP. Open complex formation requires both transcription factors A and B2, although only B2 is required after open complex formation. Interestingly, both factor requirements and related events early in the transcription cycle are promoter-dependent, consistent with differential regulation of the various promoters as suggested by others (18, 21, 31, 32). This study therefore provides the most detailed description of the human mitochondrial transcription to date and establishes a solid foundation for future studies of the structure, function, mechanism, and regulation of human mitochondrial transcription.
EXPERIMENTAL PROCEDURES
Materials
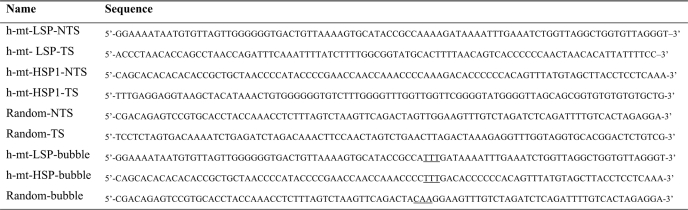
DNA oligonucleotides were from Integrated DNA Technologies (Table 1). RNA oligonucleotides were obtained from Dharmacon. Plasmids containing the human mtDNA promoters pUC18-LSP, pUC18-HSP1, and pUC18-LSP-HSP1 were a kind gift from Claes M. Gustaffson (21). Restriction endonucleases were from New England Biolabs. T4 polynucleotide kinase was from United States Biochemical Corp. RQ1 DNase was from Promega. Ultrapure NTP solutions were from GE Healthcare. [α-32P]UTP (3000 Ci/mmol) and [α-32P]ATP (3000 Ci/mmol) were from PerkinElmer Life Sciences. [γ-32P]ATP (7000 Ci/mmol) was from MP Biomedical. HPLC-purified [γ-32P]ATP (6000 Ci/mmol) was from MP Biomedical. KMnO4, tRNA, and piperidine were from Sigma. A 10-bp DNA ladder was from Invitrogen. Heparin (average molecular weight of 6000 Da) was from Sigma. All other reagents were of the highest grade available from Sigma, Fisher, or VWR Scientific.
TABLE 1.
Oligonucleotides used in this study
Recombinant Human Mitochondrial Transcription Proteins
The proteins used to perform this study can be obtained from Enzymax, LLC (Lexington, KY). Expression and purification of h-mtRNAP (amino acids 42–1230 of accession number NM_005035.2) will be published elsewhere. Expression and purification of h-mtTFA (amino acids 42–246 of accession number NM_003201.1) have been reported (30, 33). Expression and purification of h-mtTFB2 (amino acids 31–396 of accession number NM_022366.1) followed a strategy similar to that for h-mtRNAP, the details of which will be published elsewhere. A related strategy was also recently reported (27).
Purification of DNA Oligonucleotides
DNA oligonucleotides were purified by denaturing PAGE as described previously (34). Briefly, oligonucleotides were resolved by denaturing PAGE on 6% gels (37:3, acrylamide:bisacrylamide ratio). The oligonucleotide ladder was visualized by UV shadowing; a gel slice containing only the full-length oligonucleotide was removed, and the nucleic acid was electroeluted from the gel in 1× TBE (89 mm Tris base, 89 mm boric acid, and 2 mm EDTA) by using an Elutrap apparatus (Schleicher & Schuell). Oligonucleotides were desalted on Sep-Pak columns (Millipore) as specified by the manufacturer and suspended in 10 mm Tris-HCl, 0.1 mm EDTA. Concentrations were determined by measuring the absorbance at 260 nm using a Nanodrop spectrophotometer and the appropriate calculated extinction coefficient.
5′-32P Labeling of DNA and RNA Oligonucleotides
DNA oligonucleotides were end-labeled by using [γ-32P]ATP (7000 Ci/mmol) and T4 polynucleotide kinase. Reactions, typically 50 μl, contained 1× T4 polynucleotide kinase buffer provided by the manufacturer, 1 μm [γ-32P]ATP, 50 μm DNA oligonucleotide, and 0.4 unit/μl T4 polynucleotide kinase. Reactions were incubated at 37 °C for 60 min and then placed at 65 °C for 10 min to heat-inactivate T4 polynucleotide kinase. The dinucleotide RNA pApA was end-labeled by using HPLC-purified [γ-32P]ATP (6000 Ci/mmol) and T4 polynucleotide kinase by using an exchange reaction. Reactions, typically 20 μl, contained 1× exchange buffer (50 mm imidazole, pH 6.4, 12 mm MgCl2, 1 mm 2-mercaptoethanol, and 70 μm ADP), 0.36 μm HPLC-purified [γ-32P]ATP, 10 μm pApA, and 0.4 unit/μl T4 polynucleotide kinase. Reactions were incubated at 37 °C for 60 min and then placed at 65 °C for 10 min to heat-inactivate T4 polynucleotide kinase. Labeling of the DNA ladder was performed by using [γ-32P]ATP and T4 polynucleotide kinase as specified by Invitrogen.
Annealing of DNA Oligonucleotides
For each pair of oligonucleotides, the nontemplating and templating strand (top and bottom strand) were annealed at 25 μm in 10 mm Tris-HCl, pH 8.0, 0.1 mm EDTA, and 50 mm NaCl by using a Progene Thermocycler (Techne). Annealing reactions were heated to 90 °C for 1 min and slowly cooled to 10 °C at a rate of 5 °C/min. Annealing was confirmed by native 6% PAGE.
Linearization of pUC18 Plasmids
Linearization reactions contained 1–5 μg of pUC18 plasmid DNA. Digestions were performed as specified by New England Biolabs. After linearization, reactions were placed at 65 °C for 10 min to heat-inactivate the restriction endonuclease prior to use.
In Vitro Transcription Reactions Using Plasmids as Templates
Reactions contained 1× reaction buffer (10 mm HEPES, pH 7.5, 20 mm NaCl, 10 mm MgCl2, 1 mm DTT, and 0.1 μg/μl bovine serum albumin), 4 nm linearized pUC18 plasmid DNA, 1× NTP mix (400 μm ATP, 150 μm CTP, 150 μm GTP, 10 μm UTP, 0.2 μCi/μl [α-32P]UTP), 100 nm h-mtTFA, 20 nm h-mtTFB2, and 20 nm h-mtRNAP (final concentrations). Reactions were performed by incubating linearized plasmid DNA in reaction buffer at 32 °C for 5 min and then adding in the following order: h-mtTFA, h-mtTFB2, and h-mtRNAP. Between each addition of protein to the reaction, there was an incubation time of 1 min. After addition of h-mtRNAP, the reaction was allowed to incubate at 32 °C for 5 min and then initiated by addition of NTP mix. The reaction was allowed to proceed for 30 min and then quenched by addition of stop buffer (35% formamide, 0.0125% bromphenol blue, 0.0125% xylene cyanol, and 50 mm EDTA final). Products were resolved by denaturing 5% (37:3, acrylamide:bisacrylamide ratio) PAGE. Proteins were diluted immediately prior to use in 10 mm HEPES, pH 7.5, 1 mm DTT, and 20% glycerol. The volume of protein added to any reaction was always less than or equal to one-tenth the total volume. Any deviations from the above are indicated in the appropriate figure legend.
In Vitro Transcription Using Oligonucleotides as Templates
Reactions were performed essentially as described above for using plasmids as templates, except for the following modifications. Reactions contained 10 mm HEPES, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 1 mm DTT, 0.1 μg/μl bovine serum albumin, 2.5 μm DNA oligonucleotide duplex, 500 μm ATP, 500 μm CTP, 500 μm GTP, 500 μm UTP, 0.2 μCi/μl [α-32P]ATP, 2.5 μm h-mtTFA, 2.5 μm h-mtTFB2, and 2.5 μm h-mtRNAP (final concentrations). Where indicated, reactions contained 10 μm 32P-labeled pApA dinucleotide primer and/or 10 μm heparin as trap. In reactions that contained 32P-labeled pApA, [α-32P]ATP was not included. Reactions were assembled, initiated as described above, and quenched at various times by addition of stop buffer (35% formamide, 0.0125% bromphenol blue, 0.0125% xylene cyanol, and 50 mm EDTA final). Products were resolved by denaturing 28% (25:3, acrylamide:bisacrylamide ratio) PAGE. Proteins were diluted immediately prior to use in 10 mm HEPES, pH 7.5, 1 mm DTT, and 20% glycerol. The volume of protein added to any reaction was always less than or equal to one-tenth the total volume. Any deviations from the above are indicated in the appropriate figure legend.
DNase I Footprinting
Reactions were assembled as described above for in vitro transcription reactions using oligonucleotides as templates but instead using 2.5 μm 32P-labeled DNA oligonucleotide duplex. The 32P-labeled DNA duplex contained only one of the DNA strands labeled in a given reaction (template or nontemplate strand). Reactions were assembled as described above and then initiated by addition of RQ1 DNase (0.002 units/μl) and CaCl2 (1 mm). The reaction was allowed to proceed for 2 min and then quenched by stop/trap buffer (35% formamide, 0.0125% bromphenol blue, 0.0125% xylene cyanol, 50 mm EDTA final) and 25 μm trap strand final. The trap strand is unlabeled DNA oligonucleotide that is of the same sequence of the 32P-labeled DNA oligonucleotide in the reaction. This is necessary for improved resolution of the DNA fragments during PAGE (35). Quenched reactions were heated to 90 °C for 1 min and slowly cooled to 10 °C at a rate of 5 °C/min using a Progene Thermocycler prior to PAGE. Products were resolved by denaturing PAGE on 8% gels (37:3, acrylamide:bisacrylamide ratio). DNase I patterns were quantified using the line tool in ImageQuant 5.2. The data were normalized to the intensity of the band corresponding to position −40 and plotted using Excel.
Potassium Permanganate (KMnO4) Footprinting
Reactions were assembled as described above for DNase I footprinting, in a buffer modified to contain only 0.1 mm DTT. Reactions were initiated by addition of KMnO4 (30 mm), allowed to proceed for 2 min at 32 °C, and then quenched by addition of KMnO4 stop/quench buffer (100 mm β-mercaptoethanol, 0.1 μg/μl tRNA, and 0.3 m sodium acetate final concentrations). The quenched reactions were ethanol-precipitated by adding 200 μl of 100% ethanol, placed on dry ice for 10 min, and centrifuged 5 min at 16,000 × g. The pellet was washed with 200 μl of 70% ethanol, centrifuged again, dried, and suspended in 30 μl of 1 m piperidine. The piperidine cleavage reaction was performed at 90 °C for 30 min using a Progene Thermocycler. After 30 min, 50 μl of 100% butanol was added and centrifuged for 5 min at 16,000 × g. The organic phase was discarded, and the aqueous phase was ethanol-precipitated as described above. The pellet was suspended in 90% formamide, 0.0125% bromphenol blue, and 0.0125% xylene cyanol and resolved by denaturing 8% (37:3, acrylamide:bisacrylamide ratio) PAGE.
Product Analysis, Denaturing PAGE and Quantitation
The quenched reaction mixtures were heated to 70 °C for 5 min prior to loading 5 μl on a denaturing polyacrylamide gel containing 1× TBE (89 mm Tris base, 89 mm boric acid, 2 mm EDTA) and 7 m urea. Electrophoresis was performed in 1× TBE at 85 watts. Gels were exposed to a PhosphorImager screen (GE Healthcare) and scanned using a Typhoon 9410 (GE Healthcare). Gels were visualized by using a phosphorimager and quantitated by using the ImageQuant software (GE Healthcare). The amount of RNA product was determined by dividing the volume of each product by the total volume in each lane and multiplying by the total concentration (labeled and unlabeled) of the 32P-labeled species in the reaction. Bar graphs and time courses were produced by using GraphPad Prism version 4 (GraphPad software). Each graph is representative of at least two independent experiments.
RESULTS AND DISCUSSION
Human Mitochondrial Transcription Proteins Produced in E. coli Support Promoter- and Factor-dependent Transcription in Vitro
Our current understanding of the biochemistry of human mitochondrial transcription derives primarily from studies performed in cell-free extracts (10–12). Reconstitution of human mitochondrial transcription by using purified proteins has only been reported for proteins produced in a eukaryotic system, the baculovirus/insect cell expression system (21), although active human mitochondrial transcription factor A (h-mtTFA, also known as TFAM) can be produced in bacteria (30, 33). Eukaryotic expression systems are generally more costly and produce less proteins than prokaryotic systems. Our success with producing the human mitochondrial DNA-dependent RNA polymerase (h-mtRNAP, also known as POLRMT) in E. coli motivated us to pursue production of transcription factors A, B1, and B2 (h-mtTFA, also known as TFAM; h-mtTFB1, also known as TFB1M; h-mtTFB2, also known as TFB2M) in E. coli. All three proteins can be isolated from E. coli in a soluble form (Fig. 1a) (data not shown) free of contaminants such as nucleases and phosphatases that would interfere with biochemical analysis of transcription. Transcription was not supported by mtTFB1 (data not shown). Additional studies will be required to verify that the protein is appropriately folded, so this factor was not studied further.
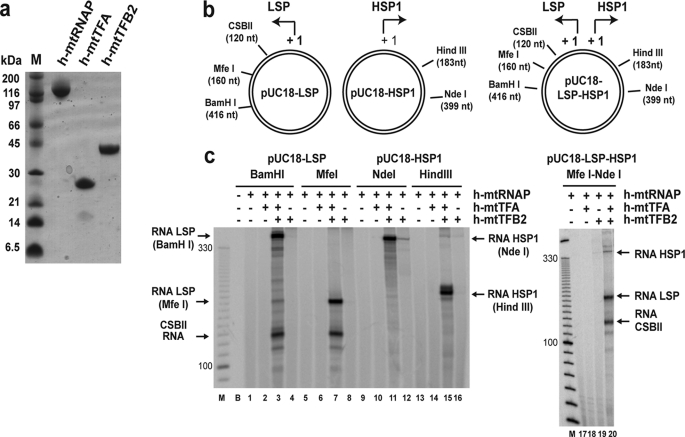
FIGURE 1.
Human mitochondrial transcription machinery produced in E. coli supports promoter-dependent transcription on established plasmid-based templates in vitro. a, recombinant human mitochondrial transcription proteins used in this study. Proteins (5 μg) were resolved by SDS-PAGE on a 10% gel and visualized by Coomassie staining. b, plasmid-based templates for in vitro transcription. Schematic of pUC18-LSP, pUC18-HSP1, and pUC18-LSP-HSP1 plasmids used in this study (21). The position of unique restriction sites and the sizes (in nucleotides, nt) of the corresponding run-off transcripts are indicated. A 120-nt transcript will be produced from LSP regardless of the restriction enzyme used because of premature termination caused by the conserved sequence block II (CSBII). c, transcription assays. Reactions were performed by combining h-mtTFA (100 nm) and h-mtTFB2 (20 nm) with linearized plasmid DNA (4 nm) in reaction buffer containing NTP mix (400 μm ATP, 150 μm CTP, 150 μm GTP, 10 μm UTP, and 0.2 μCi/μl [α-32P]UTP) at 32 °C, initiated by addition of h-mtRNAP (20 nm), and quenched after 30 min by addition of stop buffer. Products were resolved by denaturing PAGE on 5% gels and visualized by phosphorimaging. The size of run-off transcripts produced from the different linearized plasmid templates is indicated. The size of selected bands from a 10-bp DNA ladder (M) is indicated as a reference.
We were concerned that proteins produced in E. coli might lack post-translational modifications that are critical for function, given that the only success to date has required use of a baculovirus/insect cell expression system (21). Therefore, the objective of our initial experiments was to validate the function of proteins by using well established templates for transcription (21, 29). Plasmids containing human mitochondrial LSP and/or HSP1 were linearized by restriction enzyme digestion at unique sites (Fig. 1b). Mitochondrial transcription proteins were combined with the linearized transcription template, and in the presence of NTPs, run-off transcripts of the expected length were produced in all cases (Fig. 1c).
We first used the pUC18-LSP template digested with BamHI (418-nt product) or MfeI (160-nt product). In both cases, when h-mtRNAP was incubated in the reaction together with h-mtTFA and h-mtTFB2, a run-off transcript of the expected size was observed (Fig. 1c, lanes 3 and 7), suggesting that the bacterially expressed proteins are capable of recognizing the LSP promoter and the appropriate start site therein. The primer for mtDNA replication is produced by termination of transcripts initiated from LSP in a region referred to as CSBII (14). The CSBII-terminated product (120 nt) was observed independent of the site of template linearization (Fig. 1c, lanes 3 and 7), suggesting that the bacterially produced proteins are capable of sensing and responding to the CSBII termination signal. Transcription from LSP was dependent on the polymerase and both transcription factors as omission of any one of these proteins prevented production of detectable levels of either CSBII-terminated or run-off transcript (Fig. 1c, compare lane 3 with lanes 1, 2, and 4 and lane 7 with lanes 5, 6, and 8), suggesting that bacterially produced polymerase exhibits the appropriate factor dependence for recognition of and initiation and elongation on LSP.
We also validated the human mitochondrial proteins produced in E. coli on HSP1. Consistent with the results obtained for LSP, run-off transcription products of the expected size were made by the polymerase on HSP1 (Fig. 1c, lanes 11 and 15) in a manner that was most efficient when both transcription factors were present (Fig. 1c, compare lane 11 with lanes 9, 10, and 12 and lane 15 with lanes 13, 14, and 16). In the absence of h-mtTFA, some run-off product was reproducibly detected (Fig. 1c, lanes 12 and 16). This observation is quite intriguing and has been further explored as part of another study.3
To rule out the remote possibility that the observations made above were coincidental, we utilized a plasmid lacking the mitochondrial promoter sequences as DNA template for in vitro transcription. This template failed to produce products similar to those obtained using LSP or HSP1, although some RNA products could be produced by the polymerase in a transcription factor-dependent manner (supplemental Fig. S1).
There has been some suggestion that LSP is a stronger promoter than HSP1 (18, 21). The data presented above do not reveal a substantial difference in utilization of the two promoters. Previous studies that have observed preferential utilization of LSP have been performed in the context of dual promoter constructs. Therefore, we utilized a transcription template that contained LSP, HSP1, and the natural inter-promoter DNA sequence (Fig. 1b, pUC18-LSP-HSP1) (21). When transcription templates were produced by digesting the dual promoter construct with MfeI (120- and 160-nt products from LSP) and NdeI (399-nt product from HSP1), the three RNA transcripts of the predicted size were observed (Fig. 1c, lane 20), the synthesis of which required both transcription factors (Fig. 1c, compare lane 20 with lanes 17–19). In this context, utilization of LSP is substantially more efficient (Fig. 1c, lane 20), suggesting that the bacterially produced proteins recapitulate all of the phenomena reported to date regarding transcription initiation from LSP and HSP1. The basis for the enhanced efficiency of LSP relative to HSP1 is unclear but may be important in terms of the differential regulation of these promoters in vivo (10, 21, 36).
It is important to note that conditions in which our experiments were performed were nearly identical to the conditions reported for the proteins produced using the baculovirus/insect cell expression system (21). It is important to note that the concentrations of proteins used are at least 5-fold higher than that for plasmid DNA template. The requirement for an excess of protein relative to promoter is likely due to the abundance of nonspecific DNA. To a first approximation, the yield of RNA was equivalent to that observed previously. Therefore, the activity of our proteins is likely identical to proteins derived from the eukaryotic system. We conclude that post-translational modifications are not required for assembly or function of the transcription components, at least under the conditions employed here, which essentially provide a long period of time for assembly of the initiation complex.
Transcripts Produced under Standard Assay Conditions Derive from Multiple Rounds of Transcription Initiation
Our understanding of human mitochondrial transcription initiation derives from biochemical experiments that monitor products of transcription elongation formed, most likely, after multiple rounds of transcription initiation (21, 29, 30, 32, 37). Therefore, we sought a more direct approach to interrogate initiation of mitochondrial transcription.
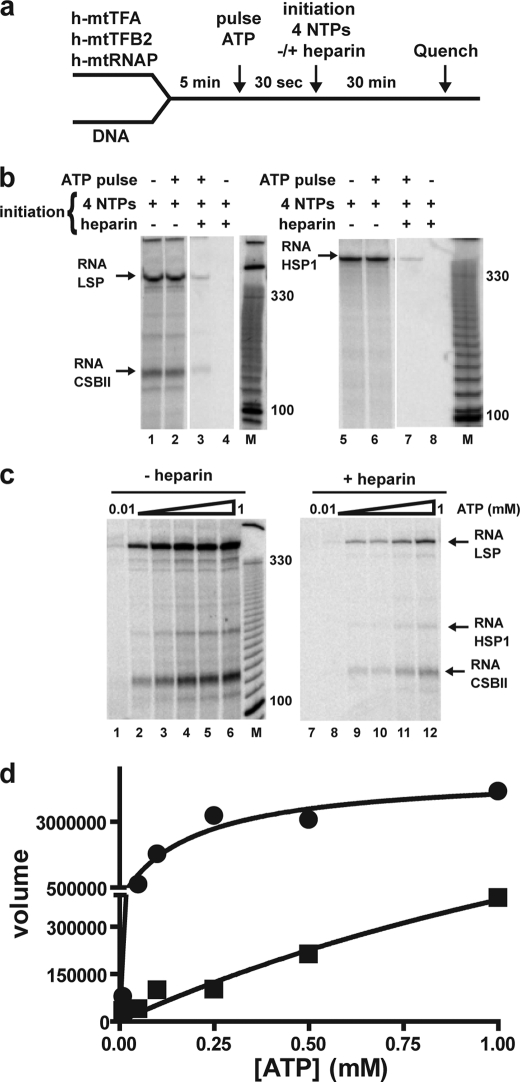
We first asked whether the transcription products observed in a standard run-off transcription experiment derive from a single or multiple rounds of transcription initiation. To do so, we performed the experiments described above in the presence or absence of heparin. Heparin is a negatively charged polysaccharide that is an efficient trap for polymerases and other nucleic acid-binding proteins when not bound to nucleic acid. After a single initiation cycle, a polymerase that terminates and disengages from the template should be trapped by heparin. Thus, heparin prevents new initiation events therefore permitting the study of a single round of transcription. We determined experimentally that the concentration of heparin that completely inhibits initiation is in the 0.5 to 1.0 μm range (supplemental Fig. S2). We refer to this type of experiment as a “pretrap” experiment as heparin is present during the assembly of the initiation complex. The subsequent experiments employed 1 μm heparin.
Our experimental design is illustrated in Fig. 2a. We initiated the transcription reaction by pulsing with ATP, which is both the first, second, and third nucleotide utilized during initiation, for 30 s. Next, we chased the initiated product with NTPs in the presence or absence of heparin. LSP transcripts of the expected size and quantity were observed in the absence (Fig. 2b, lane 1) or presence (Fig. 2b, lane 2) of an ATP pulse as long as the four NTP chase was performed in the absence of heparin. However, an ATP pulse was required to form product when the four NTP chase was performed in the presence of heparin (Fig. 2b, compare lane 3 with lane 4), suggesting that binding of the initiation nucleotide(s) and/or formation of di- or trinucleotide products was required to form an initiation complex that is resistant to heparin. The amount of transcript produced when the chase contained heparin (that reports on a single round of transcription) is substantially lower than the amount produced in the chase without heparin, indicating that most of the transcripts observed in the experiments performed above and by others (18, 21, 28–30, 32, 36–39) derive from multiple rounds of transcription initiation. A similar result was obtained when the transcription reaction was directed by HSP1 (Fig. 2b, lanes 5–8).
FIGURE 2.
Formation of an initiation complex that is resistant to heparin is dependent on ATP concentration. a, experimental design. Reactions were performed by combining h-mtTFA (100 nm), h-mtTFB2 (20 nm), and h-mtRNAP (20 nm) to linearized plasmid DNA (4 nm) in reaction buffer at 32 °C for 5 min. Some reactions were pulsed with ATP (400 μm) for 30 s prior to initiation of transcription with all four NTPs (400 μm ATP, 150 μm CTP, 150 μm GTP, 10 μm UTP, and 0.2 μCi/μl [α-32P]UTP) in the absence or presence of heparin (1 μm). Reactions were quenched after a 30-min incubation. b, ATP is required to form heparin-resistant complexes. Products from reactions on LSP (left) or HSP1 (right) were resolved by denaturing PAGE on 5% gels and visualized by phosphorimaging. In the absence of heparin, products were formed independent of an ATP pulse on LSP (lanes 1 and 2) and HSP1 (lanes 5 and 6). In the presence of heparin, products were formed on LSP and HSP1 when pulsed with ATP (lanes 2 and 7) but not in the absence of the ATP pulse (lanes 4 and 8). The size of selected bands from a 10-bp DNA ladder (M) is indicated as a reference. c, ATP concentration dependence for transcription in the absence and presence of heparin. The reaction was performed in the absence (lanes 1–6) or presence (lanes 7–12) of heparin as described in b; however, the concentration of ATP used in the pulse was varied from 0.01 to 1 mm. d, formation of heparin-resistant (initiation) complexes requires high concentration of ATP. The RNA product data (volume reported by ImageQuant software) were plotted as a function of ATP concentration. Transcription efficiency reaches a plateau at 1 mm ATP only in the absence of heparin, suggesting that concentrations in excess of 1 mm are required for initiation. When the data are fit to a hyperbola, K0.5(ATP) values for ATP of 0.2 and 2 mm are obtained in the absence and presence of heparin, respectively.
Formation of Heparin-resistant Transcription-Initiation Complexes Requires the Initiating Nucleotide (ATP) and Is Dependent on Its Concentration
The ability to form heparin-resistant, transcription-initiation complexes provided the opportunity to determine the requirements for formation of this complex. Other NTPs, in particular GTP and UTP, did not support significant accumulation of the heparin-stable complex (supplemental Fig. S3b), suggesting that the initiation nucleotide is required. Because the first three nucleotides templated are adenylate residues, it was not clear if ATP binding was sufficient to form the heparin-stable complex. In the T7 transcription system, the first nucleotide can be replaced by the nucleoside and its mono- and diphosphorylated derivatives (40). Some heparin-stable complex was formed on LSP (supplemental Fig. S3c) and HSP1 (supplemental Fig. S3d) by using ADP and AMP as the initiating nucleotide. However, we were unable to exclude the presence of trace ATP contaminants. We suggest that the ability of the pulsed nucleotide to undergo catalysis to form the di- and/or trinucleotide products enhances resistance of the transcription initiation complex to heparin.
Next, we evaluated the ATP concentration dependence of the reaction in the presence and absence of heparin. We reasoned that if the values for the concentration of ATP required for half-maximal transcription (K0.5) were the same under both conditions, then the rate-limiting step (initiation versus elongation) may also be the same. In the absence of heparin, a lower concentration of ATP was required for maximal transcription (Fig. 2c, lanes 1–6) relative to its absence (Fig. 2c, lanes 7–12). Indeed, there was at least a 10-fold difference in the observed K0.5 values measured (Fig. 2d). Clearly, in the presence of heparin, initiation is being measured so it is not surprising that the K0.5 value for ATP is so high. The low K0.5 value in the absence of heparin may suggest that elongation is being measured.
Regulation of transcription likely occurs at all stages of transcription as follows: preinitiation, initiation, post-initiation, elongation, and termination. Generally, the rate-limiting sub-step of a step in a multistep pathway is the most likely point of regulation. Obtaining information on these rate-limiting sub-steps requires that, preferably, a single step is being interrogated. The experiments presented above suggest that all of the studies performed to date that use a plasmid-based run-off transcription template to study human mitochondrial transcription cannot distinguish between transcriptional effects arising from problems with initiation, elongation, or reinitiation, especially if different from initiation (21, 29, 30, 32, 37). The use of heparin limits these templates to a single round of transcription. The high K0.5 values suggest that nucleotide binding and/or di- or trinucleotide product formation may be the rate-limiting step under these conditions. Having ATP as the initiating nucleotides for LSP and HSP1 makes initiation of these promoters sensitive to ATP concentration as suggested for human and yeast mitochondrial transcription (41–43).
Oligonucleotide-based DNA Templates for the Study of Human Mitochondrial Transcription in Vitro
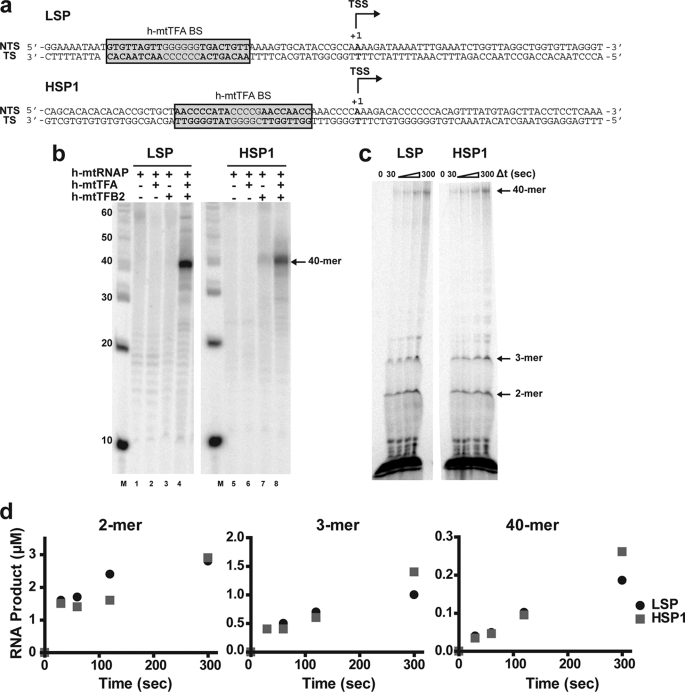
The sensitivity of the plasmid-based transcription systems is insufficient to permit transcription initiation to be interrogated fully and its rate-limiting sub-steps to be discovered. Therefore, we developed 90-bp DNA oligonucleotides representing LSP and HSP1 (Fig. 3a). Each 90-bp template contains the following: 1) 50 bp upstream of the site of transcription initiation (also referred to as the TSS and denoted as +1), which includes the h-mtTFA-binding site (18); and 2) 40 bp of template.
FIGURE 3.
DNA oligonucleotide-based templates for the study of human mitochondrial transcription in vitro. a, double-stranded DNA oligonucleotides used in this study. DNA oligonucleotides (90 bp) for LSP and HSP1 contained 50 bp upstream of the TSS (+1) and a 40-bp templating region. The reported h-mtTFA DNA-binding sites (h-mtTFA-BS) are highlighted (38). The top strand is the nontemplating strand (NTS); the bottom strand is the templating strand (TS). b, utilization of the LSP and HSP1 oligo templates by the polymerase requires h-mtTFA and h-mtTFB2. Reactions were performed by combining one or more of the following: h-mtTFA (0.5 μm), h-mtTFB2 (0.5 μm), and h-mtRNAP (0.5 μm) with LSP or HSP1 oligo (0.5 μm) in reaction buffer at 32 °C for 5 min prior to initiating transcription by addition of NTP mix (400 μm ATP, 150 μm CTP, 150 μm GTP, 10 μm UTP, and 0.2 μCi/μl [α-32P]UTP). Reaction was quenched after 10 min. Products were resolved by denaturing PAGE on 15% gels and visualized by phosphorimaging. c, direct observation of early (abortive) initiation products. Reactions were performed by combining h-mtTFA (2.5 μm), h-mtTFB2 (2.5 μm), and h-mtRNAP (2.5 μm) with DNA oligonucleotide duplex (2.5 μm) in reaction buffer at 32 °C for 5 min prior to initiating transcription by addition of NTPs (500 μm each) containing 0.2 μCi/μl [α-32P]ATP. Reactions were quenched at various times by addition of stop buffer. Product analysis was as described in b; however a 23% gel was used. d, production of full-length RNA is inefficient. The yield of 2-, 3-, and 40-mer RNA was determined on LSP and HSP1 oligos as described under “Experimental Procedures.”
We utilized these DNA oligonucleotides as template for in vitro transcription as described above, with the exception that the complex was assembled such that each component of the transcription initiation complex (h-mtTFA, h-mtTFB2, h-mtRNAP, and DNA) was present at 0.5 μm. A 40-nt run-off transcript could be produced from both the LSP and HSP1 templates (Fig. 3b, lanes 4 and 8). Production of the 40-nt product using the LSP oligo required both transcription factors A and B2 (Fig. 3b, compare lane 4 with lanes 1–3). Production of the 40-nt product using the HSP1 oligo required h-mtTFB2 (Fig. 3b, compare lanes 7 and 8 with lanes 5 and 6), but some product was detected in the absence of h-mtTFA (Fig. 3b, lane 7) as observed with plasmids. The yield of the 40-nt product using LSP appeared greater than the yield from HSP1 (Fig. 3b, compare lane 4 with lane 8), and part of this difference derives from the difference in the specific activity of the two products. LSP-derived transcripts contain 13 uridylate residues, and HSP1-derived transcripts contain only 11 uridylate residues (Fig. 3a).
Together, these results confirm that DNA oligonucleotide-based transcription templates support accurate initiation that is dependent on the presence of transcription factors as observed for plasmid-based transcription templates. We conclude that the minimal requirements for specific transcription initiation are contained within these 90-bp fragments. The ability of the human mitochondrial transcription machinery to assemble productively on DNA oligonucleotides is not surprising as this observation has been made for other transcription systems, including the phage T7 (44–46), yeast mitochondrial (47, 48), and bacterial (49) transcription systems.
Transcription on LSP and HSP1 Oligonucleotide Templates Yields Abortive Products and Sub-stoichiometric Amounts of Full-length RNA
All of the experiments performed thus far used [α-32P]UTP to label the RNA product. This selection was driven by previous studies of human mitochondrial transcription in vitro (21). As alluded to above, the first three nucleotides templated by both promoters are ATP. Therefore, the earliest products of transcription initiation cannot be observed using [α-32P]UTP. Indeed, labeling with UTP will only permit products greater than or equal to 6 or 18 nt to be observed from LSP or HSP, respectively (Fig. 3a). To address this problem, we modified our experimental design to perform labeling with [α-32P]ATP. Given that a high concentration of initiating nucleotide is required for efficient transcription, the concentration of ATP employed was 500 μm. We also elevated the concentration of the other nucleotides at this level. To increase the signal for products produced at such a low specific activity of [α-32P]ATP, the components of the transcription initiation complex were elevated by a factor of 5 to 2.5 μm.
The transcription machinery was incubated with either LSP or HSP1 oligos at 32 °C for 5 min. The reaction was initiated by addition of the NTP mix containing [α-32P]ATP. Reaction progress was monitored from 30 s to 5 min as described above; RNA product accumulation was linear over the entire 5-min time course. From the first time point, short dinucleotide (2-mer) and trinucleotide (3-mer) RNAs were produced from LSP and HSP1 and continued to accumulate over the entire 5-min reaction period (Fig. 3c). Later, full-length RNA (40-mer) was observed (Fig. 3c). Very little RNA accumulated in the 4–39-nt range, although some tetranucleotide RNA was evident late in the reaction (Fig. 3c). Interpretation of these data in the context of the more established transcription systems (50–54) leads us to suggest that the di- and trinucleotide RNAs are abortive products and that the transition into late-initiation or elongation complex may occur with the production of a 4-nt RNA.
We quantified the amount of each product (2-, 3-, and 40-mer) produced after initiation of the reaction from LSP and HSP1; the data for the 5-min time course are shown in Fig. 3d. First, initiation from both promoters was similar, with initiation from LSP exceeding that from HSP1 by no more than 2-fold (Fig. 3d). Production of abortive products exceeded the production of full-length RNA by 10-fold, suggesting that the transition into a mode competent for 40-mer production is inefficient (Fig. 3c). These experiments were performed under conditions in which the transcription initiation complex should have been present at the 2.5 μm level. Therefore, if all complexes were competent for transcription initiation and elongation at the start of the reaction, then at least 2.5 μm 40-mer RNA should have been rapidly produced, especially given that the rate of nucleotide addition by the polymerase is on the order of 30 s−1.4 Over the 5-min time course, no more than 10% of the expected 2.5 μm 40-mer RNA was observed. This low value of 40-mer RNA could not be elevated by changing protein stoichiometry or time of incubation prior to addition of nucleotides (data not shown).
Assembly of Human Mitochondrial Transcription Machinery on LSP and HSP1 Oligos
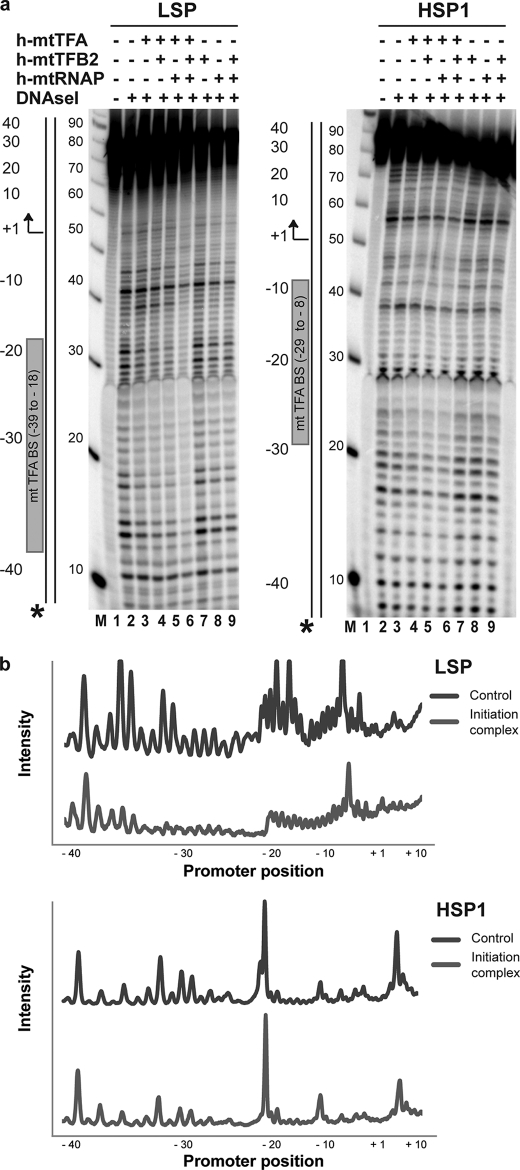
To begin to delimit the myriad possible explanations for sub-stoichiometric production of full-length RNA using LSP and HSP1 oligo templates, we used DNase I footprinting to analyze the extent of assembly of the transcription machinery on these templates. DNase I footprinting has been used extensively to characterize the human mitochondrial transcription initiation complex on plasmid-based templates (18, 32). The ability to assess promoter occupancy should provide some information on the binding competence of the transcription components, especially h-mtTFA (18, 28, 36, 38) and h-mtRNAP (32).
LSP and HSP1 oligo templates were labeled with 32P on the 5′ end of the nontemplating strand (Fig. 4a). Seventy two percent protection of LSP was observed when the nontemplating strand was labeled, and all three protein components were present (Fig. 4, a, LSP−, lane 6, and b, LSP). Protection of HSP1 was reduced 5-fold relative to LSP (Fig. 4a, HSP1−, lane 6, and b, HSP1). As a control for specificity, an end-labeled oligo was used whose sequence was a scrambled version of LSP (referred to as random in supplemental Fig. S4). Essentially no protection was observed on the random oligo (supplemental Fig. S4). Additional changes in the pattern of protection were not observed by increasing the amount of one or more of the protein components by a factor of 2 or 3 (data not shown). With the exception of h-mtTFA on one labeled version of LSP (Fig. 4a, LSP−, lane 3), individual proteins and two-protein combinations failed to produce substantial protection.
FIGURE 4.
DNase I footprinting provides evidence for assembly of initiation complexes on LSP and HSP1 oligo templates. DNase I footprinting was performed using LSP and HSP1 DNA oligonucleotide templates. Proteins were assembled on the indicated 32P-labeled DNA oligo (2.5 μm) by combining one or more of the following components: h-mtTFA (2.5 μm), h-mtTFB2 (2.5 μm), and h-mtRNAP (2.5 μm), as indicated, in reaction buffer at 32 °C. DNA cleavage was initiated by addition of RQ1 DNase (0.002 unit/μl) and CaCl2 (1 mm) and quenched after 2 min by addition of stop/trap buffer. Products were resolved by denaturing PAGE on 8% gels and visualized by phosphorimaging. The locations/orientations of the h-mtTFA-binding site and transcription start site are indicated for the LSP and HSP1 promoters. A 10-bp ladder was used as a size marker (M). a, footprinting was performed on LSP and HSP1 containing 32P in the 5′ end of the templating strand. b, quantification of selected lanes of the gels shown in a. The intensities were normalized to the −40 position of the control (lane 2 of LSP or HSP1 in a). Initiation complex refers to lane 6 of LSP or HSP1 in a. Asterisk refers to the location of the 32P label.
All of the observations described above are in good agreement with available footprinting data obtained using plasmid-based templates (32, 38). The protection pattern observed here on the nontemplating strand-labeled LSP permits us to conclude that 1 eq of all protein components relative to template is sufficient to protect the appropriate region of the template. In solution, h-mtTFA is a monomer (33). However, h-mtTFA is thought to form dimers on DNA (33). Additional studies will be required to determine whether DNA-induced dimerization of h-mtTFA is obviated by the presence of one or more of the other components of the transcription machinery. It is likely that all proteins are folded in a conformation that permits them to interact specifically with each other and with DNA. Protection of HSP1 is 5-fold lower than observed for LSP, but only a 50% reduction in yield of full-length RNA from HSP1 relative to LSP was observed (Fig. 3, b and d). Unlike for LSP, HSP1 protection does not correlate directly with occupancy. The specific binding activity observed for LSP does not prove complete functional competence but is suggestive thereof. Therefore, it is possible that the less-than-stoichiometric yield of full-length product from LSP (and by inference HSP1) is related to a defect that occurs after assembly of the protein components on the promoter.
Use of a Dinucleotide Primer Does Not Overcome the Block to Efficient Transcription on LSP and HSP1 Oligo Templates
Our initial characterization of transcription products synthesized from LSP and HSP1 oligo templates revealed the accumulation of di- and trinucleotide RNA products (Fig. 3c). Whether or not these products were truly abortive, remained bound, or could be extended was not clear. To address these questions, we performed a pulse-chase experiment (supplemental Fig. S5). The transcription complex was assembled and incubated with [α-32P]ATP for 10 s, and products were chased into full-length RNA by addition of all four NTPs (supplemental Fig. S5a). As a control, the labeling reaction was allowed to proceed in the absence of added nucleotide to show that synthesis of full-length, labeled product depended on NTP addition (supplemental Fig. S5a). Di- and trinucleotide products did not accumulate in the presence of added NTPs (pulse-chase) (supplemental Fig. S5b). In the absence of added NTPs (pulse quench), di- and trinucleotide accumulated, but full-length RNA was not produced (supplemental Fig. S5b). Curiously, a poly(rA) ladder was produced at longer times under these conditions, perhaps due to slippage synthesis as described previously for T7 RNA polymerase (51, 55). In both the T7 and bacterial transcription systems, abortive products accumulate over the entire course of a reaction (51, 53, 56, 57). This was not observed for the human mitochondrial transcription system (see pulse-chase in supplemental Fig. S5b), suggesting that the transition from initiation to elongation may be more efficient relative to these other transcription systems.
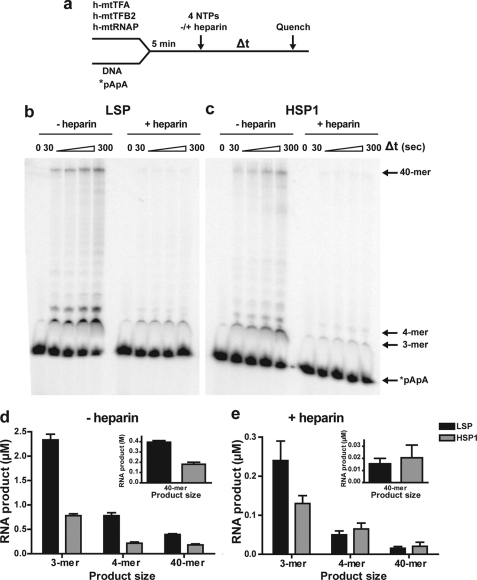
The apparent ability of the transcription complex to efficiently and rapidly utilize di- and trinucleotide products provided the opportunity to ask whether or not formation of these products contributed to the less-than-stoichiometric yield of full-length RNA relative to transcription template. For these experiments, the transcription complex was assembled on LSP or HSP1 in the presence of a 32P-labeled synthetic dinucleotide primer (*pApA) (Fig. 5a). The reaction was initiated by addition of all four NTPs in the absence or presence of heparin, and reaction progress was monitored for 5 min (Fig. 5a). Product formed in the presence of heparin should be a more accurate reflection of the quantity of active initiation complexes.
FIGURE 5.
Initiation of RNA synthesis by using a dinucleotide RNA primer does not increase transcription efficiency relative to initiation de novo on LSP or HSP1. a, experimental design. Reactions were performed by combining h-mtTFA (2.5 μm), h-mtTFB2 (2.5 μm), and h-mtRNAP (2.5 μm) with DNA oligonucleotide duplex (2.5 μm) and 32P-labeled dinucleotide (*pApA) RNA (10 μm) in reaction buffer at 32 °C for 5 min. The reaction was initiated by addition of NTPs (500 μm each) in the absence or presence of heparin (10 μm) and then quenched at various times by addition of stop buffer. Reaction products were resolved by denaturing PAGE on 25% gels and visualized by phosphorimaging. b, representative gel of an experiment using LSP as template. c, representative gel of an experiment using HSP1 as template. d, comparison of the average amount of the indicated RNA product formed at 120 s on LSP and HSP1 in the absence of heparin. e, comparison of the average amount of the indicated RNA product formed at 120 s on LSP or HSP1 in the presence of heparin.
In the absence of heparin, the dinucleotide was converted to 3-mer, 4-mer, and full-length 40-mer RNAs using LSP (Fig. 5b, −heparin) or HSP1 (Fig. 5c, −heparin). In the presence of heparin, the same product distribution was observed, but the yield of RNA product was reduced substantially (+heparin in Fig. 5, b and c). Quantitation of the products formed revealed that, in the absence of heparin, the yield of RNA products from a dinucleotide primer on LSP and HSP1 (Fig. 5d) was equivalent to that produced by using ATP on these promoters (Fig. 3d). We conclude that formation of the dinucleotide, and likely other short initiation products, does not limit formation of full-length RNA. This conclusion was not changed by increasing the concentration of pApA in the reaction (data not shown). In the presence of heparin, the yield of RNA products was reduced by a factor of 10 (Fig. 5e). This observation was striking in that it suggested that only 10% of the complexes were functioning. Therefore, it was again possible that under conditions in which multiple turnovers were allowed, only a very small fraction of the input proteins was functioning.
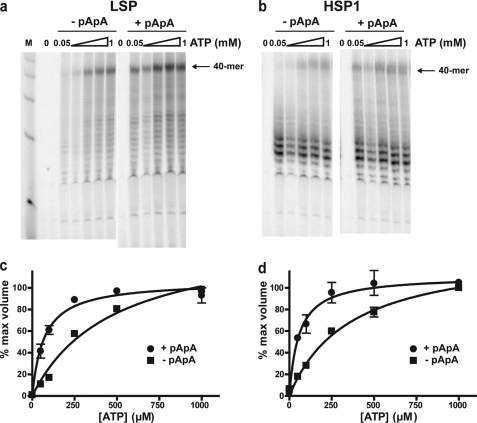
When a labeled dinucleotide was employed, products originating from initiation de novo would be missed. To rule out a preference for initiation de novo and to further define differences between dinucleotide-primed and de novo RNA synthesis, an experiment was performed in which [α-32P]UTP was again used for labeling in the presence or absence of the dinucleotide primer and under conditions in which the ATP concentration was varied (Fig. 6). This experiment showed that the dinucleotide primer stimulated synthesis of full-length RNA from both LSP (Fig. 6a) and HSP1 (Fig. 6b) at all concentrations of ATP employed. Quantitative evaluation of the ATP concentration dependence of 40-mer synthesis by de novo initiation yielded a K0.5 value in the 400–500 μm range for both promoters (Fig. 6, c and d). In contrast, the K0.5 value was at least 5-fold lower (70–90 μm) when the dinucleotide primer was used (Fig. 6, c and d). Such a high K0.5 value for the initiating nucleotide(s) is consistent with observations made in other transcription systems, including the yeast mitochondrial transcription system (43, 46, 58, 59). The efficiency of human transcription should therefore be responsive to the cellular levels of ATP as observed for the human and the yeast mitochondrial transcription systems (41, 43).
FIGURE 6.
RNA synthesis initiated using a dinucleotide RNA primer requires lower concentrations of ATP for maximal transcriptional efficiency than RNA synthesis initiated de novo on LSP or HSP1. Reactions were performed by combining h-mtTFA (0.5 μm), h-mtTFB2 (0.5 μm), and h-mtRNAP (0.5 μm) with LSP or HSP1 oligo templates (0.5 μm) in the absence (−pApA) or presence (+pApA) of dinucleotide primer (10 μm) in reaction buffer at 32 °C for 5 min. Transcription was initiated by adding NTPs (150 μm GTP, 150 μm CTP, 50 μm UTP, and 0.2 μCi/μl [α-32P]UTP) with the concentration of ATP ranging from 50 to 1000 μm. Reactions were quenched after 2 min by addition of stop buffer. Products were resolved by denaturing PAGE on 25% gels and visualized by phosphorimaging. a, representative gel of an experiment using LSP as template. b, representative gel of an experiment using HSP1 as template. c, RNA product data obtained on LSP in absence (−pApA, ■) or presence (+pApA, ●) of dinucleotide primer were expressed as a fraction of that observed at 1 mm ATP and plotted as a function of ATP concentration. The data were fit to a hyperbola, yielding K0.5(ATP) values for ATP of 500 ± 90 μm and 70 ± 10 μm in the absence or presence of dinucleotide, respectively. d, RNA product data obtained on HSP1 in absence (−pApA, ■) or presence (+pApA, ●) of dinucleotide primer were expressed as a fraction of that observed at 1 mm ATP and plotted as a function of ATP concentration. The data were fit to a hyperbola, yielding K0.5(ATP) values for ATP of 400 ± 60 and 60 ± 10 μm in the absence or presence of dinucleotide, respectively.
Formation of an Open Complex Is a Rate-limiting Step for Initiation of Transcription on LSP and HSP1 Oligo Templates
At this point of the study, it was still not clear if our protein preparations were only 10% active or if there were rate-limiting events after assembly of the transcription complex but prior to synthesis of the dinucleotide product. It is well established that formation of an open complex, i.e. melting of the double-stranded DNA surrounding the transcription start site, can be a rate-limiting step for transcription initiation (42, 59–62). To determine whether an open complex was forming on LSP and HSP1 oligos after assembly of the transcription machinery, we used KMnO4 reactivity (63, 64). KMnO4 will readily oxidize thymine bases of single-stranded DNA and sensitize the phosphodiester backbone to piperidine cleavage at the sites of modification (64). This approach is well suited to the study of the LSP and HSP1 promoters given the number of thymidines surrounding the transcription start site (Fig. 3a). When this experiment was performed, we were unable to detect any KMnO4 reactivity on LSP (supplemental Fig. S6a) or HSP1 (supplemental Fig. S6b) in the absence or presence of any combination of factors.
The inability to quantitatively form an open complex would explain the low yield of RNA from the LSP and HSP1 oligo templates. To bypass the need for open complex formation, we created “bubble” templates in which the region from +1 to +3 of the template was artificially melted due to the presence of noncomplementary nucleotides at these positions of the nontemplating strand. The presence of the bubble was confirmed by KMnO4 footprinting for the LSP (supplemental Fig. S6a) and HSP1 (supplemental Fig. S6b) bubble templates. Importantly, the presence of the transcription machinery had no significant impact on the reactivity of the bubble templates with KMnO4, consistent with the absence of an open complex on the “normal” templates (supplemental Fig. S6).
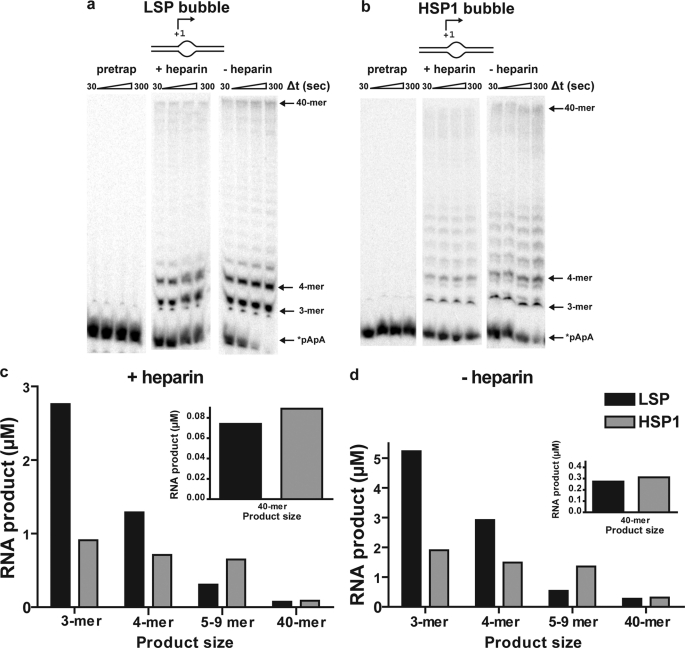
Experiments using the bubble templates were designed as follows. A pretrap experiment, in which heparin was present at the time of transcription complex assembly, was performed to show that the concentration of heparin used was sufficient to prevent reinitiation events from occurring. Experiments were then performed in which the transcription complex was assembled on the bubble template in the presence of end-labeled dinucleotide, and the reaction was initiated by addition of all four NTPs in the presence or absence of heparin. In the presence of heparin, the concentration of active complexes should be revealed. In the absence of heparin, the ability of these complexes to recycle would be revealed.
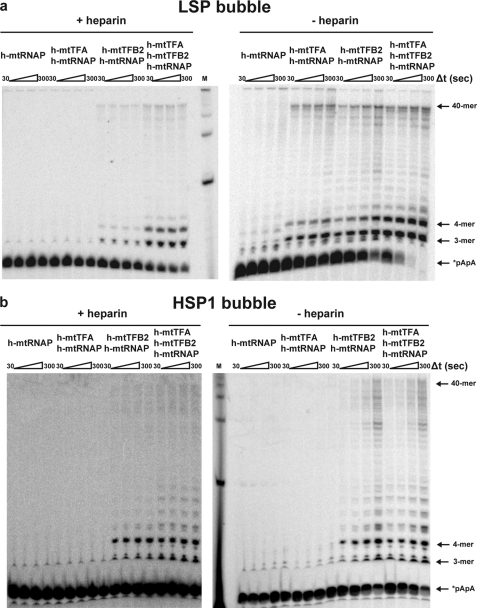
Heparin at a concentration of 10 μm was sufficient to prevent transcription on LSP (Fig. 7a, pretrap) and HSP1 (Fig. 7b, pretrap) bubble templates. When heparin was added along with the NTPs after complex assembly, a burst of product formation was observed on both promoters as fast as we could measure by manual quenching (+heparin in Fig. 7, a and b). On LSP, the primary products were 3–6 and 40 nts (Fig. 7a, +heparin). On HSP1, the primary products were 3–9 and 40 nts (Fig. 7b, +heparin). The amount of product formed in the presence of heparin did not change substantially over time on either promoter, with the final yield of extended dinucleotide being 5 μm on LSP and 3 μm on HSP1 (Fig. 7c). The product yield is now on the order of the value expected based on the concentration of assembled complex. Indeed, for LSP, this value exceeds that of the complex by a factor of 2, likely because multiple rounds of abortive synthesis are possible prior to complex dissociation. The yield from HSP1 is lower than that from LSP, suggesting that this complex is less stable as reported by others (21). The differences in stability of the two transcription complexes may reflect differences in organization of the complex imposed by differences in the positions of the TFA-binding sites relative to the transcription start site (Fig. 3a). We now conclude that the transcription machinery is fully active and that formation of an open complex is a major obstacle to transcription on linear, double-stranded oligo templates and likely linear, plasmid-based templates as well. We speculate that the use of a relaxed, circular template more like mtDNA may permit strain induced by the binding and/or bending of h-mtTFA to be harnessed for promoter melting.
FIGURE 7.
Open complex formation is a rate-limiting step for transcription initiation on LSP and HSP1 revealed by using bubble templates. Reactions were performed by combining h-mtTFA (2.5 μm), h-mtTFB2 (2.5 μm), and h-mtRNAP (2.5 μm) with LSP or HSP1 bubble templates (2.5 μm) and 32P-labeled dinucleotide (*pApA, 10 μm) in reaction buffer at 32 °C for 5 min. The reaction was initiated by addition of NTPs (500 μm each) in the presence or absence of heparin (10 μm) and then quenched at various times by addition of stop buffer. For the pretrap experiment, heparin (10 μm) was included during the assembly of proteins on DNA. a, representative gel of an experiment using LSP bubble template. b, representative gel of an experiment using HSP1 bubble template. c, comparison of the average amount of the indicated RNA product formed at 120 s on LSP and HSP1 bubble templates in the presence of heparin. d, comparison of the average amount of the indicated RNA product formed at 120 s on LSP and HSP1 in the absence of heparin.
The reason for the promoter-dependent differences in the observed lengths of the intermediate products (+heparin in Fig. 7, a and b) is not clear and has not been observed previously. This observation may suggest that differences exist in the regulation of LSP relative to HSP1 after open complex formation in vitro. The “stuttering” observed on HSP1 occurs in a GC-rich stretch of the templating strand (Fig. 3a). The high GC content of CSBII is thought to contribute to the termination activity of that sequence (14).
In the absence of heparin, all of the dinucleotide primer is consumed and more full-length RNA is produced (−heparin in Fig. 7, a and b), suggesting that the complexes can recycle. Dinucleotide primer is consumed faster on LSP than on HSP (compare Fig. 7, a, −heparin, with b, −heparin). This difference in primer consumption again suggests differences in regulation of LSP relative to HSP. Reiterative synthesis of abortive products 3–4 nt in length may occur on LSP without the necessity for polymerase to escape the promoter and may be driven by dissociation of the abortive products from the polymerase active site. In contrast, reiterative synthesis on HSP may be less facile because the products 3–9 nt in length may be retained longer in the polymerase active site, akin to pausing observed by multisubunit RNA polymerases (65–68). Although additional studies will be required to sort out the mechanistic basis for these differences, these studies provide the first indication that the early events of transcription initiation on LSP differ substantially from those on HSP1. Full-length product produced on HSP1 is on par with that produced on LSP (see 40-mer on Fig. 7d), despite the differences in complex assembly, stability, and initiation efficiency discussed above. Perhaps the reduced complex stability on HSP1 leads to a greater efficiency of promoter clearance. In neither case does the amount of full-length product approach a level stoichiometric with the template (see 40-mer in Fig. 7d). This observation may suggest that promoter clearance and transition into a fully elongation competent transcription complex may represent another rate-limiting step during the human mitochondrial transcription cycle. The reason that abortive products are not used (Fig. 7a), as predicted by the pulse-chase experiments (supplemental Fig. S5), is not known but could reflect a difference in the size of the open complex on the bubble templates relative to the size of the open complex on double-stranded templates and/or loss of sequence-specific interactions with the nontemplating strand caused by creation of the artificial bubble. Additional studies will be required to resolve this issue.
One final control was performed to show that the observations discussed above were specified by the promoter sequence and not just the presence of a bubble. When a bubble template was constructed by using the LSP-scrambled (random) sequence, products were not formed by the polymerase in the absence or presence of transcription factors when RNA synthesis was initiated by using a dinucleotide primer (supplemental Fig. S7a) or de novo (supplemental Fig. S7b).
Use of Bubble Templates Relaxes Transcription Factor Dependence, Further Distinguishes LSP from HSP1, and Reveals the Existence of a Post-initiation Step That Limits the Efficiency of Full-length RNA Synthesis
The ability of a bubble template to bypass the primary, rate-limiting step for transcription initiation in vitro on linear templates provided the opportunity to study the contributions of the different transcription factors before and after open complex formation. Experiments were performed on the LSP bubble template (Fig. 8a) and the HSP1 bubble template (Fig. 8b) in the presence of heparin. Production of full-length RNA was no longer as dependent on the presence of h-mtTFA as observed on the templates lacking a bubble (Fig. 3b), suggesting that h-mtTFA functions primarily at steps that lead to formation of the open complex. In most cases, h-mtTFB2 was required, suggesting that h-mtTFB2 functions before and after open complex formation, consistent with a recent report from Temiakov and co-workers (27). However, bubble templates did not require any transcription factors in that study (27). This difference may reflect the larger bubble and/or much longer reaction times employed (27). In all cases, the presence of all three protein components led to an increase in the amount of overall transcription (Fig. 8c, +heparin) likely because the combination of all three protein components is more stable on the promoter, consistent with DNase I footprinting experiments (Fig. 4a) and, consequently, also more resistant to heparin.
FIGURE 8.
Unique factor dependence of LSP relative to HSP1 when bubble templates are used. Proteins were assembled on the LSP1 or HSP1 bubble template (2.5 μm) by combining one or more of the following components: h-mtTFA (2.5 μm), h-mtTFB2 (2.5 μm), and h-mtRNAP (2.5 μm), as indicated, and 32P-labeled dinucleotide primer (*pApA, 10 μm) in reaction buffer at 32 °C for 5 min. The reaction was initiated by addition of NTPs (500 μm each) in the presence or absence of heparin (10 μm) and then quenched at various times by addition of stop buffer. Products were resolved by denaturing PAGE on 25% gels and visualized by phosphorimaging. a, representative gel of an experiment using LSP bubble template. b, representative gel of an experiment using HSP1 bubble template.
Experiments performed on the bubble templates in the absence of heparin reveal a substantial difference in the intrinsic properties of the two promoters. On LSP, polymerase in combination with either h-mtTFA or h-mtTFB2 is sufficient to initiate (Fig. 8a, −heparin) and produce amounts of full-length RNA equivalent to that produced in the presence of both factors (Fig. 8c, −heparin). In contrast, on HSP1, polymerase and h-mtTFA were essentially inactive (Fig. 8b, −heparin). It has been shown recently for the yeast mitochondrial RNA polymerase that a region of the polymerase termed the specificity loop interacts with the promoter (47). The same is likely true for the human mitochondrial polymerase. The need for an additional factor on the LSP bubble template may suggest that the polymerase alone is not retained at the transcription start site long enough to do catalysis. Consistent with this possibility is the observation that either h-mtTFA or h-mtTFB2 function equivalently on the LSP bubble template. Clearly, the roles for these two factors in transcription are mechanistically distinct. As discussed earlier, h-mtTFA is thought to bind and bend DNA to facilitate open complex formation (11, 12, 30, 33). However, h-mtTFB2 is thought to interact with the templating strand in the vicinity of the transcription start site, as well as the initiating nucleotide(s) (27). This assay will prove useful in determining the physical basis for functional interaction of h-mtTFA with h-mtRNAP and h-mtTFB2 with h-mtRNAP. The reason that only h-mtTFB2 is sufficient to activate use of the HSP1 bubble template is not at all clear, but this observation further highlights the likelihood that LSP and HSP1 are regulated differently.
Finally, there is a lack of correspondence between the efficiency of initiation (the amount of 3- and 4-nt product) and the amount of full-length RNA produced. The lack of correspondence on LSP is shown best by the experiment performed in the absence of heparin (Fig. 8a), where the dinucleotide primer is completely converted to 3- and 4-mer by the polymerase in the presence of both transcription factors and is only partially converted in the presence of h-mtTFA or h-mtTFB2, but the amount of 40-mer produced is essentially the same in all cases. The lack of correspondence on HSP1 is shown best by the experiment performed in the presence of heparin, where the same amount of 3- and 4-mer is produced by the polymerase in the presence of both transcription factors and in the presence of only h-mtTFB2, but the amount of 40-mer produced is different in these two situations. Together, these observations lead us to propose that a step after initiation exists that leads to formation of a competent elongation complex. Moreover, we suggest that the transit efficiency through this step, and perhaps regulation of this step, differs between the two promoters, consistent with observations made by considering the differences in the nature of the intermediate RNA products observed between the two promoters (Fig. 7, a and b).
Conclusions
We have reconstituted human mitochondrial transcription in vitro using protein components produced in E. coli, permitting us to conclude that post-translational modifications are not required for efficient and specific initiation in vitro. The utility of these proteins has been validated by using conventional, plasmid-based assay systems developed by others to study human mitochondrial transcription in cell-free extracts and with proteins produced using a baculovirus/insect cell expression system (Figs. 1 and 2 and supplemental Figs. S1–S3) (21). To date, all studies of human mitochondrial transcription have employed full-length elongation products as a surrogate for transactions that lead to assembly and function of the transcription initiation complex, regardless of the nature of the templates used (21, 27, 29, 32). We have succeeded in assembling transcription initiation complexes on DNA oligonucleotide templates representing LSP and HSP1 at concentrations that permit, for the first time, direct analysis of both transcription initiation and transcription elongation (Figs. 3–8 and supplemental Figs. S5–S7). The major findings of this study have been integrated into the context of a model for the human mitochondrial transcription cycle shown in Fig. 9.
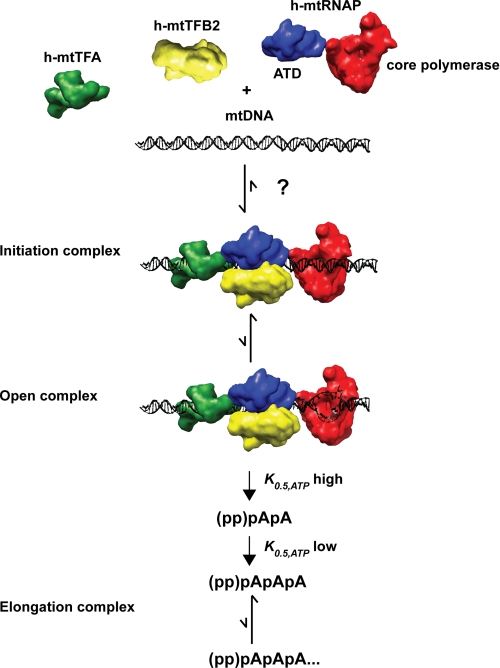
FIGURE 9.
Hypothetical model for the human mitochondrial transcription cycle. The images used of the proteins and DNA should be considered schematics but have been drawn to scale. Interactions of h-mtTFA with h-mtTFB2 and h-mtTFB2 with h-mtRNAP occur through carboxyl- or amino-terminal tails of h-mtTFA or h-mtTFB2, respectively, and are not shown explicitly (27, 29). The data presented in this study are consistent with the following model. Transcription factors and polymerase assemble on DNA to form an initiation complex by some unknown mechanism. Once formed, the initiation complex is quite stable. Open complexes do not accumulate based on potassium permanganate reactivity, creating a significant barrier to transcription initiation that can be bypassed by using bubble templates. Formation of the first phosphodiester bond requires high concentrations of ATP; therefore, the efficiency of this step will be controlled by the availability of ATP. Once a dinucleotide is formed, the dependence of the reaction on the concentration of ATP is reduced by a factor of 10. The transition into an elongation complex occurs at some point after formation of the trinucleotide, occurs inefficiently, and occurs differently on the two promoters.
The polymerase and transcription factors A and B2 assemble on the LSP or HSP1 promoter. The order of assembly, if any, was not addressed by this study. However, we were able to determine the stoichiometry of the transcription complex for the first time. We observed that 1 eq of each protein relative to DNA is sufficient to assemble the maximal level of transcription complex on the promoter (Fig. 4 and data not shown), as well as to produce the maximal amount of RNA (Fig. 3 and data not shown). The initiation complex is more stable on LSP than HSP1 (Fig. 4), consistent with previous studies (21, 38). The organization and interactions between the factors are not known, although it is clear that h-mtRNAP interacts functionally with both h-mtTFA and h-mtTFB2 (Fig. 8) (21, 32) and that h-mtTFA interacts physically with h-mtTFB2 (29). The unique amino-terminal domain of the polymerase is shown as a bridge of the core polymerase domain to the factors because this domain appears to be present in all T7-like RNA polymerases that employ transcription factors (11, 12, 29, 69). Once the initiation complex forms on linear templates, only 10% of these complexes are competent for initiation (Fig. 5). Our data suggest that formation of an open complex is a rate-limiting step for transcription on linear templates as the use of bubble templates bypasses this step (Fig. 7). The requirement for h-mtTFB2 is not alleviated on bubble templates, consistent with this factor being required before and after open complex formation. In contrast, the requirement for h-mtTFA is bypassed on bubble templates (Fig. 8), suggesting that this factor contributes to open complex formation. It is possible that the inability of h-mtTFA to shift the equilibrium from closed to open complex reflects the linear nature of the template. Mitochondrial DNA is circular, and bending of even a relaxed circular DNA will lead to more local unwinding of the DNA double helix than bending of a linear DNA. Initiation of RNA synthesis de novo from the open complex requires high concentrations of ATP, given a K0.5 value on the order of 1 mm (Fig. 6). Di- and trinucleotides are the major products when linear templates are used (Fig. 3c), and these products chase into full-length RNA (supplemental Fig. S5). These products are likely abortive products based on a yield that is greater than the polymerase concentration, even under conditions in which dissociated enzyme would be inhibited (Fig. 7). Utilization of the dinucleotide product diminished K0.5 value for ATP by at least 5-fold (Fig. 6), consistent with observations in other transcription systems that the Km value for elongating nucleotides is lower than that for initiating nucleotides (43, 46, 58, 59, 70). Production and utilization of abortive products are different on the artificial, 3-bp open complex relative to that on the double-stranded templates. The basis for this difference is unknown and requires further investigation. Finally, once efficient initiation is possible, a second rate-limiting step comes into view, namely production of a competent elongation complex (Fig. 8). Transition into an elongation complex appears to be promoter-dependent (Fig. 8), giving rise to accumulation of intermediate RNAs of different lengths (Figs. 7 and 8).
Rate-limiting steps of biochemical pathways are often those that are targeted for regulation. Like the more complex transcription systems of the eukaryotic nucleus (65, 71, 72), prokaryotes, and their phage (57, 70, 73–77), multiple steps may exist for the control of human mitochondrial transcription, including assembly of the initiation complex, formation, and/or stabilization of the open complex, initiation of the first products of RNA synthesis, and transition into an elongation complex. The system described herein sets the stage for detailed studies not only of the regulation of human mitochondrial transcription in vitro but also of the structure, function, and mechanism of the complexes that catalyze the various stages of human mitochondrial transcription.
Supplementary Material
Acknowledgments
We thank Scott Nelson, Joe Reese, Gerry Shadel, and Eric Smidansky for comments on the manuscript and Claes Gustafsson for pUC18 plasmids containing human mitochondrial promoters.
This work was supported in part by the Paul Berg Endowment of the Eberly College of Science, Pennsylvania State University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
T. E. Shutt, M. F. Lodeiro, J. Cotney, C. E. Cameron, and G. S. Shadel, submitted for publication.
E. Smidansky and C. E. Cameron, unpublished results.
- LSP
- light strand promoter
- mt
- mitochondrial
- h
- human
- h-mtRNAP
- human mitochondrial RNA polymerase
- h-mtTFA
- human mitochondrial transcription factor A
- h-mtTFB2
- human mitochondrial transcription factor B2
- HSP
- heavy strand promoter
- CSBII
- conserved sequence box II
- TSS
- transcription start site
- NTP
- nucleotide triphosphate
- oligo
- oligonucleotide
- DTT
- dithiothreitol.
REFERENCES
- 1.Wallace D. C. (2005) Annu. Rev. Genet. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallace D. C. (2008) Genetics 179, 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace D. C. (2007) Annu. Rev. Biochem. 76, 781–821 [DOI] [PubMed] [Google Scholar]
- 4.Shadel G. S. (2004) Trends Genet. 20, 513–519 [DOI] [PubMed] [Google Scholar]
- 5.DiMauro S., Schon E. A. (2008) Annu. Rev. Neurosci. 31, 91–123 [DOI] [PubMed] [Google Scholar]
- 6.Schon E. A., DiMauro S. (2007) Novartis Found Symp. 287, 214–233 [DOI] [PubMed] [Google Scholar]
- 7.Chan S. S., Copeland W. C. (2009) Biochim. Biophys. Acta 1787, 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Copeland W. C. (2008) Annu. Rev. Med. 59, 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsson N. G., Clayton D. A. (1995) Annu. Rev. Genet. 29, 151–178 [DOI] [PubMed] [Google Scholar]
- 10.Clayton D. A. (2000) Hum. Reprod. 15, Suppl. 2, 11–17 [DOI] [PubMed] [Google Scholar]
- 11.Bonawitz N. D., Clayton D. A., Shadel G. S. (2006) Mol. Cell 24, 813–825 [DOI] [PubMed] [Google Scholar]
- 12.Falkenberg M., Larsson N. G., Gustafsson C. M. (2007) Annu. Rev. Biochem. 76, 679–699 [DOI] [PubMed] [Google Scholar]
- 13.Martin M., Cho J., Cesare A. J., Griffith J. D., Attardi G. (2005) Cell 123, 1227–1240 [DOI] [PubMed] [Google Scholar]
- 14.Pham X. H., Farge G., Shi Y., Gaspari M., Gustafsson C. M., Falkenberg M. (2006) J. Biol. Chem. 281, 24647–24652 [DOI] [PubMed] [Google Scholar]
- 15.Rodeheffer M. S., Boone B. E., Bryan A. C., Shadel G. S. (2001) J. Biol. Chem. 276, 8616–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parisi M. A., Clayton D. A. (1991) Science 252, 965–969 [DOI] [PubMed] [Google Scholar]
- 17.Thomas J. O., Travers A. A. (2001) Trends Biochem. Sci. 26, 167–174 [DOI] [PubMed] [Google Scholar]
- 18.Fisher R. P., Parisi M. A., Clayton D. A. (1989) Genes Dev. 3, 2202–2217 [DOI] [PubMed] [Google Scholar]
- 19.Larsson N. G., Oldfors A., Holme E., Clayton D. A. (1994) Biochem. Biophys. Res. Commun. 200, 1374–1381 [DOI] [PubMed] [Google Scholar]
- 20.Ekstrand M. I., Falkenberg M., Rantanen A., Park C. B., Gaspari M., Hultenby K., Rustin P., Gustafsson C. M., Larsson N. G. (2004) Hum. Mol. Genet. 13, 935–944 [DOI] [PubMed] [Google Scholar]
- 21.Falkenberg M., Gaspari M., Rantanen A., Trifunovic A., Larsson N. G., Gustafsson C. M. (2002) Nat. Genet. 31, 289–294 [DOI] [PubMed] [Google Scholar]
- 22.McCulloch V., Seidel-Rogol B. L., Shadel G. S. (2002) Mol. Cell. Biol. 22, 1116–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adán C., Matsushima Y., Hernández-Sierra R., Marco-Ferreres R., Fernández-Moreno M. A., González-Vioque E., Calleja M., Aragón J. J., Kaguni L. S., Garesse R. (2008) J. Biol. Chem. 283, 12333–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushima Y., Adán C., Garesse R., Kaguni L. S. (2005) J. Biol. Chem. 280, 16815–16820 [DOI] [PubMed] [Google Scholar]
- 25.Matsushima Y., Garesse R., Kaguni L. S. (2004) J. Biol. Chem. 279, 26900–26905 [DOI] [PubMed] [Google Scholar]
- 26.Metodiev M. D., Lesko N., Park C. B., Cámara Y., Shi Y., Wibom R., Hultenby K., Gustafsson C. M., Larsson N. G. (2009) Cell Metab. 9, 386–397 [DOI] [PubMed] [Google Scholar]
- 27.Sologub M., Litonin D., Anikin M., Mustaev A., Temiakov D. (2009) Cell 139, 934–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fisher R. P., Clayton D. A. (1985) J. Biol. Chem. 260, 11330–11338 [PubMed] [Google Scholar]
- 29.McCulloch V., Shadel G. S. (2003) Mol. Cell. Biol. 23, 5816–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dairaghi D. J., Shadel G. S., Clayton D. A. (1995) J. Mol. Biol. 249, 11–28 [DOI] [PubMed] [Google Scholar]
- 31.Fernández-Silva P., Enriquez J. A., Montoya J. (2003) Exp. Physiol. 88, 41–56 [DOI] [PubMed] [Google Scholar]
- 32.Gaspari M., Falkenberg M., Larsson N. G., Gustafsson C. M. (2004) EMBO J. 23, 4606–4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gangelhoff T. A., Mungalachetty P. S., Nix J. C., Churchill M. E. (2009) Nucleic Acids Res. 37, 3153–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnold J. J., Cameron C. E. (1999) J. Biol. Chem. 274, 2706–2716 [DOI] [PubMed] [Google Scholar]
- 35.Arnold J. J., Ghosh S. K., Bevilacqua P. C., Cameron C. E. (1999) BioTechniques 27, 450–456 [DOI] [PubMed] [Google Scholar]
- 36.Fisher R. P., Clayton D. A. (1988) Mol. Cell. Biol. 8, 3496–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Cotney J., Shadel G. S. (2007) J. Biol. Chem. 282, 12610–12618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher R. P., Topper J. N., Clayton D. A. (1987) Cell 50, 247–258 [DOI] [PubMed] [Google Scholar]
- 39.Chang D. D., Fisher R. P., Clayton D. A. (1987) Biochim. Biophys. Acta 909, 85–91 [DOI] [PubMed] [Google Scholar]
- 40.Martin C. T., Coleman J. E. (1989) Biochemistry 28, 2760–2762 [DOI] [PubMed] [Google Scholar]
- 41.Gaines G., Rossi C., Attardi G. (1987) J. Biol. Chem. 262, 1907–1915 [PubMed] [Google Scholar]
- 42.Amiott E. A., Jaehning J. A. (2006) J. Biol. Chem. 281, 34982–34988 [DOI] [PubMed] [Google Scholar]
- 43.Amiott E. A., Jaehning J. A. (2006) Mol. Cell 22, 329–338 [DOI] [PubMed] [Google Scholar]
- 44.Martin C. T., Coleman J. E. (1987) Biochemistry 26, 2690–2696 [DOI] [PubMed] [Google Scholar]
- 45.Diaz G. A., Raskin C. A., McAllister W. T. (1993) J. Mol. Biol. 229, 805–811 [DOI] [PubMed] [Google Scholar]
- 46.Bandwar R. P., Jia Y., Stano N. M., Patel S. S. (2002) Biochemistry 41, 3586–3595 [DOI] [PubMed] [Google Scholar]
- 47.Nayak D., Guo Q., Sousa R. (2009) J. Biol. Chem. 284, 13641–13647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang G. Q., Paratkar S., Patel S. S. (2009) J. Biol. Chem. 284, 5514–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daube S. S., von Hippel P. H. (1992) Science 258, 1320–1324 [DOI] [PubMed] [Google Scholar]
- 50.Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. (1987) Nucleic Acids Res. 15, 8783–8798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin C. T., Muller D. K., Coleman J. E. (1988) Biochemistry 27, 3966–3974 [DOI] [PubMed] [Google Scholar]
- 52.Tintut Y., Wang J. T., Gralla J. D. (1995) J. Biol. Chem. 270, 24392–24398 [DOI] [PubMed] [Google Scholar]
- 53.Gong P., Martin C. T. (2006) J. Biol. Chem. 281, 23533–23544 [DOI] [PubMed] [Google Scholar]
- 54.Gnatt A. L., Cramer P., Fu J., Bushnell D. A., Kornberg R. D. (2001) Science 292, 1876–1882 [DOI] [PubMed] [Google Scholar]
- 55.Macdonald L. E., Zhou Y., McAllister W. T. (1993) J. Mol. Biol. 232, 1030–1047 [DOI] [PubMed] [Google Scholar]
- 56.Hsu L. M., Vo N. V., Kane C. M., Chamberlin M. J. (2003) Biochemistry 42, 3777–3786 [DOI] [PubMed] [Google Scholar]
- 57.Goldman S. R., Ebright R. H., Nickels B. E. (2009) Science 324, 927–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuzmine I., Gottlieb P. A., Martin C. T. (2003) J. Biol. Chem. 278, 2819–2823 [DOI] [PubMed] [Google Scholar]
- 59.Villemain J., Guajardo R., Sousa R. (1997) J. Mol. Biol. 273, 958–977 [DOI] [PubMed] [Google Scholar]
- 60.Jia Y., Kumar A., Patel S. S. (1996) J. Biol. Chem. 271, 30451–30458 [DOI] [PubMed] [Google Scholar]
- 61.Bandwar R. P., Patel S. S. (2002) J. Mol. Biol. 324, 63–72 [DOI] [PubMed] [Google Scholar]
- 62.Davis C. A., Capp M. W., Record M. T., Jr., Saecker R. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Place C., Oddos J., Buc H., McAllister W. T., Buckle M. (1999) Biochemistry 38, 4948–4957 [DOI] [PubMed] [Google Scholar]
- 64.Holstege F. C., Timmers H. T. (1997) Methods 12, 203–211 [DOI] [PubMed] [Google Scholar]
- 65.Price D. H. (2008) Mol. Cell 30, 7–10 [DOI] [PubMed] [Google Scholar]
- 66.Seila A. C., Core L. J., Lis J. T., Sharp P. A. (2009) Cell Cycle 8, 2557–2564 [DOI] [PubMed] [Google Scholar]
- 67.Gilmour D. S. (2009) Chromosoma 118, 1–10 [DOI] [PubMed] [Google Scholar]
- 68.Kireeva M. L., Kashlev M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8900–8905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asin-Cayuela J., Gustafsson C. M. (2007) Trends Biochem. Sci. 32, 111–117 [DOI] [PubMed] [Google Scholar]
- 70.Rutherford S. T., Villers C. L., Lee J. H., Ross W., Gourse R. L. (2009) Genes Dev. 23, 236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wade J. T., Struhl K. (2008) Curr. Opin. Genet. Dev. 18, 130–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuda N. J., Ardehali M. B., Lis J. T. (2009) Nature 461, 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uptain S. M., Kane C. M., Chamberlin M. J. (1997) Annu. Rev. Biochem. 66, 117–172 [DOI] [PubMed] [Google Scholar]
- 74.Landick R. (2006) Biochem. Soc. Trans. 34, 1062–1066 [DOI] [PubMed] [Google Scholar]
- 75.Nudler E. (2009) Annu. Rev. Biochem. 78, 335–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murakami K. S., Darst S. A. (2003) Curr. Opin. Struct. Biol. 13, 31–39 [DOI] [PubMed] [Google Scholar]
- 77.Kapanidis A. N., Margeat E., Ho S. O., Kortkhonjia E., Weiss S., Ebright R. H. (2006) Science 314, 1144–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.